Abstract
In mitotically dividing cells, the cohesin complex tethers sister chromatids, the products of DNA replication, together from the time they are generated during S phase until anaphase. Cohesion between sister chromatids ensures accurate chromosome segregation, and promotes normal gene regulation and certain kinds of DNA repair. In somatic cells, the core cohesin complex is composed of four subunits: Smc1, Smc3, Rad21 and an SA subunit. During meiotic cell divisions meiosis-specific isoforms of several of the cohesin subunits are also expressed and incorporated into distinct meiotic cohesin complexes. The relative contributions of these meiosis-specific forms of cohesin to chromosome dynamics during meiotic progression have not been fully worked out. However, the localization of these proteins during chromosome pairing and synapsis, and their unique loss-of-function phenotypes, suggest non-overlapping roles in controlling meiotic chromosome behavior. Many of the proteins that regulate cohesin function during mitosis also appear to regulate cohesin during meiosis. Here we review how cohesin contributes to meiotic chromosome dynamics, and explore similarities and differences between cohesin regulation during the mitotic cell cycle and meiotic progression. A deeper understanding of the regulation and function of cohesin in meiosis will provide important new insights into how the cohesin complex is able to promote distinct kinds of chromosome interactions under diverse conditions.
Keywords: cell cycle, cohesin complex, cohesin regulation, meiosis
Introduction
In somatic cells, sister chromatids are held together from the time they are made until cell division by a protein complex called cohesin. The cohesin complex and its regulators were originally identified in genetic screens in yeast for genes required for maintenance of sister chromatid association during mitotic arrest [1–3]. Subsequent isolation of vertebrate orthologs from frog egg extracts confirmed that cohesion between sister chromatids is mediated by a highly conserved complex of proteins, with readily identifiable orthologs from yeast to man [4].
The cohesin complex is essential for normal growth and development at the cellular and organismal level [5]. In mitotically growing cells, cohesion between sister chromatids promotes cell cycle progression and accurate chromosome segregation at anaphase, and mediates DNA repair through mechanisms that depend on homologous recombination (reviewed in [6]). In addition, cohesin promotes normal gene regulation through control of chromosome structure [7]. Aberrant cohesion causes severe developmental disorders, such as Roberts syndrome and Cornelia de Lange syndrome, and is correlated with chromosome instability in tumors [8–11].
During meiotic cell divisions, a number of unique cohesin subunits are expressed and incorporated into meiosis-specific cohesin complexes. Interestingly, these meiotic cohesins, which are essential for normal meiotic chromosome dynamics and thus fertility, have non-overlapping localization and unique regulation during meiotic progression. Here, I summarize our understanding of the contributions of meiotic cohesin to proper meiotic chromosome dynamics, and explore the analogies that can be drawn between mitotic and meiotic cohesin regulation. To this end, I begin with a brief overview of cohesin regulation in mitotically dividing cells. (More comprehensive reviews of this material can be found in [6,12,13].) I then delve into how mitotic events compare and contrast with meiotic cohesin regulation.
The cohesin cycle
Genetic, biochemical and cell biological studies have contributed to our understanding of cohesin structure, regulation and function during mitotic cell cycle progression (Fig. 1). The core cohesin complex is composed of four subunits: two elongated proteins, called Smc1 and Smc3 (for structural maintenance of chromosomes), and two non-SMC subunits, called Scc1 or Mcd1 in budding yeast (Rad21 in vertebrates and fission yeast) and Scc3 (SA1 or SA2 in vertebrates) (Fig. 2). All SMC proteins share a similar structure: they fold back on themselves to form anti-parallel coiled-coils, with their N and C termini in close proximity. Smc1 and Smc3 interact at two places: the Smc1 and Smc3 head groups interact to form a pair of inter-molecular ABC type ATPases [14], and the hinge domains formed between the coil-forming regions also interact [15]. Rad21 forms a bridge between the head groups of the SMC proteins, and a fourth subunit, Scc3 (either SA1 or SA2 in vertebrates), binds to the complex through Rad21. The core cohesin complex shares sequence and structural homology with other SMC-protein-containing complexes, such as condensin and the Smc5/6 complex, which similarly promote chromatin–chromatin interactions in different contexts (reviewed in [16,17]).
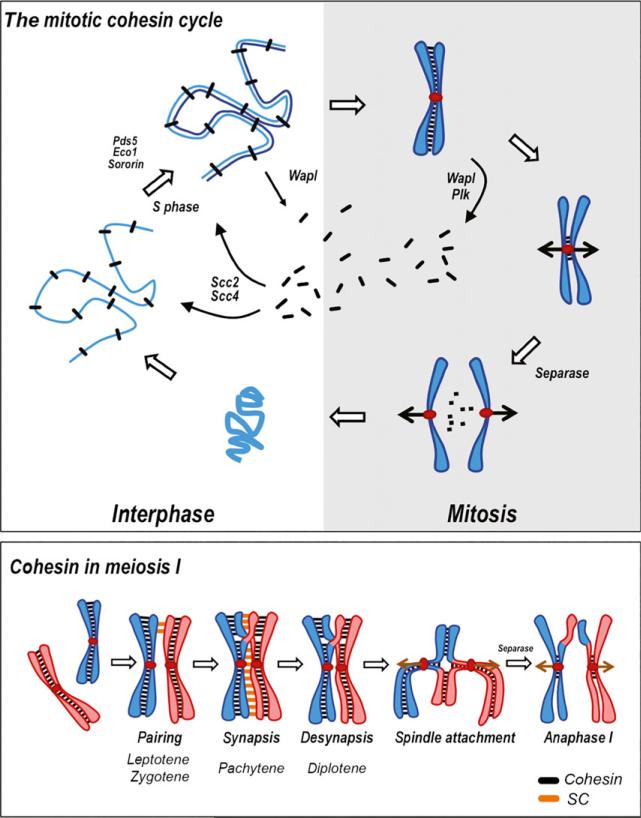
Fig. 1.
Upper panel: the mitotic cohesin cycle. Shown is the vertebrate cohesin cycle. Cohesin (black bars) is loaded onto chromosomes as cells enter interphase by the Scc2/Scc4 loader complex. Following Pds5 loading cohesin is acetylated by Eco1 during S phase, which stimulates Sororin binding and stabilization of cohesin. Throughout interphase, cohesin is kept largely dynamic by the activity of Wapl. During mitotic entry the bulk of cohesin is removed from chromosomes by both Wapl and Plk-dependent phosphorylation, although some is retained at the centromere. At anaphase onset, the site-specific protease called separase is activated and cleaves the Rad21 subunit of cohesin, allowing separation of sister chromatids. Cohesin reloads as chromosomes decondense in telophase, and the cycle begins again. Lower panel: cohesin in meiosis. This drawing illustrates the presence of cohesin during meiosis I chromosome dynamics. Cohesin (black bars) is loaded onto chromosomes during pre-meiotic S phase (not shown), tethering sister chromatids together. This cohesin is retained during homolog pairing and synapsis, and is required for the proper assembly of the synaptonemal complex (SC; orange). Following desynapsis in diplotene, cohesion between sister chromatids distal to the point of crossover provides resistance to spindle pulling forces (brown arrows) at metaphase I arrest. At anaphase, separase-dependent cleavage of cohesin on chromosome arms allows arm and homolog separation at meiosis I anaphase. At this time, cohesin is protected from cleavage at the centromere by Sgo (not shown), preventing sister separation.
Fig. 2.
Cohesin complexes in mitosis and meiosis. Cohesin is composed of four core subunits assembled as shown (top). These include two Smc proteins that fold back on themselves to form an ATPase head domain and a central hinge. A dimer of Smc proteins interacts with a kleisin subunit and an Scc3/SA protein to form the core four-subunit complex. The subunit composition of the sole mitotic cohesin complex is shown (bottom left), and the various meiotic complexes are also indicated (bottom right). All known cohesin complexes contain the same Smc3 protein. Rec8-containing cohesin is found in most eukaryotic meiocytes, while the remaining meiotic cohesins are found only in higher eukaryotes. Additional meiosis-specific subunits of cohesin include the Smc1α-like subunit called Smc1β, as well as the meiosis-specific kleisins Rec8 and Rad21L. In vertebrates one of two SA subunits, SA1 or SA2, is found in mitotic cohesin. In meiotic cohesin this is substituted with a meiosis-specific ortholog called STAG3.
