Abstract
Intramembrane proteases catalyze the signal-generating step of various cell signaling pathways, and continue to be implicated in diseases ranging from malaria infection to Parkinsonian neurodegeneration1–3. Despite playing such decisive roles, it remains unclear whether or how these membrane-immersed enzymes might be regulated directly. To address this limitation, we sought intramembrane proteases containing domains known to exert regulatory functions in other contexts, and focused on rhomboid proteases that harbor calcium-binding EF-hands. We found calcium potently stimulates proteolysis by endogenous rhomboid-4 in Drosophila cells, and, remarkably, when rhomboid-4 was purified and reconstituted in liposomes. Interestingly, deleting the amino-terminal EF-hands activated proteolysis prematurely, while residues in cytoplasmic loops connecting distal transmembrane segments mediated calcium stimulation. Rhomboid regulation was not orchestrated by either dimerization or substrate interactions. Instead, calcium increased catalytic rate by promoting substrate gating. Substrates with cleavage sites outside the membrane could be cleaved but lost the capacity to be regulated. These observations indicate substrate gating is not an essential step in catalysis, but instead evolved as a mechanism for regulating proteolysis inside the membrane. Moreover, these insights provide new approaches for studying rhomboid functions by investigating upstream inputs that trigger proteolysis.
Cell membranes are both controlled borders with the outside world as well as dynamic platforms for organizing cell signaling, metabolic pathways, and ultrastructure assembly. All of these key events rely on enzymes that reside directly within the cell membrane, yet achieving a mechanistic understanding of how these specialized enzymes function within this environment has proven challenging.
Intramembrane proteases catalyze the committed, signal-generating step of several key signaling pathways by cleaving transmembrane proteins within the membrane1–3. Their importance is underscored by repeated implication in disease. γ-secretase generates the amyloid-β peptide in Alzheimer’s disease4,5, but more recently has been successfully targeted in a spectrum of cancers6, because its activating cleavage of the Notch receptor triggers signaling2. Site-2 protease family metalloenzymes liberate transcription factors from the membrane to control cholesterol and fatty acid composition of membranes1, and signaling circuits that control virulence in pathogenic bacteria7. Rhomboid serine proteases are a family of master regulators that initiate epidermal growth factor (EGF) signaling during Drosophila development3,8, but more recently have been implicated in cleaving adhesins during malaria invasion9, and regulating mitochondrial quality control to guard against Parkinson’s disease10.
Since peptide bond cleavage is irreversible in the cell, precise regulation of protease activity is paramount. Yet it’s generally thought that intramembrane proteases are constitutively active enzymes over which the cell cannot exert direct regulation11. Instead, two mechanisms control activity. The first is transcriptional, as exemplified by Drosophila rhomboid-1: the constitutively active protease is made only when and where needed3. This mechanism has historically served as a beautiful atlas of EGF signal initiation during development. The second mechanism is centered on controlling access to substrate by segregating it from protease11. Malaria, for example, sequesters adhesins in secretory organelles before invasion, while their secretion onto the surface leads to the first encounter with an active rhomboid protease7.
The key property missing from these two mechanisms is the ability to respond rapidly to changing conditions: transcriptional and cell localization changes are ill-adapted to provide immediate responses that are hallmarks of cell signaling. Moreover, it’s essentially unprecedented for proteases to be devoid of direct enzymatic regulation in the cell, raising the possibility that this apparent discrepancy reflects our lack of understanding rather than absence of a regulatory mechanism.
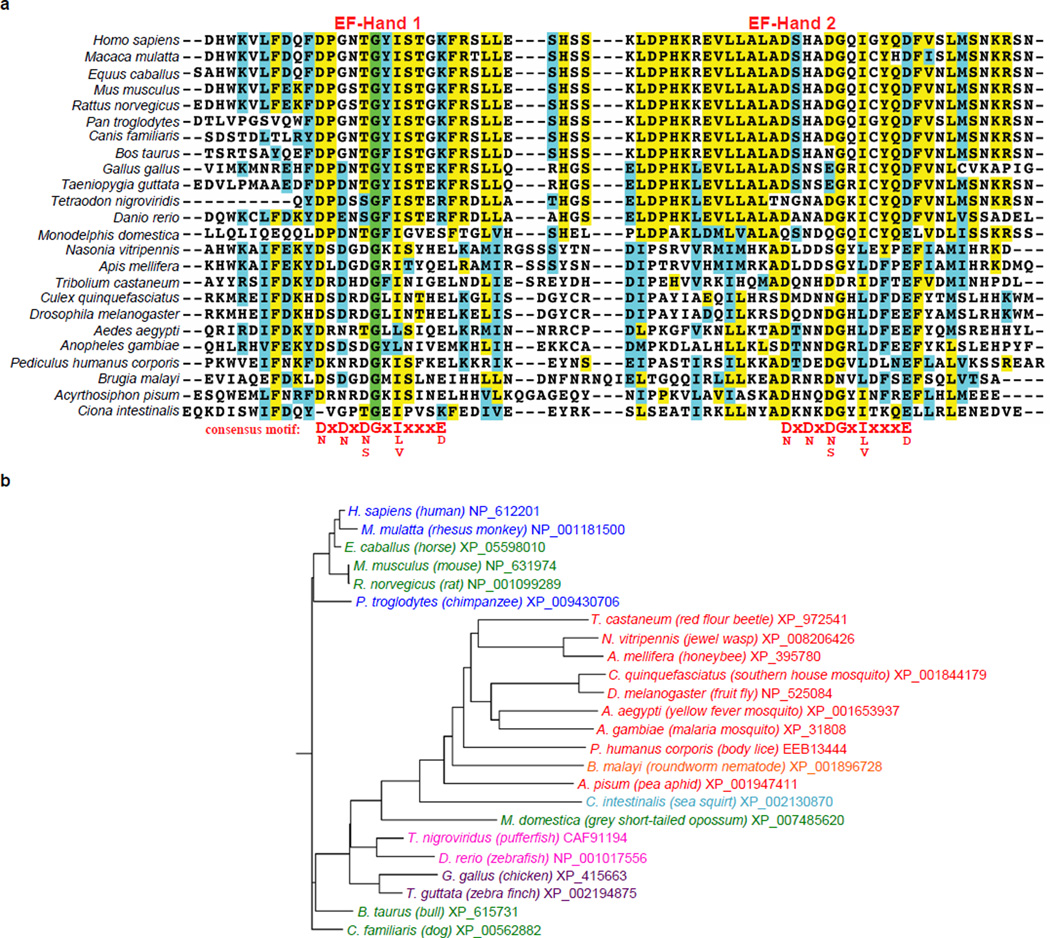
Although Escherichia coli rhomboid protease GlpG has served as a tractable model for studying the structure-function of intramembrane proteolysis12, no information is available on its cellular role. This knowledge gap prohibits deciphering regulatory mechanisms. Instead, as a new approach to this question, we searched for rhomboid proteins that contain additional domains with precedent for regulating protein activity and focused on a conserved subset of over two dozen animal rhomboid enzymes with EF-hand domains appended to their cytosolic N-termini (Fig. 1a and Extended Data Fig. 1). EF-hands are helix-loop-helix motifs in which calcium binding at the loop serves either a structural or regulatory role. In the latter, calcium binding separates the helices and exposes a new surface for binding a regulatory partner13. EF-hands typically occur in pairs to form a stable helical bundle, perhaps the best characterized of which are the EF-hands of calmodulin13.
