Abstract
Focal adhesions (FAs) link the extracellular matrix (ECM) to the actin cytoskeleton to mediate cell adhesion, migration, mechanosensing and signaling. FAs have conserved nanoscale protein organization, suggesting that the position of proteins within FAs regulates their activity and function. Vinculin binds different FA proteins to mediate distinct cellular functions, but how vinculin’s interactions are spatiotemporally organized within FA is unknown. Using interferometric photo-activation localization (iPALM) super-resolution microscopy to assay vinculin nanoscale localization and a FRET biosensor to assay vinculin conformation, we found that upward repositioning within the FA during FA maturation facilitates vinculin activation and mechanical reinforcement of FA. Inactive vinculin localizes to the lower integrin signaling layer in FA by binding to phospho-paxillin. Talin binding activates vinculin and targets active vinculin higher in FA where vinculin can engage retrograde actin flow. Thus, specific protein interactions are spatially segregated within FA at the nano-scale to regulate vinculin activation and function.
Introduction
Integrin-mediated focal adhesions (FAs) are plasma membrane-associated organelles that physically connect the actin cytoskeleton to the extracellular matrix (ECM), providing specific adhesion between cells and their surroundings to mediate tissue formation and immune responses. FAs also serve as signaling hubs where cells sense biochemical and physical cues in their environment that inform cell decision-making in the cell cycle, differentiation and death. Additionally, they serve as sites of force transmission between the cytoskeleton and the surroundings to drive tissue morphogenesis, cell movement, and ECM remodeling. These diverse functions of FAs are reflected in their biochemical complexity. FAs contain hundreds of different proteins and their composition changes in response to physical stimuli, making them important sites of mechano-transduction1–3. Thus, FAs are multifaceted organelles that mediate an array of functions involving biochemical and physical interactions between the cell and its environment.
Although FAs are functionally and biochemically complex, they have conserved dynamics and structure4. FAs form during protrusion of the cell edge as small (<250 nm) nascent FAs containing clustered integrins, FAK, and paxillin5. Nascent FAs undergo a process of actomyosin-dependent maturation in which they grow to several microns in length and change molecular composition6. Mature FAs exhibit variations in protein composition along their length, with phosphorylated paxillin concentrating at their distal tips facing the cell periphery7, and actin binding proteins such as vinculin, VASP, and α-actinin concentrating at their proximal tips where they attach to actin stress fibers4,8,9. In addition, super-resolution microscopy recently revealed that FA proteins exhibit differential nano-scale localization along the axial dimension of their 200 nm thickness4. This showed that proteins localize to three general FA nano-domains: A membrane-proximal integrin signaling layer (ISL) containing FAK and paxillin located within ~10–20 nm of the plasma membrane; An actin regulatory layer (ARL) containing α-actinin, VASP, and zyxin that initiates ~50–60 nm from the membrane and extends upwards into the stress fiber; And a force transduction layer (FTL) containing the rod domain of talin that spans between the ISL and the ARL4. However, the functional consequence of this organized structure has yet to be explored. The nano-scale segregation of proteins into different axial FA domains could sterically limit the possible protein-protein interactions, which in turn could dictate specific downstream functional effects. Furthermore, whether this architecture is altered to mediate distinct FA functions, or if it evolves dynamically during FA maturation is not known.
Vinculin is an essential protein required for multiple FA functions, including stabilizing and strengthening FAs and promoting their maturation10–14, ECM mechanosensing15, regulating actin cytoskeletal dynamics16, and signaling to control cell death17. Vinculin has over 14 putative binding partners at FA including talin18, actin19, paxillin20, PIP221, Arp2/322 and vinexin23, and specific vinculin-protein interactions have been ascribed to distinct FA functions. For example, vinculin interaction with paxillin mediates FA mechanosensing15, its interaction with actin is required for regulation of lamellipodial actin dynamics,16 and talin binding by vinculin mediates FA strengthening11. Thus, the spatio-temporal regulation of different vinculin interactions likely regulates cellular function. Additionally, vinculin’s interaction with its binding partners is regulated by an auto-inhibitory, high-affinity intramolecular interaction between its head and tail domains24,25, and release of auto-inhibition is believed to require simultaneous binding of multiple ligands26. However, how vinculin activation and protein interactions are spatio-temporally regulated during FA formation and maturation is not known.
In this study, we sought to understand how distinct molecular interactions regulate vinculin activation and function within the context of the three-dimensional FA nano-architecture. Using super-resolution microscopy to assay vinculin nano-scale organization and a FRET biosensor to assay vinculin activation, we found that inactive vinculin associates with the lower ISL by binding to phospho-paxillin, while talin binding is required to activate vinculin and target active vinculin to higher FA layers where vinculin binds actin. Furthermore, we show that vinculin distribution shifts from the ISL to the FTL and ARL as myosin-II contractility promotes FA maturation. These results suggest that nanometer-scale changes in vinculin position during FA maturation facilitate vinculin activation and mechanical reinforcement of FA.
Results
Vinculin is distributed throughout the three FA nano-domains
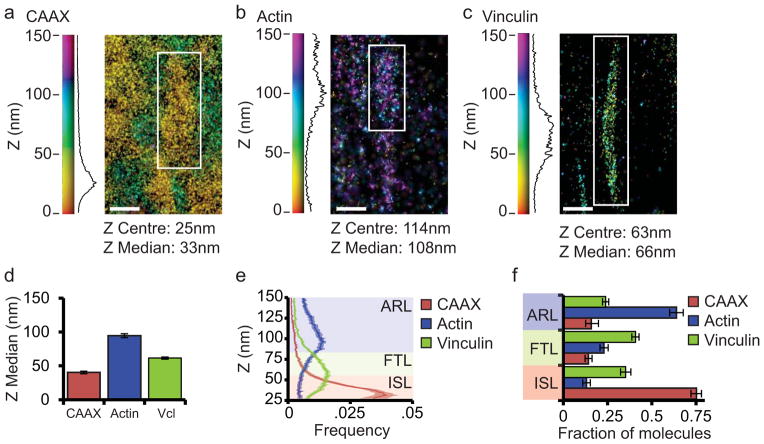
We hypothesize that the organization of vinculin into axial FA nano-domains is regulated by specific protein-protein interactions. To serve as a conceptual framework for our analysis of vinculin axial distribution within FA, we quantitatively defined the ISL and ARL layers in human foreskin fibroblasts (HFFs) by iPALM imaging of a plasma membrane marker (CAAX fused to tandem Eos (CAAX-tdEos)) and actin (actin fused to monomeric Eos2 (actin-mEos)), respectively, and defined the FTL as the intervening region. Fluorescent protein fusions were transiently expressed in cells plated on fibronectin-coated coverslips and fixed for iPALM imaging. For display, representative FA were rendered with colours indicating the axial (Z) position of each localized molecule relative to the position of the coverslip, and the Z-distribution of all the molecules in the rendered FA plotted in the accompanying histogram (Fig. 1a–b). Determination of the medians of the Z distributions (Zmed) showed that CAAX-tdEos and actin-mEos localized to ~40nm and ~95nm, respectively, similar to previous reports4 (Fig. 1d, Supplementary Table 1–2). Then, based on visual comparison of averaged 1 nm-binned histograms of Z-distributions of each probe (Fig. 1e), we arbitrarily defined the ISL as 25–54nm above the coverslip, where most (75%) of CAAX-tdEos and little (13%) of the actin-mEos localized. Similarly, we defined the ARL as 85–150nm above the coverslip where most (64%) actin-mEos and little CAAX-tdEos resided (16%), and the intervening FTL as 55–84nm above the coverslip (Fig. 1f). Thus, for the purpose of our analysis, the ISL consists of the 30nm region adjacent to the plasma membrane, the ARL extends from 60nm above the membrane into the stress fiber, with the FTL in the intervening 30nm.
Figure 1. Vinculin is distributed throughout the three FA nano-domains.
(a, b, c) Representative iPALM renderings from HFFs expressing CAAX-tdEos to label the plasma membrane (a), actin-mEos2 to label actin (b), or vinculin-N-tdEos (c). In (a–c) the colourscale represents Z-position (nm), FAs oriented with the distal tip facing up, scale bar = 1 micron. Histograms of the Z-position of molecules within individual FAs (white boxes in a–c) displayed next to the colourscale. The measured Gaussian Z-centre and Z-median of the distribution is shown. (d) Mean of Z-median measurements from individual FAs. (e) Averaged Z-position frequency histograms of molecules within FAs. Solid line, mean frequency; Shaded region, bootstrapped 95% confidence intervals. (f) Mean fraction of molecules localized to each of the three FA layers. Colouring in (e, f) is used to highlight the three FA layers. ISL: integrin signaling layer (red, 25–54 nm above the coverslip), FTL: force transduction layer (green, 55–84 nm above the coverslip), ARL: actin regulatory layer (purple, 85–150 nm above the coverslip). Graphs in (d–f) represent measurements of n=27 FAs from 5 CAAX-tdEos expressing cells, n=56 FAs from 8 actin-mEos2 expressing cells and n=115 FAs from 8 vinculin-N-tdEos expressing cells Data in all bar graphs are represented as mean ± bootstrapped 95% confidence intervals.
We next characterized the distribution of vinculin within these defined axial FA nano-domains. Transient expression of vinculin fused to tdEos at its N-terminus (WT-vinculin-N-tdEos) followed by iPALM analysis showed that WT-vinculin-N-tdEos consistently exhibited a Zmed of ~62nm (Fig. 1c,d, Supplementary Figure 1). Using the above definitions, we found that 35% of the vinculin molecules localized in the ISL, 41% localized in the FTL, and 24% localized in the ARL (Fig. 1e–f). Thus, vinculin is distributed throughout the three FA nano-domains, with the majority of molecules residing in the ISL and FTL.
