Abstract
The first cell fate decisions during mammalian development establish tissues essential for healthy pregnancy. The mouse has served as a valuable model for discovering pathways regulating the first cell fate decisions, because of the ease with which early embryos can be recovered and an arsenal of classical and emerging methods for manipulating gene expression. Here we summarize the major pathways that govern the first cell fate decisions in mouse development. This knowledge serves as a paradigm for exploring how emergent properties of a self-organizing system can dynamically regulate gene expression and cell fate plasticity. Moreover, it brings to light the processes that establish healthy pregnancy and embryonic stem (ES) cells. We also describe unsolved mysteries and new technologies that could help overcome experimental challenges in the field.
Keywords: emergent properties, preimplantation, stem cells, reproduction, gene networks, pluripotency
Spotlight on the mouse blastocyst
The past ten years have met with an explosion of literature characterizing the molecular and cellular events leading to formation of the mouse blastocyst. This tiny structure has been pushed into the spotlight for several reasons. First, formation of the blastocyst, which occurs during the first few days after fertilization, proceeds through a sequence of events that are morphologically highly similar between mice and humans. Defects in the preimplantation period can cause pregnancy loss, and recurrent pregnancy loss impacts 5% of couples trying to conceive [1]. Therefore, the mouse provides an excellent model for reproductive biology. Second, the blastocyst is the source of embryonic stem (ES) cells, a cell type famous for its abilities to produce unlimited quantities of itself while maintaining capacity for forming tissues and organs, and thus the blastocyst is an important tool in stem cell research. Third, newly improved methods for single-cell gene expression, live imaging, and confocal microscopy have enabled exquisite access to all cells of the embryo throughout the preimplantation period. Together with a variety of tools for genetic manipulation and in vitro culture of preimplantation stages, these approaches allow for precise resolution of the temporospatial roles of genes and signaling pathways in blastocyst formation. Here we highlight some of the studies that have contributed to our current understanding of how patterned gene expression is initiated and maintained during the first cell fate decisions in mammalian development.
The importance of timing
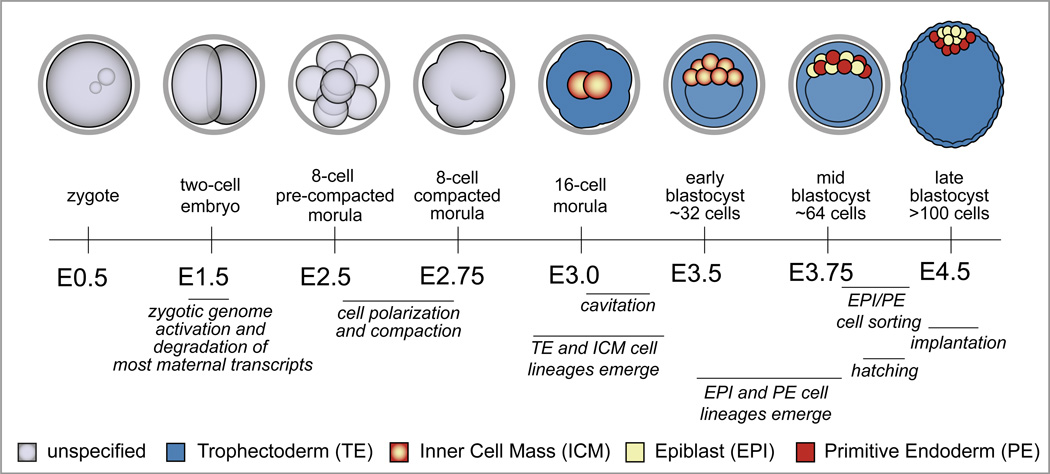
To understand current models of the molecular mechanisms of blastocyst formation, it is important to first clarify the conventions used for properly staging preimplantation embryos. In mice, embryos of desired developmental stages are produced by timed matings. Mice are assumed to have mated around midnight (E0.0) if a solid, seminiferous plug is detected within the vagina the following morning. Thus, litters of embryos at discrete developmental stages, such as zygote, morula, and blastocyst (Glossary), can be recovered at predictable time points (Fig. 1). Notably these developmental stages are based on gross morphological features and are, therefore, relatively broad and can include embryos undergoing diverse developmental events. In addition, cleavage divisions are asynchronous, both within and among embryos in a litter, and mutations, as well as genetic background [2], can alter developmental timing. Therefore, it is now standard to report embryo staging both in terms of days since fertilization and cell number (e.g., E3.5 and ~32 cells). By the same argument, it is important to compare experimental and control embryos that are stage-matched, in terms of cell number, and not just time since fertilization. This increased level of accuracy has allowed for more precise resolution of the dynamic gene regulatory events occurring during the first cell fate decisions in mammalian development.
Figure 1. Developmental milestones prior to implantation.
Cell fates are color-coded according to the legend at the bottom of the figure, and E refers to the number of days after fertilization, which occurs at E0.0. Zygotic genome activation occurs at the two-cell stage, concurrent with degradation of the maternally provided transcriptome. Cell polarization and compaction occur at the late eight-cell stage. TE and ICM cell fates emerge during the eight to sixteen cell transition, and then the blastocoel forms by cavitation, starting between E3.0 and E3.25. During blastocyst expansion, the TE exists as a polarized epithelium, while ICM cells are nonpolar. As the ICM matures, EPI and PE cell fates emerge in the absence of an apparent spatial pattern. Eventually, EPI and PE cells occupy discrete layers within the ICM. Implantation occurs around E4.5, after the embryo hatches from the zona pellucida (grey ring encircling the cleavage stage embryos).
