Abstract
We present a robot that enables high-content studies of alert adult Drosophila by combining operations including gentle picking, translations and rotations, characterizations of fly phenotypes and behaviors, micro-dissection or release. To illustrate, we assessed fly morphology, tracked odor-evoked locomotion, sorted flies by sex, and dissected the cuticle to image neural activity. The robot's tireless capacity for precise manipulations enables a scalable platform for screening flies’ complex attributes and behavioral patterns.
Biologists increasingly rely on automated systems to improve the consistency, speed, precision, duration and throughput of experimentation with small animal species. For nematodes, zebrafish, and larval or embryonic flies, fluidic systems can sort and screen individual animals based on phenotypes or behavior1-5. Unlike animals living in aqueous media, adult fruit flies have largely eluded automated handling. Given the fly's prominence in multiple research fields, automated handling would have a major impact.
Video tracking systems can classify some fly behaviors6-9 but active manipulation, sorting, micro-dissection and many detailed assessments have evaded automation due to the delicacy and complexity of the required operations. To clear this challenge, we created a robotic system that is programmable for diverse needs, as illustrated here by sex sorting; analyses of fly morphology; micro-injection; fiber-optic light delivery; locomotor assessments; micro-dissection; and imaging of neural activity. Users can program other applications as sequences of existing operations or by adding machine vision analyses. The robot has a similar footprint as a laptop computer, uses affordable parts (<$5000 total) and is scalable to multiple units.
Aspects of our instrumentation existed previously but not for handling flies. Robots with parallel kinematic architectures can make fast precise movements, and machine-vision-guided robots can capture fast-moving objects10,11. Thus, automated capture of flies seemed feasible, but a robot needs unusual precision and speed to gently capture a fly (thorax ~1 mm across; walking speed ~3 cm/s; ref. 12). To avoid causing injury the machine must exert ultra-low, milliNewton forces to carry a fly (~1.0 mg) and overcome flight or walking forces (~0.1–1 mN) (refs.13,14). To manipulate flies biologists typically anesthetize them, but anesthesia affects the insect nervous system15,16 and necessitates a recovery period to restore normal behavior and physiology17. We avoided anesthesia. By integrating machine vision into the robot's effector head, our device captures, manipulates, mounts, releases and dissects non-anesthetized flies.
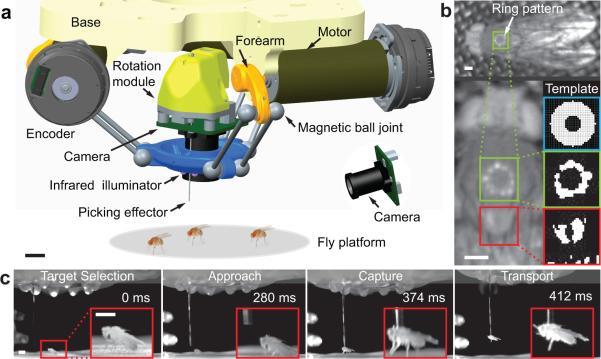
The robot has parallel kinematic chains that connect a base platform to the effector that picks the fly (Fig. 1a, Supplementary Fig. 1). Compared to cascaded serial chains, parallel chains offer superior rigidity and keep the moving mass lightweight, key advantages for precise execution of rapid movements. Three rotary motors drive the effector's three-dimensional translations. To implement this, we invented magnetic ball joints. Magnetic forces hold the joint and allow greater angular range than conventional ball-and-sockets while minimizing backlash and improving precision. As a safety mechanism, the joints detach during an accidental collision. To rotate the picked fly in yaw, another rotary motor turns the picking effector, which holds the fly by suction.
Fig. 1. A high-speed robot that uses real-time machine vision guidance to identify, capture and manipulate non-anesthetized adult flies.
a. Computer-assisted design schematic of the robot. Scale bar: 10 mm. Flies are not to scale.
b. Top: The robot targets the thorax by locating the reflection from the robot's infrared ring illuminator. Left: Magnified image shows the ring reflection off the thorax (green box) and for comparison a neighboring region (red box). Right: Using a ring template (blue box), an image-analysis algorithm matches the template to a binary-segmented image of the thorax (green box), but not to the neighboring subfield (red box). Scale bars: 0.5 mm.
c. High-speed videography reveals the speed at which the robot tracks and grabs the fly. Insets show close-ups in each frame. Scale bars: 1.5 mm.