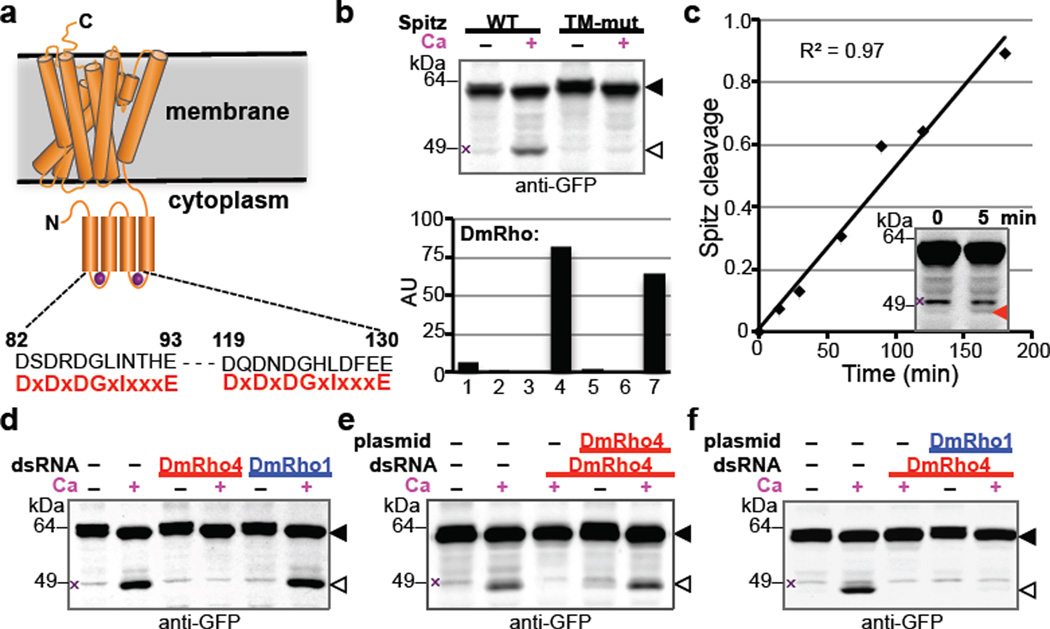
Figure 1. Calcium rapidly stimulates intramembrane proteolysis in Drosophila cells by endogenous DmRho4.
a Diagram comparing the predicted calcium-binding loop residues of DmRho4 to an EF-hand consensus (in red). b Calcium ionophore treatment of Drosophila S2R+ cells induced cleavage of GFP-Spitz, but not its cleavage-site mutant, by endogenous DmRho4. Graph shows expression levels of Drosophila rhomboid genes in S2R+ cells (RNAseq data from modENCODE, modencode.org). c Ionophore-induced Spitz cleavage was detectable within 5 min (red triangle) and linear for 3h. d RNAi knockdown of DmRho4 but not of DmRho1 abrogated calcium-induced cleavage of GFP-Spitz. e Plasmid expression of DmRho4 rescued calcium-induced cleavage of GFP-Spitz in S2R+ cells undergoing RNAi. f Calcium-stimulated Spitz cleavage abolished by DmRho4 RNAi could not be rescued by DmRho1 overexpression. All images are anti-GFP western analyses, with substrate and cleavage bands denoted by black or open triangles, respectively, and non-specific bands marked by × (see Fig. 3d for untransfected cells).
Since rhomboid function is best understood in Drosophila, we sought to study the EF-hand containing Drosophila rhomboid-4 (DmRho4) under physiological conditions by searching for cell lines that endogenously express DmRho4. We focused on the well-characterized S2R+ cell line, which also expresses the housekeeping mitochondrial rhomboid and low amounts of DmRho1, but no other rhomboid. Treating S2R+ cells with a calcium ionophore potently stimulated processing of the EGF ligand Spitz by >50 fold, but not its transmembrane mutant (Fig. 1b), and processing was rapid, becoming detectable within 5 minutes (Fig. 1c). Targeting DmRho4 with RNAi removed this stimulation completely, while parallel RNAi against DmRho1 had no effect whatsoever (Fig. 1d). Importantly, exogenous expression of DmRho4 lacking the RNAi target sequence fully rescued calcium stimulation (Fig. 1e). Finally, calcium activated DmRho4 proteolysis directly and not other steps such as Spitz trafficking, because even high levels of DmRho1 could not substitute for removing DmRho4 during calcium stimulation (Fig. 1f). Calcium therefore triggers potent and rapid proteolysis by DmRho4 under physiological conditions.
Our goal was to study direct regulation of rhomboid enzymes. We therefore next tested the unlikely possibility that calcium directly regulates pure DmRho4 expressed and purified from bacteria and reconstituted into liposomes. Remarkably, addition of calcium directly stimulated intramembrane proteolysis of DmRho4 by >10-fold, but not a panel of other rhomboid proteases, without any other protein factors present (Fig. 2a). This stimulation was selective since other divalent metal ions failed to have this effect.
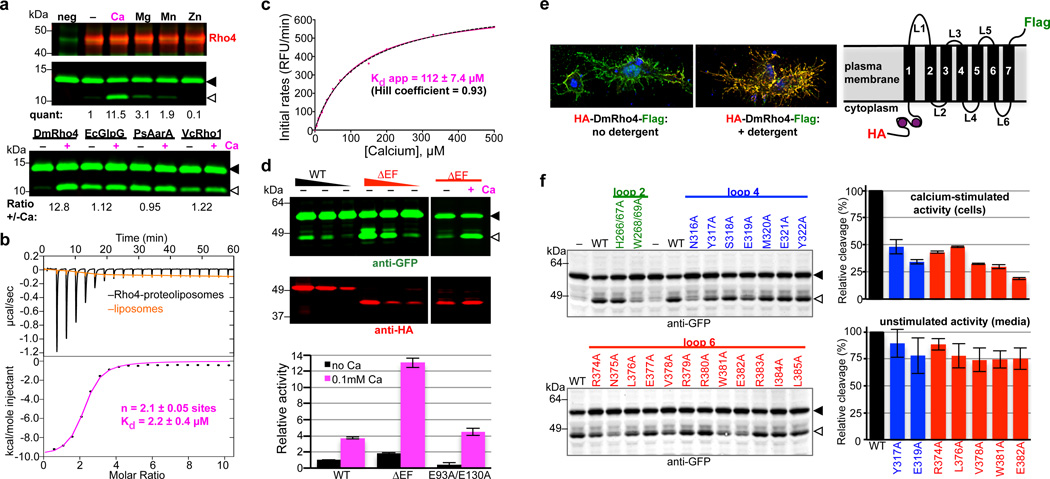
Figure 2. Calcium directly regulates intramembrane proteolytic activity of DmRho4.
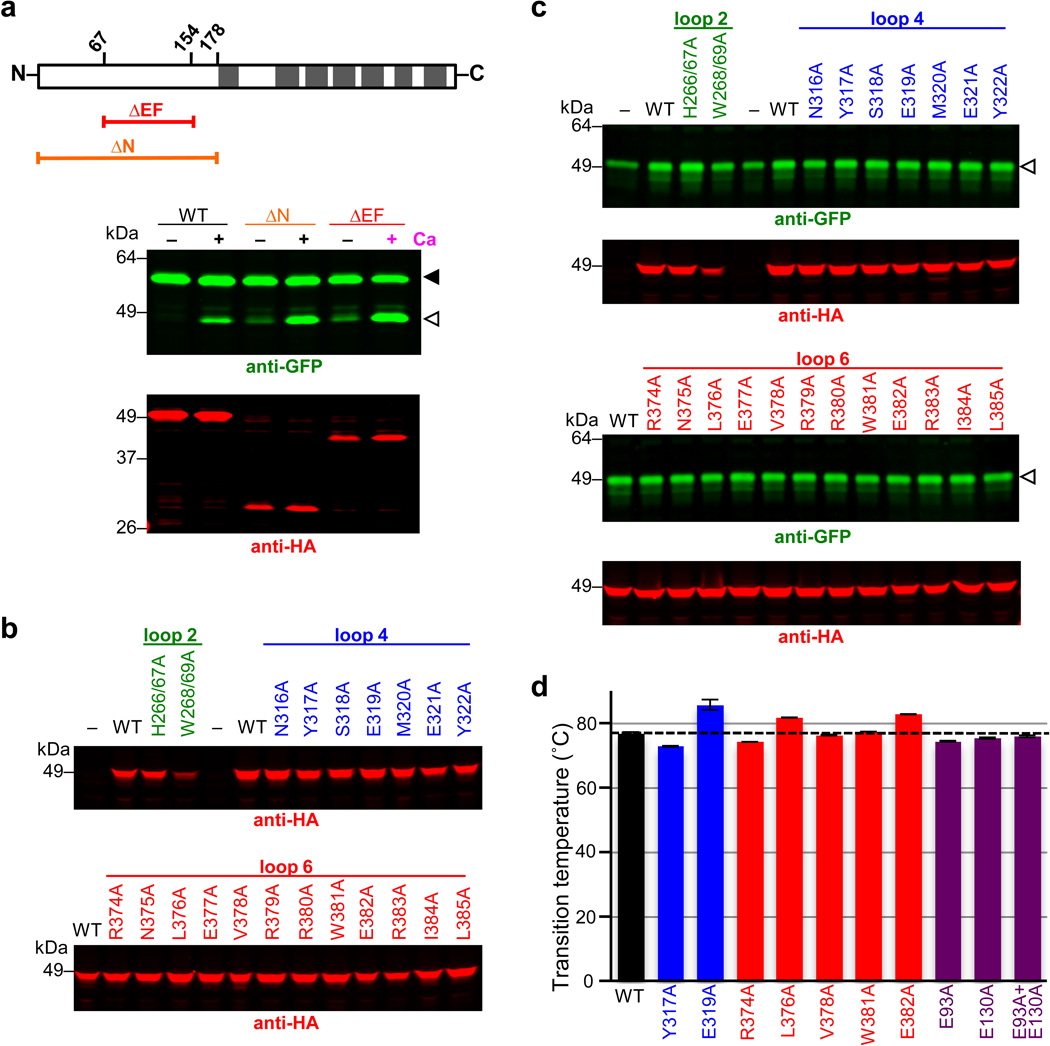
a Proteolysis assay with pure reconstituted DmRho4 ± a panel of 1mM divalent metal ions (upper panels), and four different rhomboid enzymes reconstituted into proteoliposomes ± calcium (lower panel). EcGlpG is from Escherichia coli, PsAarA is from Providencia stuartii, and VcRho1 is from Vibrio cholerae. b Analysis of calcium binding to DmRho4 in proteoliposomes by isothermal titration calorimetry (upper graph shows the thermograms, lower graph is the liposome-subtracted quantification). c Calcium titration analysis of DmRho4 proteolysis using an inducible real-time reconstitution assay16. Black dashed line shows an alternate fit with an optimal Hill coefficient. d Titration of wildtype (WT) and ∆EF DmRho4 in S2R+ cells comparing basal, unstimulated Spitz cleavage (left panel) and Spitz cleavage by DmRho4-∆EF ± calcium ionophore (right panel; also see Extended Data Set 2a). Lower graph shows in vitro activity of WT and EF-Hand mutants of DmRho4 in proteoliposomes (error bars indicate standard deviation for experimental replicates). e Topology of 3xHA-DmRho4-Flag