The interaction of cohesin with chromosomes is regulated by a number of proteins, which collectively ensure proper cohesin dynamics in response to cell cycle progression (Table 1). This modulation of the association of cohesin with chromatin in response to cell cycle progression is often referred to as the cohesin cycle. This cycle refers to the binding of cohesin to chromatin, the tethering together of sister chromatids, and finally release of chromatids at cell division. The cycle can be broken down into three distinct molecular steps, described here.
Table 1.
Cohesin regulators in mitosis and meiosis. Summary of cohesin regulatory proteins and their localization and phenotypes in meiosis, if known. In the case of Wapl, plant data were included because there were few data available from yeast or vertebrate models.
| Cohesin regulators |
Meiotic roles? |
|||
|---|---|---|---|---|
| Yeast protein | Function | Metazoan form(s) | Meiotic localization | Meiotic phenotype |
| Eco1 (Sc) Eso1 (Sp) | Cohesin acetylation and function | Esco1 Esco2 | Along chromosome axes, LE | Fission yeast: Smc3 acetylation Budding yeast and vertebrates: ND |
| Scc2 Scc4 | Cohesin loading complex | Nipbl Mau2 | Along chromosome axes into early pachytene | Loss of cohesion in yeast. Not tested in vertebrates |
| Rad61 | Destabilization of cohesion, blocked by Smc3 acetylation | ND | ND | |
| Wapl (vertebrates) | Along chromosome axes in pachytene | ND | ||
| Wapl1, Wapl2 (plants) | ND | Cohesin not properly removed in meiosis, broken chromosomes, uneven segregation | ||
| Pds5 | Scaffold for cohesin regulation | Pds5A Pds5B | Pds5A absent, Pds5B on axial cores | Pds5A: no apparent phenotype Pds5B: germ cell failure |
| Esp1 ‘Separase’ | Site specific protease, cleaves Rad21 at anaphase | Separase | ND | Cleaves Rec8 and Rad21. Rad21L cleavage not tested |
| Not found | Inhibits Wapl, stabilizes cohesion | Sororin | Along chromosome axes after synapsis | ND |
Cohesin loading
Cohesin is loaded onto chromosomes by a heterodimeric complex composed of the Scc2 and Scc4 proteins [18]. In vertebrates cohesin is first loaded to chromosomes at mitotic exit [4,19], while in budding yeast cohesin loading occurs somewhat later, during S phase. In all cases cohesin loading requires the activity of the Scc2/Scc4 complex, which becomes dispensable for cohesion after DNA replication is complete in G2 [18]. Loading of cohesin onto chromosomes requires nucleotide hydrolysis by the SMC head domains [14,20], which is stimulated by the Scc2/Scc4 complex [21]. This hydrolysis is thought to lead to transient opening of the cohesin ring at the hinge interaction domain, and topological entrapment of DNA within [22]. Because they are thought to ensure this entrapment by forming a third component of a tripartite cohesin ring, proteins in the Rad21 family of SMC-interacting proteins have been dubbed ‘kleisins’ from the Greek word for closure [23]. Once cohesin is loaded onto chromatin, the cohesin-interacting protein Pds5 associates with the complex. Interestingly, the interaction between cohesin and Pds5 is weak in solution, and may depend upon a particular conformation of cohesin that is achieved during chromatin association [19].
Cohesion establishment
The requirement for cohesin loading prior to the completion of DNA replication reflects the fact that cohesin must be present on chromatin during DNA replication in order to be activated to tether sister chromatids together [24]. This activation process is often referred to as ‘cohesion establishment’. The precise molecular details of this activation are poorly understood, but modification of cohesin by members of the Eco1 family of acetyltransferases is critical. The Eco enzymes (called Esco1 and Esco2 in vertebrates) interact directly with components of the replication machinery and modify the Smc3 subunit of cohesin [25–29]. In the absence of Eco/Esco-dependent modification in both yeast and vertebrates, cohesin binds to chromosomes, but fails to tether sister chromatids together [30,31]. How does acetylation promote cohesion establishment? The simplest model is that acetylation of Smc3 disrupts interaction of the Wapl protein with the cohesin complex. Wapl (Rad61 in budding yeast) is the major cohesion-disrupting activity in the cell, thought to promote opening of the Rad21–Smc3 gate and unload cohesin from chromatin [32–35]. Wapl is thus sometimes referred to as an ‘anti-establishment’ factor. When Wapl activity is blocked cohesin's interaction with chromatin becomes more stable [36]. Acetylation of Smc3 by Eco1 may also have effects on the intrinsic chromatin binding activity of cohesin, independent of Wapl [37].
Metazoans express an additional essential cohesion regulator, not found in fungal systems, called Sororin [38]. This protein binds to cohesin in a replication- and acetylation-dependent manner, and is required for cohesion maintenance from the time of DNA replication until anaphase [39,40]. Sororin is thought to antagonize the activity of Wapl, perhaps by competing with it for interaction with the cohesin complex [41]. Interestingly, Sororin binds to cohesin only following cohesion establishment [26]. Thus Sororin's interaction with cohesin provides a biochemical mark for active cohesion. Understanding the nature of the Sororin binding site promises to provide critical information about the nature of the establishment mechanism.
The activities of Wapl, Eco and Sororin are all critically dependent on a cohesin-interacting protein called Pds5, which interacts with the core cohesin complex through both the SA and Rad21 [36,42–46]. Pds5 is therefore essential for both the establishment of cohesion during S phase and for cohesion maintenance in G2 [47,48]. In vertebrates, Pds5 binds cohesin when it is chromatin bound and is required for cohesin's acetylation by the Esco enzymes [19,42]. In addition, destabilization of cohesion by Wapl occurs through Pds5 [36,46]. Thus Pds5 serves a scaffold function to integrate pro- and anti-cohesion activities during cell cycle progression.
Interestingly, in vertebrates only a small pool of cohesin is stabilized on chromatin during DNA replication [49]. The bulk of cohesin associates dynamically with chromatin, constantly loading and unloading, perhaps to fulfill additional roles in modulating chromosome architecture. Through its effect on chromosome structure cohesin plays significant roles in gene regulation and programmed gene rearrangements (such immunoglobulin and T cell receptor gene rearrangements) that are likely to be independent of sister tethering per se [50–53]. In fact, chromatin cohesin is found associated with particular chromosomal loci throughout the cell cycle, even in G1 when there are not yet two sister chromatids to be tethered together [54]. The precise role of ‘non-cohesive’ cohesin in interphase cells is not clear and the subject of current investigation by many laboratories [55,56].
Cohesin release
Following cohesin loading and DNA replication-dependent cohesion establishment, sister chromatids remain tethered together by cohesin until the metaphase–anaphase transition. In budding and fission yeast cohesin remains largely associated with chromosomes until anaphase [57]. In vertebrate cells, the bulk of cohesin is removed from chromosome arms during mitotic entry by phosphorylation and by the activity of the Wapl protein [36,46,58]. Protection of cohesion at the centromere regions is accomplished by specific recruitment of a phosphatase, PP2A, to the centromeric region of the chromosomes by Sgo1 [59,60]. PP2A is thought to resist cohesin release by maintaining centromeric cohesin in its dephosphorylated state [61–63]. In metazoans, PP2a also prevents phosphorylation-dependent removal of Sororin from the centromeric region of chromosomes, thus protecting cohesin from Wapl-dependent removal in metaphase [64].
In the final step of the cohesin cycle, cleavage of the Rad21 subunit of cohesin by a site-specific protease called separase releases the cohesin complex and allows anaphase separation of chromosomes (reviewed in [65]). Separase is activated at the metaphase–anaphase transition both by degradation of an inhibitory protein called Securin and through loss of inhibitory phosphorylation on separase itself as mitotic kinases are inactivated [66,67]. In vertebrate cells, only a small fraction of cohesin remains associated with chromosomes and is thus cleaved at the metaphase to anaphase transition [68]. The bulk of cohesin remains intact, can be redeployed in telophase as nuclei are reforming, and is thought to play a significant role in chromosome architecture in G1 prior DNA replication. In budding yeast, in contrast, virtually all Rad21 is cleaved at anaphase and does not accumulate again until the next S phase [69].