The first two cell fate decisions in development
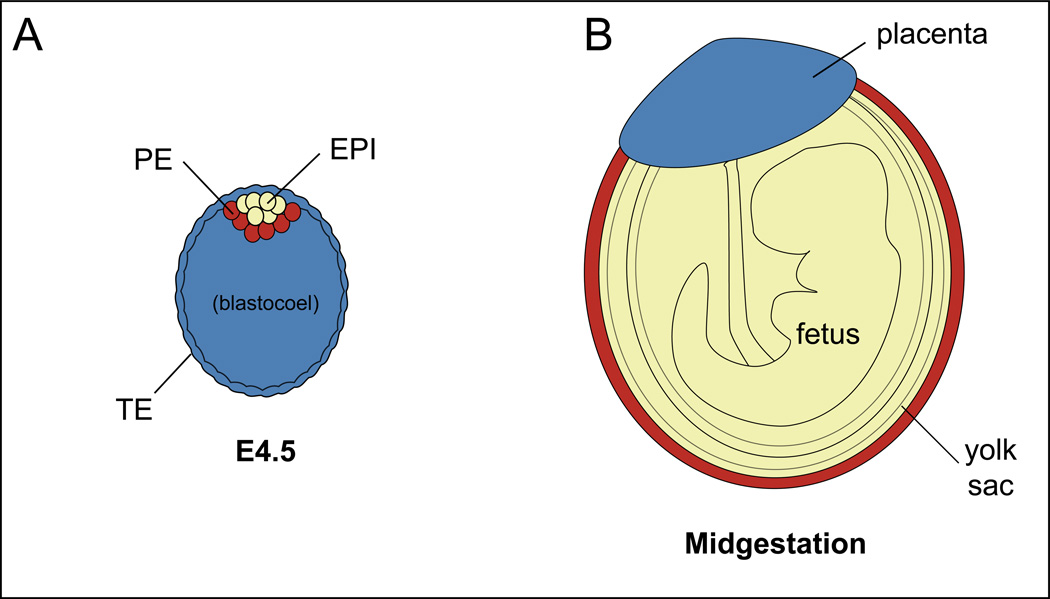
During the first three days after fertilization, the goal of the embryo is to establish three lineages: one fetal [epiblast (EPI)] and two extraembryonic [trophectoderm (TE) and primitive endoderm (PE)] (Fig. 2A). These three lineages, EPI, TE, and PE, will go on to contribute to fetus, placenta, and yolk sac, respectively (Fig. 2B), and are established by two cell fate decisions. The first cell fate decision segregates the TE from the inner cell mass (ICM), beginning around the 16-cell stage (E3.0) (Fig. 1). The second cell fate decision segregates the ICM into EPI and the PE cell types, beginning around the 64-cell stage (E3.5-E3.75). The EPI cells are the progenitors of ES cells, and, therefore, uncovering the molecular mechanisms by which EPI cells are established and maintained can teach us about the origins of pluripotency. However, it is important to recognize that ES cells are not present in the embryo at any stage, but are grown from EPI cells over the course of days in culture. Moreover, ES cell progenitors not fully established until E4.5 [3], which is a relatively late stage of blastocyst development. Therefore, the mechanisms maintaining expression of pluripotency genes in ES cells not necessarily identical to the mechanisms by which pluripotency gene expression is first established in the embryo. These observations highlight the importance of understanding the mechanisms of gene regulation in the embryo in addition to important lessons from stem cell lines derived from the blastocyst [4, 5].
Figure 2. Destinies of the blastocyst lineages.
A) At E4.5, the blastocyst contains three cell types, corresponding to three major lineages: one fetal, and two extraembryonic. B) The three major lineages are color-coded in the later mouse embryo, according to their cell type of origin in the blastocyst.
Although diverse pathways and mechanisms regulate the first two cell fate decisions, a common theme unites both decisions. For both decisions, there is first a round of cell signaling, which selects cell fates (TE versus ICM or EPI versus PE), and then these cell fates are reinforced by lineage-specific transcription factors. Next, we discuss details of the roles and regulation of the signaling pathways and transcription factors that regulate the first two cell fate decisions.
The first cell fate decision: establishment of TE and ICM cell types prior to blastocyst formation
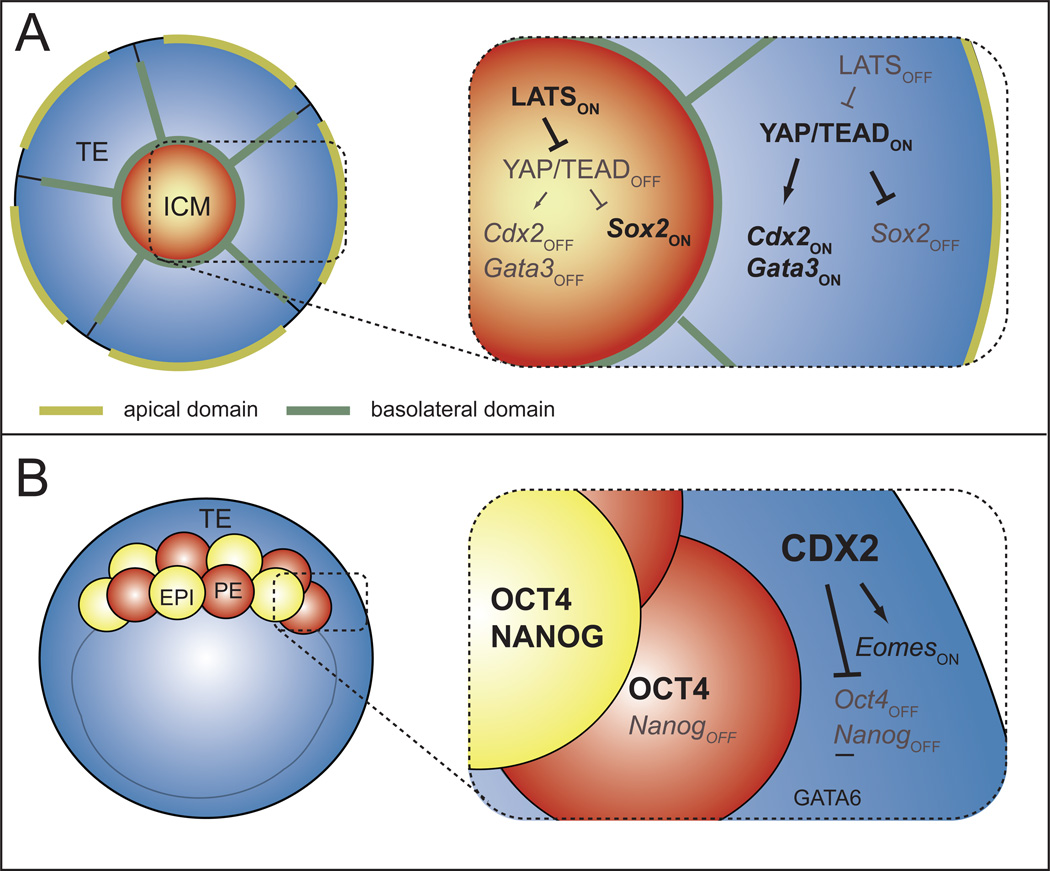
The first cell fate decision, segregation of TE and ICM cell fates occurs around the 16-cell stage, prior to blastocyst formation, when inside and outside cell populations are first established. Prior to the 16-cell stage, blastomeres are equivalent in terms of polarity, position, and degree of cell-cell contact. However, at the 16-cell stage, 1 or 2 cells come to occupy the inside of the embryo, and thus two populations emerge, each with distinct properties. Inside cells are apolar and make contact with cells on all sides, while outside cells are polarized along the radial axis of the embryo and lack cell contacts on their outer surface. Because cell polarization is one of the earliest recognized differences in these two populations, mechanisms regulating polarization of blastomeres are an intense area of research [6–10]. In addition, cell polarity has been shown to regulate the activity of the HIPPO signaling pathway [11–15], and the HIPPO pathway establishes the patterned expression of transcription factors that will later reinforce TE and ICM fates after blastocyst formation (Fig. 3A).
Figure 3. Initiation and maintenance of TE and ICM cell fates.
A) Beginning around the 16-cell stage, the HIPPO pathway is active in the ICM, where it represses expression of TE genes, such as Cdx2 and Sox2. The HIPPO pathway is inactive in the TE, enabling the transcriptional regulation of both ICM and TE genes in parallel (WWTR1 omitted for simplicity, please see text for details). B) At later stages, CDX2 reinforces the first cell fate decision by regulating expression of later-acting TE and ICM genes, such as Eomes and Oct4/Nanog, and GATA6 also helps repress Nanog in the TE. Simultaneously, OCT4 helps repress TE genes in some ICM cells. Please see text for relevant citations.