in S2R+ cells as assessed by deconvolution immunofluorescence. The N-terminal HA-tag (red) was inaccessible while the C-terminal Flag tag (green) was accessible in the absence of detergent, indicating that the N-terminus is cytosolic while the C-terminus of DmRho4 is extracellular (blue marks nuclei). f Ability of DmRho4 cytosolic loop mutants to cleave GFP-Spitz in response to calcium ionophore stimulation in S2R+ cells was quantified by anti-GFP western analysis (also see Extended Data Set 2b for DmRho4 levels). Graphs show activity of selectively compromised loop 4 and 6 mutants under calcium-stimulated conditions in cells (upper graph) versus unstimulated conditions (lower graph, measured as cleavage product accumulation in culture media after 24 hours, also see Extended Data Fig. 2c). Error bars indicate standard deviation for experimental replicates. Black triangles and open triangles denote substrate and cleavage bands, respectively, throughout.
We used isothermal titration calorimetry (ITC) to characterize the thermodynamic basis of calcium binding to the EF hand domain (Fig. 2b). The resulting thermograms revealed two sites for calcium binding with similar Kds of 2.1 ± 0.05 µM. Interestingly, a calcium titration revealed stimulation of DmRho4 protease activity (Fig. 2c) could be fit with a binding isotherm with an apparent Kd of 112 ± 7.4 µM, implying that calcium binding to at least a third, lower affinity site (beyond detection by ITC) is required for enzyme activation. To assess this further, we deleted the EF domain entirely (ΔEF), and found DmRho4 became dysregulated in cells, with its basal activity elevated by ~10-fold (Fig. 2d). This effect was direct because pure DmRho4ΔEF also displayed elevated activity in vitro. However, calcium still stimulated proteolysis of the E93A+E130A mutant that abrogates binding of calcium to EF-hands, DmRho4ΔEF, and DmRho4ΔN missing its entire N-terminus (Fig. 2d and Extended Data Set 2a). These observations further indicate that another calcium-binding site outside its N-terminal domain is the basis of DmRho4 stimulation, while the EF-hand domain functions to limit activation.
To begin mapping the calcium-binding site(s) responsible for stimulating proteolysis, we first examined the topology of DmRho4 in S2R+ cells (Fig. 2e). An antibody accessibility approach revealed the N-terminus of DmRho4 resides in the cytosol while the C-terminus is extracellular, indicating that loops 2, 4 and 6 are cytosolic and thus candidates for calcium binding. We mutated all 24 residues in these three cytosolic loops to alanine and assessed their activity in Drosophila cells (Fig. 2f and Extended Data Fig. 2b, c). The mutants fell into 3 classes: most did not affect activity, three perturbed both stimulated and unstimulated activity and were therefore likely structural mutants, and seven selectively compromised calcium-stimulated proteolysis but not DmRho4 structure (Extended Data Fig. 2d), as revealed in a sensitive thermostability assay14. The latter localized to loops 4 and 6, indicating that calcium binding at a site formed by loops 4 and 6 specifically activates the intramembrane enzyme core, but how?
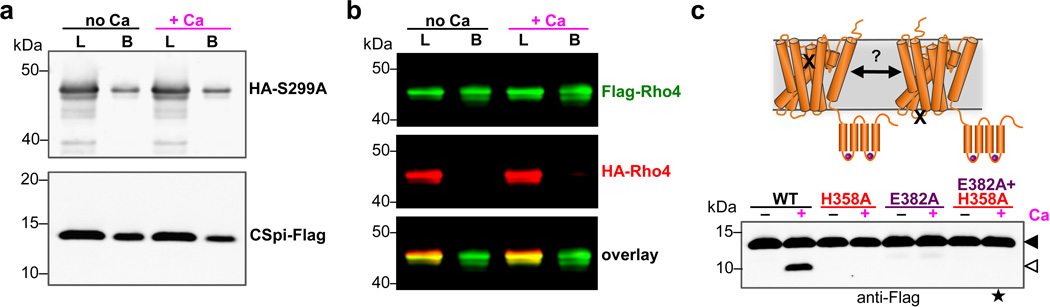
We used complementary biochemical and enzymatic approaches to decipher the mechanism of calcium stimulation. Although calcium binding could expose an exosite for substrate binding15, we did not detect any increased interaction between DmRho4 and substrate in the presence of calcium by coimmunoprecipitation analysis from either Drosophila cells (Fig. 3a) or proteoliposomes (Extended Data Fig. 3a). This was also functionally corroborated by kinetic analysis using an inducible real-time reconstitution assay for studying catalysis directly within the membrane16; calcium stimulated the catalytic rate of DmRho4 at least 6-fold, but did not increase affinity of protease for substrate (Fig. 3b).
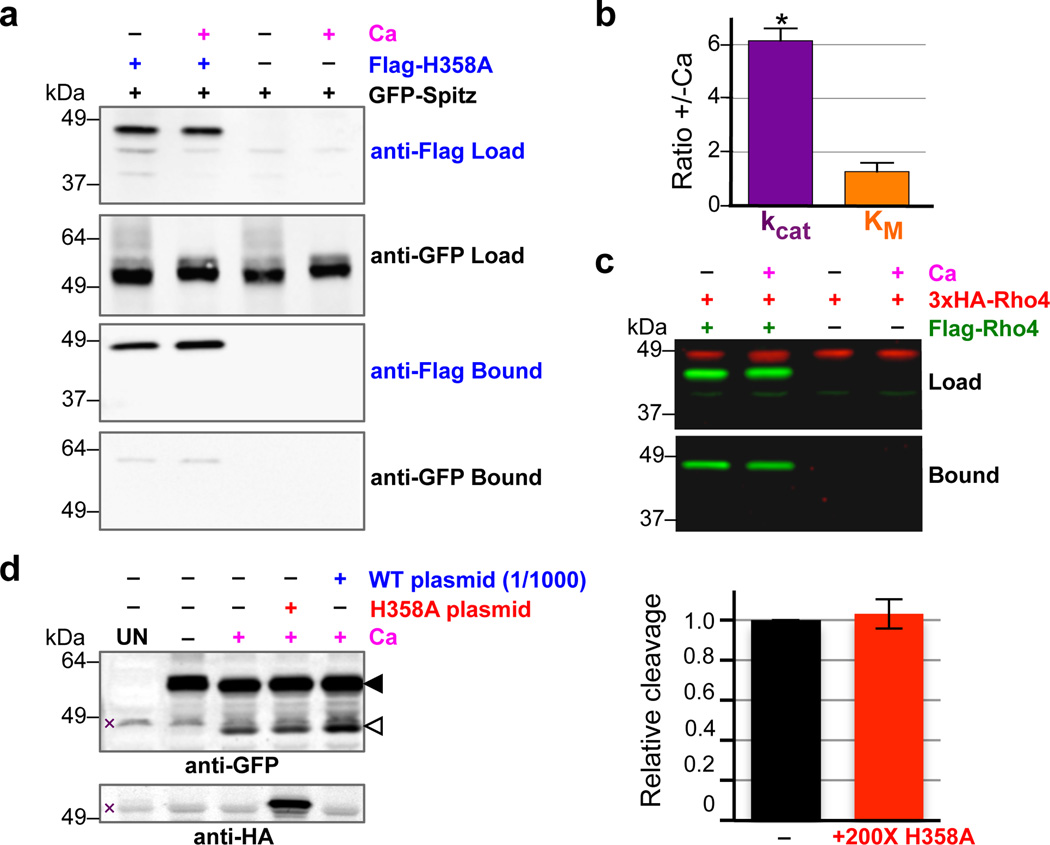
Figure 3. Intermolecular interactions do not mediate DmRho4 regulation by calcium.