In summary, during transit through the cohesin cycle the cohesin ring is thought to open in three distinct ways: at the hinge region during loading onto chromosomes, at the Smc3–Rad21 interface during unloading by Wapl, and by cleavage of the Rad21 subunit at anaphase. These activities of cohesin are controlled by several proteins, including Sororin, Pds5, Eco acetyltransferases and Wapl, that collectively ensure proper sister chromatid cohesion.
Cohesion and the DNA damage response
In budding yeast, cohesion between sister chromatids is increased both locally and throughout the nucleus in response to DNA double strand breaks and is critical for DNA double strand break repair [70–73]. A number of the proteins that promote cohesion establishment during cell cycle progression also promote cohesion establishment in response to DNA damage signaling. In response to DNA damage these proteins, including the Scc2/Scc4 cohesin loader and Eco1 acetyltransferase, act downstream of the ATM/Tel1 and ATR/Mec1 checkpoint kinases and phosphorylation of the histone variant H2AX at the sites of DNA damage [73,74]. Mutation or decreased expression of cohesin subunits or cohesin regulators, such as Scc2/Scc4, Wapl, Pds5, Eco1/Esco1/2, separase, and in vertebrates Sororin, all confer increased sensitivity to DNA damage [72,74–78]. Sister chromatid cohesion is thought to promote double strand break repair by ensuring the close proximity of an undamaged sister chromatid as a template for repair of the damaged sister chromatid through a recombination-based mechanism. Such a mechanism must necessarily be restricted to S phase or G2 cells, in which two sister chromatids are present. Precisely how this restriction occurs is not clear, and may differ between yeast and higher eukaryotes [74,79].
The meiotic cohesins
During meiotic prophase I chromosomes undergo an elaborate series of movements and structural rearrangements, in which homologous chromosomes sort themselves out, pair and synapse (see accompanying reviews by Sansam and Pezza [80], Kurdzo and Dawson [81]). Pairing refers to the initial DNA homology-dependent interactions between homologous chromosomes, while synapsis is the process by which the homologous chromosome pairs become intimately associated with each other through the synaptonemal complex (SC), allowing for progression of homologous recombination (reviewed in Sansam and Pezza, this issue [80]). During synapsis, a proteinaceous bridge, called the SC, is formed between homologous chromosomes (see Kurdzo and Dawson, this issue [81]). The first SC element to form is the axial element (AE), which assembles on each sister chromatid pair, called a univalent, prior to synapsis. As the chromosome cores become more tightly apposed during synapsis, additional proteins are recruited to form the transverse filaments and the central element. At this time, called the pachytene stage, the AE proteins constitute part of the lateral elements (LEs) of the mature tripartite zipper-like SC structure. Together, one pair of homologous chromosomes that are synapsed is referred to as a bivalent.
The alignment, pairing, synapsis and recombination of homologous chromosomes in meiotic prophase present an enormously complicated set of steric and topological challenges, and proper progression of these rearrangements is critically dependent upon the cohesin complex. Interestingly, the subunit composition of cohesin is altered in meiosis, with meiosis-specific paralogs of various cohesin subunits expressed and incorporated into the holocomplex (Fig. 2 and Table 2). These cohesins, which include meiosis-specific kleisin subunits called Rec8 or in vertebrates Rad21L, as well as a meiosis-specific SMC protein in some species (Smc1β; Fig. 2), are essential for normal meiotic chromosome structure and dynamics. While many of the mitotic functions of cohesin are likely to be conserved during meiosis, the regulation of cohesin during meiotic progression is complex and not fully understood.
Table 2.
Cohesin in mitosis and meiosis. Summary of cohesin subunits in yeast and mammalian mitosis and meiosis. The meiotic phenotypes are summarized in the last two columns. See text for details. DSB, double strand break.
| Mitotic cohesin |
Meiotic cohesins (mammalian) |
|||||
|---|---|---|---|---|---|---|
| Yeast protein | Metazoan form(s) | Function | Meiosis specific isoform | Meiotic localization | Meiotic arrest point | Other phenotypes? |
| Smc1 | Smc1 | Core cohesin complex | Smc1β | Chromosome axes | Pachytene-like | Females: complete error-prone meiosis. Males: incomplete synapsis and meiotic failure. Shortened axes |
| Smc3 | Smc3 | Core cohesin subunit. | – | |||
| Scc1 (Rad21, Mcd1) | Rad21 | ‘Kleisin’ subunit. Cleaved for cohesin release at anaphase | Rec8 | Chromosome axes, alternating with Rad21L | Zygotene-like | Meiotic failure. Some pairing, failure to repair DSBs. Shortened axes |
| Rad21L | Chromosome axes, alternating with Rec8 | Zygotene-like | Meiotic failure. Pattern on homologs similar prior to synapsis. Failure to repair DSBs | |||
| Scc3 | SA-1 SA-2 |
Linker to Pds5, other | Stag3 | Chromosome axes | Zygotene-like | Meiotic failure. No DSB repair. Loss of Smc1β, Rec8 and Rad21L on cores, shortened axes |
During meiosis cohesion is released in two steps (Fig. 1). In anaphase of meiosis I, cohesin is removed from chromosome arms, allowing separation of homologs (disjunction) by the site-specific protease called separase. At centromeres, cohesion is protected from removal in meiosis I by the activity of the Shugoshin protein Sgo1 (Japanese for ‘guardian spirit’) which recruits PP2A phosphatase to the centromere [59,60]. As phosphorylation of cohesin is required for separase-dependent cleavage, centromeric cohesion is protected from cleavage by the presence of PP2A. In meiosis II, cohesion at the centromere is no longer protected from separase and sister chomatids are thus unlinked and segregated during the second meiotic division. The mechanism by which centromeric cohesion is lost in meiosis II anaphase is controversial and may vary between species.
The geometry of bivalents after desynapsis is such that the resistance to spindle pulling forces in meiosis I is provided principally by cohesion that is distal to the sites of crossing over (Fig. 1 and [82]). In human females, cohesion established in meiosis I must have the remarkable ability to persist for up to four decades, as meiotic prophase and cohesion establishment occur before birth and cohesion must be maintained until meiosis I release at ovulation (reviewed in [83]). This has led to the proposal that failure of meiotic cohesion contributes to the age-related increase in aneuploidy seen in women [84], a hypothesis that has been experimentally validated in flies and mice [85,86]. In naturally aging female mice, centromeric cohesion declines with age and females that are genetically deficient in certain forms of meiotic cohesin, such as cohesin containing Rad21L (see below), while born fertile, show an accelerated decline in fertility as they age [87]. When their oocytes are examined, this reduced fertility is seen to be accompanied by migration of chiasmata (the cytological manifestations of crossovers) towards the distal end of chromosomes and the appearance of unpaired homologs, as would be predicted if cohesion loss were the underlying mechanism of meiotic failure [88]. Recent work in Drosophila suggests that cohesin is renewed throughout meiotic prophase, and that this renewal is necessary for chiasma maintenance and accurate chromosome segregation, although chiasma formation may also be compromised in this model due to premature SC disassembly [89]. The mechanism of age-dependent cohesion loss in oocytes may be analogous to the ‘cohesion fatigue’ seen in somatic cells during sustained metaphase arrest, although the absence of spindle pulling forces during meiotic arrest might suggest alternative mechanisms [90,91]. As aneuploidy is a major contributor to birth defects and spontaneous abortion, understanding the mechanisms involved in the establishment, maintenance and loss of meiotic cohesion is of great interest.
Meiotic kleisins: Rec8 and Rad21L
Virtually all eukaryotic species express a meiosis-specific paralog of Rad21 called Rec8. Rec8 is found in cohesin complexes present during meiotic prophase and is localized to the chromosome core prior to chromosome synapsis (Figs 2 and 3). Rec8 was originally identified in Schizosaccharomyces pombe in a screen for genes that are required for meiotic recombination [92]. Subsequent work done in numerous model organisms has demonstrated a conserved essential function for Rec8 in meiotic chromosome dynamics. Rec8-containing cohesin complexes are loaded onto chromosomes during pre-meiotic S phase. In budding yeast, mutations in Rec8 result in complete failure of sister chromatid cohesion as well as failure of homologous chromosome pairing [93]. In mice, the absence of Rec8 causes germ cell failure and sterility in both males and females. In Rec8–/– mice the initial stages in homologous chromosome pairing occur, but chromosomes do not synapse, and recombination fails [94,95].