The system tracks and picks individual flies by machine vision, using infrared illumination as flies generally do not initiate flight in visible darkness (Fig. 1; Supplementary Figs. 1–3; Videos 1,2). After screwing a vial of flies into a loading chamber, flies climb the vial walls and emerge onto the picking platform (Supplementary Fig. 4; Video 3). A stationary camera provides coarse locations of all available flies (up to ~50) and guides selection of one for picking (Fig. 1a; Supplementary Fig. 5). We exchanged vials as needed, allowing one-by-one studies of ~1,000 flies in ~10 h.
The robot head has a camera and a ring of infrared LEDs that move over the chosen fly to determine its location and orientation. The ring creates a stereotyped reflection off the thorax (Fig. 1b) that scarcely varies with the fly's orientation or position, allowing the robot to reliably identify it for real-time tracking of fly motion (Supplementary Figs. 1–3,6; Video 1). Given the <20% size variability of the thorax and the mechanical compliance of fly legs, it sufficed for the robot to pick flies at a fixed height above the picking platform. Unlike humans, the robot has sufficient speed (maximum: 22 cm/s) and precision to connect the fly to the picking effector.
A tubular suction effector gently holds the thorax (Videos 1,2,4,5). Suction gating engages or disengages the holding force (~4.5 mN across 0.53 mm2 of thorax), and a pressure sensor detects a good connection. The robot can identify and pick a still fly in <2 s (Supplementary Table 1, Videos 1,2,6,7). For an ambulatory fly, the robots tracks it until it pauses, then gently lifts it upward (Videos 1,2). The robot can rotate the fly in yaw, translate it in three dimensions, bring it to an inspection camera, tether or deliver it elsewhere (Videos 4–7).These maneuvers are flexibly combinable to serve many applications.
To benchmark handling speed, the robot continuously transferred flies back and forth across a divided platform (Video 6). After selecting a fly the robot attempted a pick; whenever the pressure sensor detected success the robot delivered the fly to the platform's opposite side. During continuous iteration (Video 7), the robot picked a fly every 8.4 ± 3.2 s (s.d.), capturing 84 ± 5% (s.d.) of these on the thorax (Supplementary Table 2). The robot returned the 16% captured elsewhere on the body to the platform for another try.
Human visual inspection is often key to establishing fly matings, but computer-vision should provide comparable reliability. By carrying a fly to a high-magnification camera (Video 8, Supplementary Fig. 7) the robot classified the sex by the number of dark abdominal segments [99 ± 1% (s.d.) sex accuracy in ~20 ms computation] (Fig. 2a, Supplementary Table 2). Total time to pick and sort each fly was ~20 s.
Fig. 2. Machine vision based assessments of fly phenotypes.
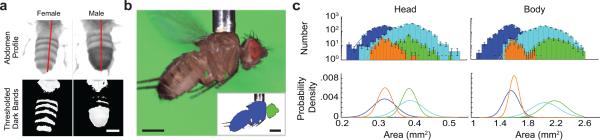
a. The robot discriminated flies by sex to 99% accuracy. Top: Guided by real-time machine vision the robot rotated the fly to view the abdomen. Bottom: Another algorithm counted the abdominal bands. Scale bar: 0.5 mm.
b. After sex determination, flies underwent analyses of body morphology. Top: Raw image of a fly held by the robot's picker. Inset: Segmentation into head (green) and body (blue) regions. Scale bars: 0.5 mm.
c. Top: Histograms (logarithmic y-axis) and Gaussian fits (solid lines) of head and body areas determined as in b. Male Oregon-R (blue); male inbred (orange); female Oregon-R (cyan); female inbred (green). For head and body areas, the size distributions for males and females are markedly distinct for both fly groups. The two genotypes had distinguishable distributions for head and body areas, for males and females (P-values: 10−5–0.05 for all four comparisons between Oregon-R and inbred; Kolmogorov-Smirnov test). Errors are s.d., estimated as counting errors. Bottom: Gaussian fits (linear y-axis), normalized to unity area to highlight the differences in the corresponding statistical distributions.