a Anti-Flag coimmunoprecipitation analysis of catalytically inactive Flag-DmRho4-H358A and GFP-Spitz from S2R+ cells untreated or treated with calcium ionophore. b Effect of calcium on the steady-state kinetic parameters of intramembrane proteolysis by reconstituted DmRho4 (mean ± standard deviation of 3 independent experiments, compared using a paired t-test). c Anti-Flag coimmunoprecipitation of Flag-DmRho4 and HA-DmRho4 co-expressed in S2R+ cells untreated or treated with calcium ionophore. d Overexpressing catalytically-inactive DmRho4 (H358A) had no effect on the calcium-stimulated activity of endogenous DmRho4 in S2R+ cells (compare cleavage bands, denoted by open triangle, in lanes 3 versus 4, and quantified in graph on right). Expressing low levels of wildtype 3xHA-DmRho4 (1/1,000 amount of input plasmid) was used to quantify the level of endogenous DmRho4 (by comparing protease activity) relative to 3xHA-DmRho4-H358A expression (by comparing anti-HA signals). UN indicates S2R+ cells not transfected with GFP-Spitz (x denotes non-specific bands).
Recently rhomboid proteases have been postulated to exist as dimers17. In contrast to this model, co-expressing two DmRho4 molecules carrying different epitope tags under calcium signaling conditions in Drosophila cells (Fig. 3c) or co-reconstituted into proteoliposomes (Extended Data Fig. 3b) did not result in dimerization. Functionally, we also did not observe calcium stimulation of protease activity in trans, which is a classical test of allostery resulting from oligomerization: a catalytically-inactive rhomboid enzyme that can still bind calcium could not stimulate the activity of a DmRho4 enzyme carrying a mutation that compromised calcium binding (Extended Data Fig. 3c).
These observations collectively suggest that DmRho4 is not regulated by interaction with any other proteins. To test this further in Drosophila cells, we overexpressed inactive DmRho4 to outcompete the endogenous enzyme for any binding partners (Fig. 3d). Remarkably, ~200-fold more inactive DmRho4 had no effect whatsoever on the ability of the endogenous DmRho4 to process Spitz. This observation, in particular, indicates that rhomboid regulation under physiological conditions is not mediated by dimerization, substrate affinity, or additional factors (although factors that fine tune responses in different contexts remain possible).
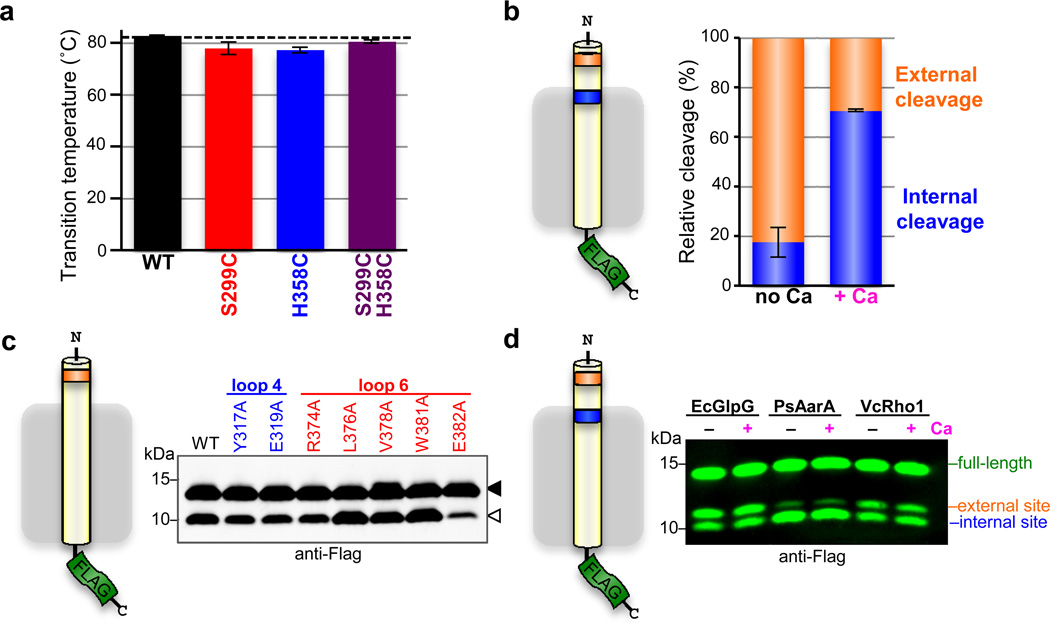
In contrast to intermolecular target binding, calcium could directly stimulate the activity of a single DmRho4 enzyme through intramolecular allostery. Calpains are the precedent for this type of activation, with calcium binding resulting in a conformation change that aligns the catalytic residues18. Since structure of a eukaryotic rhomboid enzyme has never been solved, we used a biochemical cross-linking approach to test whether calcium aligns the catalytic residues of DmRho4. Cysteines installed at the catalytic serine and histidine positions had no effect on the structural stability of DmRho4 (Extended Data Fig. 4a) and could readily and reversibly be oxidized to form a disulfide bridge (Fig. 4a). Importantly, calcium did not affect the amount of cross-linking, revealing that the DmRho4 catalytic residues are pre-aligned with no influence from calcium binding.
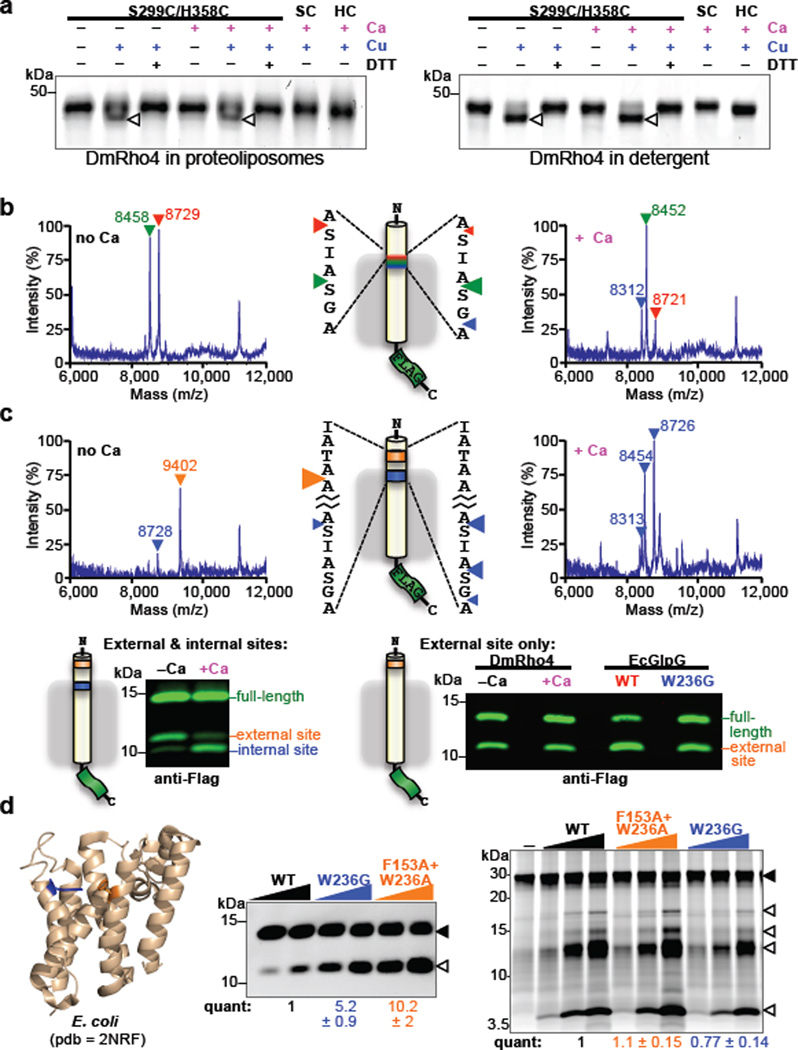
Figure 4. Regulation of intramembrane proteolysis by lateral substrate gating.