The HIPPO signaling pathway is well known for regulating cell-cell interactions in diverse developmental and homeostatic contexts [16, 17]. In the preimplantation mouse embryo, HIPPO pathway signaling negatively regulates activity of a transcriptional complex that includes YAP, WWTR1, and TEAD4 [9, 11, 12, 14, 18–20]. Since HIPPO signaling is repressed in outside cells [11–13, 18], the YAP/WWTR1/TEAD4 complex is thought to be active in outside cells, where it promotes expression of TE genes, including Cdx2 and Gata3 [18, 21, 22], and represses expression of the ICM gene Sox2 prior to blastocyst formation [23] (Fig. 3A). Importantly, CDX2 and SOX2 do not regulate each other’s expression [21, 23], and Gata3 null embryos survive beyond the blastocyst stage [24]. Therefore TE and ICM genes are independently regulated by the HIPPO pathway prior to blastocyst formation. Notably, other pluripotency genes, such as OCT4 and NANOG are expressed ubiquitously until after blastocyst formation, and are not influenced by HIPPO signaling prior to blastocyst formation [19, 20, 25–27]. Therefore, CDX2 and SOX2 are the earliest fate markers that provide readouts of HIPPO signaling prior to blastocyst formation.
The mechanisms that produce the spatially restricted HIPPO pathway signaling have been examined. Surprisingly, the mouse HIPPO orthologues Mst1/2 do not regulate YAP/WWTR1/TEAD4 activity or TE/ICM cell fate during preimplantation stages [19]. Rather, the most upstream components of the HIPPO pathway identified to function in TE/ICM specification are proteins encoded by the Angiomotin (Amot) family and Nf2 genes [11, 12, 19]. These proteins have been proposed to act as a switch in the HIPPO pathway, turning the pathway on in nonpolar inside cells and off in polarized outside cells, through differential recruitment and activation of LATS kinases [11–13, 19], which directly regulate activation of the YAP/WWTR1/TEAD4 complex [18]. Interestingly, HIPPO pathway members also promote ICM fates after blastocyst formation, and this role is described later.
Recently, an alternative role for TEAD4 was proposed [28], based on the observation that Tead4 was dispensable for blastocyst formation and CDX2 expression in embryos cultured in hypoxic conditions. This led to the proposal that TEAD4 regulates Cdx2 expression indirectly, by maintaining cell survival in conditions of oxidative stress, such as normoxia. However, TEAD4 can regulate Cdx2 expression directly [22, 29]. Thus it is also possible that in hypoxic conditions, Tead4 paralogues rescue Tead4 loss of function, but this hypothesis has not yet been tested. In addition, loss of Tead4 could be rescued by NOTCH signaling, which was recently reported to act synergistically with HIPPO signaling [29]. This was surprising because embryos completely lacking the NOTCH signaling effector RBPJ lack an obvious preimplantation phenotype [30]. However, defects in TE development were observed when NOTCH pathway components were examined in a genetic background sensitized by mutations in the HIPPO pathway [29]. Thus NOTCH signaling appears to potentiate the differentiative signals generated by the HIPPO pathway. These observations are consistent with programs of genetic redundancy to ensure a robust developmental program, and highlight the need to evaluate the roles of not just individual genes, but interacting genes and pathways.
In addition to HIPPO-mediated regulation of TE and ICM gene expression, other mechanisms for assigning TE/ICM cell fates may also be in play. For example, it has been proposed that Cdx2 is differentially partitioned into cells by oriented cell divisions, and that CDX2 then drives cells to an outside position [31, 32]. However, oriented cell divisions have been shown not to correlate with TE/ICM cell fate in the early embryo [33]. Moreover, while overexpressed Cdx2 is sufficient to alter cell position [32], endogenous Cdx2 expression is a consequence, rather than a cause, of cell position in the blastocyst [34–36]. These observations therefore support the idea that regulators of cell fate normally act downstream of cell position, and differential inheritance of Cdx2 could help reinforce outside cell fate, which is already established by polarity-dependent HIPPO signaling.
The first cell fate decision: maintenance of TE and ICM cell types after blastocyst formation
After blastocyst formation, TE and ICM fates are then maintained by lineage-specific transcription factors. In the TE, CDX2 plays two essential roles, both occurring after blastocyst formation (after E3.5) (Fig. 3B). One role of CDX2 is to promote expression of TE genes in the TE, such as Eomes [27, 35]. The other role of CDX2 is to repress expression of ICM genes in the TE, such as Oct4 and Nanog [27]. Therefore, in the TE, CDX2 transcriptionally promotes expression of TE genes and represses expression of ICM genes after blastocyst formation.
Whether ICM genes reciprocally repress expression of TE genes in the ICM has also been investigated. Curiously, although Sox2 expression localizes to the ICM well before Oct4 and Nanog (E3.0 versus E3.5-E4.0) [23, 37], Sox2 is not required for restricting expression of TE genes to the TE [23]. One report suggested that NANOG represses Cdx2 in the ICM, as weakly elevated levels of CDX2 were detected in the ICM of Nanog null blastocysts [38]. However, this phenotype was not observed in other studies [39, 40]. By contrast, there is consensus that OCT4 represses expression of multiple TE genes in the ICM [21, 41]. Curiously, however, the requirement for Oct4 is not absolute because TE genes are absent in some cells of Oct4 null blastocysts [21, 41, 42 1998]. Although the ICM is a mixture of EPI and PE cell types at this stage, OCT4 functions in both cell types at this stage [42, 43]. Because TE genes are only partially derepressed in ICM cells of Oct4 null embryos, this suggests that OCT4 must regulate expression of TE genes indirectly in the ICM. This hypothesis is consistent with the observation that OCT4 regulates expression of TE genes indirectly in ES cell lines [44].
The second cell fate decision: establishment of EPI and PE cell types at the mid-blastocyst stage
Similar to the first cell fate decision, the second cell fate decision is also initiated by cell signaling. During the second cell fate decision, FGF4/MAPK signaling selects PE and EPI cell fate by promoting expression of PE genes and repressing expression of EPI genes within an initially homogenous ICM [45–48]. At the time of blastocyst formation (E3.5), ICM cells co-express Oct4, Nanog, Sox2, and Gata6 [23, 37, 42, 45]. Soon thereafter, FGF4/MAPK signaling in some ICM cells leads to repression of Nanog and Sox2 and elevation of Gata6 expression in PE cells [23, 45–48]. By contrast, OCT4 expression is maintained in both EPI and PE cells at equivalent levels [37, 49], and its expression is not influenced by FGF4/MAPK signaling [48]. Around the time that PE and EPI cells acquire distinct gene expression profiles, additional PE genes are expressed de novo in PE cells, including Sox17, Gata4, and Pdgfra [50–53]. Therefore, around the 64-cell stage (E3.75), the ICM contains a mixture of EPI and PE cells, arranged in a salt and pepper fashion.