Vinculin is required to maintain talin in a vertically extended conformation in FA
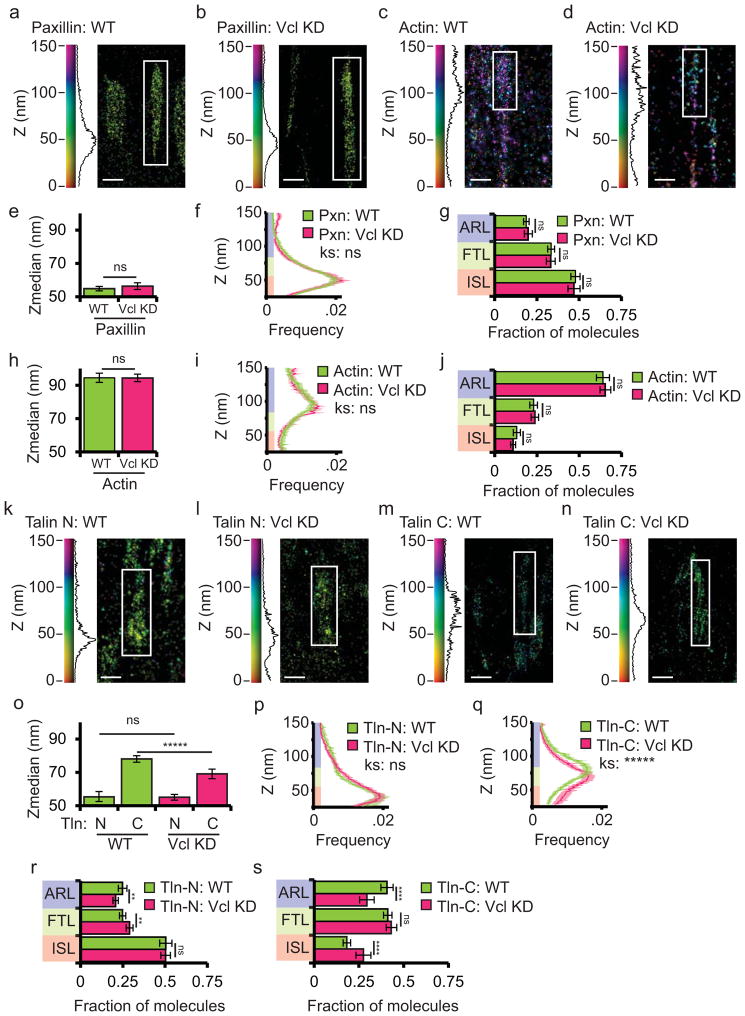
We next sought to determine the requirement for vinculin in the nano-scale organization of its binding partners and the molecular architecture of FA. We performed iPALM analyses of cells in the presence (wild type, WT) and absence (knock down, KD) of vinculin, examining the distribution of the ISL protein paxillin, the ARL protein actin, and talin, an extended protein that spans the FTL by binding integrin tails27 in the ISL and actin28 in the ARL4,29. Small interfering RNAs (siRNA) targeting the 3′UTR knocked down >95% of vinculin in HFFs (Vcl KD, Supplementary Figure 2a). We transiently expressed paxillin-tdEos (paxillin with n-terminally fused tdEos), actin-mEos, talin-N-tdEos (talin with n-terminally fused tdEos) and talin-C-tdEos (talin with c-terminally fused tdEos) in either WT or vinculin KD background. Comparison of WT and vinculin KD cells by two-sampled Kolmogorov-Smirnov test and ANOVA analyses showed that vinculin had an insignificant effect on the localization of paxillin (Fig. 2a–b), with ~50% of paxillin-tdEos localized in the ISL under both conditions (Fig. 2e–g). Similarly, the loss of vinculin had an insignificant effect on the localization of actin or the n-terminus of talin in FA, with ~75% of actin-mEos2 localized in the ARL and ~50% of talin-N-tdEos localized in the ISL (Fig. 2c,d, h–j, k,l, o,p,r). In contrast, compared to WT cells, in vinculin KD, the Zmed of talin-C-tdEos was significantly decreased by ~10nm, the fraction of molecules in the ARL was reduced, and there was a concomitant increase in the fraction of molecules in the ISL (Fig. 2m–o, q, s). Furthermore, talin-C-tdEos nanoscale position was rescued by the overexpression of vinculin-mTurquoise (Supplementary Figure 2). Thus, in the presence of vinculin, the talin tail is localized ~23nm above the head, while in the absence of vinculin, the tail only extends ~14nm above the head. We conclude that while vinculin is not required for paxillin or actin nanoscale positioning within FA, vinculin is required for talin to be maintained in a maximal vertically extended conformation.
Figure 2. Vinculin is required to maintain talin in a vertically extended conformation.
(a, c, k, m): Representative iPALM renderings from wild-type (WT) HFFs expressing paxillin-tdEos (Pxn, a), actin-mEos2 (c), N-terminally tagged talin-N-tdEos (k) or C-terminally tagged talin-C-tdEos (m). (b, d, l, n): Representative iPALM rendering from HFFs treated with siRNAs targeting vinculin (Vcl KD) and additionally expressing paxillin-tdEos (b), actin-mEos2 (d), talin-N-tdEos (l) or talin-C-tdEos (n). In (a–d and k–n), the colourscale represents Z-position (nm), FAs oriented with the distal tip facing up, scale bar = 1 micron. Histograms of the Z-position of molecules within individual FAs (white boxes in a–d, k–n) displayed next to the colourscale. Actin-mEos2 WT data is duplicated from figure 1 for comparison purposes. (e, h, o) Mean of Z-median measurements from individual FAs. (f, i, p, q) Averaged Z-position frequency histograms of molecules within FAs. Solid line, mean frequency; Shaded region, bootstrapped 95% confidence intervals. Significance tested with two-sample KS test. (g, j, r, s) Mean fraction of molecules localized to the three FA layers. Colouring in (f,g, i, j, and p–s) is used to highlight the FA layers as in Fig. 1. Graphs in (e–g) represent measurements of n=120 FAs from 7 WT cells and n=117FAs from 6 Vcl KD cells. Graphs in (h–j) represent measurements of n=56 FAs from 8 WT cells and n=50 FAs from 6 Vcl KD cells. Graphs in (o–s) represents data from n=75 FAs from 7 TlnN WT cells, n=107 FAs from 5 TlnN KD cells, n= 95 FAs from 5 TlnC WT cells, and n=66 FAs from 5 TlnC KD cells. Data in all bargraphs are represented as mean± 95% bootstrapped confidence intervals and significance is tested with one way ANOVA. For statistical tests: * p<0.05, ** P<0.01, *** p<0.001, ****p<0.0001, *****p<0.00001 ns: not significant.
Vinculin is oriented in FAs with the tail above the head
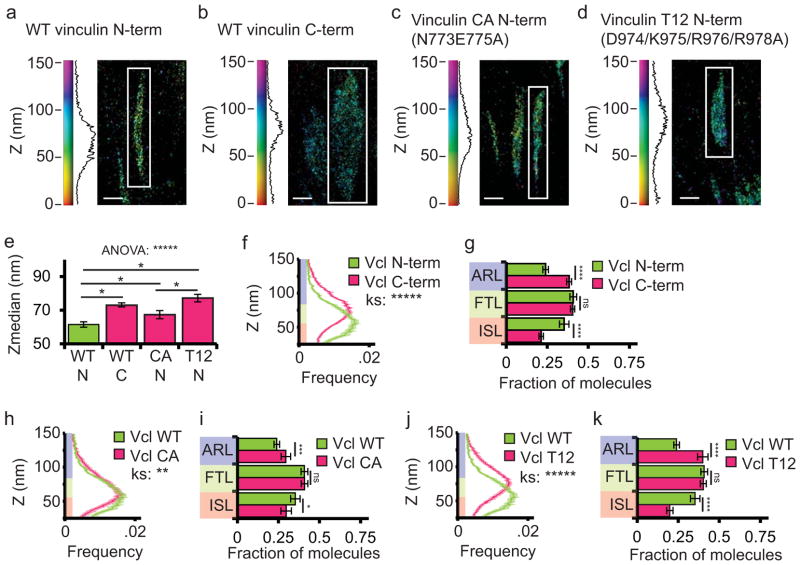
We next sought to determine if vinculin exhibits a preferred orientation in FA by comparing the localization of the N-and C-termini of WT vinculin. We performed iPALM on WT-vinculin-N-tdEos and WT-vinculin-C-tdEos (Fig. 3a,b). Similar to a previous study,30 we found that WT-vinculin-C-tdEos localized significantly higher than WT-vinculin-N-tdEos (Fig. 3e) and had a significantly different Z-distribution (Fig 3f), with more WT-vinculin-N-tdEos localized in the ISL and more WT-vinculin-C-tdEos localized in the ARL (Fig. 3g). Since the N-terminus is adjacent to the vinculin head and the C-terminus is adjacent to the vinculin tail31, this suggests that the majority of vinculin molecules are oriented in FAs with the tail above the head.
Figure 3. Vinculin is oriented in FAs with the tail above the head, and activation promotes association of vinculin with the force transduction and actin regulatory layers.
(a–d) Representative iPALM rendering from HFFs expressing N-terminally tagged wild-type (WT) vinculin-N-tdEos (a, data duplicated from figure 1 for comparison purposes), WT C-terminally tagged vinculin-C-tdEos (b), constitutively active (CA, N773/E775A) vinculin-N-tdEos (c), or constitutively active (T12, D974/K975/R976/R978A) vinculin-N-tdEos (d). In (a–d) the colourscale represents Z-position (nm), FAs oriented with the distal tip facing up, and scale bar = 1 micron. Histograms of the Z-position of molecules within individual FAs (white boxes in a–d) displayed next to the colourscale. (e) Mean of Z-median measurements of the position of molecules from individual. Significance tested with one way ANOVA, (* difference is significant at p<0.05, determined by post-hoc Tukey test) (f, h, j) Averaged Z-position frequency histograms of molecules within FAs. Solid line, mean frequency; Shaded region, bootstrapped 95% confidence intervals. Significance tested with two-sample KS test. (g, i, k) Mean fraction of molecules localized to the three FA layers. Significance is tested with one way ANOVA. Colouring in (f–k) used to highlight the FA layers as in Fig 1. Graphs in (e–k) represent measurements of n=115FA from 8 WT vinculin-N-tdEos expressing cells, n=137 FAs from 5 WT vinculin-C-tdEos expressing cells, n=82 FAs from 8 CA-vinculin-N-tdEos cells and n=103 FAs from 6 T12-vinculin-N-tdEos expressing cells. Data in all bar graphs are represented as mean± 95% bootstrapped confidence intervals. For statistical tests: * p<0.05, ** P<0.01, *** p<0.001, ****p<0.0001, *****p<0.00001 ns: not significant).
Activation promotes association of vinculin with the FTL and ARL
Because vinculin activation affects its ability to interact with multiple binding partners including talin and actin26, we next sought to determine if vinculin activation influences its nano-scale organization within FA. We utilized iPALM analysis to measure the effect of activating point mutations (Vinculin N773/E775A (“CA Vinculin”) and Vinculin D974/K975/R976/R978A (“T12 Vinculin”)) on the Z-position of N-and C-terminally tagged vinculin-tdEos fusion constructs in FA16,32,33 (Fig. 3c–d). We found that both CA-vinculin-N-tdEos and T12-Vinculin-N-tdEos localized significantly higher in FA and had a higher percentage of molecules localized in the ARL compared with WT-vinculin-N-tdEos (Fig. 3e,h–k). Additionally, CA-vinculin-C-tdEos localized significantly higher in FA than CA-vinculin-N-tdEos, suggesting that activated vinculin is oriented in FA with the tail above the head (Supplementary Figure 3). We conclude that activation promotes an upwards shift in the distribution of vinculin within FA.
Talin binding is required for vinculin activation in FA and promotes localization of active vinculin to the FTL and ARL
The talin α-helical rod domain contains several mechano-sensitive vinculin binding sites spanning the FTL that mediate vinculin activation in vitro34 and strengthening of the integrin-actin link in cells35. We thus sought to determine the role of talin in vinculin activation and nanoscale localization in FA. To assay activation in cells, we utilized a vinculin activation FRET biosensor36 in which donor (mTurquoise) and acceptor (NeonGreen37) fluorophores were inserted into the vinculin protein such that FRET occurs when vinculin tail interacts with the head in the auto-inhibited, closed conformation, and FRET decreases when auto-inhibition is relieved and vinculin is activated. Unlike positive and negative FRET controls (Supplementary Figure 4a–c), WT-vinculin-FRET exhibited a significantly lower FRET ratio at FA compared with the cytosol (Supplementary Figure 4d–e), consistent with previous reports36. In contrast, when we introduced the N773/E775A activating mutation, CA-vinculin-FRET exhibited a FRET ratio at the same low level as FA-localized WT-vinculin-FRET, independent of localization. (Supplementary Figure 4d–e). Together, these results confirm previous reports36 indicating that vinculin activation is selectively increased at FA.