a Calcium did not enhance disulfide crosslinking of cysteines installed at the catalytic serine and histidine positions of DmRho4 (triangles indicate crosslinked products). b Shift in substrate cleavage site generated by reconstituted DmRho4 ± calcium as analyzed by mass spectrometry following anti-Flag immunoaffinity capture. Masses and triangles indicating cleavage sites are color-matched. c Processing of a substrate carrying intramembrane (blue) and external (orange) cleavage sites in APP-Flag that was co-reconstituted into liposomes with DmRho4, and assayed ± calcium. Reactions were analyzed by mass spectrometry (top) and quantitative western blotting (lower panels). Processing of a substrate carrying only the external cleavage site was compared for DmRho4 ± calcium, and for EcGlpG versus its gate-open mutant W236G. d Quantitative cleavage analysis of wild type and gate-open mutants of EcGlpG for the APP-Spi7-Flag transmembrane substrate and a soluble BODIPY-casein substrate. Full-length substrate (solid triangles) and cleavage products (open triangles) are indicated.
Stimulation of the catalytic rate constant kcat by calcium was strikingly reminiscent of the increase in kcat we measured for gate-open mutants of GlpG16. This is an attractive parallel, because gate-opening is the rate-limiting step for rhomboid intramembrane proteolysis, and the loops that we predict bind calcium also connect a transmembrane gating helix to the rest of the enzyme. One consequence of gate-opening is that substrates can enter deeper into the protease active site, which is reflected in a shift of cleavage site deeper into the substrate transmembrane segment19. Similarly, calcium-stimulated proteolysis shifted the cleavage site 3 residues deeper into the transmembrane segment for DmRho4 but not other rhomboid proteases, consistent with calcium specifically stimulating DmRho4 proteolysis by facilitating gate-opening (Fig. 4b).
To explore the functional consequence of this shift, we examined proteolysis of a series of transmembrane substrates that we engineered to have cleavage sites inside the membrane, outside the membrane, or both in the same molecule. Remarkably, the external site was used very well in the absence of calcium, while the intramembrane site was barely cleaved (Fig. 4c and Extended Data Fig. 4b). Moreover, loop 4 and 6 mutants that compromised calcium regulation in cells also readily cleaved the external site (Extended Data Fig. 4c), which independently confirms that the catalytic residues are competent for catalysis in the absence of calcium. However, addition of calcium shifted the cleavage site from the external site almost exclusively to the intramembrane site for DmRho4 while having no effect on the cleavage site selection of other rhomboid enzymes (Fig. 4c and Extended Data Fig. 4d).
Although no information is available on how E. coli GlpG is regulated, we also extended these analyses to this widely-studied enzyme by comparing wildtype versus gate-open mutants20. Both wildtype and gate-open GlpG cleaved the external site with similar efficiency (Fig. 4c), while proteolysis at the intramembrane site was specifically stimulated by gate-open mutants (Fig. 4d). Moreover, although gate-open mutants of E. coli GlpG stimulated proteolysis of transmembrane substrates by ~10-fold (Fig. 4d), a soluble casein substrate that approaches the active site from above (not laterally from the membrane) was not proteolyzed at any higher level (Fig. 4d). Therefore, cleavage outside the membrane by rhomboid proteases is readily possible, but this type of cleavage is difficult to regulate. A similar analysis of DmRho4 will require identifying gate-open mutants, which should no longer respond to calcium. Lack of a DmRho4 structure and <12% sequence identity with GlpG has hindered this approach.
In summary, we discovered that calcium binding directly, rapidly, and potently stimulates DmRho4 intramembrane proteolysis by triggering gate-opening. In addition to revealing, to our knowledge, the first mechanism for directly regulating any intramembrane protease, these observations resolve the long-standing mystery of why these proteases gate substrate entry. Gating has been controversial, because rhomboid proteases can be made to process substrates without lateral gate opening21, and cleavage can be moved to external sites in substrates22. In doing so, it was questioned why intramembrane gate-opening occurs, if at all. Our observations now reveal that intramembrane substrate gating is a means of enzyme regulation, not a necessary step in catalysis: cleavage outside the membrane is possible, but cannot be tightly regulated directly.
The ability of cytosolic regions to regulate substrate gating at a distant, intramembrane site also suggests deeper organizational features within rhomboid architecture that are only beginning to be studied23. Interestingly, the ‘non-canonical’ mode of calcium binding to DmRho4 loops is reminiscent of synaptotagmin activation, which also involves loops that are placed into close contact with lipid molecules. In fact, rhomboid activation may also involve lipids, which might explain the apparent high calcium levels needed for full enzyme activation in vitro: synaptotagmin exhibits an intrinsic Kd of 530 µM for calcium at the C2 site that decreases to 3–4 µM when appropriate lipids are provided24. Although ultimately structural analysis is required to reveal the precise architecture of calcium binding, likely involvement of lipid, and impact on gating in DmRho4, so far no rhomboid enzyme with an intact extramembraneous domain has produced well-diffracting crystals. Our studies provide incentive to move beyond GlpG and focus structural biology efforts on these more complex rhomboid proteases.
A particularly exciting implication of these enzymatic properties is that rhomboid proteases can directly integrate upstream signals from other signaling pathways. In fact, this may have medical implications, since Ventrhoid/RHBDL3, a human rhomboid that contains potential calcium-binding residues in its EF-hands and cytoplasmic loops, is expressed in the nervous system25 and may be linked to a mental retardation syndrome26. In this light, studying upstream regulation provides a powerful new approach towards revealing biological functions of rhomboid proteases that have evaded discovery. In fact, prior efforts could have missed important roles because they were studying rhomboid functions under unstimulated conditions. Activation is not limited to calcium signaling, since a diversity of recognizable domains have been appended to different rhomboid proteins including Zn fingers, β-propellers, and tetracopeptide repeats27. It should be noted that not all extramembraneous domains necessarily serve direct regulatory functions. For example, trafficking signals have been found in the cytosolic domains of parasitic rhomboid enzymes28.
Finally, while we focused our studies on rhomboid proteases, substrate gating has been proposed for other intramembrane proteases29,30, raising the possibility that gating may be a general mechanism for directly regulating intramembrane proteolysis.
ONLINE METHODS
Rhomboid Expression Constructs
The Drosophila rhomboid-4 open reading frame (ORF) was subcloned from SD06923 (Drosophila Genomics Research Center) into pGEX-6P-1 or pET21 for bacterial expression, or into pRmHa3 for expression in Drosophila S2R+ cells. An N-terminal 3xHA-tag was introduced to allow detection of DmRho4 by anti-HA western analysis. Residue substitutions were introduced by site-directed mutagenesis using Pfu Ultra in a Stratagene 96 Gradient Robocycler (Agilent Technologies) and were confirmed by sequencing the entire ORF. The EF-hand domain of DmRho4 (residues 68–154) or its entire N-terminal domain (residues 1–176) were deleted by site-directed mutagenesis to generate the ∆EF and ∆N mutants, respectively.