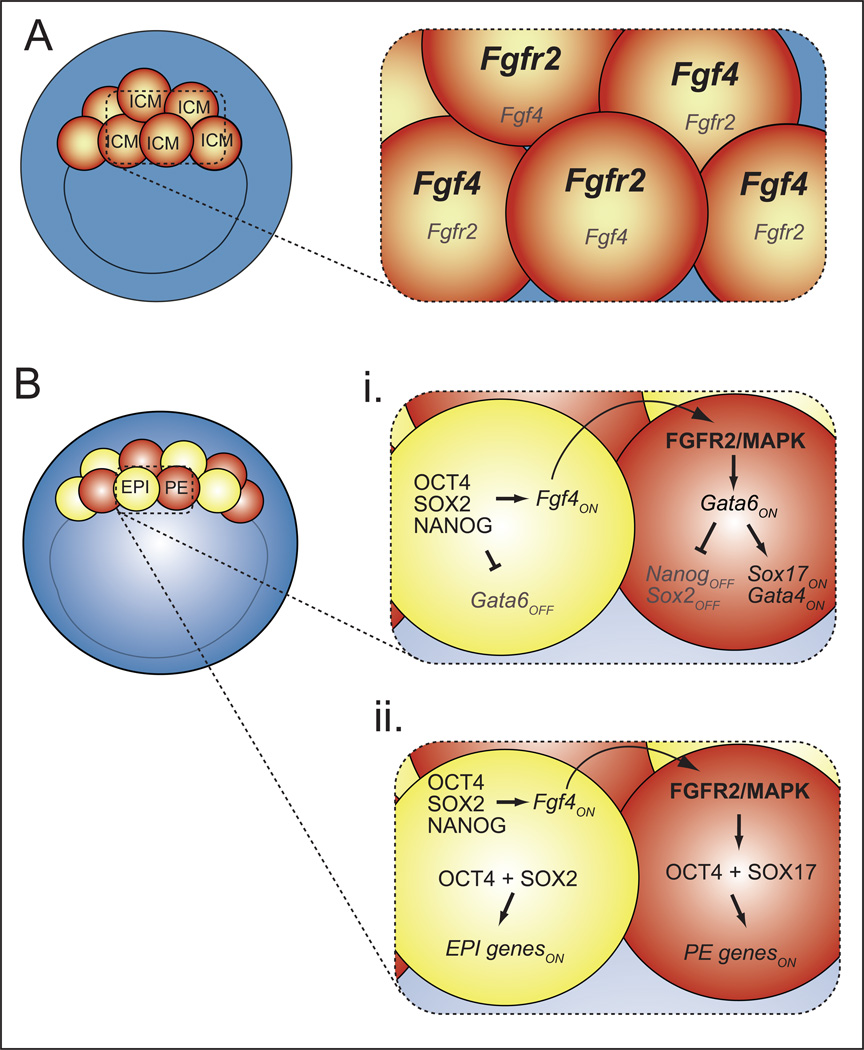
Given the importance of FGF4/MAPK signaling in the second cell fate decision, the timing and localization of the expression of FGF pathway components has been examined. These studies have shown that, initially, transcripts encoding the FGF4 ligand and its receptor FGFR2 are present in a mutually exclusive pattern in ICM cells as early as the 32-cell stage (Fig. 4A) [37, 54]. Prior to blastocyst formation, GATA6 and NANOG are still expressed ubiquitously, and in an FGF4-independent manner [46, 47]. Thus, the regulated expression of FGF pathway components is the earliest, and most upstream, molecular difference among ICM cells that has yet been identified. These observations raise several questions about the mechanisms by which FGF4/MAPK signaling regulates the second cell fate decision.
Figure 4. Origins of cellular heterogeneity in the ICM.
A) At the onset of blastocyst formation, Fgf4 and Fgfr2 transcripts exhibit heterogeneous distribution within the ICM. Bi) Around the 64-cell stage (E3.75), EPI factors promote expression of FGF4, which is received and transduced by FGFR2/MAPK. FGF4/MAPK signaling increases expression of GATA6, which represses NANOG and promotes expression of PE genes in PE cells. In EPI cells, NANOG represses expression of GATA6. Bii) In both EPI and PE cells, OCT4 is transcriptionally active, but the choice of targets depends on the presence of either SOX2 or SOX17. Please see text for relevant citations.
The first question raised is, how could an FGF ligand, which should be capable of signaling over many cell diameters, establish heterogeneous gene expression among cells in close proximity? One model is that the restricted expression of the receptor, encoded by Fgfr2 [37], limits which cells receive the signal and adopt a PE fate. However, high doses of exogenously supplied FGF4 are sufficient to convert all ICM cells to PE cells [47], and, therefore, all cells are capable of responding to FGF4. This observation suggests that the amount of Fgf4 expressed in the ICM must also be limiting. This proposal is supported by the observation that an intermediate dose of exogenous Fgf4 can re-create a salt and pepper pattern of EPI and PE cells in Fgf4 null blastocysts [55].
The second question raised is, what establishes the initially heterogeneous expression of Fgf4 and Fgfr2 in the early ICM? There is currently no evidence for a role for NOTCH signaling in a lateral inhibition type of mechanism at this stage [29]. Rather, other models have been proposed. One model is that the stage at which new ICM cells are formed can influence cell fate and/or expression of FGFR2 [51, 56, 57]. However, there is evidence that ICM cell origins do not influence ICM fates [45, 47]. An alternative model is that the salt-and-peppering of ligand/receptor expression is initially stochastic [54, 58]. Differences in the expression levels of FGF4 ligand and its receptor might then be maintained by a feed-forward mechanism. For example, expression of NANOG, and SOX2 are both restricted to EPI cells by FGF4 signaling, where they help maintain expression of Fgf4 in EPI cells after E3.75 (Fig. 4B) [23, 39, 42, 45, 47, 48]. This model is consistent with reports of ICM cell plasticity [59, 60].
The second cell fate decision: transcription factor maintenance of EPI and PE cell types
During the second cell fate decision, transcription factors reinforce EPI and PE cell fates. However, until recently, it was unclear whether transcription factors acted upstream or downstream of FGF/MAPK signaling to promote PE cell fate. Several models were consistent with the observations that FGF4/MAPK signaling is necessary and sufficient to induce PE cell fate at the expense of EPI cell fate. For example, FGF4/MAPK signaling could upregulate expression of Gata6, and GATA6 could repress expression of Nanog in PE cells. Alternatively, FGF4/MAPK signaling could repress expression of Nanog and, if NANOG normally represses expression of Gata6, then Gata6 would become upregulated in PE cells. These models have been resolved by examining the epistatic relationship between FGF4/MAPK signaling and Gata6 and Nanog mutants.
Analyses using null alleles has revealed that FGF/MAPK acts at the top of the PE gene hierarchy (Fig. 4Bi), where it first increases expression of Gata6 in PE cells, and GATA6, in turn, represses expression of Nanog in PE cells [61, 62]. Notably, GATA6, like NANOG, is expressed ubiquitously before it becomes upregulated in PE cells, and it is unclear what drives the initial expression of Gata6 in the morula [27, 39, 46]. Surprisingly, GATA6 also appears to repress Nanog in TE cells [61], indicating that GATA6 is active in multiple cell types of the blastocyst. Conversely, NANOG represses expression of Gata6 in EPI cells [39]. Importantly, however, Gata6 is the only PE gene known to be ectopically expressed in EPI cells of Nanog null embryos. Other PE genes, such as Gata4, Sox17, and others, show markedly reduced expression in Nanog null embryos, owing to reduced expression of the PE-promoting signal Fgf4 [39, 40]. These observations and others suggest that GATA6 promotes PE gene expression and represses EPI genes in parallel, downstream of FGF4/MAPK (Fig. 4Bi).