We used machine-vision algorithms to measure cross-sectional head and body areas of 1046 Oregon-R flies (Supplementary Fig. 8). The robot picked each fly, classified its sex, and took 4894 images total in ~10 h (Fig. 2b). Males and females had distinct head and body areas [Heads: males, 0.32 ± 0.03 mm2; females, 0.39 ± 0.04 mm2; Bodies: males, 1.6 ± 0.2 mm2; females, 2.0 ± 0.2 mm2; mean ± s.d.; n = 1288 male and 1796 female images]. Size distributions closed matched Gaussian fits across two orders of magnitudes of statistical frequency (Fig. 2c).
To demonstrate fine morphological discriminations, the robot examined Drosophila melanogaster derived and inbred from native populations in the eastern United States (Online Methods). Initially, head areas [males: 0.33 ± 0.03 mm2 (mean ± s.d.); females: 0.39 ± 0.03 mm2] and body areas [males: 1.6 ± 0.1 mm2 (mean ± s.d; females: 2.2 ± 0.2 mm2] of these flies (n = 67) seemed indistinguishable from those of Oregon-R flies. However, the robot's dataset of 1113 flies revealed plain differences between the populations (Fig. 2c) [Kolmogorov-Smirnov tests comparing male and female body sizes (both P < 10−5), male heads (P = 4·10−5), and female heads (P = 0.05) between inbred and Oregon-R]. Inbred lines had larger median body areas (males: 4% larger than Oregon-R; females: 7%) and finer differences in median head areas that would surely elude human inspection (males: 1.3% larger than Oregon-R; females: 0.3%) (Fig. 2c).
After picking a fly the robot can tether it for behavioral studies, microsurgery or brain imaging. After locating the neck the robot can glue it to a stationary fiber or detachably insert the proboscis into a suction tube (Supplementary Figs. 9–11, Video 4). Like flies tethered manually18,19, those tethered robotically exhibited flying and walking behaviors. The robot placed flies on a trackball for automated determinations of odor-evoked locomotor responses (Fig. 3a–c, Supplementary Fig. 12, Video 9). Comparisons of locomotor responses revealed no speed differences between flies handled manually or robotically (n = 10 trials per fly; n = 4 flies per group; Wilcoxon rank sum test; P = 0.12–0.66 for sideways, forward and angular speeds). Flies held by suction can be released and saved for later experimentation (Videos 4,6,7,9).
Fig. 3. Automated assessments of sensory-evoked behavioral responses, programmable microsurgery, and two-photon imaging of olfactory neural dynamics.
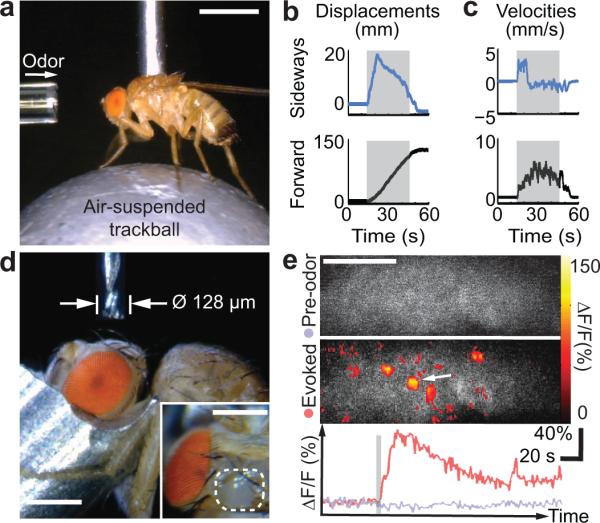
a. To test odor-induced locomotion, the robot holds a fly on an air-suspended trackball. A glass capillary delivers benzaldehyde, a repulsive odor, to the antennae. Scale bar: 1 mm.
b, c. Total displacements, b, and velocities, c, of the fly's sideways and forward locomotor responses for a representative trial (30 s odor stimulation; gray bar). Positive values denote rightward and forward walking.