Drosophila Cell Culture and Manipulation
Drosophila S2R+ cells (purchased from the Drosophila Genomics Research Center) were cultured at 25°C in Schneider’s insect medium (Invitrogen) supplemented with 10% fetal bovine serum (FBS, Sigma). For transfection experiments, S2R+ cells were seeded into 6-well plates and transfected with a total of ~2 µg of plasmid DNA (0.5 µg each of pRmHa3-GFP-Spitz and pRmHa3-Star plus 1 µg pBluescript, and 5–25 ng of pRmHa3-3xHA-DmRho4 if applicable) and 5 µl of XtremeGENE HP (Roche). Transfection complexes were formed at room temperature in 100 µl of DMEM for 15 minutes before being added dropwise to cells. Expression from the metallothionine promoter was induced the following day with 0.5 mM CuSO4 for ~24 hours. For ionophore experiments, cells were then washed with Insect Saline (10 mM HEPES pH 7.3, 120 mM NaCl, 5 mM KCl, 32 mM sucrose, 8mM MgCl2, 2 mM CaCl2), and incubated with 6 µM ionomycin (Sigma) in Insect Saline for 2–3 hours, unless otherwise indicated. Cells were lysed in reducing Laemmli buffer, the lysates resolved on 4–20% tris-glycine SDS polyacrylamide gels, and electrotransferred to nitrocellulose. GFP-Spitz (or its transmembrane mutant SGA→LLL) and 3xHA-DmRho4 were detected by anti-GFP and anti-HA western analysis, respectively, and fluorescence quantified with an Odyssey infrared laser scanner (LiCor Biosciences). The protein level of endogenous DmRho4 in S2R+ cells was estimated by titrating wildtype 3xHA-DmRho4 by transfection: matching the level of transfected 3xHA-DmRho4 to the endogenous DmRho4 expression level resulted in a doubling of GFP-Spitz processing (Fig. 3d). To detect basal (unstimulated) DmRho4 activity, Schneider’s serum-free media was conditioned for 24 hours, and the GFP-Spitz that had been released by proteolysis was quantified by anti-GFP western analysis of media fractions.
Drosophila Cell RNA Interference
Templates for in vitro transcription were amplified by PCR with primers corresponding to the divergent N-terminal regions of DmRho1 (ORF nucleotides 1–451) and DmRho4 (ORF nucleotides 1–501) and incorporated T7 promoter sequences. RNA was generated using the RiboMAX T7 kit (Promega) according to manufacturer’s instructions, and purified using the RNeasy protocol (Qiagen). dsRNA was formed by mixing equal amounts of each strand in annealing buffer (1 mM HEPES pH 7.3, 0.5 mM EDTA) and boiling for 5 minutes, followed by slow cooling. dsRNA was analyzed by agarose gel electrophoresis. S2R+ cells seeded in 6-well plates were washed with serum-free Schneider’s media, and ~25 µg of dsRNA was added to each well containing 1mL of serum-free Schneider’s insect media. After a 1–2 hour incubation, 3 mL of Schneider’s media + 10% FBS was added, and the cells were incubated at 25°C for 3 days. On the third day, cells were assayed by transfection as described above.
Drosophila Cell Microscopy
S2R+ cells were seeded onto glass coverslips, transfected with 3xHA-DmRho4-1xFlag (which was verified to be proteolytically active and calcium regulated), induced with 0.5 mM CuSO4 for ~24 hours, and fixed in 4 % formaldehyde in PBS for 20 minutes. Cells were blocked with 1% bovine serum albumin in the presence or absence of 0.1 % TritonX-100, and stained with 1/200 anti-HA and anti-Flag antibodies overnight. The resulting immune complexes were detected with 1/500 secondary antibodies conjugated to Alexa-488 or Alexa-594 (Molecular Probes), mounted in the presence of DAPI, and imaged on a DeltaVision Elite deconvolution microscope (GE Healthcare) using 0.2 µm optical sections with a 1.4 NA 100× objective.
Recombinant Protein Production
Wild-type or engineered variants of DmRho4 were expressed as N-terminal GST- or His-tag fusions in E. coli BL21(DE3) cells. Cultures were grown in M9+ minimal media containing 100 µg/mL ampicillin at 37°C, shaking at 250 rpm. When cultures reached an OD600 of 0.5, protein expression was induced with the addition of 50 µM IPTG for 16–18h at 16°C. Bacterial cell lysates were prepared using a French pressure apparatus operating at 10,000 psi and cell membranes were pelleted by ultracentrifugation at 350,000 × g for 30 min. DmRho4 was solubilized from membranes in 2% dodecyl-β-D-maltoside (DDM) for 1h at 4°C, followed by ultracentrifugation at 350,000 × g for 30 min to remove insoluble material. GST-DmRho4 was affinity-purified on glutathione-sepharose and eluted by PreScission protease (GE Healthcare) cleavage of the GST-tag at 4°C overnight, while His-Rho4 was affinity-purified with Ni-NTA (Qiagen) or His-tag resin (Roche) and eluted using imidazole as recommended by the manufacturers. The N-terminal His-tag was removed by thrombin cleavage at 4°C overnight. Bacterial rhomboid proteases were expressed as N-terminal GST fusion proteins and purified by glutathione-sepharose affinity as described above. The APP-Spi7-Flag substrate was produced as described previously14,31,32. Briefly, expression of APP-Spi7-Flag was induced for 2–3h at 37°C in BL21(DE3) cells with 1 mM IPTG. Cells were lysed using a French pressure apparatus and lysates were cleared of cellular debris by centrifugation at 3,000 × g for 10 min. The substrate was affinity purified using anti-Flag M2 agarose (Sigma). A variant of APP-Spi7-Flag with an additional rhomboid cleavage site outside the transmembrane domain was constructed by inverse PCR to substitute residues 20–25 (FFAEDV) of APP-Spi7-Flag with the sequence IATAAF from Providencia stuartii TatA. APP-TatA-Flag had only the external TatA site.
Intramembrane Proteolysis Assays
Rhomboid proteases were co-reconstituted with substrates into liposomes using the inducible reconstitution system that we described recently16. Bacterial rhomboid proteases were reconstituted in liposomes formed from an E. coli polar lipid extract, while DmRho4 was reconstituted in liposomes formed from a yeast polar lipid extract or 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine lipids (Avanti Polar Lipids). All enzymes were assayed for 1 hour at 37°C, except DmRho4, which was assayed at 25°C for 2–4 hours. APP+Spi7-Flag reaction products were resolved by SDS-PAGE and quantified by anti-Flag western analysis using an Odyssey infrared laser scanner (LiCor Biosciences), while products for the real-time assay using FITC-TatA were quantified using a Synergy H4 Hybrid plate reader (Biotek) scanning once per minute. Proteolysis assays were supplemented with 0.5 mM calcium unless otherwise indicated. For calcium titration experiments, 10 pmol of DmRho4 and 200 pmol FITC-TatA were co-reconstituted into 30 µg yeast liposomes and total calcium was titrated from 0 to 1 mM. For kinetic analysis, 10 pmol of Rho4 was titrated against 15-600 pmol FITC-TatA substrate in the presence or absence of 0.5 mM calcium, initial reaction rates were extracted and fit to a Michaelis-Menten model using Prism software.
Casein Cleavage Assay
Wild type GlpG and engineered variants were assayed in reactions consisting of 6, 12, or 24 pmol enzyme and 50 µg/mL BODIPY FL casein (Life Technologies) in 50 mM Tris pH 7.4, 150 mM NaCl and 0.1% DDM for 1h at 37°C. Reactions were quenched by adding an equal volume of 2X Tricine sample buffer, resolved by SDS-PAGE on 16% Tricine gels (Life Technologies) and imaged using a Typhoon Imager (GE Healthcare) with settings of 488 nm for excitation and 526 nm for emission.
Isothermal Titration Calorimetry
Isothermal titration calorimetry was performed using a Microcal iTC200 instrument (GE Healthcare). The reaction cell contained 20–30 µM DmRho4 in proteoliposomes and the reference cell contained water. A motorized syringe loaded with 1 mM calcium was used to perform successive 2 µL injections into the reaction cell at 25°C. Control experiments titrating calcium against liposomes were performed to determine the heat of titrant dilution, which subtracted from the heat of reaction yielded the effective heat of calcium binding. Data were fit using Origin analysis software.
Thermostability Analysis
Wild-type and engineered variants of DmRho4 were subjected to quantitative thermostability analysis as described previously14. Briefly, pure DmRho4 was diluted to 5µM in 50mM Tris pH 7.4, 150 mM NaCl, 0.1% dodecyl-β-D-maltoside, 0.5% or 1.2% nonyl-glucoside, heated from 25°C to 85°C at a rate of 0.2°C per minute, and differential static light scattering was quantified every 0.5°C in a Stargazer-384 instrument (Harbinger Biotech, Toronto, Canada). Light scattering data were fit to a two-state Boltzmann curve using Stargazer BioActive software to derive transition temperature midpoints (Tm).