The role of EPI genes, including Oct4, Nanog, and Sox2, in maintaining the second cell fate decision has also been evaluated. None of these genes appears to be required for initial expression of each other [23, 39, 42, 63, 64]. However, because all three genes help promote expression of EPI-expressed Fgf4 [23, 39, 41, 42, 65], all three transcription factors also promote expression of PE genes non cell-autonomously [23, 39, 40, 42, 43]. In addition, OCT4 promotes expression of PE genes cell-autonomously [42, 43]. Therefore OCT4 has two activities in the ICM: promoting both EPI and PE gene expression. This observation raises the question of how OCT4 transcriptional activity is regulated so that OCT4 can regulate expression of PE and EPI-specific gene targets in a cell type-appropriate manner. An attractive model is that OCT4 transcriptional activity is regulated by cell type-specific SOX factors (Fig. 4Bii). This model is supported by the observation that SOX2 and SOX17 direct OCT4 to bind the regulatory regions of pluripotency and PE genes, respectively [66].
The second cell fate decision: the latest stages
By the late-blastocyst stage (E4.0-E4.5), EPI and PE cells sort into distinct layers, with PE cells underlying the EPI and migrating down along the mural TE. Once PE cells arrive at their ultimate hypoblast position, expression of Sox7 is induced [67]. Two mechanisms are known to regulate the process of cell sorting. First, PE cells express molecules that facilitate their sorting, including Laminin and DAB2 [68–70]. Expression of these molecules is dependent on Oct4 and Sox2, and cell-sorting defects are observed in the absence of either transcription factor [23, 42]. Second, PE cells that do not sort correctly undergo apoptosis [52], revealing that a mechanism is in place to assure that EPI and PE cells are properly localized just prior to implantation.
Several signaling pathways are thought to maintain the ICM lineages in the late blastocyst, including LIF/IL6/JAK/STAT and BMP pathways [71–73]. Interestingly, several early-expressed PE genes, such as Sox17, Pdgfra, and Sox7 are not essential until after implantation [46, 67, 74]. Therefore, even though these genes exhibit early-localized expression in PE cells, they may act redundantly with each other in the blastocyst. After blastocyst formation, the HIPPO pathway continues to function. Loss of the HIPPO pathway member Nf2 leads to gain of TE genes within the ICM at the late blastocyst stage [19]. In addition, loss of Lats leads to loss of ICM gene expression and even loss of ICM cells by late blastocyst stage [12, 25]. These observations suggest that HIPPO signaling maintains survival or position of ICM cells as blastocyst maturation proceeds. However, the mechanism of cell loss is not yet known.
It has been more difficult to determine whether genes that are required early in cell fate specification are also required later, because perdurance of the protein product of the deleted gene could mask loss-of-function phenotypes. In one case, deletion of Oct4 in the late blastocyst was achieved using the Cre/lox system [43]. Interestingly, later deletion of Oct4 did not disrupt PE cell fate in the late blastocyst. These observations suggest that Oct4 is not required for late PE cell fate, and that the reported PE defects in late Oct4 null blastocysts [41, 42] are due to an earlier requirement for Oct4. Therefore, temporal inactivation of genes can be used to pinpoint when genes are active in development. However, successful temporal gene inactivation is only feasible when appropriate Cre lines are available, and it is only informative when elimination of the target protein is confirmed to follow the dynamics of the gene deletion.
Maternal determinants do not regulate mouse development
One longstanding question is the degree to which mammalian development is regulated by cytoplasmic determinants, which are known to dictate embryo patterning in a number of model organisms. For example, fly and frog embryos inherit information about where their head and tail should be from the mother, who produces an egg with intrinsic asymmetry. Although many maternal-effect genes are important in mammals [75], no maternally produced protein that impacts cell fates of the mouse embryo has been identified. Notably, the majority of maternal transcripts as well as many maternal proteins are degraded very early in development, many days before cell fate acquisition. Moreover, the patterns of cell division that precede establishment of the EPI lineage can be highly variable among embryos [76]. For these reasons, it is difficult to imagine that the spatial distribution of a maternally deposited factor could direct cell fate decisions as a cytoplasmic determinant.
The role of maternal OCT4, and SOX2 in early cell fate decisions has been scrutinized using null alleles. Independent studies have confirmed that even though levels of maternal OCT4 and SOX2 are quite high in oocytes, maternal OCT4 and maternal SOX2 are dispensable for development [23, 42, 63, 77]. By contrast, CDX2 is not detectable in oocytes [78, 79], and the level of maternal Cdx2 detected in oocytes is ten times lower than the level detected in ES cells, where Cdx2 has no role [80, 81]. Not surprisingly, deletion of maternal Cdx2 alone does not disturb development, and deletion of maternal and zygotic Cdx2 phenocopies deletion of zygotic Cdx2 [23, 81]. In contrast to these results, another study recently reported a role for maternal Cdx2 [82]. It is not clear what is the basis for the discrepancy since both groups examined similar numbers of embryos, and used the same Cre driver to delete oocyte-expressed Cdx2. Differences between the groups included culture conditions, the Cdx2 allele used, and possibly genetic background. Therefore, the Cdx2 null phenotype may be influenced by interactions with as-yet unidentified environmental or genetic factors. This hypothesis is consistent with our recent observations that Cdx2 activity is influenced by genetic background in ES cells [83]. Regardless, both groups reported that in embryos lacking both maternal and zygotic Cdx2, cell fate allocation was similar to that in embryos lacking zygotic Cdx2, arguing that maternal Cdx2 is unimportant for cell fate decisions in the early embryo.
Forging ahead
Although technological advances in gene expression analysis and imaging have provided new insight into the genetic regulation of cell fate specification in the mouse early embryo, many mysteries are still unsolved (Outstanding Questions Box). Several of the challenges intrinsic to working with preimplantation embryos might be mitigated by emerging technology. For example, the small cell number of the blastocyst makes it difficult to identify transcription factor binding sites in the three cell types of the blastocyst. One solution involves using recently developed methods for sorting and pooling the three cell types from the blastocyst [84]. Another challenge is that genetic redundancy may mask the essential roles of genes, and breeding double and triple mutants is not considered time or cost-effective. However, emerging genome editing technology may simplify the process of introducing multiple mutant alleles in the zygote [85]. Another challenge is that gene discovery is sometimes slower in mice than in other model organisms that are better suited to forward genetic techniques. However, advances in genome sequencing technology make forward genetic approaches more feasible in the mouse. Finally, it is becoming apparent that both development and disease are governed by the interaction of multiple genes and pathways of individually small effect, and thus the real mechanisms that regulate development might be distorted by the reductionist approach of null alleles and two or three gene outputs. However, single-cell gene expression analyses have facilitated systems views of how pathways, and not just individual genes, modulate developmental processes [3, 42, 54]. Moreover, unique features of preimplantation embryos have allowed advances in techniques for live imaging gene expression changes during development [86]. With these advances in place, chances are good that the spotlight will remain on the blastocyst for many repeat performances to come!