d. With the fly head held by suction under robotic control, a 128-μm-diameter end mill executes a pre-programmed trajectory under machine-vision feedback to open the cuticle for brain imaging and remove trachea and fat bodies above the brain (Video 10). Inset: Automated microsurgery yields a clean excision (enclosed by white dashed line) of the head capsule to expose the left lobe of the mushroom body calyx. Scale bars: 0.25 mm.
e. Two-photon fluorescence imaging of odor-evoked neural Ca2+ activity following robotic microsurgery in a UAS-GCaMP3;;OK107 fly. As the robot holds the fly detachably by suction, delivery of ethyl acetate odor evokes neural Ca2+ responses. Maps of fluorescence changes (ΔF/F) show Ca2+ activity of the mushroom body Kenyon cells, before (Top) and during delivery of ethyl acetate (Middle). Time courses (Bottom) of fluorescence changes (ΔF/F) before (lavender trace) and in response (red trace) to ethyl acetate (gray bar), averaged over the area marked by the white arrow in the middle panel. Spatial scale bar: 20 μm.
After tethering a fly, our system can drill holes (~25 μm diameter or larger) for microinjection or fiber-optic light delivery (Supplementary Fig. 13). Alternatively, by transferring the fly to a three-dimensional translation stage beneath a high-speed end mill (≤ 25,000 rpm; 25–254 μm diameter) executing computer-programmable microsurgeries, the robot can open the fly's cuticle with micron-scale precision (Fig. 3d). The stage moves the mounted fly along a pre-defined cutting trajectory (Video 10), removing cuticle, trachea and fat bodies under saline immersion to keep the tissue moist and clear debris. Using these methods we prepared flies for two-photon microscopy (Supplementary Fig. 13d), and visualized odor-evoked neural Ca2+ dynamics in the mushroom body (50% success on 14 dissected flies; Fig. 3e). In flies expressing a Ca2+-indicator in Kenyon cells (Online Methods), we observed spatially sparse but temporally prolonged odor-evoked neural activation, consistent with prior studies20.
Overall, our programmable system flexibly combines automated handling, surgical maneuvers, machine-vision and behavioral assessments — without using anesthesia and while providing greater statistical power than humans can easily muster. A key virtue is the possibility for performing multiple analyses of individual flies, such as of morphological, behavioral and neurophysiological traits, for studies of how attributes interrelate. The capacity to catch and release individual flies will also allow time-lapse experiments involving repeated examinations of phenotypes across days or weeks, for studies of development, aging, or disease. Future implementations might include additional mechanical capabilities or multiple picking units (Supplementary Fig. 14). As with any new technology users will need time to explore the possibilities, but we expect a diverse library of programs will develop.
Online Methods
Fly stocks
We performed automated handling and odor-evoked locomotion studies using 3–10 day old male and female flies from the Oregon-R line (wild type). For machine vision analyses of fly morphology, we used both Oregon-R flies and recently derived, fully inbred lines from orchard populations in Pennsylvania and Maine (gift from D. Petrov and A. Bergland of Stanford University). For experiments involving two-photon imaging, we used female UAS-GCaMP3;;OK107 fly lines. We raised flies on standard cornmeal agar media under a 12-h light/dark cycle at 25°C and 50% relative humidity.
Robot construction
We actuated the robot with three DC motors (Maxon RE-25 part 118743), mounted at 120° increments upon a flat, circular base (100 mm diameter) made by laser machining in acrylic. An optical encoder with 40,000 quadrature counts (US Digital EC35) accompanying each motor sensed its angular position. Three separate position controllers, one master (Maxon 378308) controlling two slaves (Maxon 390438), synchronously drove the three motors (Supplementary Figs. 1–3).
We made the effector head (~50 mm diameter; Fig. 1a) in plastic (Objet, VeroWhitePlus) by three-dimensional printing. It held an onboard camera (Imaging Source DMM 22BUC03-ML), a board lens (f=8 mm; Edmund Optics NT-55574), the infrared ring illuminator (T1 package 880 nm LED with 17° emission angle), the picking effector (polished stainless steel hypodermic tube of 0.508 mm O.D. and 0.305 mm I.D.), and a rotation module to alter the fly's yaw.