Rhomboid-Substrate Co-Immunoprecipitation
S2R+ cells were co-transfected with catalytically inactive pRmHa3-Flag-DmRho4-H358A and pRmHa3-GFP-Spitz or, as a negative control, with pRmHa3-GFP-Spitz alone, and were either untreated or treated with 6 µM ionomycin in Insect Saline or Ca-free Insect Saline, as described above. Cell lysates were solubilised for 1h at room temperature (RT) in 25mM Tris pH 7.4, 150mM MgCl2, Complete EDTA-free protease inhibitor cocktail (Roche), 0.25% DDM in either the presence of 1mM CaCl2 or 1mM MgCl2. Cell debris was removed by centrifugation at 16,000 × g for 20 min at 4°C. Immunoprecipitations were carried out using anti-Flag M2 agarose for 1h at RT. Beads washed in the presence of 1mM CaCl2 or 1mM MgCl2 were resuspended in SDS sample buffer, then load and bound fractions were resolved by SDS-PAGE followed by anti-Flag/anti-GFP western analysis. For co-immunoprecipitation analysis in proteoliposmes, catalytically inactive HA-tagged DmRho4 (S299A) was co-reconstituted with APP-Spi7-Flag as described above in buffer consisting of 50 mM Tris pH 7.5, 150mM NaCl, and either no calcium or 0.5 mM CaCl2. Proteoliposomes were incubated for 1h at RT, solubilised with 1% DDM for 30 min at RT, and then immunoprecipitation was performed with anti-Flag M2 agarose for 1h at RT in the presence or absence of calcium. Washed beads were resuspended in SDS sample buffer and the input and bound fractions were compared by anti-HA and anti-Flag western analysis.
Rhomboid Co-Immunoprecipitation
S2R+ cells were co-transfected with expression constructs encoding triple HA-tagged and Flag-tagged DmRho4, and as a control, with HA-tagged DmRho4 alone as described above, and were then either untreated or treated with 6 µM ionomycin in Insect Saline or Ca-free Insect Saline. Cells were solubilised with 0.25% DDM (as described above) and immunoprecipitations were performed with anti-Flag M2 agarose for 1h at RT. Beads washed in the presence of 1mM CaCl2or 1mM MgCl2 were resuspended in SDS sample buffer. Load and bound fractions were detected by anti-HA and anti-Flag western analysis. Pure samples of HA-tagged and Flag-tagged DmRho4 were also co-reconstituted into proteoliposomes in the absence or presence of 0.5mM CaCl2, then solubilised with 1% DDM for 30 min at RT, immunoprecipitated with anti-Flag M2 agarose (as described above) and then subjected to anti-HA/anti-Flag western analysis.
Disulfide Cross-linking
A cysteine-less mutant of DmRho4 was generated by substituting the three amino-terminal native cysteine residues with serine residues (C104S, C150S, C176S) and the native transmembrane cysteine residue with a valine residue (C334V). Using this construct as a template, the active site residues S299 and H358 were replaced with cysteine residues, either singly, to generate S299C and H358C, or in combination, to generate the double mutant S299C/H358C. Pure cysteine-substituted proteins were reduced by treatment with 5mM TCEP for 30 min at RT and then passed through a Zeba spin column (Pierce) equilibrated in 50 mM Tris, 150 mM NaCl, 0.1% DDM. Proteins were then oxidized, either in detergent micelles or after reconstitution into proteoliposomes, by the addition of 50 or 100 µM copper phenanthroline, respectively, for 15 min at RT in the absence or presence of 0.5 mM calcium. Note that due to the random orientation of rhomboid in reconstituted proteoliposomes, only half of the DmRho4 was accessible to the oxidizing reagent. Control reactions were carried out in parallel with no copper but with 50 mM DTT to prevent spontaneous oxidation. Reactions were stopped by the addition of SDS-sample buffer. As an additional control the oxidation observed for the double mutants was reversed by the addition of 50 mM DTT for 10 min to the oxidized proteins in SDS sample buffer. Samples were analysed by SDS-PAGE and stained using either IRDye Blue Protein Stain (LiCor) or Krypton Infrared Protein Stain (Pierce).
Mass Spectrometry
Full-length substrate and C-terminal cleavage products were purified from in vitro proteolysis assays by anti-Flag immunoaffinity isolation and analyzed by MALDI-TOF mass spectrometry using sinapinic acid matrix as described previously19,32.
Extended Data
Extended Data Figure 1. The Rhomboid-4 subfamily of rhomboid proteases.
a ClustalW multiple sequence alignment of the conserved amino-terminal EF-Hand domains of 24 members of the Rhomboid-4 subfamily (generated in Biology Workbench, http://workbench.sdsc.edu). Identical residues are shaded in green, highly conserved residues in yellow and similar residues in cyan. The EF-hand calcium-binding loop consensus sequence is given below the alignment. b Rooted tree of the 24 Rhomboid-4 homologues. Genus species names are color-coded as follows: primates (blue), other mammals (green), birds (purple), fish (pink), non-vertebrate chordate (cyan), nematode (orange), and insects (red), with common names given in parenthesis, followed by NCBI accession numbers.
Extended Data Figure 2. Activity and thermostability analysis of DmRho4 mutants.
a Comparison of calcium stimulation of DmRho4 versus its EF hand domain deletion mutant (ΔEF), and a mutant lacking the entire cytosolic domain (ΔN). Upper diagram shows position of domains (demarked by residue numbers) and the corresponding deletion constructs. Transmembrane segments are shown as grey rectangles. GFP-Spitz substrate and cleavage products (green bands in the anti-GFP western) are denoted by black and white triangles, respectively. DmRho4 protein levels are shown as red bands (anti-HA western). b Analysis of DmRho4 loop 2, 4, and 6 mutant protein levels from Fig. 2f (calcium stimulation conditions). c DmRho4 loop 2, 4, and 6 mutants were assayed for cleavage of GFP-Spitz under basal (unstimulated) conditions for ~24 hours in the absence of calcium. Cleavage product (green bands, white arrowhead) was detected in media fractions for most of the mutants at levels comparable to the wild type enzyme. Corresponding DmRho4 protein levels are shown as red bands (anti-HA western analyses). d Wild type DmRho4 and engineered variants were expressed and purified from bacteria, subjected to quantitative thermal stability analysis, and transition temperature midpoints (Tms) were derived (error bars indicate the standard deviation of four experimental replicates). The thermal stability of mutant DmRho4 proteases was indistinguishable from that of wild type DmRho4.
Extended Data Figure 3. Calcium does not regulate DmRho4 through intermolecular interactions.
a Anti-Flag coimmunoprecipitation analysis of HA-DmRho4 and APP-Spi7-Flag substrate from proteoliposomes in the presence or absence of 0.5 mM calcium. An inactive mutant of DmRho4 (S299A) was used to facilitate substrate complex isolation. The amount of HA-tagged DmRo4 co-immunoprecipitated with the Flag-tagged substrate was not affected by the presence of 0.5mM calcium. L denotes ‘load’, B denotes ‘bound’. b Anti-Flag coimmunoprecipitation of Flag-DmRho4 and HA-DmRho4 from proteoliposomes. HA-tagged DmRho4 failed to coimmunoprecipitate with Flag-tagged DmRho4 in both the absence and presence of 0.5 mM calcium. c Mixing a catalytic mutant (S299A) and a calcium-binding mutant (E382A) cannot rescue calcium stimulation in trans (star indicates lane where a product would be expected with the mixed single mutants).
Extended Data Figure 4. Lateral substrate gating underlies direct regulation of intramembrane proteolysis.
a Thermostability analysis of single and double cysteine mutants of DmRho4 (error bars indicate the standard deviation of four experimental replicates). b Average relative proportions of cleavage at the external cleavage site (orange) compared to the internal cleavage site (blue) are shown for DmRho4 in the absence (no Ca) and presence (+ Ca) of 1 mM calcium (error bars indicate standard error of replicate experiments). The external site was favoured in the absence of calcium (approximately 80%) while internal cleavage was preferred in the presence of calcium (approximately 70%). c DmRho4 loop 4 and loop 6 calcium-binding site mutants retained calcium-independent cleavage of a substrate harbouring only an external cleavage site. Full-length substrate (solid triangle) and cleavage product (open triangle) are indicated. c Cleavage of a substrate with external and internal cleavage sites was compared for E. coli GlpG, P.stuartii AarA, and V. cholerae Rho1 in the absence (no Ca) or presence (+ Ca) of 0.5 mM calcium. The relative proportions of cleavage at the two sites varied between the bacterial rhomboid proteases, but in no case did calcium alter the cleavage site preference.