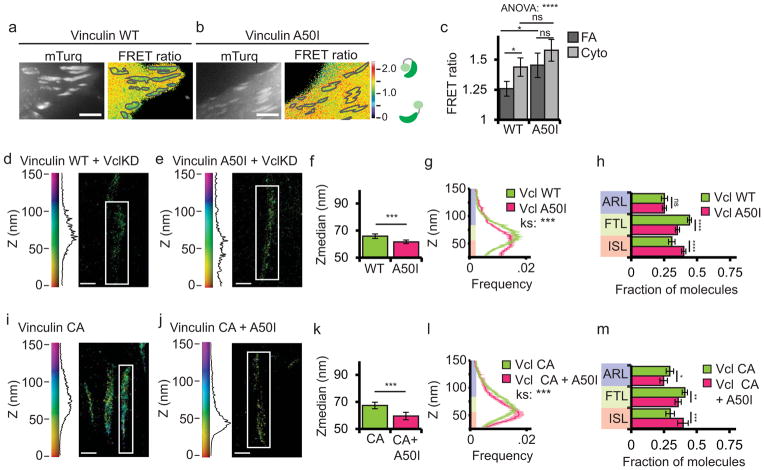
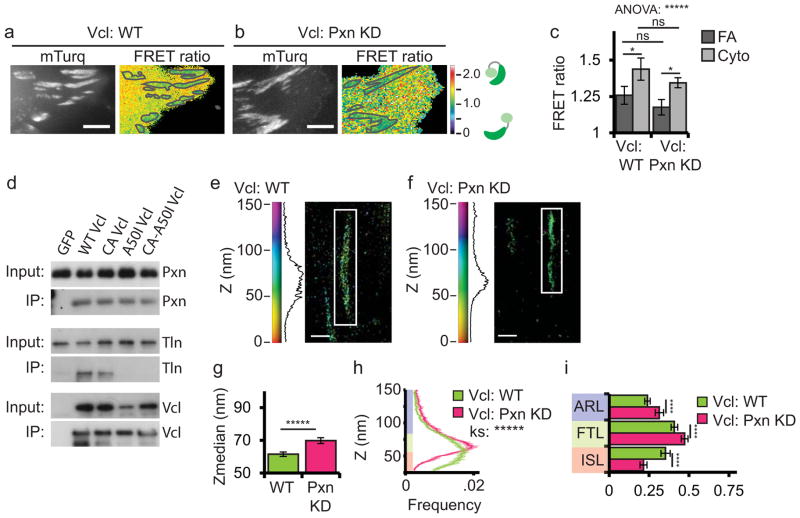
To determine the contribution of talin binding to the activation of vinculin in cells, we introduced the A50I mutation into the WT-vinculin-FRET biosensor (Fig. 4a–c). This mutation inhibits talin binding to the vinculin head in vitro33, and in cells targets to FA, but results in reduced FA number and decreased adhesion strength and traction force10. Expression of A50I-vinculin-FRET in cells showed that in contrast to WT-vinculin-FRET that exhibited reduced FRET at FA, the FRET ratio of A50I-vinculin-FRET was at the same high level in both the cytoplasm and FA (Fig. 4a–c). This suggests that talin binding is required for vinculin activation at FA.
Figure 4. Talin binding is required for vinculin activation in FA and promotes localization of active vinculin to the force transduction and actin regulatory layers.
(a, b) mTurquoise (left) and processed FRET ratio image (right) of HFFs expressing either wild-type (WT) or A50I vinculin FRET biosensor. In (a, b) the mask (grey lines) was created from the mTurq image and superimposed onto the FRET ratio image, scale bars = 5 micron. (c) Quantification of the mean FRET ratio value inside FAs (FA) and outside FAs (Cyto) from n=18 WT and n=15 A50I cells. (* difference is significant at p<0.05 cutoff, determined by Tukey test post-hoc analysis). (d, e, i, j) Representative iPALM renderings from HFFs treated with siRNAs targeting vinculin (KD) and additionally expressing vinculin-N-tdEos (d); or A50I-vinculin-tdEos (e); and WT HFFs expressing constitutively active (CA, N773/E775A) CA-vinculin-tdEos (i, data duplicated from figure 3 for comparison purposes); or CA-A50I-vinculin-tdEos (j). In (d, e, i, j) the colourscale represents Z-position (nm), FAs oriented with the distal tip facing up, scale bar = 1 micron. Histograms of the Z-position of molecules within individual FAs (white boxes in a–d) displayed next to colourscale. (f, k) Mean of Z-median measurements from individual FAs. (g, l) Averaged Z-position frequency histograms of molecules within FAs. Solid line, mean frequency; Shaded region, bootstrapped 95% confidence intervals. Significance tested with two-sample KS test. (h, m) Mean fraction of molecules localized to the three FA layers. Colouring in (g, h, l, and m) used to highlight the FA layers as in Fig. 1. Graphs in (f–h) represent measurements of n=133 FA from 7 WT-vinculin-N-tdEos and n=102 FA from 7 A50I-Vinculin-tdEos cells. Graphs in (k–m) represent measurements of n=82 FA from 8 CA-vinculin-N-tdEos and n=129 FA from 6 CA-A50I-Vinculin-tdEos cells. Data in all bar graphs are represented as mean± 95% bootstrapped confidence intervals with significance tested by one way ANOVA. * p<0.05, ** P<0.01, *** p<0.001, ****p<0.0001, *****p<0.00001 ns: not significant.
Next, we examined the role of talin binding in regulating vinculin nano-scale localization. We knocked down vinculin and rescued with WT-vinculin-N-tdEos or A50I-vinculin-N-tdEos (Fig. 4d,e). iPALM analysis showed that compared to WT-vinculin-N-tdEos, A50I-vinculin-N-tdEos localized significantly lower in FA than WT-vinculin-N-tdEos and had a significantly different Z-distribution, with reduced localization to the FTL and increased localization to the ISL (Fig. 4f–h). We conclude that talin binding promotes a shift in vinculin localization from the ISL to the FTL.
Since talin binding is required for vinculin activation in vitro26 and at FAs (Fig. 4a–c), we next determined if talin binding was required for vinculin localization to the FTL, or whether activation was sufficient to localize vinculin to the FTL independent of talin binding. We introduced the A50I mutation into CA-vinculin-N-tdEos (A50I-CA-vinculin-N-tdEos) and expressed this construct in WT cells (Fig. 4j). iPALM analysis showed that compared to CA-vinculin-N-tdEos (Fig. 4i), A50I-CA-vinculin-N-tdEos localized significantly lower in FA, with significantly decreased localization to the FTL and ARL and increased localization to the ISL (Fig. 4k–m). Thus, reducing the head-tail affinity is not sufficient to rescue the localization defect of A50I-vinculin-N-tdEos. Together, these FRET and iPALM results show that talin binding is required for activation of vinculin in FA and promotes an upward shift in localization of active vinculin from the ISL to the FTL and ARL.
Paxillin is not required for vinculin activation but promotes vinculin localization to the ISL
Because paxillin binding is critical to vinculin’s function in force transmission and mechanosensation15, we determined the role of paxillin in vinculin activation and nanoscale localization in FA. We used siRNA to knock down >95% of endogenous paxillin and ~50% of the paxillin homologue HIC-5 (paxillin KD, Supplementary Figure 5a), expressed WT-vinculin-FRET, and performed FRET analysis in control versus paxillin KD cells (Fig. 5a–c). This showed that loss of paxillin had no effect on either vinculin targeting to FA38 or FRET ratios, with WT-vinculin-FRET exhibiting significantly lower FRET in FA than the cytosol in both controls and paxillin KD (Fig. 5c). We conclude that vinculin activation at FA does not require paxillin.
Figure 5. Paxillin is not required for vinculin activation but promotes vinculin localization to the integrin signaling layer.
(a, b) mTurquoise (left) and processed FRET ratio image (right) of WT or paxillin KD HFFs additionally expressing a vinculin (Vcl) FRET biosensor. In (a, b) the mask (grey lines) was created from the mTurq image and superimposed onto the FRET ratio image, scale bars = 5 micron. WT-vinculin FRET data duplicated from figure 4 for comparison purposes. (c) Quantification of mean FRET ratio value inside FAs (FA) and outside FAs (Cyto) from n=18 WT and n=16 PxnKD cells. (* difference is significant at p<0.05 cutoff, determined by Tukey test post-hoc analysis) (d) Western blots of immunoprecipitations (IP) with anti-GFP antibodies of GFP-tagged vinculins. Input lysates and IP fractions blotted for paxillin (pxn), talin (tln) and vinculin (vcl). See also Supplemental Figure 5. (e, f) Representative iPALM rendering from HFFs expressing vinculin-N-tdEos in WT (e, data duplicated from figure 1 for comparison purposes) or paxillin KD backgrounds (f). In (e, f) the colourscale represents Z-position (nm), FAs oriented with the distal tip facing up, scale bar = 1 micron. Histograms of the Z-position of molecules within individual FAs (white boxes in a–d) displayed next to the colourscale. (g) Mean of Z-median measurements from individual FAs (h) Averaged Z-position frequency histograms of molecules within FAs. Solid line, mean frequency; Shaded region, bootstrapped 95% confidence intervals. Significance tested with two-sample KS test. (i) Mean fraction of molecules localized to the three FA layers. Colouring in (h, i) used to highlight the FA layers as in Fig. 1. Graphs in (g–i) represent measurements of n=115 FA from 8 WT-vinculin-N-tdEos and n=109 FA from 6 Paxillin-Hic5 siRNA cells. Data in all bar graphs are represented as mean± 95% bootstrapped confidence intervals with significance tested by one way ANOVA. * p<0.05, ** P<0.01, *** p<0.001, ****p<0.0001, *****p<0.00001 ns: not significant.
The above results show that paxillin has no effect on vinculin activation in FAs, and that a talin binding mutant of vinculin localizes to FAs in an inactive conformation, begging the question of whether paxillin may recruit inactive vinculin to FA. To address this, we assayed the ability of talin and paxillin to co-immunoprecipitate (co-IP) with GFP-tagged vinculin mutants (Fig. 5d, Supplementary Figure 5b–c). We knocked down endogenous vinculin and rescued with WT-vinculin-GFP, CA-vinculin-GFP, A50I-vinculin-GFP or the double mutant A50I-CA-vinculin-GFP. Probing the immunoprecipitates for talin or paxillin showed that both proteins co-IPed with WT and CA-vinculin, suggesting that the N773/E775A mutation does not strongly enhance binding. In contrast, while A50I-vinculin-GFP or A50I-CA-vinculin-GFP did not co-IP talin as expected13, paxillin was able to co-IP with both of these constructs (Fig. 5d). Consistent with our observation that paxillin is not required for vinculin activation at FA, these results show that paxillin is able to bind vinculin independently of talin binding, and supports the notion that paxillin binding could target inactive vinculin to FAs.
To determine the role of paxillin in vinculin nanoscale organization, we performed iPALM analysis of WT-vinculin-N-tdEos in the presence and absence of paxillin (Fig 5e–f). We found that compared to control, paxillin KD resulted in WT-vinculin-N-tdEos localizing significantly higher in the FA with a significantly altered Z-distribution and decreased localization of molecules in the ISL (Fig. 5g–i), suggesting that paxillin is required to efficiently target vinculin to the ISL.
Binding to phospho-paxillin promotes vinculin localization to the ISL
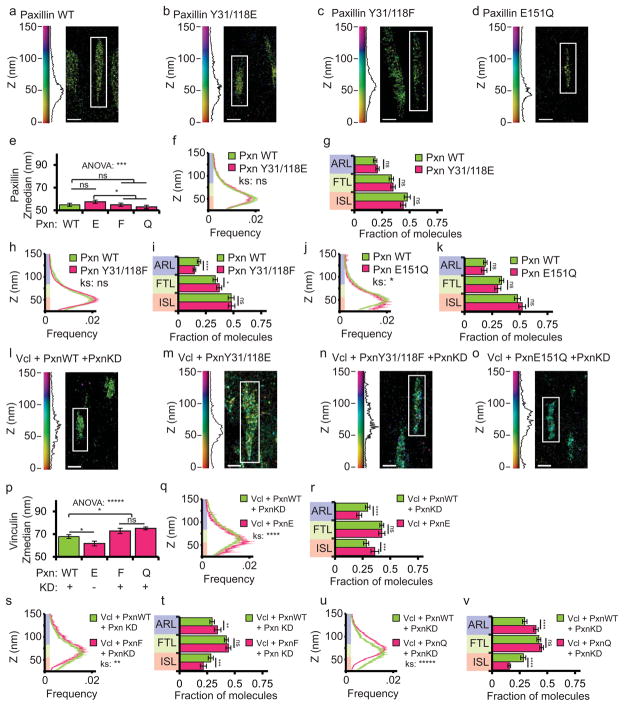
We next sought to examine the role of paxillin binding in vinculin nanoscale localization in FA. The vinculin-paxillin interaction requires paxillin phosphorylation on Y31 and Y118 by FAK in response to myosin contractility38. Therefore, phosphomimetic paxillin (Y31/118E-paxillin) will bind to vinculin, while non-phosphorylatable paxillin (Y31/118F-paxillin) does not38. Additionally, E151Q-paxillin is a point mutant specifically defective in vinculin binding15,39 All three paxillin-GFP mutants co-localize with WT paxillin at FA and have no effect on FA size15. To confirm that neither the vinculin-paxillin interaction nor these mutations affected paxillin nano-scale localization within FA, we introduced the mutations into paxillin-tdEos constructs and performed iPALM analysis (Fig. 6a–d). This showed that all three paxillin-tdEos constructs exhibited Z-positions and Z-distributions that were not significantly different from WT-paxillin-tdEos (Fig. 6e–k). Therefore, vinculin binding and paxillin phospho-regulation are not required to target paxillin to the ISL within FAs.