The effector head connected to the three actuating motors through plastic (Objet, VeroWhitePlus) forearms and parallelograms. The forearms were three-dimensionally printed shapes of 20 mm length (Fig. 1a), within which we press fit cylindrical Neodymium magnets that had hollow cores. These magnets plus stainless steel (Type 440C) balls allowed us to form the four ball joints of each parallelogram. Each ball was connected at one end of a stainless steel rod (62 mm long), creating two barbell-like structures along the long edges of each parallelogram (Fig. 1a).
The rotation module had a gear assembly and pager motor from a servo (Hitec nano HS-35HD) to drive the picking effector rotation, via a flexible tube coupling, with position sensing from an optical encoder with 1200 quadrature counts (US Digital E4). We drove the pager motor with a position controller (Maxon 390438 in slave mode). The picking effector was connected to a suction source (–28” Hg of pressure) and a differential pressure sensor (Honeywell HSC TruStabiity rated at 1 psi), and was electronically gated by a solenoid valve (Humphreys H010E1).
We activated the picking suction and the LEDs and read the pressure sensor through a microcontroller (PIC32MX460 on a UBW32 board from Sparkfun) (Supplementary Fig. 2). We set the intensity of the LEDs by controlling the input voltage to an LED current driver (LuxDrive 1000mA BuckPuck) through a Digital-to-Analog Converter (MCP4725).
Picking platforms for non-anesthetized flies
In our initial work, we aspirated flies onto a metal mesh (100 × 100 openings per inch) picking platform or gently tapped them out of a vial. Weak negative air pressure across the mesh ensured that the flies fell inside the picking workspace. Once the flies were inside the picking platform, we disengaged the platform suction so the flies could upright themselves. In later work, we replaced the mesh platform with the rapid loading platform (Supplementary Fig. 4), which accepts standard vials of flies. To keep the flies inside either platform until all automatic handling tasks were done, we operated the robot with only near-infrared illumination, to sharply curtail the flies’ escapes by flight. A thin layer of silicone grease (Bayer) surrounding the platform served as a barrier to impede the flies from escaping the robot's picking workspace (Supplementary Fig. 4b). We arranged LEDs (880 nm emission) around the boundary of the platform to provide illumination for the stationary localization camera (Imaging Source DMM 22BUC03-ML) (Fig. 1c).
This localization camera yielded the approximate location of all flies in the picking platform. We mounted this camera at a suitable position and angle so as not to obstruct the robot's motion and adjusted the camera lens (f=16 mm, Edmund Optics NT-64108) to capture scenes of the entire picking platform. Flies appeared in these images as dark objects against the bright infrared illumination coming through the wire mesh.
To identify flies within these images (Supplementary Fig. 5), we first identified candidate flies by taking pixels that were darker than in a reference image of the platform without any flies, and then we binarized the images based on this distinction. We transformed the coordinates of the objects in the binarized images to their actual locations on the picking platform by applying a homographic transform (Supplementary Fig. 5). We identified individual flies among the candidate objects by accepting only binarized objects of >50 pixels, and we estimated the location of each fly by the centroid of the binarized object.
Automated picking
After randomly selecting a fly from among those identified on the platform, the robot moved the onboard camera over this fly to track it. Using the illumination from the platform LEDs, the robot acquired an image from the onboard camera and binarized the pixel values in an attempt to find a fly. If a fly was present, the robot turned on the ring illuminator and triggered the onboard camera again.
To locate the ring reflection on the fly thorax (Supplementary Fig. 6), an image analysis algorithm compared a binarized version of the image from the onboard camera to a 33 × 33 pixel template region (Fig. 1b) that corresponded to 0.53 mm × 0.53 mm on the thorax. Across all possible displacements between the centroid of the template and the image center, we calculated an overlap score between the template and a binarized image region equal in size: bright image pixels within the ring template scored positively whereas bright pixels outside the ring template scored negatively. We detected the presence of the ring if the net score exceeded a threshold; the position of the ring was determined from the template displacement that yielded the highest score. We estimated the vectorial orientation of the picked fly by examining the first principal component of all segmented pixels and then identifying the position of the head by its proximity to the ring reflection. To assess picking performance, we instructed the robot to serially pick and release flies continually for 23 min (Video 7 and Supplementary Table 2).