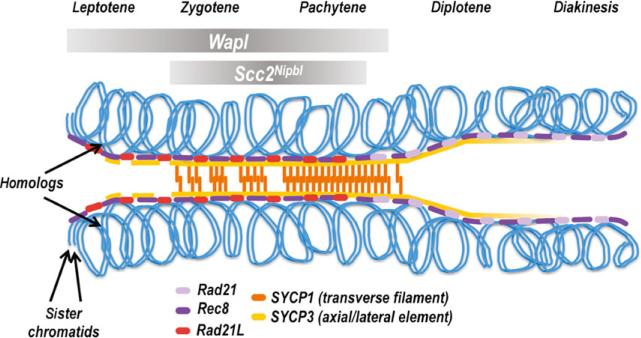
Fig. 3.
Meiotic cohesins during meiosis I prophase. The relative timing of loading of the different kleisin subunits during mammalian meiotic prophase is indicated. Rec8 and Rad21L are thought to be loaded during pre-meiotic S phase; Rad21-containing complexes are not present early and decorate the chromosome axes beginning in late pachytene. Rec8 and Rad21L have non-overlapping patterns of localization along the chromosome axes. For simplicity, the timing with which the cohesin regulators Wapl and Scc2Nipbl are associated with the chromosome axes is indicated by the grey bars at the top. Precise localization of these regulatory proteins relative to the SC components has not been determined.
In mice, loss of Rec8 leads to the aberrant assembly of axial structures, suggesting that Rec8-dependent cohesion controls proper development and homeostasis of SC structures [94]. Indeed, in Rec8–/– mice SYCP1, a component of the central region of mature SC, is occasionally observed between loosely associated sister chromatids (rather than between homologous partners), and there is also ultrastructural evidence of central element structures between sister chromatids. Interestingly, chromosomes with these abnormal SC structures often contain numerous breaks and discontinuities in the AEs as revealed by silver staining. In these meiocytes, recombination is initiated but does not progress beyond the initial steps [96].
Although Rad21-containing complexes can be found in meiotic cells in some organisms, several lines of evidence suggest that the tethering of sister chromatids together during meiosis occurs exclusively through Rec8 cohesin. In budding yeast, Rec8 is seen decorating meiotic chromosomes, while Rad21 is greatly diminished, and loss of Rec8 function leads to premature separation of sister chromatids prior to the first meiotic division, in a manner indistinguishable from that seen with the loss of the core cohesin subunit Smc3 [93]. In mouse oocytes, despite the presence of Rad21 cohesin complexes, the artificial cleavage of Rec8 with an engineered protease site causes premature separation of both homologs and sister chromatids in meiosis I, demonstrating unambiguously that sister chromatids are tethered exclusively by Rec8 in these cells [97].
In addition to the Rec8 protein, vertebrates express an additional Rad21 ortholog called Rad21L, also only during meiosis. Interestingly Rad21L is found on chromosome cores in a non-overlapping, alternating punctate pattern with Rec8, suggesting that Rad21L and Rec8 perform unique functions in meiotic chromosome development [98,99] (Fig. 3). In the mouse, Rad21L mutants arrest, like Rec8 mutants, in a diplotene-like state, with persistence of early-mid DNA repair markers suggesting impaired recombination and repair. In the absence of both Rec8 and Rad21L, mice are completely unable to form AE or LE structures [96]. This phenotype is reminiscent of the Rec8 phenotype in yeast, suggesting that meiosis-specific cohesin is essential for AE assembly and that Rec8-containing complexes are sufficient in yeast, while two distinct meiotic kleisins, Rec8 and Rad21L, contribute to this function in vertebrates.
The localization of Rec8 and Rad21L is consistent with a model of combined, non-redundant function for the two proteins in axis formation and synapsis. Interestingly, the patterns of Rec8 and Rad21L on the not-yet synapsed regions of homologous chromosomes in leptotene are remarkably similar between homologs. This observation has led to the proposal that Rad21L and Rec8 patterns on univalents may serve as a kind of bar code to enhance homolog recognition and thus pairing and synapsis [99]. There is some evidence for physical interaction of Rad21L, but not Rec8 or Rad21, with the transverse filament protein SYCP1 [98]. One possible model is that unique patterns of SYCP1 recruitment by Rad21L contribute to the efficiency of homolog recognition and nucleation of SC assembly. Indeed, when double strand breaks are prevented by mutation of Spo11, initial interactions between homologous chromosomes depend upon Rad21L, consistent with the model that Rad21L-dependent chromosome structure underlies initial homolog recognition [100].
Rad21L and Rec8 are clearly not functionally redundant. Unlike Rec8, Rad21L staining of the chromosome cores does not persist past pachytene [98,99,101]. Additionally, Rec8 is essential for centromere cohesion, while Rad21L is not found on the metaphase centromere and does not directly impact centromere cohesion.
The nematode Caenorhabditis elegans also expresses meiosis-specific kleisins, including REC-8 and two seemingly redundant additional kleisins called COH-3 and COH-4 [102,103]. Analysis of mutant phenotypes provides compelling evidence that these meiotic kleisins, as well as the SCC-1 kleisin (also present during meiosis), confer distinct functional properties to the cohesin complexes containing them. For example, REC-8 cohesin is required to establish cohesion during DNA replication, while COH-3 and COH-4 (COH-3/4) containing complexes are induced to become cohesive in response to meiotic DNA double strand breaks, well after pre-meiotic DNA replication is complete [104]. It is possible that Rad21L fulfills a function similar to COH-3/4 in vertebrates, as it is seen to load at a comparably late time during meiotic progression [87,98]. Similarly, the kleisin subunits may also determine the mechanism(s) by which particular complexes are removed from meiotic chromosomes during meiotic progression.
Additional meiotic cohesin subunits: Smc1β and STAG3
In vertebrate meiosis, not only are meiotic kleisin subunits expressed and incorporated into cohesin complexes, but meiosis-specific core subunits are also found in the complex (Fig. 2). A meiosis-specific form of Smc1 called Smc1β is found in some cohesin complexes in mammals [105]. (To avoid confusion the originally identified form is now referred to as Smc1α.) Smc1β-containing cohesin is found along chromosome axes throughout meiotic prophase, including through desynapsis in diplotene, and accumulates around centromeres in diakinesis [105]. Extensive immunoprecipitation analysis indicates that Smc1β is found in distinct separate complexes with each of the three kleisins present in meiosis: Rad21, Rad21L and Rec8 [98,101].
In the absence of Smc1β the meiotic chromosome axis–loop structure is abnormal in prophase, with chromosomes being ~ 50% shorter than their wild-type counterparts [106]. In males, meiosis arrests at a pachytene-like state, suggesting that pairing and synapsis are largely functional. Recombination fails and the animals of both sexes are sterile. Smc1β is not present until leptotene, when it accumulates on chromosome cores and remains there until anaphase of meiosis I. Smc1β remains associated with centromeres until anaphase II, suggesting that Smc1β cohesin prevents sister chromatid separation in anaphase of meiosis I [107].
Vertebrates also express a meiosis-specific SA subunit, called STAG3 (Fig. 2). Like its mitotic counterparts SA1 and SA2, STAG3 interacts with the kleisin subunits of cohesin. It appears that STAG3 is the one invariant subunit of meiotic cohesin complexes: there is no evidence that STAG3 interacts with SMC1α-containing cohesin, and in Rec8/Rad21L double mutants STAG3 is no longer found along chromosome axes. There is conflicting evidence as to whether the mitotic SA proteins SA1 and SA2 are found in complexes with the meiotic kleisins Rad21L and Rec8 [98,99,101].