Figure 6. Binding to phospho-paxillin promotes vinculin localization to the integrin signaling layer.
(a–d, l–o) Representative iPALM renderings from HFFexpressing either wild-type (WT) paxillin-tdEos (a, Pxn, duplicated from figure 2 for comparison purposes), paxillin Y31/118E-tdEos (b), paxillin Y31/118F-tdEos (c), paxillin E151Q-tdEos (d), WT-vinculin-tdEos (Vcl) and WT-paxillin-mCerulean in a paxillin/hic5 siRNA (paxillin KD) background (l); WT-vinculin-tdEos and paxillin Y31/118E-mCerulean (m), WT-vinculin-tdEos and paxillin Y31/118F-mCerulean in a paxillin KD background (n) or WT-vinculin-tdEos and paxillin E151Q-mCerulean in a paxillin KD background (o). In (a–d, l–o) the colourscale represents Z-position (nm), FAs oriented with the distal tip facing up, scale bar = 1 micron. Histograms of the Z-position of molecules within individual FAs (white boxes in a–d) displayed next to the colourscale. (e, p) Mean of Z-median measurements from individual FAs. (* difference is significant at p<0.05, determined by post-hoc Tukey test) (f, h, j, q, s, u) Averaged Z-position frequency histograms of molecules within FAs. Solid line, mean frequency; Shaded region, bootstrapped 95% confidence intervals. Significance tested with two-sample KS test. (g, i, k, r, t, v) Mean fraction of molecules localized to the three FA layers. Colouring in (f–k and q–v) used to highlight the FA layers as in Fig. 1. Graphs in (e–k) represent measurements of n=120 FA from 7 WT-paxillin-tdEos expressing cells, n=150 FA from 6 paxillin Y31/118E expressing cells, n=152 FA from 6 paxillin Y31/118F expressing cells, and N=79 FA from 8 paxillin E151Q-tdEos expressing cells. Graphs in (p–v) represent measurements of n=110 FA from 5 WT-paxillin-mCer expressing cells, n=92 FA from 8 paxillin Y31/118E-mCer expressing cells, n=117 FA from 6 paxillin Y31/118F-mCer expressing cells, and N=175 FA from 6 paxillin E151Q-mCer expressing cells. Data in all bar graphs are represented as mean± 95% bootstrapped confidence intervals with significance tested by one way ANOVA. * p<0.05, ** P<0.01, *** p<0.001, ****p<0.0001, *****p<0.00001 ns: not significant.
To determine if vinculin targeting to the ISL required the vinculin-paxillin interaction, we introduced Y31/118E, Y31/118F and E151Q mutations into paxillin-mCerulean (WT-paxillin-mCer) for co-expression with WT-vinculin-N-tdEos (Fig. 6l–o). Co-expression of WT-paxillin-mCer with WT-vinculin-N-tdEos in either WT or paxillin KD HFFs resulted in similar vinculin Z-postion measurements (Supplementary Figure 5d–h). When Y31/118E-paxillin-mCer was co-expressed with vinculin-N-tdEos in paxillin KD cells to promote the vinculin-paxillin interaction, vinculin localized significantly lower in the FA with significantly more molecules in the ISL compared to vinculin-N-tdEos when it was co-expressed WT-paxillin-mCer (Fig. 6p–r). In contrast, vinculin-N-tdEos localized significantly higher in FAs and had less molecules in the ISL when paxillin KD was rescued with either Y31/118F-paxillin-mCer or E151Q-paxillin-mCer to disrupt the paxillin-vinculin interaction (Fig. 6p,s–v). We conclude that paxillin binding mediated by Y31 and Y118 phosphorylation promotes efficient targeting of vinculin to the ISL. Together, these results show that the vinculin-phospho-paxillin interaction promotes association of vinculin with the ISL independent of vinculin activation.
Actin binding does not regulate vinculin activation or nano-scale localization in FA
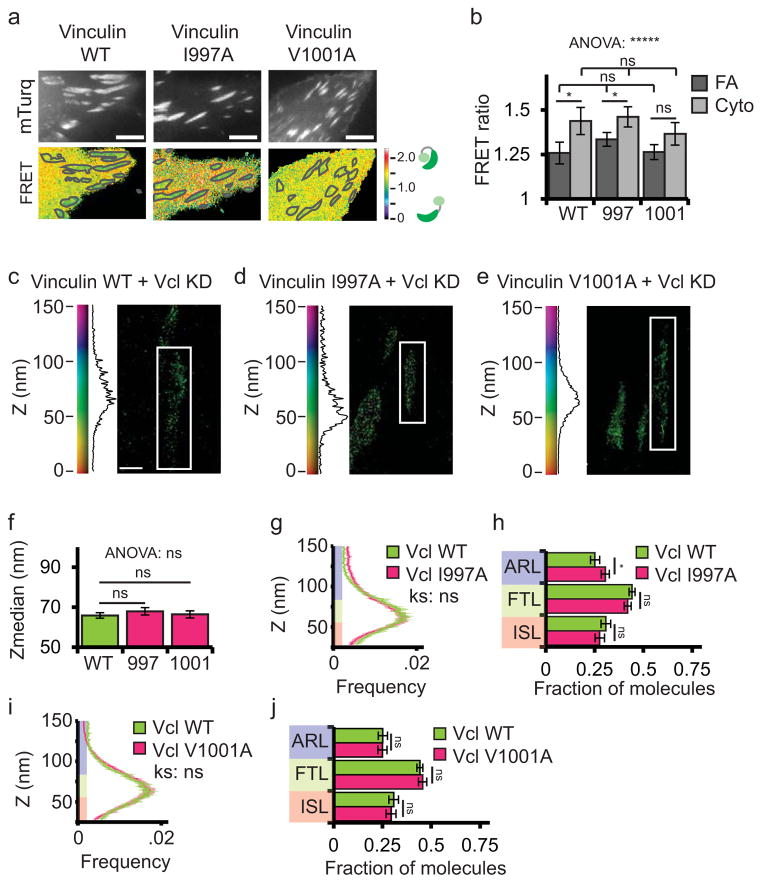
Because actin binding is critical for vinculin’s function in regulating actin and adhesion dynamics16, we examined the role of actin in regulating vinculin activation and nanoscale localization at FA. Two independent amino acid point mutations (I997A and V1001A) have been shown to reduce actin binding to the vinculin tail without perturbing vinculin auto-inhibition, and result in decreased mechanical response to force in cells16,40. To determine the role of actin binding in regulating vinculin activation, we introduced these mutations into the vinculin-FRET biosensor (I997A-vinculin FRET and V1001A-vinculin-FRET, Fig. 7a–b). Analysis of FRET images showed that neither I997A-vinculin-FRET nor V1001A-vinculin-FRET had significantly different FRET values in FAs or the cytoplasm compared with WT-vinculin-FRET (Fig. 7b). Thus, actin binding is not required for vinculin activation at FAs.
Figure 7. Actin binding does not regulate vinculin activation or nano-scale localization.
(a) mTurquoise (top) and processed FRET ratio (bottom) images of wild type (WT) HFFs expressing WT or actin-binding mutant (I997A and V1001A) vinculin FRET biosensor. Scale bar = 5 micron. The mask (grey lines) was created from the mTurq image and superimposed onto FRET ratio image. WT data is duplicated from figure 4 for comparison purposes. (b) Quantification of the mean FRET ratio value inside FAs (FA) and outside FAs (Cyto) from n=18 WT, n=21 I997A, and n=16 V1001A cells. (* difference is significant at p<0.05, determined by post-hoc Tukey test). (c–e) Representative iPALM renderings from HFFs expressing vinculin-N-tdEos (c), I997A-vinculin-tdEos (d), or V1001A-vinculin-tdEos (e) in a vinculin siRNA (vinculin KD) background. In (c–e) the colourscale represents Z-position (nm), FAs oriented with the distal tip facing up, scale bar = 1 micron. Histograms of the Z-position of molecules within individual FAs (white boxes in a–d) displayed next to the colourscale. WT-Vinculin data duplicated from figure 4 for comparison purposes. (f) Mean of Z-median measurements from individual FAs. (* difference is significant at p<0.05, determined by post-hoc Tukey test) (g, i) Averaged Z-position frequency histograms of molecules within FAs. Solid line, mean frequency; Shaded region, bootstrapped 95% confidence intervals. Significance tested with two-sample KS test. (h, j) Mean fraction of molecules localized to the three FA layers. Colouring in (f–k and q–v) used to highlight the FA layers as in Fig. 1. Graphs in (f–j) represent measurements of n=133 FA from 7 WT-vinculin-N-tdEos, n=155 FA from 6 I997A-vinculin-tdEos cells, and n=182 FA from 8 V1001A-Vinculin-tdEos cells. Data in all bar graphs are represented as mean± 95% bootstrapped confidence intervals with significance tested by one way ANOVA. * p<0.05, ** P<0.01, *** p<0.001, ****p<0.0001, *****p<0.00001 ns: not significant.
To determine the effect of actin binding on vinculin nanoscale localization within FA, we rescued vinculin-KD with WT-vinculin-N-tdEos, I997A-vinculin-N-tdEos or V1001A-vinculin-N-tdEos (Fig. 7c–e) and performed iPALM analysis. Surprisingly, we found no significant difference in z-position or z-distribution between WT-vinculin and the actin-binding mutants (Fig. 7f–j). Thus, while actin binding is important for regulating actin dynamics and force transduction16,40, it is not required for vinculin activation or nanoscale organization at FAs.
Vinculin exhibits a gradient in activation and axial position across single FAs
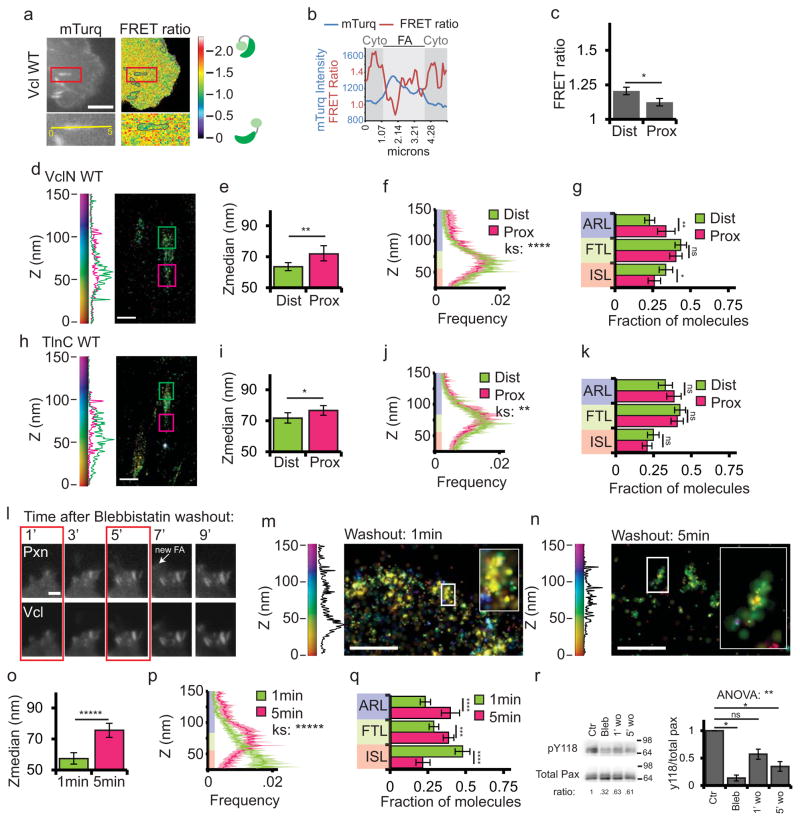
Individual FAs vary in biochemical composition and distribution of traction forces across their length. Phospho-paxillin7 and high ECM traction forces15,41 are concentrated at the distal tip of FAs near the leading edge, while actin-associated proteins4,8,9 and lower forces15,41 are located at the proximal FA tip facing the cell center. Therefore, we sought to determine whether vinculin activation or axial position varied along the length of individual FA. To determine if vinculin activation spatially varied within FA, we measured the mean FRET ratio in the distal and proximal thirds of FAs (Fig. 8a–c). Neither the FA-targeted control probe nor CA-vinculin-FRET exhibited significant differences in FRET ratio between the distal and proximal thirds of FAs (Supplementary Figure 6a–f). In contrast, WT-vinculin-FRET exhibited significantly higher FRET ratio values in the distal third of FAs compared with the proximal third, although both FA regions had lower FRET ratio values than the cytoplasm (compare Fig. 4c and 8c). This suggests that active vinculin is concentrated in the proximal FA tip.