For high-magnification inspection of individual flies, the robot brought each picked fly to a camera (Imaging Source DMM 22BUC03-ML) equipped with a high magnification lens (f=16 mm, Edmund Optics NT-83107).
Automated handling routines
We wrote the robot control software in the Visual C++ software development environment. Elemental operations include fly tracking, picking, release, three-dimensional translation and yaw rotation of the picked fly, each operation implemented as a separate routine. Users may program new fly handling routines by specifying a sequence of operations and the desired time delays between successive steps. Through a graphical user interface, users can initiate execution of a pre-defined routine with a single button press, and can modify destination coordinates and rotations angles during run-time through text inputs.
If adaptive handling routines are desired, the user may also implement novel machine vision algorithms, using images or video acquired from the robot's onboard camera or accessory high-resolution cameras, to guide automated robotic navigation or to execute decision trees within the programmed sequence of operations. Inputs from other sensors can readily be accommodated as well.
To illustrate the interactions between sensing and decision-making in an adaptive handling routine, we describe here the process of transferring a picked fly to a head tether. Utilizing image feedback from the onboard camera, a first algorithm calculates the position and orientation of the tether. A second algorithm navigates the robot to a predefined inspection location, identifies the neck location of each picked fly from images taken by a static camera and then commands the translational and rotational trajectory of the robot to guide the fly head onto the tether. If the fly is incorrectly picked, the algorithm will decide to release the fly into the platform rather than tether it. Users may build a library of similar adaptive handling routines for different inspection tasks and behavioral assays and then combine these routines to automate increasingly complex experiments.
Automated neck detection algorithm
To head-fix a fly for microsurgery and imaging, our machine vision algorithm determined the position of the apse of the fly's neck, where it would be tethered (Supplementary Figs. 9–11). The algorithm had two main steps, which involved identifying the: (1) fly's yaw orientation, to help align the fly to the head holder; (2) position of the neck apse using the segmented and average silhouette of the fly, to help precisely tether the fly's head to the tether.
Automated sex sorting algorithm
The robot brought each picked fly to a stream of air to induce the fly to fly, so that the wings did not occlude the abdomen. We rotated the yaw of the picked fly so that the abdomen was optimally visible to the inspection camera (Fig. 2a, Supplementary Fig. 7), which was mounted at a ~45° angle to the picking platform.
Once the fly was optimally oriented, an image analysis algorithm checked for the presence of two distinct features: >2 dark bands, to indicate that we were looking at the abdomen; the presence of a dark-colored segment at the posterior of the male abdomen, to discriminate between male and female flies (Supplementary Fig. 7).
Determination of body and head cross-sectional areas
In images of single flies acquired from a sideways view and with the animal's long-axis parallel to the plane of the image, we first segmented the image of the fly from the background area. We then further distinguished the head and body regions, and computed their respective areas. Please see Supplementary Fig. 8 for details.
Microsurgery
The robot attached the head-fixed fly to a head holder mounted on a kinematic magnetic base, which provided micron-scale reproducible mating to a three-dimensional translational stage (Sutter Microsystems MP-285). This stage sat under a stationary rotary tool (Proxxon 38481) with a micro-endmill (Performance Micro Tool). We limited the movements of the fly using a custom-made dome placed over the thorax to prevent the fly from accidentally cutting itself on the rotating micro end-mill (Video 10). We maneuvered the fly's head to the rotating end-mill by moving the translational stage. Once the endmill punctured the cuticle, we applied adult hemolymph-like solution (AHLS)21 to the opening from a local reservoir above the dome. The three-dimensional translation stage then automatically moved the fly's head according to a pre-programmed trajectory under the rotating mill to cut cuticle, trachea and fat bodies above the brain. The AHLS kept the exposed brain hydrated and prevented surgically removed debris from adhering to the cutting surfaces or clogging the endmill's flutes. After automated surgery was done, we immediately replenished the AHLS to ready the fly for brain imaging.
High-speed videography
We acquired high-speed movies of the robot picking a fly using a Phantom v1610 camera (500 Hz frame rate) under 880-nm-illumination from LEDs.