Stag3–/– mice have the most severe meiotic phenotype of all the single meiosis-specific cohesin mutants [108–110]. This phenotype is reminiscent of the Rad21L–/–/Rec8–/– double mutant [108,110]. AEs entirely fail to develop and some markers of incomplete DNA repair such as γH2AX are present throughout chromatin. In addition, both centromeric and telomeric sister chromatid cohesion are compromised. Interestingly, in the absence of STAG3 there is still some limited recruitment of mitotic cohesins to the chromosome axes, which are grossly shortened [110]. Double strand breaks are generated, although recombination does not progress. In addition, the total level of Rec8 cohesin is reduced, suggesting that STAG3 in some way contributes to the expression or stability of these complexes [109].
Fission yeast also expresses a meiosis-specific Scc3 ortholog called Rec11. Rec11, like Rec8, is required for meiotic sister chromatid cohesion. Unlike many other meiosis-specific subunits of cohesin, this protein shows preferential localization to the chromosome arms, in association with Rec8 kleisin, while at the centromere Rec8 is found associated with the Scc3 subunit (Psm3) that is active in both mitosis and meiosis [111].
Sexual dimorphism
In mice that are compromised for meiotic cohesin expression, the meiotic phenotypes can differ between male and female animals, with the males generally being more severely compromised [112]. It is possible that the mechanisms are somehow distinct between males and females. An alternative explanation for this is that checkpoint activation in male meiosis leads to apoptotic degradation of defective cells at an earlier stage in meiotic progression. Indeed, failures in synapsis that occur due to loss of meiotic cohesion in males prevent meiotic sex chromosome inactivation as well as silencing of non-paired regions in somatic chromosomes, leading to ectopic expression of pro-apoptotic genes (reviewed in [113]). Whatever the cause, meiotic progression is generally more compromised in male meiotic cohesin mutant mice (e.g. Smc1β–/– or Rad21L–/–) than in female [87,106].
Meiotic cohesion and chromosome structure
Meiotic cohesin promotes normal chromosome structure during meiotic progression. The effects may indeed fully explain the contributions of these cohesins in pairing and synapsis. In vertebrates, meiotic cohesin regulates normal axis–loop structures and thus chromosome length [94,95,106,110,114]. When meiotic cohesion is disrupted, the chromosome axes become shorter and the cloud of chromatin loops around the core is enlarged. It is easy to see how this might present a problem with pairing and ultimately synapsis, with most univalents being shorter and puffier than their wild-type counterparts. Cohesin at the core of the sister chromatid pair ensures the proper chromatin loop size and somehow resists the condensation caused by assembly of the SC. Smc1β, Stag3, Rec8 and Pds5 all contribute to normal chromosome loop–axis geometry: chromosomes are shorter in their absence.
Cohesin regulation in meiosis
Although important conclusions have been drawn based on the localization and mutant phenotypes of the meiotic cohesins, relatively little is known about how these complexes are regulated. In mitotic cells, cohesion establishment is stimulated both during DNA replication and in response to DNA damage (reviewed in [115,116]), and there is significant mechanistic overlap in these events. As described above, several proteins modulate cohesin's association with chromatin in response to mitotic cell cycle progression (Table 2). A number of these proteins have specific and interesting localization on meiotic chromosomes that suggests that they play specific roles in meiosis (Fig. 3).
The cohesin loader: Scc2/4
In mitotically growing cells, the Scc2/Scc4 complex is required both for DNA replication-mediated cohesion establishment and DNA damage-induced cohesion. In vertebrates, during mitotic cell cycle progression the association of this complex with chromatin is controlled by Dbf4-dependent kinase and is thus dependent on replication licensing, at least in the early embryo [117–119]. In budding yeast Scc2/Scc4 is recruited to nucleosome-free regions of chromatin, which are generated by the activity of the RSC chromatin remodeling complex [120]. It is required both for Rec8 expression and meiotic cohesion [121].
In Drosophila meiocytes, Scc2Nipbl colocalizes with the Smc1 and Smc3 subunits of cohesin along the arms of meiotic chromosomes in pachytene, and perhaps earlier [122] (Fig. 3). Similarly in mouse Scc2Nipbl is found associated with chromosome axes from zygotene until chromosomes are fully synapsed in late pachytene [123]. Interestingly, this localization suggests that cohesin loading by the Scc2Nipbl protein may occur well past pre-meiotic S phase, when the cohesin complexes containing Rec8 and Rad21L are loaded onto chromatin. The presence of the cohesin loader at this late stage (pachytene) is consistent with the observation that Rad21-containing cohesin complexes load onto chromosomes at this time. Thus, cohesion may be stabilized on chromosomes in meiosis by mechanisms that are distinct from the well-understood replication-dependent events in mitotic cells. It is possible that some fraction of meiotic cohesin loading is more analogous to damage-induced cohesion establishment, stimulated by signaling events downstream of meiotic DNA double strand breaks [70]. Consistent with this model, in C. elegans SCC2 is required for cohesin loading and for normal processing of meiotic double strand breaks [124]. In plants and insects, cohesin subunits have also been shown to bind meiotic chromosomes in a manner that suggests they are independent of DNA replication [125,126].
In humans, the Scc2Nipbl gene is haplo-insufficient: loss of one allele results in Cornelia de Lange syndrome, a pervasive developmental disorder [9]. In a mouse model of Cornelia de Lange syndrome (Scc2Nipbl+/–), although there is no direct reporting of meiotic phenotype, the frequency of mutant offspring is well below the expected Mendelian ratios and fertility is reduced, consistent with defects in gametogenesis [127]. Understanding the precise role of Scc2/Scc4 in meiosis is confounded by the pleiotropic effects that partial loss of function have on gene expression and thus growth and health throughout development.
The cohesin stability network in meiosis
As described above, several proteins modulate the stability of the interaction between cohesin and chromatin during mitotic progression. These proteins include the Eco/Esco enzymes, Wapl, Sororin and Pds5. While our mechanistic understanding of the roles that these proteins play in cohesin regulation is based on studies of mitotic cohesion, localization patterns and mutant phenotypes suggest that they play equally critical roles in meiotic chromosome dynamics.
Eco/Esco enzymes
During mitotic cell cycles, acetylation of the Smc3 subunit of cohesin by members of the Eco family of acetyltransferases is essential for the establishment of cohesion between sister chromatids [27–29]. Eco-dependent acetylation of Smc3 during DNA replication renders the cohesin complex resistant to the destabilizing activity of Wapl [41]. All vertebrates express two distinctly different Eco enzymes, called Esco1 and Esco2; both are capable of acetylating the Smc3 subunit of cohesin [26,30]. The activity of Eco1 in budding yeast is promoted through interaction of Eco with the replication machinery [128]. In contrast, the vertebrate enzymes can acetylate cohesin in a replication-independent manner, although only the acetylation during DNA replication is productive for cohesion establishment [26]. Cohesion establishment in meiosis does not precisely mimic the S phase reactions seen in mitotic cohesion establishment [26,28,31,128]. The meiosis-specific cohesin subunits Smc1β and STAG3 are both expressed in leptotene, after pre-meiotic S is complete, suggesting that their loading occurs independently of replication-dependent acetylation.
Localization of Esco enzymes has not proved informative about their roles in meiosis. The staining pattern for Esco2 during meiotic prophase is diffuse and not localized to particular structures. The exception is in the pachytene stage of male meiosis, in which the paired sex chromosomes, a structure called the sex body, stain intensely for Esco2 throughout the unpaired chromatin ([129]; S. R. and R. Pezza, unpublished). The sex body also stains intensely for markers of DNA damage signaling, including the phosphorylated histone variant H2AX (γH2AX) [130]. In budding yeast, Eco1 activity is downstream of H2AX phosphorylation during the DNA damage response [74,131]. It is possible that Esco2 localization to the sex body reflects the unique damage-signaling properties of this chromatin [132]. The expression and localization during meiosis of the other vertebrate cohesin acetylating protein Esco1 have not yet been reported.