Figure 8. Vinculin activation and nano-scale localization are spatiotemporally regulated.
(a) Top: mTurquoise (mTurq) and processed mTurquoise/NeonGreen FRET ratio images of an HFF cell expressing the vinculin FRET biosensor. Bottom: Zoom of outlined area. FA mask (grey lines) created from the mTurq image and superimposed onto FRET ratio image. (b) mTurquoise intensity and FRET ratio along the 5um long line in the zoom in (a). Dark shaded areas are cytosolic (cyto) regions adjacent to the FA (light shaded area) (c) Mean FRET ratio quantification of the distal (Dist, facing cell edge) 1/3 and proximal (Prox, facing cell center) 1/3 of FA linescans. (d, h) Representative iPALM renderings of HFFs expressing vinculin-N-tdEos (d) or talin-C-tdEos (h). Histograms of the Z-position of molecules within Dist and Prox thirds displayed next to the colourscale. (e, i) Mean of Z-median measurements from Dist or Prox thirds (f, j) Averaged Z-position frequency histograms of molecules. Solid line, mean frequency; Shaded region, bootstrapped 95% confidence intervals. (g, k) Mean fraction of molecules localized to the three FA layers. (l) Time-lapse TIRFM images of FAs during blebbistatin washout. HFFs expressing paxillin-mCherry (top) and vinculin-EGFP (bottom) treated with 50uM blebbistatin for 2 hours. Blebbistatin washout at t=0 min, Arrow: newly formed FA. Boxed areas: time points for iPALM analysis. Scale bar= 2μm. (m, n) Representative iPALM renderings of vinculin-N-tdEos at 1min (m) and 5min (n) after blebbistatin washout. Histograms of Z-positions of molecules within individual FAs (boxes in m-n) displayed next to the colourscale. (o) Mean Z-median measurements from individual FAs (p) Averaged Z-position histograms of molecules within FAs. Solid line, mean frequency; Shaded region, bootstrapped 95% confidence intervals. (q) Mean fraction of molecules localized to the three FA layers. (r) Left: Representative immunoblots of HFFs after 2h of DMSO (Ctr) or 50uM blebbistatin treatment (Bleb), or 1min or 5min after blebbistatin washout (1′ wo, 5′ wo), probed with antibodies to total or pY118 paxillin. Ratio: ratio of pY118:total paxillin, normalized to control. Right: Quantification of ratios. Graphs in (c) represent measurements of n=17 FA from 6 cells, (e–g) n=40 FA from 10 cells, (i–k) n=33 FA from 5 cells, (o–q) n=61 FA from 6 cells and n=52 FA from 7 cells fixed 1 and 5 min after blebbistatin washout respectively, (r) n=3 experiments. Colouring in (f, g, j, k, p, q) highlights FA layers as in Fig. 1. Data in (e, g, i, k, o, q) are mean± 95% bootstrapped confidence interval, in (c,r) mean±S.E.M, significance tested by one way ANOVA. Significance tested with two-sample KS test in (f, j, p) and with Tukey post-hoc Tukey in (r). * p<0.05, ** P<0.01, *** p<0.001, ****p<0.0001, *****p<0.00001 ns: not significant).
To determine if vinculin nano-scale localization varied along the length of FAs, we quantified the axial position of WT-vinculin-N-tdEos in the distal and proximal thirds of FAs. iPALM analysis showed that vinculin localized significantly higher and had a significantly increased ARL localization in the proximal FA third compared to the distal FA third (Fig. 8d–g). Together these results show that vinculin exhibits gradients of low-to-high activation and axial position from the distal tip to proximal tip of single FAs.
Because our results indicate that vinculin activation and nano-scale localization are regulated by interactions with talin, we hypothesized that the talin binding could mediate the observed gradient in vinculin activation and axial position across single FA. Previous studies showed that talin and vinculin preferentially interact in the proximal tip of FA42. To determine if talin vertical extension could mediate the localization of vinculin to the FTL and ARL in the proximal FA tip, we compared the axial position of talin-C-tdEos in the distal and proximal thirds of FA (Fig. 8h–k). This showed that the talin C-terminus was significantly higher in the proximal than in the distal FA tip (Fig. 8i), indicating that maximum talin vertical extension spatially corresponds with higher vinculin axial position within individual FA.
Vinculin is recruited initially to the ISL and later to the FTL and ARL during myosin II-mediated FA maturation
Previous reports showed that vinculin activation increases during FA dynamics and growth36, which together with our current results suggest that vinculin nano-scale localization and binding partners may evolve during FA maturation. To obtain a synchronized population of FAs at different stages of maturation, we performed washouts of the myosin-II ATPase inhibitor, blebbistatin, and performed iPALM analysis of vinculin axial position during contractility-induced FA growth (Fig. 8l–q). We first characterized the kinetics of contractility-induced FA maturation in cells co-expressing paxillin-mCherry and vinculin-GFP. Blebbistatin treatment (50uM, 2h) inhibited FA maturation and reduced vinculin localization at FAs.38,43 Replacement of blebbistatin-containing with drug-free media during live-cell TIRF revealed that immature, diffraction-limited FAs at the cell edge underwent coordinated growth, with vinculin recruited to maturing FA as early as 1min after washout (Fig. 8l). By 5min post-washout, FA were elongated and the cell edge had advanced, and by 7min, a new row of nascent FA formed. Thus, we analyzed vinculin at 1 and 5min after blebbistatin washout to ensure that all FAs had begun maturation simultaneously.
To determine the nano-scale localization of vinculin during FA maturation, we expressed WT-vinculin-N-tdEos in cells, treated them with blebbistatin, and fixed at 1min or 5min after blebbistatin washout (Fig. 8m–n). iPALM analysis showed that at 1 min after washout, vinculin localized ~57 nm above the coverslip, with half of the molecules localized to the ISL (Fig. 8o–q), but by 5 min after induction of FA maturation, vinculin was localized ~20 nm higher with a majority of molecules in the FTL and ARL (Fig. 8p–q). Thus, vinculin localizes to the ISL at the onset of myosin II activity, and vinculin distribution shifts upward as FA undergo contractility-induced maturation.
To determine if paxillin phosphorylation could be responsible for recruitment of vinculin to the ISL in nascent FA, we characterized the dynamics of paxillin phosphorylation on Y118 (pY118-paxillin) during myosin II-induced FA maturation (Fig. 8r). Western blot analysis showed that, as reported38, blebbistatin treatment significantly reduced pY118 paxillin. One minute after blebbistatin washout, pY118 paxillin increased more than twofold with no additional increase at 5min post-washout. Therefore, consistent with earlier findings44, contractility-dependent phosphorylation of paxillinY118 occurs within the first minute of FA maturation, mirroring the kinetics of vinculin localization to the ISL during contractility-induced FA maturation. Together, our findings demonstrate that vinculin nano-scale localization evolves during myosin II-dependent FA maturation and suggest that vinculin localizes first to the ISL by interaction with phospho-paxillin in nascent FAs. As FAs grow, vinculin molecules exhibit increased localization to the FTL and ARL, likely by talin-mediated activation.
Discussion
This study provides the first demonstration of protein activity and function regulated by the nano-scale molecular architecture of FAs. While vinculin has many characterized molecular interactions that regulate distinct FA functions, it was previously unknown how vinculin’s protein interactions are spatiotemporally regulated within FAs during maturation. Using super-resolution microscopy and a FRET biosensor in combination with point-mutants with well-characterized in vitro protein binding defects and cellular phenotypes, we dissected the roles of paxillin, talin, and actin in vinculin’s activation, orientation, and nano-scale position within FA. Our work highlights the mind-blowing power of light microscopy for determining protein activity and interactions with nanometer precision within organelles in intact cells. Our results suggest that inactive vinculin is recruited to the membrane-proximal ISL by binding to phospho-paxillin. Talin binding promotes vinculin activation and targeting of active vinculin to the FTL where vinculin can engage actin in the ARL to modulate retrograde flow and strengthen adhesion. Thus, the spatial segregation of paxillin, talin, and actin along the z-axis of the FA regulates vinculin recruitment, activation, and function during FA maturation.
Based on our study and the work of others, we propose a model in which upwards redistribution of vinculin within FA structure during maturation facilitates vinculin activation and mechanical reinforcement of FA (Supplementary Figure 7). At the leading edge, integrins are activated in the protruding lamellipodium by talin-mediated45 engagement of actin retrograde flow46,47 to form nascent FAs and bind ECM. FAK and paxillin are quickly recruited5, and tension on engaged integrins48 promotes FAK activation38 and rapid tyrosine phosphorylation of paxillin7,44. Paxillin phosphorylation in nascent FA mediates the recruitment of inactive vinculin to the ISL7,38, although other pathways for recruitment are possible. Interaction with phospho-paxillin puts vinculin into proximity with its activating ligands, including PIP2 in the membrane21, talin32, and actin26, where it acts as a coincidence detector26,31. As tension builds, talin stretches to unmask vinculin binding sites35, allowing talin and a second ligand to mediate vinculin activation and promote vinculin association with the upper FA layers where it binds actin to engage retrograde flow16 and bolster the force-transmitting link between talin, integrin, and the ECM11,16. Vinculin binding stabilizes talin in a vertically extended conformation, allowing for further extension of the talin rod, exposing additional cryptic vinculin binding sites49 and further recruitment of active vinculin, allowing for positive-feedback to mechanically reinforce FA. In mature FA, inactive vinculin remains associated with phospho-paxillin in the ISL in the distal FA tip, and active vinculin is distributed in the FTL and ARL in the proximal FA tip.
Our results suggest that talin is a master regulator of vinculin nanoscale localization and function. We show that talin binding is required for activation and localization to the FTL and ARL in vivo, while surprisingly, actin binding is not. Functionally, talin binding is critical for vinculin’s ability to strengthen FA and promote FA growth10,11,13, suggesting that vinculin performs these jobs in the upper layers of the FA. Talin’s capacity to regulate vinculin’s association with the ARL also suggests it controls vinculin’s ability to engage retrograde actin flow and mechanically reinforce FAs16. Furthermore, while in vitro experiments have shown that vinculin binding prevents refolding of stretched talin,34,49 our results provide the first in vivo evidence that vinculin binding is required to maintain talin in a vertically extended conformation.