Measurements of odor-evoked locomotion
The trackball setup closely resembled that of our prior work22. Two optical pen mice (Finger System, Korea) were directed at the equator of an air-suspended, hollow high-density polyethylene ball (6.35 mm diameter; ~80 mg mass; Precision Plastic Ball Co.). The pen mice were 2.3 cm away from the ball and tracked its rotational motion at a readout rate of 120 Hz. We converted the digital readouts to physical units and thereby computed the fly's forward, sideways, and turning velocities as described previously18, using custom software written in MATLAB (Mathworks). Velocities shown in Fig. 3 and Supplementary Fig. 12 are smoothed over a 1 s time window. For calculations of the average odor-evoked velocities, stationary states (<1 mm s−1 forward velocity) were excluded from the analysis. Calculations of mean sideways and rotational speeds were based on the absolute values of velocity values.
Two hours before behavioral experiments, we transferred 2–3 d old female Oregon-R flies from culture vials to empty vials using wet Kimwipes paper. For the flies in the manual handled group, we anesthetized the animals by inserting the vials into ice and then transferring them to a cold iron platform for mounting. We glued a fine syringe needle to the middle of the fly's dorsal thorax using ultraviolet-cured glue. We positioned the glued flies atop the trackball by using a three-dimensional translation stage. For the flies in the robotic handling group, we loaded the animals onto the picking platform and instructed the robot to pick them as needed. Once a fly was well picked, we exchanged the picking platform for the trackball setup. The robot held the fly ~0.7 mm above the trackball, allowing the fly to walk in place. A custom-built olfactometer delivered constant air flow (30 mL/min) and switched between clean air and air with odor. The flow exited a glass capillary (0.35 mm inner diameter) ~1 mm in front of the fly's antenna. We presented 10 trials of 2% benzaldehyde (a strong repulsive odor to flies), each lasting 30 s. Between each trial we presented clean air for 180 s.
Two-photon microscopy
We used a two-photon microscope (Prairie Technologies) equipped with a 0.95 NA water-immersion objective 20× (Olympus XLUMPFL). We imaged the Kenyon cells of the mushroom body (4 Hz frame rate) using ultra-short pulsed illumination of 920 nm, with 10-20 mW of power at the specimen plane. We delivered continuous air flow (142 mL/min) to the fly antennae through a stainless steel tube (0.3 mm inner diameter). We presented odor to the fly by re-directing the flow of air through a chamber containing filter paper dipped in ethyl acetate oil for 2 s intervals. Each fly was allowed at least 1 min of recovery time after each 2 s of odor presentation.
Analysis of neural Ca2+ dynamics
We aligned images acquired from two-photon microscopy with the Turboreg23 (http://bigwww.epfl.ch/thevenaz/turboreg/) plugin for NIH ImageJ software. We spatially filtered each aligned image with a Gaussian kernel (5 × 5 pixels; σ=2 pixels). We computed the baseline fluorescence of each pixel, F0, by averaging the pixel intensity across a 12.5 s interval before odor delivery. To extract the temporal waveform of cell's Ca2+ dynamics, we selected pixels from the chosen neuron and computed the relative changes in fluorescence as (F(t) – F0)/F0. We averaged the resulting time traces to obtain the final waveform estimate (Fig. 3e).
Statistical analyses
We performed all statistical analyses using MATLAB (Mathworks) software. Sample sizes were chosen using our own and published empirical measurements to gauge effect magnitudes. There was no formal randomization procedure, but flies were informally chosen in a random manner for all studies. No animals were excluded from analyses. The experimenters were not blind to each fly's genotype. For analyses of head and body areas, we excluded images of flies in which the wings occluded the body or in which the animals were incorrectly positioned. All statistical tests were non-parametric, to avoid assumptions of normal distributions or equal variance across groups.
Code availability
Readers interested in the software code for our analyses should please write the corresponding authors.
Supplementary Material
Acknowledgements
We thank T.R. Clandinin, L. Liang, L. Luo, G. Dietzl, S. Sinha and T.M. Baer for advice, A. Janjua, and J. Li for technical assistance, and D. Petrov and A. Bergland for providing inbred fly lines. We thank the W.M. Keck Foundation, the Stanford Bio-X program, and an NIH Director's Pioneer Award for research funding (M.J.S.), and the Stanford-NIBIB Training program in Biomedical Imaging Instrumentation (J.R.M.).