The meiotic phenotype of loss of Eco/Esco activity has not been thoroughly investigated. In fission yeast meiosis, the homolog of Eco1, called Eso1, is essential for cohesion between sister chromatids, which is in turn required for mono-polar orientation of sister kinetochores [133,134]. In the absence of cohesin or Eso1 activity sister chromatids undergo premature separation and thus are pulled away from each other in meiosis I. The meiotic function of Eso1 occurs primarily through acetylation of the Smc3 subunit of cohesin, called Psm3, although there may be additional acetylation substrates important for proper meiotic chromosome dynamics [133]. Esco2-deficient mice have been developed, though their meiotic phenotypes have not been described [135]. Similarly, Esco2 morphant zebrafish have been reported to have global cell cycle defects, although meiotic effects were not specifically reported [136]. Human patients with Roberts syndrome, which is caused by loss of Esco2 function, generally do not survive to sexual maturity and the effect of Esco2 on human gametogenesis has not been reported. Pregnancies have been reported among the small number of female patients who survive to adulthood, suggesting that fertility is not fully compromised [137].
Wapl and Sororin
In the absence of Wapl, mitotic chromosomes in vertebrate cells show an apparent over-cohesion phenotype, with excessive cohesin associated with interphase and mitotic chromosomes [36,46]. Although mitotic progression appears relatively normal, aneuploidy and micronuclei develop over time, suggesting that Wapl is essential for genome stability [138]. Cohesin association with chromatin throughout interphase is largely dynamic, and this dynamicity is critically dependent on Wapl [36]. In meiosis, Wapl shows specific localization to the chromosome axes both prior to and during synapsis [139,140]. The presence of Wapl on meiotic chromosome axes may suggest that cohesin's association with chromatin is dynamic throughout meiotic prophase.
In somatic cells, Sororin binds to cohesin after it is activated during DNA replication and inhibits the activity of Wapl, thereby stabilizing cohesion. Sororin recruitment requires acetylation of the cohesin complex by the Esco proteins [39,141]. In meiosis, Sororin is found on chromosome axes, but only after synapsis, and is retained at centromeres in diplotene (S. R. and R. Pezza, in preparation). This staining pattern suggests that during meiosis Sororin stabilizes cohesion after synapsis. This is remarkably different from the mitotic mechanism, in which Sororin is recruited during DNA replication to stabilize active cohesion [39], and suggests that it may be essential that cohesin remain dynamic to accommodate the complex chromosome behaviors of meiotic prophase.
Pds5
Given its critical importance to mitotic cohesion, and the fact that it promotes Eco, Wapl and Sororin function, it is not surprising that Pds5 also plays a critical role in meiosis. In budding yeast, Pds5 is required for meiotic cohesion and is found along chromosome cores in meiotic prophase, colocalizing with, and dependent upon, Rec8 [142]. In contrast, Rec8 loading does not require Pds5. At anaphase of meiosis I, Pds5 is retained on centromeres, much like the meiotic cohesin complexes containing Rec8 and Smc1β. In the absence of Pds5, sister chromatids separate prematurely. Interestingly, much like Rec8 in vertebrate meiosis, Pds5 both promotes homolog pairing and inhibits ectopic synapsis between sister chromatids [143]. Although the data are still limited, it appears that the role of Pds5 in meiosis is consistent with its role in mitotic cohesion, as was originally proposed in Sordaria [144]. Vertebrates express two orthologs of Pds5, called Pds5A and Pds5B [38,45]. It is still not entirely clear what the functional differences are between these two proteins, although in mouse models there are some phenotypic differences. Pds5A is not required for gametogenesis [145], while Pds5B is highly expressed in the mouse testis and during meiotic prophase is found along chromosome axes prior to and during synapsis, in a manner similar to that of yeast Pds5 [146]. These data all suggest that Pds5 interacts with meiotic cohesin as it does with mitotic cohesins, promoting cohesin activity, most probably by integrating the activities of cohesin regulators such as Wapl and Sororin.
Cohesin phosphorylation
In vertebrates, the association of cohesin with chromatin is modified during mitotic entry by phosphorylation, particularly by the Polo-like kinase (Plk1) [58]. Phosphorylation promotes removal of cohesin from chromatin, thus promoting sister chromatid resolution and mitotic condensation in prophase I. In yeast meiosis, phosphorylation of Rec8 cohesin by both casein kinase 1 and Dbf4-dependent kinase promote its cleavage by separase, leading to separation of homologs in meiosis I [61–63].
Questions remaining
Much of what we understand about cohesin function comes from studies in model organisms, particularly budding yeast, and with a few exceptions the regulatory steps appear to be conserved throughout eukaryotic phylogeny. Currently, our understanding of the functional differences between meiotic and mitotic cohesin complexes is very limited. What, for example, are the biochemical differences between the meiotic and mitotic cohesins? Does their unique biochemistry affect their interaction with regulatory proteins such as the Eco enzymes, the Scc2/Scc4 loader complex, Pds5 or Wapl? Does STAG3 interact with Pds5 in the same manner as the mitotic counterparts SA1 and SA2?
One very interesting puzzle is the nature of cohesin dynamics throughout meiotic prophase. Smc1β is not present during pre-meiotic S phase and yet is found on chromosome cores starting in leptotene. The cohesin loaded during pre-meiotic S exclusively contains Smc1α (not Smc1β). Similarly, Rad21, which is largely absent in early meiotic prophase, is found to load onto chromosome cores in late pachytene, while Rad21L is removed. It is difficult to draw analogies with replicative mitotic cohesion establishment because this occurs exclusively during DNA replication. Does the mechanism of cohesion establishment in meiotic prophase more closely resemble replicative cohesion establishment or damage-induced cohesion? What is the role of the Scc2 cohesin loader on synapsed chromosomes?
It is possible that cohesin contributes to meiotic chromosome dynamics not only through its canonical role in tethering pieces of chromatin together in replication-dependent reactions, but also by promoting higher order interactions between meiotic structures. Experiments indicate that, at least in some systems, cohesin and separase play important roles in centrosome dynamics [147–150]. The events are thought to be DNA independent, leading to the notion that cohesin might in fact be a multi-purpose intracellular glue that can be deployed in very different circumstances. Might meiotic cohesins tether together not only sister chromatids but also other meiotic chromosome components such as elements of the SC? Should the term ‘cohesion establishment’ be reserved for replication-dependent tethering of sister chromatids, and do we need a new term to explain some of the reactions that occur in meiotic prophase? A deeper understanding of cohesin regulation during meiotic progression, as well as the development of meiotic models in which cohesin can be conditionally inactivated, will help us address this exciting question. Biochemical analyses of meiotic cohesins and the identification of interacting proteins might contribute to a fuller understanding of meiotic cohesin function. Finally, experiments in which the stability of association of various cohesin subunits with meiotic chromosomes, such as fluorescence recovery after photobleaching analysis, would be extremely helpful in determining whether meiotic cohesin undergoes an ‘establishment’-like transition during progression through meiotic prophase.
Acknowledgements
I apologize to all of the people whose work was not cited directly or sufficiently due to space limitations. I am grateful to Dean Dawson and Roberto Pezza for critical reading of the manuscript, and all members of the Program in Cell Cycle and Cancer Biology for helpful discussions. The author's laboratory is funded by grant R01GM101250 from the National Institutes of Health and the Oklahoma Center for Adult Stem Cell Research.
Abbreviations
- AE
axial element
- LE
lateral element
- SC
synaptonemal complex
Footnotes
Author contribution
Susannah Rankin wrote the review in its entirety.