Our data suggests that vinculin dynamics are spatiotemporally regulated by distinct protein-interactions to mediate ECM mechanosensing. Since the vinculin-phospho-paxilin interaction promotes traction force oscillations in FA that facilitate mechanosensing and durotaxis,15 the dynamics of vinculin recruitment and nanoscale localization mediated by this interaction are also likely critical for mechanosensing. FRAP studies suggests that the vinculin phospho-paxilin interaction is very labile, while activated vinculin is more stably associated with FAs.33,38 Paxillin and vinculin have also been observed to interact in both the FA and the cytoplasm50, which could allow rapid loading of preassembled complexes into the FA from the cytoplasm. Thus, dynamic traction oscillations in FA could be mediated by rapid activation and redistribution of the labile, paxillin-bound pool of inactive vinculin to the upper FA layers where it reinforces FA traction.
In conclusion, the distinct vertical distributions of paxillin, talin and actin allow for the segregation of vinculin-protein interactions into nano-domains that spatio-temporally regulate vinculin activation and function during FA maturation. We suggest that spatial segregation of different protein-protein interactions into molecularly distinct nano-domains within organelles could be a broader mechanism for regulating protein activity in cells.
MATERIALS AND METHODS
Cell culture and transfection
HFF cells were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA) and maintained at 37°C at 5% CO2 in DMEM supplemented with 15% FBS (Gibco, Grand Island, NY, USA). Transfections of 2μg of plasmid DNA were performed using a Nucleofector (NHDF solution, program U-020. Lonza, Basal, Switzerland). For FRET experiments, cells were plated for 6–8hrs prior to imaging on 22 × 22mm #1.5 coverslips pre-coated with 15μg/ml fibronectin (2hr at 37°C, Millipore, Billerica, MA, USA). Imaging was performed in growth media without phenol red and supplemented with 20mM HEPES and 30 units/mL Oxyrase (Mansfield, OH, USA). For iPALM experiments, fluorescent fiducials are affixed to the coverslip to provide a constant internal reference for calibration, alignment, and spatial drift correction. 80 to 100 nm gold (Au) nanoparticles were sparsely adsorbed (~2,000 per mm2) to the coverglass surface and immobilized by 30–50nm of sputtered SiO2, as previously described4. Prior to experiments, fiducialed coverslips were UV-sterilized for 15 min, rinsed with Dulbecco’s phosphate buffered saline (DPBS, Invitrogen, Grand Island, NY, USA), coated with 15 μg/ml fibronectin (2hr at 37°C, Millipore, Billerica, MA, USA), and incubated with 1% heat-inactivated bovine serum albumin (1h, 37°C, Sigma-Aldrich. St. Louis, MO, USA) before a final rinse with DPBS. For iPALM experiments, cells were transfected for ~36h and then replated onto fibronectin coated coverslips for 6–8h, allowing time for cells to spread and migrate with minimal ECM fibrillogenesis. Cells were fixed with 4% PFA in PHEM buffer (PIPES 60mM, HEPES 25mM, EGTA 10mM, MgCl2 2mM, pH 6.9) for 15min. Imaging was performed in PHEM buffer supplemented with 2mM Trolox (Calbiochem, Billerica, MA, USA) and 0.04mg/mL Catalase (Sigma-Aldrich. St. Louis, MO, USA) to scavenge oxygen and inhibit photodamage and bleaching. We imaged focal-adhesion-containing lamella areas, typically no greater than 20μm from the cell edge, containing at least three fiducials for calibration and drift correction.
Pharmacological treatments
50μM blebbistatin was used to inhibit myosinII ATPase activity (Torronto Research Chemicals, North York, Ontario, Canada).
Plasmids
Plasmids encoding actin-mEos2, talin-N-tdEos, talin-C-tdEos, paxillin-N-tdEos, WT-vinculin-N-tdEos, CAAX-tdEos, vinculin-EGFP and paxillin-mCherry were previously described4,16,38. All fluorescent protein expression vectors were generated using a C1 or N1 (Clontech-style) cloning vector backbone. To construct the fluorescent protein cloning vectors, the cDNA encoding mCerulean, tdEos, mNeonGreen, and mTurquoise was amplified with a 5′ primer encoding an AgeI site and a 3′ primer encoding a BspEI site (C1) or a NotI site (N1) for insertion into the corresponding vector backbone. The resulting PCR products were purified, digested, and ligated into similarly treated EGFP-C1 and EGFP-N1 vector backbones, yielding C1 and N1 cloning vectors for mCerulean, tdEos, mNeonGreen, and mTurquoise.
To generate the chicken paxillin (NM_204984.1) vector, an advanced EGFP variant mEmerald (wtGFP + F64L, S65T, S72A, N149K, M153T, I167T, A206K), was initially used to characterize the fusion. To produce a paxillin fusion with a 22 amino acid linker separating the C terminus of paxillin from the fluorescent protein, paxillin cDNA was PCR amplified with a 5′ primer encoding an EcoRI site and a 3′ primer encoding a NotI site. The resulting PCR product and mEmerald-N1 were both digested by the appropriate restriction enzymes, gel purified and ligated to yield mEmerald-Paxillin-22. Upon sequence verification of the vector, mEmerald-Paxillin-22 and mCerulean-N1 were digested by EcoRI and NotI, gel purified, and ligated to yield mCerulean-Paxillin-22.
To prepare the vinculin fusion, human vinculin (NM_003373.3) was PCR amplified with a 5′ primer encoding a NheI site and a 3′ primer encoding an EcoRI site. The resulting PCR product and mEmerald-N1 were digested by the appropriate restriction enzymes, gel purified and ligated to yield mEmerald-Vinculin-21. Upon sequence verification of the vector, mEmerald-Vinculin-21, tdEos-N1, mNeonGreen-N1, and mTurquoise-N1 were digested with NheI and EcoRI, gel purified, and ligated to yield tdEos-Vinculin-21 (WT-vinculin-C-tdEos), mNeonGreen-Vinculin-21, and mTurquoise-Vinculin-21.
Dr. Susan W. Craig generously provided cDNAs of ECFP-EYFP vinculin FRET probes36. To generate the mTurquoise-mNeonGreen vinculin tail probe, the ECFP-EYFP vinculin tail probe was PCR amplified with AgeI forward and BspEI reverse primers (Supplementary Table 3). The resulting PCR product was digested, gel purified, and ligated into a similarly treated C1 cloning vector. Upon sequence verification, mTurquoise was PCR amplified with KpnI forward and XhoI reverse primers (Supplementary Table 3). The PCR fragment was digested, purified, and ligated to the similarly treated ECFP-EYFP-vinculin tail probe described above. This yielded the mTurquoise-EYFP vinculin tail probe intermediate. Following sequence verification, mNeonGreen was PCR amplified with NotI forward and NotI reverse primers (Supplementary Table 3). The PCR fragment was digested, purified, and ligated to a similarly treated mTurquoise-EYFP vinculin tail probe intermediate. This resulted in the mTurquoise-mNeonGreen vinculin tail probe.
To generate the vinculin control probe, HindIII forward and KpnI reverse primers (Table S3) were used to PCR amplify ECFP and amino acids (1–419) of vinculin. The resulting PCR product was purified, digested, and ligated to a similarly treated mNeonGreen C1 cloning vector. This resulted in an ECFP-mNeonGreen vinculin control intermediate. Following sequence verification, mTurquoise was PCR amplified using HindIII forward and EcoRI reverse primers (Supplementary Table 3). The PCR fragment was digested, purified, and ligated to a similarly treated ECFP-mNeonGreen vinculin control probe intermediate. This resulted in the mTurquoise-mNeonGreen vinculin control probe.
To construct the mTurquoise-mNeonGreen-FRET-10 probe, NheI forward and AgeI reverse primers were used to PCR amplify mNeonGreen. The resulting PCR product was digested, gel purified, and ligated to a similarly treated mTurquoise-N1 cloning vector, generating a 10 amino acid linker (SGLRSPPVAT) separating the fluorescent proteins. This yielded mTurquoise-mNeonGreen FRET-10.
All fusion DNA was purified using the Plasmid Maxi kit (QIAGEN, Valencia, CA, USA) and sequence verified (The Florida State University Bioanalytical and Molecular Cloning DNA Sequencing Laboratory, Tallahassee, FL, USA). To ensure proper localization, all fusion proteins were characterized by transfection in HeLa cells (CCL2 line; ATCC, Manassas, VA, USA) using Effectene (QIAGEN) and ~1μg plasmid DNA. Transfected cells were grown on coverglass in DMEM/F12, fixed after 48h, and mounted with Gelvatol.
Mutagenesis to generate vinculin and paxillin mutants
The vinculin and paxillin point mutants were generated from the WT constructs using the Quick-change II site-directed mutagenesis kit (Agilent Technologies, Santa Clare, CA, USA) and the forward primers listed in Supplementary Table 3 and their reverse complements.
siRNA Knockdown of endogenous protein
To inhibit paxillin expression in HFF cells, we used the ON_TARGETplus SMART-pool for Human PXN (Cat# L-005163-00-0005), and to inhibit Hic-5 expression we used the ON_TARGETplus SMART-pool for Human TGFB1/1(Cat# L-006565-00-0005, Dharmacon, Lafayette, CO, USA). To inhibit vinculin expression we pooled two custom siRNAs targeted to the 3′UTR of vinculin (Sequences in Supplementary Table 3). (Dharmacon, Lafayette, CO, USA). siRNA was transfected into cells using the Nucleofector and knockdown was achieved after 48h (paxillin and Hic5) or 72h (vinculin).
Western blot
Cells lysed in 2X SDS Protein Gel Loading Solution (Quality Biological, Gaithersburg, MD, USA) were separated by SDS-PAGE and electro-transferred to Immobilon-P PVDF membrane (Millipore, Billerica, MA, USA). Membranes were blocked with 3% non-fat dry milk in TBS-T buffer (20mM Tris pH 7.6, 137mM NaCl2, 0.1% (vol/vol) Tween-20) incubated with primary antibody overnight at 4°C, washed with TBS-T, incubated with appropriate HRP-conjugated secondary antibodies and visualized with Immobilon chemiluminescent HRP Substrate (Millipore, Billerica, MA, USA). Antibodies and dilutions: mouse anti-paxillin (1:1000, BD Transduction Labs cat# 610051, Franklin Lakes, NJ, USA). Mouse anti-Talin (1:1000, Abcam cat# ab11188, Cambridge, MA, USA). Mouse anti-vinculin (1:1000, Sigma-Aldrich cat# V4505. St. Louis, MO, USA), rabbit anti-gfp (1:5,000. Abcam cat# ab290, Cambridge, MA, USA), rabbit anti-Y118 paxillin (1:500, Invitrogen cat# 44-722G, Grand Island, NY, USA). HRP-conjugated anti-mouse (cat# 715-035-150) or anti-rabbit (cat# 711-035-152), 1:5,000 (Jackson ImmunoResearch Laboratories, Inc, West Grove, PA). To quantify paxillin phosphorylation levels, unsaturated exposures of the western blots were acquired with a myECL Imager (ThermoFischer, Waltham, MA, USA). The background-corrected mean intensity was measured for each band using the myECL software. The phosphopaxillin intensity was divided by the total paxillin intensity, and values were normalized relative to control treatment.
Immunoprecipitation
HFF cells were transfected with vinculin siRNA for 60h. Cells were then transfected with GFP (non-specific control) or vinculin-GFP constructs, plated in 10cm tissue culture plates precoated with 15μg/ml fibronectin, and supplemented with 15% FBS for 12h. Cells were harvested in lysis buffer (50mM Tris pH 8.0, 150mM NaCl, 5mM EDTA, 5% glycerol, 1% Triton-X + complete mini-EDTA free protease inhibitor cocktail and PhosSTOP phosphatase inhibitor (Roche Diagnostics, Indianapolis, IN, USA) for 20min at 4°C. Lysates were clarified by 20min centrifugation at 14,000 rpm. Supernatants were incubated with rabbit anti-GFP antibody (1:300, Abcam cat# ab290, Cambridge, MA, USA) and rotated overnight at 4°C. The following day, the solution was incubated with 30μl proteinA Dynabeads (Invitrogen, Grand Island, NY) with rotation at 4°C for 1h. Beads were washed two times with lysis buffer, 1 time with DPBS and resuspended in 30 μl 2X SDS Protein Gel Loading Solution (Quality Biological, Gaithersburg, MD, USA). Samples were boiled for 10min and analyzed by Western blotting using the appropriate antibodies.