Footnotes
Author Contributions
J.S., E.T.W.H. contributed equally and innovated all hardware and software for the robotic system and performed all experiments. C.H. performed and analyzed the odor-evoked trackball experiments. J.R.M. created and performed the machine vision analyses of head and body areas. M.J.S. conceived and supervised the research program. All authors wrote the paper.
Competing Financial Interests
The authors declare no competing financial interest.
References
- 1.Albrecht DR, Bargmann CI. Nature methods. 2011;8:599–605. doi: 10.1038/nmeth.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chalasani SH, et al. Nature. 2007;450:63–70. doi: 10.1038/nature06292. [DOI] [PubMed] [Google Scholar]
- 3.Yanik MF, Rohde CB, Pardo-Martin C. Annual review of biomedical engineering. 2011;13:185–217. doi: 10.1146/annurev-bioeng-071910-124703. [DOI] [PubMed] [Google Scholar]
- 4.Chung K, et al. Nature methods. 2011;8:171–176. doi: 10.1038/nmeth.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crane MM, et al. Nature methods. 2012;9:977–980. doi: 10.1038/nmeth.2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kabra M, Robie AA, Rivera-Alba M, Branson S, Branson K. Nature methods. 2013;10:64–67. doi: 10.1038/nmeth.2281. [DOI] [PubMed] [Google Scholar]
- 7.Dankert H, Wang L, Hoopfer ED, Anderson DJ, Perona P. Nature methods. 2009;6:297–303. doi: 10.1038/nmeth.1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Branson K, Robie AA, Bender J, Perona P, Dickinson MH. Nature methods. 2009;6:451–457. doi: 10.1038/nmeth.1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Straw AD, Branson K, Neumann TR, Dickinson MH. Journal of the Royal Society, Interface / the Royal Society. 2011;8:395–409. doi: 10.1098/rsif.2010.0230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaumette F, Hutchinson S. Robotics & Automation Magazine, IEEE. 2006;13:82–90. [Google Scholar]
- 11.Higashimori M, Kaneko M, Namiki A, Ishikawa M. The International Journal of Robotics Research. 2005;24:743–753. [Google Scholar]
- 12.Mendes CS, Bartos I, Akay T, Márka S, Mann RS. eLife. 2013;2:e00231. doi: 10.7554/eLife.00231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dickinson M, Gotz KJ. Exp. Biol. 1996;199:2085–2104. doi: 10.1242/jeb.199.9.2085. [DOI] [PubMed] [Google Scholar]
- 14.Zumstein N, Forman O, Nongthomba U, Sparrow JC, Elliott CJ. J Exp Biol. 2004;207:3515–3522. doi: 10.1242/jeb.01181. [DOI] [PubMed] [Google Scholar]
- 15.Barron AB. Journal of Insect Physiology. 2000;46:439–442. doi: 10.1016/s0022-1910(99)00129-8. [DOI] [PubMed] [Google Scholar]
- 16.Nicolas G, Sillans D. Annual Review of Entomology. 1989;34:97–116. [Google Scholar]
- 17.Jean David R, et al. Journal of Thermal Biology. 1998;23:291–299. [Google Scholar]
- 18.Seelig JD, et al. Nature methods. 2010;7:535–540. doi: 10.1038/nmeth.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maimon G, Straw AD, Dickinson MH. Nat Neurosci. 2010;13:393–399. doi: 10.1038/nn.2492. [DOI] [PubMed] [Google Scholar]
- 20.Honegger KS, Campbell RA, Turner GC. Journal of neuroscience. 2011;31:11772–11785. doi: 10.1523/JNEUROSCI.1099-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilson RI, Turner GC, Laurent G. Science. 2004;303:366–370. doi: 10.1126/science.1090782. [DOI] [PubMed] [Google Scholar]
- 22.Clark DA, Bursztyn L, Horowitz MA, Schnitzer MJ, Clandinin TR. Neuron. 2011;70:1165–1177. doi: 10.1016/j.neuron.2011.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thevenaz P, Ruttimann UE, Unser M. IEEE Transactions on Image Processing. 1998;7:27–41. doi: 10.1109/83.650848. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.