Highlights.
The first two cell fate decisions in mammalian development create embryonic stem (ES) cell progenitors and two extraembryonic cell types essential for fetal development
During blastocyst formation, cell fates are initially specified by cell signaling, and are then reinforced by lineage-specific transcription factors
Technological advances in imaging and gene expression analysis enables new insights into mechanisms of cell fate specification in the mouse early embryo
Outstanding Questions Box.
What regulates the timing of developmental milestones events? Halved two-cell embryos form a blastocyst structure after the same number of days as intact two-cell embryos, even though halved embryos contain half the number of cells as their intact counterparts. Therefore, a mechanism exists for measuring time, which is independent of the number of cell divisions. This mechanism has not been identified.
What regulates the self-organizing properties of the embryo? Cells of the early embryo spontaneously acquire differences in cell polarity, which plays an essential role in directing cell fate. Disassembled embryos can reassemble and reestablish this polarity, even though cells may have changed their original position within the embryo. The pathways that dynamically regulate cell polarity are unknown.
How is cellular heterogeneity established in the embryo? The inner cell mass spontaneously subdivides into two cell types: one pluripotent and one extraembryonic, present in a 1:1 ratio. How these two cell types arise in balanced numbers is still unclear.
Why are some genes essential but others are not? Some genes with specific expression patterns are essential for early mouse development, whereas other genes, with identical expression patterns, are not. Therefore early embryonic processes seem protected against loss-of-function mutations in some genes, but not others. It is unclear how these observed patterns modulate the robustness of regulatory signaling networks involved in early embryogenesis, or how this complex developmental program evolved in placental mammals.
Acknowledgements
We apologize to colleagues whose work could not be discussed due to word limitations. We thank members of the Ralston, Floer, Knott, Smith, and Latham Labs at Michigan State University for discussions. This work is supported by National Institutes of Health grant R01 GM104009 to A.R.
Glossary
- Blastocoel
the fluid-filled cavity of the blastocyst
- Blastocyst
the embryo after formation and expansion of the blastocoel (32-cell stage to implantation)
- Cavitation
formation of a cavity
- Cleavage
a special type of cell division that decreases cell size by halving the cell
- Cleavage stages
preimplantation development
- Compaction
the process by which totipotent blastomeres of the mouse embryo increase cell-cell contact and polarize along the radial axis of the embryo. Compaction creates apical (outside) and basolateral (inside) polarity and occurs at the late 8-cell stage
- Epiblast (EPI)
the fetal lineage of the embryo, present in the late blastocyst
- Implantation
the process of embryo embedding within the stroma of the uterus
- Inner Cell Mass (ICM)
the inside cells of the blastocyst, which contain progenitors of ES cells or fetus and primitive endoderm
- Morula
the embryo from the 8-cell stage until blastocyst formation, when the embryo morphologically resembles a mulberry
- Preimplantation
the period of embryonic development after fertilization and prior to uterine implantation
- Primitive Endoderm (PE)
an extraembryonic lineage present in the blastocyst that will contribute to the yolk sac endoderm
- Trophectoderm (TE)
the extraembryonic lineage present in the blastocyst that will differentiate to become the trophoblast of the placenta
- Zona pellucida
the thick glycoprotein shell that surrounds the preimplantation embryo until hatching
- Zygote
the 1-cell embryo
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Stephenson M, Kutteh W. Evaluation and management of recurrent early pregnancy loss. Clin Obstet Gynecol. 2007;50:132–145. doi: 10.1097/GRF.0b013e31802f1c28. [DOI] [PubMed] [Google Scholar]
- 2.Molls M, et al. A comparison of the cell kinetics of pre-implantation mouse embryos from two different mouse strains. Cell Tissue Kinet. 1983;16:277–283. [PubMed] [Google Scholar]
- 3.Boroviak T, et al. The ability of inner-cell-mass cells to self-renew as embryonic stem cells is acquired following epiblast specification. Nat Cell Biol. 2014;16:516–528. doi: 10.1038/ncb2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parfitt DE, Shen MM. From blastocyst to gastrula: gene regulatory networks of embryonic stem cells and early mouse embryogenesis. Philos Trans R Soc Lond B Biol Sci. 2014;369 doi: 10.1098/rstb.2013.0542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ralston A, Rossant J. Genetic regulation of stem cell origins in the mouse embryo. Clin Genet. 2005;68:106–112. doi: 10.1111/j.1399-0004.2005.00478.x. [DOI] [PubMed] [Google Scholar]
- 6.Posfai E, et al. Mechanisms of pluripotency in vivo and in vitro. Curr Top Dev Biol. 2014;107:1–37. doi: 10.1016/B978-0-12-416022-4.00001-9. [DOI] [PubMed] [Google Scholar]
- 7.Choi I, et al. Transcription factor AP-2γ is a core regulator of tight junction biogenesis and cavity formation during mouse early embryogenesis. Development. 2012;139:4623–4632. doi: 10.1242/dev.086645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao Z, et al. Transcription factor AP-2gamma incudes early Cdx2 expression and represses HIPPO signaling to specify trophectoderm lineage. Development. 2015 doi: 10.1242/dev.120238. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kono K, et al. Inhibition of RHO-ROCK signaling enhances ICM and suppresses TE characteristics through activation of Hippo signaling in the mouse blastocyst. Dev Biol. 2014;394:142–155. doi: 10.1016/j.ydbio.2014.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alarcon VB. Cell polarity regulator PARD6B is essential for trophectoderm formation in the preimplantation mouse embryo. Biol Reprod. 2010;83:347–358. doi: 10.1095/biolreprod.110.084400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leung CY, Zernicka-Goetz M. Angiomotin prevents pluripotent lineage differentiation in mouse embryos via Hippo pathway-dependent and -independent mechanisms. Nat Commun. 2013;4:2251. doi: 10.1038/ncomms3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirate Y, et al. Polarity-dependent distribution of angiomotin localizes Hippo signaling in preimplantation embryos. Curr Biol. 2013;23:1181–1194. doi: 10.1016/j.cub.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirate Y, Sasaki H. The role of angiomotin phosphorylation in the Hippo pathway during preimplantation mouse development. Tissue Barriers. 2014;2:e28127. doi: 10.4161/tisb.28127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anani S, et al. Initiation of Hippo signaling is linked to polarity rather than to cell position in the pre-implantation mouse embryo. Development. 2014;141:2813–2824. doi: 10.1242/dev.107276. [DOI] [PubMed] [Google Scholar]
- 15.Lorthongpanich C, et al. Developmental fate and lineage commitment of singled mouse blastomeres. Development. 2012;139:3722–3731. doi: 10.1242/dev.086454. [DOI] [PubMed] [Google Scholar]
- 16.Varelas X. The Hippo pathway effectors TAZ and YAP in development, homeostasis and disease. Development. 2014;141:1614–1626. doi: 10.1242/dev.102376. [DOI] [PubMed] [Google Scholar]