TIRF Microscopy
Dual-color time-lapse TIRF microscopy of EGFP-Vinculin and mCherry paxillin in living cells was performed at 37°C using an Apo TIRF 100x 1.49 NA oil immersion objective lens (Nikon Instruments, Melville, NY, USA) on an inverted Eclipse Ti microscope system (Nikon Instruments, Melville, NY, USA51). Pairs of EGFP (using 488nm laser illumination, Coherent, Santa Clara, CA, USA) and mCherry (using 561nm laser illumination, Coherent, Santa Clara, CA, USA) images were captured in rapid succession at 20sec intervals using a CCD (coolsnap HQ2; Photometrics, Tuscon, AZ, USA) operated in the 5 MHz readout mode. Blebbistatin washout during high resolution imaging was performed using an RC-30 perfusion chamber (Warner Instruments, Hamden, CT, USA) as previously described 44.
FRET Imaging and Analysis
Widefield epifluorescence imaging of FRET biosensors in living cells was performed at 37°C using an 60x 1.40 NA PlanApo oil immersion objective lens (Nikon Instruments, Melville, NY, USA) on an inverted TE2000-E microscope system (Nikon Instruments, Melville, NY, USA). Illumination was provided by a solid state white light (Lumencor SOLA SE, Beaverton, OR, USA) using the CFP/YFP/HcRed-3XA Sedat filter set with a 444/521/608 dichroic mirror (Semrock, Rochester, NY, USA). The following filters were used: mTurquoise image: Excitation 427/10, Emission 472/30; FRET image: Excitation 427/10, Emission 542/27; NeonGreen image: Excitation 504/12, Emission 542/27. Cells were located and brought into focus with mTurquoise filters to prevent bleaching of the acceptor fluorophore. Images were captured in rapid succession (mTurquoise, then FRET, then NeonGreen) using a CCD (Coolsnap HQ2, Photometrics, Tuscon, AZ, USA). For each channel, ten images were acquired in a cell-free area to use for shade correction. Ten dark-current images were acquired using mTurquoise filters without illumination.
The raw mTurqouise, FRET and Neon Green images were aligned using the rigid body method of the StackReg ImageJ plugin52. The FRET ratio was calculated using MATLAB to run the Danuser Lab Biosensors processing software 2.1(available for download: http://lccb.hms.harvard.edu/software.html). Briefly, images were manually thresholded to create a binary mask of the cell. Dark-current images were filtered with a 3×3 median filter, averaged, and subtracted from each raw image. Shade correction images were filtered with a 3×3 median filter and averaged. The dark-current-corrected images were divided by the averaged shade correction image. Using the mask of the cell to define the background, the average background intensity was subtracted from the image. Bleed-through of mTurquoise and NeonGreen into the FRET channel was corrected with experimentally determined bleed-through coefficients. The FRET ratio image was created by dividing the corrected FRET image by the corrected mTurquoise image. The mask of the cell was applied so that the ratio value outside the cell was zero.
For each cell, FAs were segmented using a combination of Otsu and Rosin thresholding of the raw mTurquoise image. The resulting mask was applied to the processed FRET ratio image to determine the mean FRET ratio value within FAs (inside the masked regions) and in the cytoplasm (outside the masked regions). To compare the distal and proximal regions of FAs, a line-scan was manually drawn along the long axis of individual FAs. The FA was defined as the pixels along the line-scan where the intensity was >2*standard deviation of the adjacent cytoplasm intensity. The FA line-scan was divided into thirds, and the mean FRET value in the distal 1/3 and proximal 1/3 were computed.
iPALM imaging
Instrumentation associated with the iPALM method has been described in detail53. The sample was mounted between two dual-opposed infinity-corrected objectives (Nikon CFI Apochromat TIRF 60x, NA=1.49, Nikon Instruments Inc., Melville, NY) with index matching oil (Cargille type DF, Cargille Laboratories, Cedar Grove, NJ). We used light from a 50mW 405nm diode laser (Coherent Inc., Santa Clara, CA), attenuated by neutral density filters, to achieve single fluorophore-level Eos activation and a 150mW 561nm diode pumped solid-state laser (CrystaLaser, Reno, NV) for excitation of activated fluorophores. Narrow bandwidth laser line filters (MaxLine©LL01-561-12.5 and MaxDiode©LD01-405/10-12.5, Semrock Inc., Rochester, NY) were placed before the objectives to reject emission noise from the laser and autofluorescence generated in the optical path. The activation and excitation beams entered the back focal planes of the objectives through slots in custom turning mirrors, and the radial position of the activation and excitation beams were adjusted to produce near total internal reflection condition. The emission fluorescence signal was collected by the two objective lenses, and mirrors were used to direct the emitted light from the objectives into the custom designed beam-splitter (Rocky Mountain Instruments, Lafayette, CO). The output beams from the 3-way beam splitter were then focused on three separate EMCCD cameras (Andor iXon DU-897, Andor Technology, Belfast, Northern Ireland) via achromatic lenses (01LAO799, CVI Melles Griot, Albuquerque, NM). Long-pass and band-pass optical filters (RazorEdge©LP02-568RU-25 and BrightLine©FF01-588/21-25, Semrock Inc., Rochester, NY) were placed in front of the cameras to reject the excitation light. Typical iPALM data acquisition consisted of 20,000–40,000 images acquired with three Andor iXon EMCCD cameras operated in EM gain, frame-transfer mode with a 50ms exposure per image. Data acquisition was carried out using software written in LABVIEW (National Instruments). Data analysis, image processing, and rendering was performed using software written in IDL (ITT Visual Information Solutions) and run on a Linux computational cluster at HHMI Janelia Farm Research Campus.
25–100nm gold (Au) nanoparticles were used for drift correction, image registration, and interferometry calibration. For each iPALM image set, the optics were aligned using localization of a single fiducial. The focal plane was found with the bottom objective, which was then held constant, while the top objective was adjusted to the same focal plane and lateral position. To properly align the optics for interference, the sample holder was continuously oscillated along the z-axis over a range of 400nm. The beam-splitter was then translated up or down until the intensity of the fiducial oscillated due to the interferometric effect. A calibration curve was acquired by translating the sample in discrete z steps of 1 nm over a range of 250nm. The intensity of the fiducial would then oscillate with a similar period but with phase differences between the cameras. Fine alignment of the position and tilt of the bottom mirror in the beamsplitter was performed to obtain ~120° phase difference between each camera. Once alignment was achieved and a final calibration curve was taken, the acquisition of raw image sets could proceed.
After image acquisition was complete, the raw datasets were processed to localize individual molecules and extract their X, Y, and Z coordinates, as described previously4,53. First the individual camera images were aligned and added together to form a sum image. Individual fluorescent particles in the sum image were fit to a two-dimensional Gaussian by nonlinear least square fitting to obtain x and y coordinates. The peak amplitude of fluorescent particles in each individual camera image was determined with two-dimensional Gaussian fit. These amplitudes were used to extract the z-coordinate of each fluorescent molecule from the calibration curve. Sample drift was corrected by tracking the position of fiducials. Lateral sample drift was typically less than 5 nm and vertical (z-coordinate) sample drift varied between 10nm and 60nm. iPALM images were rendered from the processed list of three-dimensional molecular coordinates. The position of the x-y localization was represented by a normalized two-dimensional Gaussian with a width proportional to lateral localization uncertainty, while the position of the Z-coordinate was represented by colour.
Analysis of processed iPALM data
The measured molecular X, Y, and Z coordinates were exported into a .txt file for analysis in MATLAB. Processed iPALM datasets included the coordinates of fluorescent molecules localized in FAs and the surrounding cytoplasm as well as auto-fluorescent molecules inside and outside the cells. To quantify the spatial distribution of the proteins specifically residing within individual FA regions, we manually segmented FAs from the top-view X-Y iPALM images such that each rectangular ROI contained a single FA and the immediately adjacent space. For each individual FA, the vertical Z coordinates contained in the ROI was plotted in a 1nm binned histogram. As described previously4, in addition to the main peak representing molecules in the FA, each histogram contained a smaller peak of auto-fluorescent molecules from the substrate surface. We determined the centre vertical positions (Z-centre) and width parameter (σ) of each peak using a Guassian fit. We subtracted the coverslip Z-centre from the FA Z-centre, so that FA Z-centre measurements represent the distance from the local coverslip. We previously found that integrin cytoplasmic tails are ~25–30nm above the coverslip and the FAs extend ~150nm above the coverslip4. Therefore, molecules with X–Y coordinates within the ROI and Z coordinates between 25–150 nm above the local coverslip Z-centre were considered “FA-associated”. For each ROI, we calculated the median Z-position of FA-associated molecules and subtracted the coverslip Z-centre from this value to obtain the Z-median relative to the coverslip. Finally, for each segmented FA we calculated the percent of FA-associated molecules in each of the defined FA layers (ISL: 25–54nm, FTL: 55–84nm, ARL: 85–150nm). Thus, we are able to calculate the Z-centre, Z-median, and Z-distribution for each individual FA in the cell.
To compare different conditions, we calculated the average Z-median, average Z-distribution and average percent of molecules in each FA layer, and we computed 95% confidence intervals by resampling our datasets 10,000 times using the Bootstrapping method. Confidence intervals are displayed as error bars on bar graphs or as lightly shaded regions on the average Z-distribution plots.
Statistical analysis
Differences between iPALM Zmedian, percent localization to each layer, and mean FRET ratios were determined by an analysis of variance (ANOVA) followed by Tukey post-hoc anlaysis and differences were considered significant at p<0.05. Average iPALM Z-distributions were statistically compared using a two-sample Kolmogorov-Smirnov test. Unless otherwise stated, all data are presented as mean ± 95% confidence interval of the mean.
Supplementary Material
Acknowledgments
The authors thank Susan Craig (Johns Hopkins University) for the cDNA encoding the vinculin FRET activation biosensor, Gaudenz Danuser (UT Southwestern) for the FRET biosensor image analysis package, William Shin for maintenance of the Waterman Lab microscopes, Dorothy Honemond and Schwanna Thacker for administrative assistance, Tony Kanchanawong (National University of Singapore) for sharing and discussing iPALM protocols, and members of the Waterman Lab, Greg Alushin (NHLBI), Hunter Elliot (Harvard Medical School) and Justin Taraska (NHLBI) for helpful discussions. Funding: Division of Intramural Research, NHLBI (L.B.C. and C.M.W.); Howard Hughes Medical Institute (G.S. and H.F.H.); GM081764 (S.L.C.), GM080568 (S.L.C.).
Footnotes
Author Contributions
L.B.C and C.M.W. conceived the study and wrote the manuscript with input from all authors. L.B.C., C.M.W. and S.L.C. designed experiments. L.B.C. performed and analyzed most experiments. L.B.C and G.S. performed iPALM imaging. M.A.B. and M.W.D. designed novel cDNA constructs and performed cloning. G.S. and H.F.H. conceived of, built and maintained iPALM instrumentation and developed iPALM processing tools. L.B.C., M.A.B., M.W.D, and C.M.W. designed the summary cartoon.