ACKNOWLEDGEMENTS
This work was supported by NIH grant 2R01AI066025, the Howard Hughes Medical Institute, and the David and Lucile Packard Foundation. We are grateful to our colleague and friend Dr Andrew Holland for use of his deconvolution microscope.
Footnotes
The authors declare that no financial conflict of interest exists.
AUTHOR CONTRIBUTIONS
SU and RB designed the research. SU performed all Drosophila cell biology experiments, while RB performed all biochemistry experiments. SU wrote the manuscript and RB made the figures.
REFERENCES
- 1.Brown MS, Ye J, Rawson RB, Goldstein JL. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell. 2000;100:391–398. doi: 10.1016/s0092-8674(00)80675-3. [DOI] [PubMed] [Google Scholar]
- 2.De Strooper B, et al. A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature. 1999;398:518–522. doi: 10.1038/19083. [DOI] [PubMed] [Google Scholar]
- 3.Bier E, Jan LY, Jan Y. N. rhomboid, a gene required for dorsoventral axis establishment and peripheral nervous system development in Drosophila melanogaster. Genes Dev. 1990;4:190–203. doi: 10.1101/gad.4.2.190. [DOI] [PubMed] [Google Scholar]
- 4.Lichtenthaler SF, Haass C, Steiner H. Regulated intramembrane proteolysis--lessons from amyloid precursor protein processing. J. Neurochem. 2011;117:779–796. doi: 10.1111/j.1471-4159.2011.07248.x. [DOI] [PubMed] [Google Scholar]
- 5.Chavez-Gutierrez L, et al. The mechanism of gamma-Secretase dysfunction in familial Alzheimer disease. EMBO J. 2012;31:2261–2274. doi: 10.1038/emboj.2012.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Golde TE, Koo EH, Felsenstein KM, Osborne BA, Miele L. gamma-Secretase inhibitors and modulators. Biochim. Biophys. Acta. 2013;1828:2898–2907. doi: 10.1016/j.bbamem.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Urban S. Making the cut: central roles of intramembrane proteolysis in pathogenic microorganisms. Nature Reviews Microbiology. 2009;7:411–423. doi: 10.1038/nrmicro2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Urban S, Lee JR, Freeman M. A family of Rhomboid intramembrane proteases activates all membrane-tether EGF ligands in Drosophila. EMBO J. 2002;21:4277–4286. doi: 10.1093/emboj/cdf434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baker RP, Wijetilaka R, Urban S. Two Plasmodium Rhomboid Proteases Preferentially Cleave Different Adhesins Implicated in All Invasive Stages of Malaria. PLoS Pathogens. 2006;2:e113, 922–932. doi: 10.1371/journal.ppat.0020113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jin SM, et al. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J. Cell Biol. 2010;191:933–942. doi: 10.1083/jcb.201008084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morohashi Y, Tomita T. Protein trafficking and maturation regulate intramembrane proteolysis. Biochim. Biophys. Acta. 2013;1828:2855–2861. doi: 10.1016/j.bbamem.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 12.Brooks CL, Lemieux MJ. Untangling structure-function relationships in the rhomboid family of intramembrane proteases. Biochim. Biophys. Acta. 2013;1828:2862–2872. doi: 10.1016/j.bbamem.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 13.Bhattacharya S, Bunick CG, Chazin WJ. Target selectivity in EF-hand calcium binding proteins. Biochim. Biophys. Acta. 2004;1742:69–79. doi: 10.1016/j.bbamcr.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 14.Baker RP, Urban S. Architectural and thermodynamic principles underlying intramembrane protease function. Nature Chemical Biology. 2012;8:759–768. doi: 10.1038/nchembio.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fleig L, et al. Ubiquitin-dependent intramembrane rhomboid protease promotes ERAD of membrane proteins. Mol. Cell. 2012;47:558–569. doi: 10.1016/j.molcel.2012.06.008. [DOI] [PubMed] [Google Scholar]
- 16.Dickey SW, Baker RP, Cho S, Urban S. Proteolysis inside the membrane is a rate-governed reaction not driven by substrate affinity. Cell. 2013;155:1270–1281. doi: 10.1016/j.cell.2013.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sampathkumar P, et al. Oligomeric state study of prokaryotic rhomboid proteases. Biochim. Biophys. Acta. 2012;1818:3090–3097. doi: 10.1016/j.bbamem.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 18.Moldoveanu T, et al. A Ca(2+) switch aligns the active site of calpain. Cell. 2002;108:649–660. doi: 10.1016/s0092-8674(02)00659-1. [DOI] [PubMed] [Google Scholar]
- 19.Moin SM, Urban S. Membrane immersion allows rhomboid proteases to achieve specificity by reading transmembrane segment dynamics. eLife. 2012;1:e00173. doi: 10.7554/eLife.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Urban S, Baker RP. In vivo analysis reveals substrate-gating mutants of a rhomboid intramembrane protease display increased activity in living cells. Biol. Chem. 2008;389:1107–1115. doi: 10.1515/BC.2008.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xue Y, Ha Y. Large lateral movement of transmembrane helix S5 is not required for substrate access to the active site of rhomboid intramembrane protease. J. Biol. Chem. 2013;288:16645–16654. doi: 10.1074/jbc.M112.438127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strisovsky K, Sharpe HJ, Freeman M. Sequence-specific intramembrane proteolysis: identification of a recognition motif in rhomboid substrates. Mol. Cell. 2009;36:1048–1059. doi: 10.1016/j.molcel.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bondar AN, del Val C, White SH. Rhomboid protease dynamics and lipid interactions. Structure. 2009;17:395–405. doi: 10.1016/j.str.2008.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fernandez-Chacon R, et al. Synaptotagmin I functions as a calcium regulator of release probability. Nature. 2001;410:41–49. doi: 10.1038/35065004. [DOI] [PubMed] [Google Scholar]
- 25.Jaszai J, Brand M. Cloning and expression of Ventrhoid, a novel vertebrate homologue of the Drosophila EGF pathway gene rhomboid. Mech. Dev. 2002;113:73–77. doi: 10.1016/s0925-4773(01)00655-4. [DOI] [PubMed] [Google Scholar]
- 26.Venturin M, et al. Mental retardation and cardiovascular malformations in NF1 microdeleted patients point to candidate genes in 17q11.2. J. Med. Genet. 2004;41:35–41. doi: 10.1136/jmg.2003.014761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koonin EV, et al. The rhomboids: a nearly ubiquitous family of intramembrane serine proteases that probably evolved by multiple ancient horizontal gene transfers. Genome biology. 2003;4:R19. doi: 10.1186/gb-2003-4-3-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sheiner L, Dowse TJ, Soldati-Favre D. Identification of trafficking determinants for polytopic rhomboid proteases in Toxoplasma gondii. Traffic (Copenhagen, Denmark) 2008;9:665–677. doi: 10.1111/j.1600-0854.2008.00736.x. [DOI] [PubMed] [Google Scholar]
- 29.Feng L, et al. Structure of a site-2 protease family intramembrane metalloprotease. Science. 2007;318:1608–1612. doi: 10.1126/science.1150755. [DOI] [PubMed] [Google Scholar]
- 30.Li X, et al. Structure of a presenilin family intramembrane aspartate protease. Nature. 2013;493:56–61. doi: 10.1038/nature11801. [DOI] [PubMed] [Google Scholar]
REFERENCES
- 31.Urban S, Wolfe MS. Reconstitution of intramembrane proteolysis in vitro reveals that pure rhomboid is sufficient for catalysis and specificity. Proc. Natl. Acad. Sci. U. S. A. 2005;102:1883–1888. doi: 10.1073/pnas.0408306102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baker RP, Young K, Feng L, Shi Y, Urban S. Enzymatic analysis of a rhomboid intramembrane protease implicates transmembrane helix 5 as the lateral substrate gate. Proc. Natl. Acad. Sci. U. S. A. 2007;104:8257–8262. doi: 10.1073/pnas.0700814104. [DOI] [PMC free article] [PubMed] [Google Scholar]