- 17.Halder G, Johnson RL. Hippo signaling: growth control and beyond. Development. 2011;138:9–22. doi: 10.1242/dev.045500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishioka N, et al. The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev Cell. 2009;16:398–410. doi: 10.1016/j.devcel.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 19.Cockburn K, et al. The Hippo pathway member Nf2 is required for inner cell mass specification. Curr Biol. 2013;23:1195–1201. doi: 10.1016/j.cub.2013.05.044. [DOI] [PubMed] [Google Scholar]
- 20.Yagi R, et al. Transcription factor TEAD4 specifies the trophectoderm lineage at the beginning of mammalian development. Development. 2007;134:3827–3836. doi: 10.1242/dev.010223. [DOI] [PubMed] [Google Scholar]
- 21.Ralston A, et al. Gata3 regulates trophoblast development downstream of Tead4 and in parallel to Cdx2. Development. 2010;137:395–403. doi: 10.1242/dev.038828. [DOI] [PubMed] [Google Scholar]
- 22.Home P, et al. Altered subcellular localization of transcription factor TEAD4 regulates first mammalian cell lineage commitment. Proc Natl Acad Sci U S A. 2012;109:7362–7367. doi: 10.1073/pnas.1201595109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wicklow E, et al. HIPPO pathway members restrict SOX2 to the inner cell mass where it promotes ICM fates in the mouse blastocyst. PLoS Genet. 2014;10:e1004618. doi: 10.1371/journal.pgen.1004618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma G, et al. GATA-2 and GATA-3 regulate trophoblast-specific gene expression in vivo. Development. 1997;124:907–914. doi: 10.1242/dev.124.4.907. [DOI] [PubMed] [Google Scholar]
- 25.Lorthongpanich C, et al. Temporal reduction of LATS kinases in the early preimplantation embryo prevents ICM lineage differentiation. Genes Dev. 2013;27:1441–1446. doi: 10.1101/gad.219618.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nishioka N, et al. Tead4 is required for specification of trophectoderm in pre-implantation mouse embryos. Mech Dev. 2008;125:270–283. doi: 10.1016/j.mod.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 27.Strumpf D, et al. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development. 2005;132:2093–2102. doi: 10.1242/dev.01801. [DOI] [PubMed] [Google Scholar]
- 28.Kaneko KJ, DePamphilis ML. TEAD4 establishes the energy homeostasis essential for blastocoel formation. Development. 2013;140:3680–3690. doi: 10.1242/dev.093799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rayon T, et al. Notch and hippo converge on cdx2 to specify the trophectoderm lineage in the mouse blastocyst. Dev Cell. 2014;30:410–422. doi: 10.1016/j.devcel.2014.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Souilhol C, et al. RBP-Jkappa-dependent notch signaling is dispensable for mouse early embryonic development. Mol Cell Biol. 2006;26:4769–4774. doi: 10.1128/MCB.00319-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skamagki M, et al. Asymmetric localization of Cdx2 mRNA during the first cell-fate decision in early mouse development. Cell Rep. 2013;3:442–457. doi: 10.1016/j.celrep.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jedrusik A, et al. Maternally and zygotically provided Cdx2 have novel and critical roles for early development of the mouse embryo. Dev Biol. 2010;344:66–78. doi: 10.1016/j.ydbio.2010.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Watanabe T, et al. Limited predictive value of blastomere angle of division in trophectoderm and inner cell mass specification. Development. 2014;141:2279–2288. doi: 10.1242/dev.103267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McDole K, Zheng Y. Generation and live imaging of an endogenous Cdx2 reporter mouse line. Genesis. 2012;50:775–782. doi: 10.1002/dvg.22049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ralston A, Rossant J. Cdx2 acts downstream of cell polarization to cell-autonomously promote trophectoderm fate in the early mouse embryo. Dev Biol. 2008;313:614–629. doi: 10.1016/j.ydbio.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 36.Dietrich JE, Hiiragi T. Stochastic patterning in the mouse pre-implantation embryo. Development. 2007;134:4219–4231. doi: 10.1242/dev.003798. [DOI] [PubMed] [Google Scholar]
- 37.Guo G, et al. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev Cell. 2010;18:675–685. doi: 10.1016/j.devcel.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 38.Chen L, et al. Cross-regulation of the Nanog and Cdx2 promoters. Cell Res. 2009;19:1052–1061. doi: 10.1038/cr.2009.79. [DOI] [PubMed] [Google Scholar]
- 39.Frankenberg S, et al. Primitive Endoderm Differentiates via a Three-Step Mechanism Involving Nanog and RTK Signaling. Dev Cell. 2011;21:1005–1013. doi: 10.1016/j.devcel.2011.10.019. [DOI] [PubMed] [Google Scholar]
- 40.Messerschmidt DM, Kemler R. Nanog is required for primitive endoderm formation through a non-cell autonomous mechanism. Dev Biol. 2010;344:129–137. doi: 10.1016/j.ydbio.2010.04.020. [DOI] [PubMed] [Google Scholar]
- 41.Nichols J, et al. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- 42.Frum T, et al. Oct4 cell-autonomously promotes primitive endoderm development in the mouse blastocyst. Dev Cell. 2013;25:610–622. doi: 10.1016/j.devcel.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Le Bin GC, et al. Oct4 is required for lineage priming in the developing inner cell mass of the mouse blastocyst. Development. 2014;141:1001–1010. doi: 10.1242/dev.096875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Niwa H, et al. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell. 2005;123:917–929. doi: 10.1016/j.cell.2005.08.040. [DOI] [PubMed] [Google Scholar]
- 45.Chazaud C, et al. Early lineage segregation between epiblast and primitive endoderm in mouse blastocysts through the Grb2-MAPK pathway. Dev Cell. 2006;10:615–624. doi: 10.1016/j.devcel.2006.02.020. [DOI] [PubMed] [Google Scholar]
- 46.Kang M, et al. FGF4 is required for lineage restriction and salt-and-pepper distribution of primitive endoderm factors but not their initial expression in the mouse. Development. 2013;140:267–279. doi: 10.1242/dev.084996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamanaka Y, et al. FGF signal-dependent segregation of primitive endoderm and epiblast in the mouse blastocyst. Development. 2010;137:715–724. doi: 10.1242/dev.043471. [DOI] [PubMed] [Google Scholar]
- 48.Nichols J, et al. Suppression of Erk signalling promotes ground state pluripotency in the mouse embryo. Development. 2009;136:3215–3222. doi: 10.1242/dev.038893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Palmieri S, et al. Oct-4 transcription factor is differentially expressed in the mouse embryo during establishment of the first two extraembryonic cell lineages involved in implantation. Dev Biol. 1994;166:259–267. doi: 10.1006/dbio.1994.1312. [DOI] [PubMed] [Google Scholar]