References
- 1.Spencer F, Gerring S, Connelly C, Hieter P. Mitotic chromosome transmission fidelity mutants in Saccharomyces cerevisiae. Genetics. 1990;124:237–249. doi: 10.1093/genetics/124.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Strunnikov A, Larionov V, Koshland D. SMC1: an essential yeast gene encoding a putative head-rod-tail protein is required for nuclear division and defines a new ubiquitous protein family. J Cell Biol. 1993;123:1635–1648. doi: 10.1083/jcb.123.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Michaelis C, Ciosk R, Nasmyth K. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- 4.Losada A, Hirano M, Hirano T. Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes Dev. 1998;12:1986–1997. doi: 10.1101/gad.12.13.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Losada A. Cohesin in cancer: chromosome segregation and beyond. Nat Rev Cancer. 2014;14:389–393. doi: 10.1038/nrc3743. [DOI] [PubMed] [Google Scholar]
- 6.Nasmyth K, Haering CH. Cohesin: its roles and mechanisms. Annu Rev Genet. 2009;43:525–558. doi: 10.1146/annurev-genet-102108-134233. [DOI] [PubMed] [Google Scholar]
- 7.Ball AR, Chen Y-Y, Yokomori K. Mechanisms of cohesin-mediated gene regulation and lessons learned from cohesinopathies. Biochim Biophys Acta. 2014;1839:191–202. doi: 10.1016/j.bbagrm.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tonkin ET, Wang T-J, Lisgo S, Bamshad MJ, Strachan T. NIPBL, encoding a homolog of fungal Scc2-type sister chromatid cohesion proteins and fly Nipped-B, is mutated in Cornelia de Lange syndrome. Nat Genet. 2004;36:636–641. doi: 10.1038/ng1363. [DOI] [PubMed] [Google Scholar]
- 9.Krantz ID, Mccallum J, Descipio C, Kaur M, Gillis LA, Yaeger D, Jukofsky L, Wasserman N, Bottani A, Morris CA, et al. Cornelia de Lange syndrome is caused by mutations in NIPBL, the human homolog of Drosophila melanogaster Nipped-B. Nat Genet. 2004;36:631–635. doi: 10.1038/ng1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vega H, Waisfisz Q, Gordillo M, Sakai N, Yanagihara I, Yamada M, Van Gosliga D, Kayserili H, Xu C, Ozono K, et al. Roberts syndrome is caused by mutations in ESCO2, a human homolog of yeast ECO1 that is essential for the establishment of sister chromatid cohesion. Nat Genet. 2005;37:468–470. doi: 10.1038/ng1548. [DOI] [PubMed] [Google Scholar]
- 11.Barber TD, McManus K, Yuen KWY, Reis M, Parmigiani G, Shen D, Barrett I, Nouhi Y, Spencer F, Markowitz S, et al. Chromatid cohesion defects may underlie chromosome instability in human colorectal cancers. Proc Natl Acad Sci USA. 2008;105:3443–3448. doi: 10.1073/pnas.0712384105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marston AL. Chromosome segregation in budding yeast: sister chromatid cohesion and related mechanisms. Genetics. 2014;196:31–63. doi: 10.1534/genetics.112.145144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peters JM, Nishiyama T. Sister chromatid cohesion. Cold Spring Harb Perspect Biol. 2012;4:1–18. doi: 10.1101/cshperspect.a011130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arumugam P, Gruber S, Tanaka K, Haering CH, Mechtler K, Nasmyth K. ATP hydrolysis is required for cohesin's association with chromosomes. Curr Biol. 2003;13:1941–1953. doi: 10.1016/j.cub.2003.10.036. [DOI] [PubMed] [Google Scholar]
- 15.Hirano M, Hirano T. Hinge-mediated dimerization of SMC protein is essential for its dynamic interaction with DNA. EMBO J. 2002;21:5733–5744. doi: 10.1093/emboj/cdf575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirano T. At the heart of the chromosome: SMC proteins in action. Nat Rev Mol Cell Biol. 2006;7:311–322. doi: 10.1038/nrm1909. [DOI] [PubMed] [Google Scholar]
- 17.Jeppsson K, Kanno T, Shirahige K, Sjögren C. The maintenance of chromosome structure: positioning and functioning of SMC complexes. Nature Publishing Group. 2014;15:601–614. doi: 10.1038/nrm3857. [DOI] [PubMed] [Google Scholar]
- 18.Ciosk R, Shirayama M, Shevchenko A, Tanaka T, Toth A, Shevchenko A, Nasmyth K. Cohesin's binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol Cell. 2000;5:243–254. doi: 10.1016/s1097-2765(00)80420-7. [DOI] [PubMed] [Google Scholar]
- 19.Sumara I, Vorlaufer E, Gieffers C, Peters BH, Peters JM. Characterization of vertebrate cohesin complexes and their regulation in prophase. J Cell Biol. 2000;151:749–762. doi: 10.1083/jcb.151.4.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weitzer S, Lehane C, Uhlmann F. A model for ATP hydrolysis-dependent binding of cohesin to DNA. Curr Biol. 2003;13:1930–1940. doi: 10.1016/j.cub.2003.10.030. [DOI] [PubMed] [Google Scholar]
- 21.Murayama Y, Uhlmann F. Biochemical reconstitution of topological DNA binding by the cohesin ring. Nature. 2013;505:367–371. doi: 10.1038/nature12867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gruber S, Arumugam P, Katou Y, Kuglitsch D, Helmhart W, Shirahige K, Nasmyth K. Evidence that loading of cohesin onto chromosomes involves opening of its SMC hinge. Cell. 2006;127:523–537. doi: 10.1016/j.cell.2006.08.048. [DOI] [PubMed] [Google Scholar]
- 23.Schleiffer A, Kaitna S, Maurer-Stroh S, Glotzer M, Nasmyth K, Eisenhaber F. Kleisins: a superfamily of bacterial and eukaryotic SMC protein partners. Mol Cell. 2003;11:571–575. doi: 10.1016/s1097-2765(03)00108-4. [DOI] [PubMed] [Google Scholar]
- 24.Uhlmann F, Nasmyth K. Cohesion between sister chromatids must be established during DNA replication. Curr Biol. 1998;8:1095–1101. doi: 10.1016/s0960-9822(98)70463-4. [DOI] [PubMed] [Google Scholar]
- 25.Lengronne A, McIntyre J, Katou Y, Kanoh Y, Hopfner K-P, Shirahige K, Uhlmann F. Establishment of sister chromatid cohesion at the S. cerevisiae replication fork. Mol Cell. 2006;23:787–799. doi: 10.1016/j.molcel.2006.08.018. [DOI] [PubMed] [Google Scholar]
- 26.Song J, Lafont A, Chen J, Wu FM, Shirahige K, Rankin S. Cohesin acetylation promotes sister chromatid cohesion only in association with the replication machinery. J Biol Chem. 2012;287:34325–34336. doi: 10.1074/jbc.M112.400192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rolef Ben-Shahar T, Heeger S, Lehane C, East P, Flynn H, Skehel M, Uhlmann F. Eco1-dependent cohesin acetylation during establishment of sister chromatid cohesion. Science. 2008;321:563–566. doi: 10.1126/science.1157774. [DOI] [PubMed] [Google Scholar]
- 28.Zhang J, Shi X, Li Y, Kim B-J, Jia J, Huang Z, Yang T, Fu X, Jung SY, Wang Y, et al. Acetylation of Smc3 by Eco1 Is required for S phase sister chromatid cohesion in both human and yeast. Mol Cell. 2008;31:143–151. doi: 10.1016/j.molcel.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 29.Unal E, Heidinger-Pauli JM, Kim W, Guacci V, Onn I, Gygi SP, Koshland DE. A molecular determinant for the establishment of sister chromatid cohesion. Science. 2008;321:566–569. doi: 10.1126/science.1157880. [DOI] [PubMed] [Google Scholar]
- 30.Hou F, Zou H. Two human orthologues of Eco1/Ctf7 acetyltransferases are both required for proper sister-chromatid cohesion. Mol Biol Cell. 2005;16:3908–3918. doi: 10.1091/mbc.E04-12-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tóth A, Ciosk R, Uhlmann F, Galova M, Schleiffer A, Nasmyth K. Yeast cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev. 1999;13:320–333. doi: 10.1101/gad.13.3.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shintomi K, Hirano T. Releasing cohesin from chromosome arms in early mitosis: opposing actions of Wapl-Pds5 and Sgo1. Genes Dev. 2009;23:2224–2236. doi: 10.1101/gad.1844309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eichinger CS, Kurze A, Oliveira RA, Nasmyth K. Disengaging the Smc3/kleisin interface releases cohesin from Drosophila chromosomes during interphase and mitosis. EMBO J. 2013;32:656–665. doi: 10.1038/emboj.2012.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chan K-L, Roig MB, Hu B, Beckouët F, Metson J, Nasmyth K. Cohesin's DNA exit gate is distinct from its entrance gate and is regulated by acetylation. Cell. 2012;150:961–974. doi: 10.1016/j.cell.2012.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buheitel J, Stemmann O. Prophase pathway-dependent removal of cohesin from human chromosomes requires opening of the Smc3– Scc1 gate. EMBO J. 2013;32:666–676. doi: 10.1038/emboj.2013.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kueng S, Hegemann B, Peters BH, Lipp JJ, Schleiffer A, Mechtler K, Peters J-M. Wapl controls the dynamic association of cohesin with chromatin. Cell. 2006;127:955–967. doi: 10.1016/j.cell.2006.09.040. [DOI] [PubMed] [Google Scholar]