References
- 1.Kuo JCC, Han X, Hsiao CTT, Yates JR, III, Waterman CM. Analysis of the myosin-II-responsive focal adhesion proteome reveals a role for beta-Pix in negative regulation of focal adhesion maturation. Nat Cell Biol. 2011;13:383–393. doi: 10.1038/ncb2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Byron A, Humphries JD, Bass MD, Knight D, Humphries MJ. Proteomic analysis of integrin adhesion complexes. Sci Signal. 2011;4(pt2) doi: 10.1126/scisignal.2001827. [DOI] [PubMed] [Google Scholar]
- 3.Schiller HB, Friedel CC, Boulegue C, Fässler R. Quantitative proteomics of the integrin adhesome show a myosin II-dependent recruitment of LIM domain proteins. EMBO Rep. 2011;12:259–66. doi: 10.1038/embor.2011.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kanchanawong P, et al. Nanoscale architecture of integrin-based cell adhesions. Nature. 2010;468:580–4. doi: 10.1038/nature09621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi CK, et al. Actin and alpha-actinin orchestrate the assembly and maturation of nascent adhesions in a myosin II motor-independent manner. Nat Cell Biol. 2008;10:1039–50. doi: 10.1038/ncb1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zaidel-Bar R, Ballestrem C, Kam Z, Geiger B. Early molecular events in the assembly of matrix adhesions at the leading edge of migrating cells. J Cell Sci. 2003;116:4605–13. doi: 10.1242/jcs.00792. [DOI] [PubMed] [Google Scholar]
- 7.Zaidel-Bar R, Milo R, Kam Z, Geiger B. A paxillin tyrosine phosphorylation switch regulates the assembly and form of cell-matrix adhesions. J Cell Sci. 2007;120:137–48. doi: 10.1242/jcs.03314. [DOI] [PubMed] [Google Scholar]
- 8.Zamir E, et al. Molecular diversity of cell-matrix adhesions. J Cell Sci. 1999;112(Pt 1):1655–69. doi: 10.1242/jcs.112.11.1655. [DOI] [PubMed] [Google Scholar]
- 9.Wolfenson H, et al. A role for the juxtamembrane cytoplasm in the molecular dynamics of focal adhesions. PLoS One. 2009;4:e4304. doi: 10.1371/journal.pone.0004304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diez G, Auernheimer V, Fabry B, Goldmann WH. Head/tail interaction of vinculin influences cell mechanical behavior. Biochem Biophys Res Commun. 2011;406:85–8. doi: 10.1016/j.bbrc.2011.01.115. [DOI] [PubMed] [Google Scholar]
- 11.Dumbauld DW, et al. How vinculin regulates force transmission. Proc Natl Acad Sci U S A. 2013;110:9788–93. doi: 10.1073/pnas.1216209110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saunders RM, et al. Role of vinculin in regulating focal adhesion turnover. Eur J Cell Biol. 2006;85:487–500. doi: 10.1016/j.ejcb.2006.01.014. [DOI] [PubMed] [Google Scholar]
- 13.Humphries JD, et al. Vinculin controls focal adhesion formation by direct interactions with talin and actin. J Cell Biol. 2007;179:1043–57. doi: 10.1083/jcb.200703036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carisey A, et al. Vinculin regulates the recruitment and release of core focal adhesion proteins in a force-dependent manner. Curr Biol. 2013;23:271–81. doi: 10.1016/j.cub.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Plotnikov SV, Pasapera AM, Sabass B, Waterman CM. Force fluctuations within focal adhesions mediate ECM-rigidity sensing to guide directed cell migration. Cell. 2012;151:1513–27. doi: 10.1016/j.cell.2012.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thievessen I, et al. Vinculin-actin interaction couples actin retrograde flow to focal adhesions, but is dispensable for focal adhesion growth. J Cell Biol. 2013;202:163–77. doi: 10.1083/jcb.201303129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Subauste MC, et al. Vinculin modulation of paxillin-FAK interactions regulates ERK to control survival and motility. J Cell Biol. 2004;165:371–81. doi: 10.1083/jcb.200308011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burridge K, Mangeat P. An interaction between vinculin and talin. Nature. 308:744–6. doi: 10.1038/308744a0. [DOI] [PubMed] [Google Scholar]
- 19.Jockusch BM, Isenberg G. Interaction of alpha-actinin and vinculin with actin: opposite effects on filament network formation. Proc Natl Acad Sci U S A. 1981;78:3005–9. doi: 10.1073/pnas.78.5.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Turner CE, Glenney JR, Burridge K. Paxillin: a new vinculin-binding protein present in focal adhesions. J Cell Biol. 1990;111:1059–68. doi: 10.1083/jcb.111.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilmore AP, Burridge K. Regulation of vinculin binding to talin and actin by phosphatidyl-inositol-4-5-bisphosphate. Nature. 1996;381:531–5. doi: 10.1038/381531a0. [DOI] [PubMed] [Google Scholar]
- 22.DeMali KA, Barlow CA, Burridge K. Recruitment of the Arp2/3 complex to vinculin: coupling membrane protrusion to matrix adhesion. J Cell Biol. 2002;159:881–91. doi: 10.1083/jcb.200206043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kioka N, et al. Vinexin: a novel vinculin-binding protein with multiple SH3 domains enhances actin cytoskeletal organization. J Cell Biol. 1999;144:59–69. doi: 10.1083/jcb.144.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson RP, Craig SW. An intramolecular association between the head and tail domains of vinculin modulates talin binding. J Biol Chem. 1994;269:12611–12619. [PubMed] [Google Scholar]
- 25.Johnson RP, Craig SW. F-actin binding site masked by the intramolecular association of vinculin head and tail domains. Nature. 1995;373:261–4. doi: 10.1038/373261a0. [DOI] [PubMed] [Google Scholar]
- 26.Chen H, Choudhury DM, Craig SW. Coincidence of actin filaments and talin is required to activate vinculin. J Biol Chem. 2006;281:40389–98. doi: 10.1074/jbc.M607324200. [DOI] [PubMed] [Google Scholar]
- 27.Calderwood DA, et al. The Talin Head Domain Binds to Integrin Subunit Cytoplasmic Tails and Regulates Integrin Activation. J Biol Chem. 1999;274:28071–28074. doi: 10.1074/jbc.274.40.28071. [DOI] [PubMed] [Google Scholar]
- 28.Goldmann WH, et al. Examining F-Actin Interaction with Intact Talin and Talin Head and Tail Fragment using Static and Dynamic Light Scattering. Eur J Biochem. 1997;250:447–450. doi: 10.1111/j.1432-1033.1997.0447a.x. [DOI] [PubMed] [Google Scholar]
- 29.Critchley DR. Biochemical and structural properties of the integrin-associated cytoskeletal protein talin. Annu Rev Biophys. 2009;38:235–54. doi: 10.1146/annurev.biophys.050708.133744. [DOI] [PubMed] [Google Scholar]
- 30.Rubashkin MG, et al. Force Engages Vinculin and Promotes Tumor Progression by Enhancing PI3K Activation of Phosphatidylinositol (3,4,5)-Triphosphate. Cancer Res. 2014;74:4597–4611. doi: 10.1158/0008-5472.CAN-13-3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bakolitsa C, et al. Structural basis for vinculin activation at sites of cell adhesion. Nature. 2004;430:583–6. doi: 10.1038/nature02610. [DOI] [PubMed] [Google Scholar]
- 32.Cohen DM, Chen H, Johnson RP, Choudhury B, Craig SW. Two distinct head-tail interfaces cooperate to suppress activation of vinculin by talin. J Biol Chem. 2005;280:17109–17. doi: 10.1074/jbc.M414704200. [DOI] [PubMed] [Google Scholar]
- 33.Cohen DM, Kutscher B, Chen H, Murphy DB, Craig SW. A conformational switch in vinculin drives formation and dynamics of a talin-vinculin complex at focal adhesions. J Biol Chem. 2006;281:16006–15. doi: 10.1074/jbc.M600738200. [DOI] [PubMed] [Google Scholar]
- 34.Ciobanasu C, Faivre B, Le Clainche C. Actomyosin-dependent formation of the mechanosensitive talin-vinculin complex reinforces actin anchoring. Nat Commun. 2014;5:3095. doi: 10.1038/ncomms4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Del Rio A, et al. Stretching single talin rod molecules activates vinculin binding. Science. 2009;323:638–41. doi: 10.1126/science.1162912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen H, Cohen DM, Choudhury DM, Kioka N, Craig SW. Spatial distribution and functional significance of activated vinculin in living cells. J Cell Biol. 2005;169:459–70. doi: 10.1083/jcb.200410100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shaner NC, et al. A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat Methods. 2013;10:407–9. doi: 10.1038/nmeth.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pasapera AM, Schneider IC, Rericha E, Schlaepfer DD, Waterman CM. Myosin II activity regulates vinculin recruitment to focal adhesions through FAK-mediated paxillin phosphorylation. J Cell Biol. 2010;188:877–90. doi: 10.1083/jcb.200906012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brown MC, Perrotta JA, Turner CE. Identification of LIM3 as the principal determinant of paxillin focal adhesion localization and characterization of a novel motif on paxillin directing vinculin and focal adhesion kinase binding. J Cell Biol. 1996;135:1109–23. doi: 10.1083/jcb.135.4.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thompson PM, et al. Identification of an Actin Binding Surface on Vinculin that Mediates Mechanical Cell and Focal Adhesion Properties. Structure. 2014;22:697–706. doi: 10.1016/j.str.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gardel ML, et al. Traction stress in focal adhesions correlates biphasically with actin retrograde flow speed. J Cell Biol. 2008;183:999–1005. doi: 10.1083/jcb.200810060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bachir AI, et al. Integrin-Associated Complexes Form Hierarchically with Variable Stoichiometry in Nascent Adhesions. Curr Biol. 2014;24:1845–1853. doi: 10.1016/j.cub.2014.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wolfenson H, Bershadsky A, Henis YI, Geiger B. Actomyosin-generated tension controls the molecular kinetics of focal adhesions. J Cell Sci. 2011;124:1425–32. doi: 10.1242/jcs.077388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schneider IC, Hays CK, Waterman CM. Epidermal growth factor-induced contraction regulates paxillin phosphorylation to temporally separate traction generation from de-adhesion. Mol Biol Cell. 2009;20:3155–67. doi: 10.1091/mbc.E09-03-0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tadokoro S, et al. Talin binding to integrin beta tails: a final common step in integrin activation. Science. 2003;302:103–6. doi: 10.1126/science.1086652. [DOI] [PubMed] [Google Scholar]
- 46.Astrof NS, Salas A, Shimaoka M, Chen J, Springer TA. Importance of force linkage in mechanochemistry of adhesion receptors. Biochemistry. 2006;45:15020–8. doi: 10.1021/bi061566o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang G, Giannone G, Critchley DR, Fukumoto E, Sheetz MP. Two-piconewton slip bond between fibronectin and the cytoskeleton depends on talin. Nature. 2003;424:334–7. doi: 10.1038/nature01805. [DOI] [PubMed] [Google Scholar]
- 48.Friedland JC, Lee MH, Boettiger D. Mechanically activated integrin switch controls alpha5beta1 function. Science (80- ) 2009;323:642–644. doi: 10.1126/science.1168441. [DOI] [PubMed] [Google Scholar]
- 49.Yao M, et al. Mechanical activation of vinculin binding to talin locks talin in an unfolded conformation. Sci Rep. 2014;4:4610. doi: 10.1038/srep04610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hoffmann JE, Fermin Y, Stricker RL, Ickstadt K, Zamir E. Symmetric exchange of multi-protein building blocks between stationary focal adhesions and the cytosol. Elife. 2014;3:e02257–e02257. doi: 10.7554/eLife.02257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shin W, Fischer RS, Kanchanawong P, Kim Y, Lim J, Myers KA, Nishimura Y, Plotnikov SV, Thievessen I, Yarar D, Sabass B, Waterman CM. Live Cell Imaging A Lab Manual. 2. 2010. [Google Scholar]
- 52.Thévenaz P, Ruttimann UE, Unser M. A pyramid approach to subpixel registration based on intensity. IEEE Trans Image Process. 1998;7:27–41. doi: 10.1109/83.650848. [DOI] [PubMed] [Google Scholar]
- 53.Shtengel G, et al. Imaging cellular ultrastructure by PALM, iPALM, and correlative iPALM-EM. Methods Cell Biol. 2014;123:273–94. doi: 10.1016/B978-0-12-420138-5.00015-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.