- 50.Niakan KK, et al. Sox17 promotes differentiation in mouse embryonic stem cells by directly regulating extraembryonic gene expression and indirectly antagonizing self-renewal. Genes Dev. 2010;24:312–326. doi: 10.1101/gad.1833510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morris SA, et al. Origin and formation of the first two distinct cell types of the inner cell mass in the mouse embryo. Proc Natl Acad Sci U S A. 2010;107:6364–6369. doi: 10.1073/pnas.0915063107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Plusa B, et al. Distinct sequential cell behaviours direct primitive endoderm formation in the mouse blastocyst. Development. 2008;135:3081–3091. doi: 10.1242/dev.021519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Artus J, et al. A role for PDGF signaling in expansion of the extra-embryonic endoderm lineage of the mouse blastocyst. Development. 2010;137:3361–3372. doi: 10.1242/dev.050864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ohnishi Y, et al. Cell-to-cell expression variability followed by signal reinforcement progressively segregates early mouse lineages. Nat Cell Biol. 2014;16:27–37. doi: 10.1038/ncb2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krawchuk D, et al. FGF4 is a limiting factor controlling the proportions of primitive endoderm and epiblast in the ICM of the mouse blastocyst. Dev Biol. 2013;384:65–71. doi: 10.1016/j.ydbio.2013.09.023. [DOI] [PubMed] [Google Scholar]
- 56.Morris SA, et al. The differential response to Fgf signalling in cells internalized at different times influences lineage segregation in preimplantation mouse embryos. Open Biol. 2013;3:130104. doi: 10.1098/rsob.130104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krupa M, et al. Allocation of inner cells to epiblast vs primitive endoderm in the mouse embryo is biased but not determined by the round of asymmetric divisions (8 →16- and 16→32-cells) Dev Biol. 2014;385:136–148. doi: 10.1016/j.ydbio.2013.09.008. [DOI] [PubMed] [Google Scholar]
- 58.Martinez Arias A, et al. A molecular basis for developmental plasticity in early mammalian embryos. Development. 2013;140:3499–3510. doi: 10.1242/dev.091959. [DOI] [PubMed] [Google Scholar]
- 59.Xenopoulos P, et al. Heterogeneities in Nanog Expression Drive Stable Commitment to Pluripotency in the Mouse Blastocyst. Cell Rep. 2015 doi: 10.1016/j.celrep.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Grabarek JB, et al. Differential plasticity of epiblast and primitive endoderm precursors within the ICM of the early mouse embryo. Development. 2012;139:129–139. doi: 10.1242/dev.067702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schrode N, et al. GATA6 Levels Modulate Primitive Endoderm Cell Fate Choice and Timing in the Mouse Blastocyst. Dev Cell. 2014;29:454–467. doi: 10.1016/j.devcel.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bessonnard S, et al. Gata6, Nanog and Erk signaling control cell fate in the inner cell mass through a tristable regulatory network. Development. 2014;141:3637–3648. doi: 10.1242/dev.109678. [DOI] [PubMed] [Google Scholar]
- 63.Wu G, et al. Establishment of totipotency does not depend on Oct4A. Nat Cell Biol. 2013;15:1089–1097. doi: 10.1038/ncb2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Silva J, et al. Nanog is the gateway to the pluripotent ground state. Cell. 2009;138:722–737. doi: 10.1016/j.cell.2009.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Avilion AA, et al. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003;17:126–140. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Aksoy I, et al. Oct4 switches partnering from Sox2 to Sox17 to reinterpret the enhancer code and specify endoderm. EMBO J. 2013;32:938–953. doi: 10.1038/emboj.2013.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Artus J, et al. The primitive endoderm lineage of the mouse blastocyst: sequential transcription factor activation and regulation of differentiation by Sox17. Dev Biol. 2011;350:393–404. doi: 10.1016/j.ydbio.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Morris S, et al. Dual roles for the Dab2 adaptor protein in embryonic development and kidney transport. EMBO J. 2002;21:1555–1564. doi: 10.1093/emboj/21.7.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang DH, et al. Disabled-2 is essential for endodermal cell positioning and structure formation during mouse embryogenesis. Dev Biol. 2002;251:27–44. doi: 10.1006/dbio.2002.0810. [DOI] [PubMed] [Google Scholar]
- 70.Smyth N, et al. Absence of basement membranes after targeting the LAMC1 gene results in embryonic lethality due to failure of endoderm differentiation. J Cell Biol. 1999;144:151–160. doi: 10.1083/jcb.144.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Do DV, et al. A genetic and developmental pathway from STAT3 to the OCT4-NANOG circuit is essential for maintenance of ICM lineages in vivo. Genes Dev. 2013;27:1378–1390. doi: 10.1101/gad.221176.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Graham SJ, et al. BMP signalling regulates the pre-implantation development of extra-embryonic cell lineages in the mouse embryo. Nat Commun. 2014;5:5667. doi: 10.1038/ncomms6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Reyes de Mochel NS, et al. BMP signaling is required for cell cleavage in preimplantation-mouse embryos. Dev Biol. 2015;397:45–55. doi: 10.1016/j.ydbio.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wat MJ, et al. Mouse model reveals the role of SOX7 in the development of congenital diaphragmatic hernia associated with recurrent deletions of 8p23.1. Hum Mol Genet. 2012;21:4115–4125. doi: 10.1093/hmg/dds241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li L, et al. Maternal control of early mouse development. Development. 2010;137:859–870. doi: 10.1242/dev.039487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Marikawa Y, Alarcón VB. Establishment of trophectoderm and inner cell mass lineages in the mouse embryo. Mol Reprod Dev. 2009;76:1019–1032. doi: 10.1002/mrd.21057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Campolo F, et al. Essential role of Sox2 for the establishment and maintenance of the germ cell line. Stem Cells. 2013;31:1408–1421. doi: 10.1002/stem.1392. [DOI] [PubMed] [Google Scholar]
- 78.Beck F, et al. Expression of Cdx-2 in the mouse embryo and placenta: possible role in patterning of the extra-embryonic membranes. Dev Dyn. 1995;204:219–227. doi: 10.1002/aja.1002040302. [DOI] [PubMed] [Google Scholar]
- 79.Wang S, et al. Proteome of mouse oocytes at different developmental stages. Proc Natl Acad Sci U S A. 2010;107:17639–17644. doi: 10.1073/pnas.1013185107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chawengsaksophak K, et al. Cdx2 is essential for axial elongation in mouse development. Proc Natl Acad Sci U S A. 2004;101:7641–7645. doi: 10.1073/pnas.0401654101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Blij S, et al. Maternal Cdx2 is dispensable for mouse development. Development. 2012;139:3969–3972. doi: 10.1242/dev.086025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jedrusik A, et al. Maternal-zygotic knockout reveals a critical role of Cdx2 in the morula to blastocyst transition. Dev Biol. 2015;398:147–152. doi: 10.1016/j.ydbio.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Blij S, et al. Cdx2 efficiently induces trophoblast stem-like cells in naïve, but not primed, pluripotent stem cells. Stem Cells Dev. 2015 doi: 10.1089/scd.2014.0395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rugg-Gunn PJ, et al. Cell-surface proteomics identifies lineage-specific markers of embryo-derived stem cells. Dev Cell. 2012;22:887–901. doi: 10.1016/j.devcel.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang H, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nowotschin S, Hadjantonakis AK. Live imaging mouse embryonic development: seeing is believing and revealing. Methods Mol Biol. 2014;1092:405–420. doi: 10.1007/978-1-60327-292-6_24. [DOI] [PMC free article] [PubMed] [Google Scholar]