- 37.Guacci V, Stricklin J, Bloom MS, Guō X, Bhatter M, Koshland D. A novel mechanism for the establishment of sister chromatid cohesion by the ECO1 acetyltransferase. Mol Biol Cell. 2015;26:117–133. doi: 10.1091/mbc.E14-08-1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rankin S, Ayad NG, Kirschner MW. Sororin, a substrate of the anaphase-promoting complex, is required for sister chromatid cohesion in vertebrates. Mol Cell. 2005;18:185–200. doi: 10.1016/j.molcel.2005.03.017. [DOI] [PubMed] [Google Scholar]
- 39.Lafont AL, Song J, Rankin S. Sororin cooperates with the acetyltransferase Eco2 to ensure DNA replication-dependent sister chromatid cohesion. Proc Natl Acad Sci USA. 2010;107:20364–20369. doi: 10.1073/pnas.1011069107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmitz J, Watrin E, Lénárt P, Mechtler K, Peters J-M. Sororin is required for stable binding of cohesin to chromatin and for sister chromatid cohesion in interphase. Curr Biol. 2007;17:630–636. doi: 10.1016/j.cub.2007.02.029. [DOI] [PubMed] [Google Scholar]
- 41.Nishiyama T, Ladurner R, Schmitz J, Kreidl E, Schleiffer A, Bhaskara V, Bando M, Shirahige K, Hyman AA, Mechtler K, et al. Sororin mediates sister chromatid cohesion by antagonizing Wapl. Cell. 2010;143:737–749. doi: 10.1016/j.cell.2010.10.031. [DOI] [PubMed] [Google Scholar]
- 42.Chan K-L, Gligoris T, Upcher W, Kato Y, Shirahige K, Nasmyth K, Beckouët F. Pds5 promotes and protects cohesin acetylation. Proc Natl Acad Sci USA. 2013;110:13020–13025. doi: 10.1073/pnas.1306900110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vaur S, Feytout A, Vazquez S, Javerzat J-P. Pds5 promotes cohesin acetylation and stable cohesin-chromosome interaction. EMBO Rep. 2012;13:645–652. doi: 10.1038/embor.2012.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tedeschi A, Wutz G, Huet S, Jaritz M, Wuensche A, Schirghuber E, Davidson IF, Tang W, Cisneros DA, Bhaskara V, et al. Wapl is an essential regulator of chromatin structure and chromosome segregation. Nature. 2013;501:564–568. doi: 10.1038/nature12471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Losada A, Yokochi T, Hirano T. Functional contribution of Pds5 to cohesin-mediated cohesion in human cells and Xenopus egg extracts. J Cell Sci. 2005;118:2133–2141. doi: 10.1242/jcs.02355. [DOI] [PubMed] [Google Scholar]
- 46.Gandhi R, Gillespie PJ, Hirano T. Human Wapl Is a cohesin-binding protein that promotes sister-chromatid resolution in mitotic prophase. Curr Biol. 2006;16:2406–2417. doi: 10.1016/j.cub.2006.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Panizza S, Tanaka T, Hochwagen A, Eisenhaber F, Nasmyth K. Pds5 cooperates with cohesin in maintaining sister chromatid cohesion. Curr Biol. 2000;10:1557–1564. doi: 10.1016/s0960-9822(00)00854-x. [DOI] [PubMed] [Google Scholar]
- 48.Hartman T, Stead K, Koshland D, Guacci V. Pds5p is an essential chromosomal protein required for both sister chromatid cohesion and condensation in Saccharomyces cerevisiae. J Cell Biol. 2000;151:613–626. doi: 10.1083/jcb.151.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gerlich D, Koch B, Dupeux F, Peters J-M, Ellenberg J. Live-cell imaging reveals a stable cohesin-chromatin interaction after but not before DNA replication. Curr Biol. 2006;16:1571–1578. doi: 10.1016/j.cub.2006.06.068. [DOI] [PubMed] [Google Scholar]
- 50.Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hadjur S, Williams LM, Ryan NK, Cobb BS, Sexton T, Fraser P, Fisher AG, Merkenschlager M. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature. 2009;460:410–413. doi: 10.1038/nature08079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Degner SC, Wong TP, Jankevicius G, Feeney AJ. Cutting edge: developmental stage-specific recruitment of cohesin to CTCF sites throughout immunoglobulin loci during B lymphocyte development. J. Immunol. 2009;182:44–48. doi: 10.4049/jimmunol.182.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seitan VC, Hao B, Tachibana-Konwalski K, Lavagnolli T, Mira-Bontenbal H, Brown KE, Teng G, Carroll T, Terry A, Horan K, et al. A role for cohesin in T-cell-receptor rearrangement and thymocyte differentiation. Nature. 2011;476:467–471. doi: 10.1038/nature10312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wendt K, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 55.Bonora G, Plath K, Denholtz M. A mechanistic link between gene regulation and genome architecture in mammalian development. Curr Opin Genet Dev. 2014;27C:92–101. doi: 10.1016/j.gde.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dorsett D, Merkenschlager M. Cohesin at active genes: a unifying theme for cohesin and gene expression from model organisms to humans. Curr Opin Cell Biol. 2013;25:327–333. doi: 10.1016/j.ceb.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schmidt CK, Brookes N, Uhlmann F. Conserved features of cohesin binding along fission yeast chromosomes. Genome Biol. 2009;10:R52. doi: 10.1186/gb-2009-10-5-r52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sumara I, Vorlaufer E, Stukenberg PT, Kelm O, Redemann N, Nigg EA, Peters J-M. The dissociation of cohesin from chromosomes in prophase is regulated by Polo-like kinase. Mol Cell. 2002;9:515–525. doi: 10.1016/s1097-2765(02)00473-2. [DOI] [PubMed] [Google Scholar]
- 59.Riedel C, Katis V, Katou Y, Mori S, Itoh T, Helmhart W, Galova M, Petronczki M, Gregan J, Cetin B, et al. Protein phosphatase 2A protects centromeric sister chromatid cohesion during meiosis I. Nature. 2006;441:53–61. doi: 10.1038/nature04664. [DOI] [PubMed] [Google Scholar]
- 60.Kitajima TS, Sakuno T, Ishiguro K-I, Iemura S-I, Natsume T, Kawashima SA, Watanabe Y. Shugoshin collaborates with protein phosphatase 2A to protect cohesin. Nature. 2006;441:46–52. doi: 10.1038/nature04663. [DOI] [PubMed] [Google Scholar]
- 61.Katis VL, Lipp JJ, Imre R, Bogdanova A, Okaz E, Habermann B, Mechtler K, Nasmyth K, Zachariae W. Rec8 phosphorylation by casein kinase 1 and Cdc7-Dbf4 kinase regulates cohesin cleavage by separase during meiosis. Dev Cell. 2010;18:397–409. doi: 10.1016/j.devcel.2010.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rumpf C, Cipak L, Dudas A, Benko Z, Pozgajova M, Riedel CG, Ammerer G, Mechtler K, Gregan J. Casein kinase 1 is required for efficient removal of Rec8 during meiosis I. Cell Cycle. 2010;9:2657–2662. doi: 10.4161/cc.9.13.12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ishiguro T, Tanaka K, Sakuno T, Watanabe Y. Shugoshin-PP2A counteracts casein-kinase-1-dependent cleavage of Rec8 by separase. Nat Cell Biol. 2010;12:500–506. doi: 10.1038/ncb2052. [DOI] [PubMed] [Google Scholar]
- 64.Liu H, Rankin S, Yu H. Phosphorylation-enabled binding of SGO1-PP2A to cohesin protects sororin and centromeric cohesion during mitosis. Nat Cell Biol. 2012;15:40–49. doi: 10.1038/ncb2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moschou PN, Bozhkov PV. Separases: biochemistry and function. Physiol Plant. 2012;145:67–76. doi: 10.1111/j.1399-3054.2011.01550.x. [DOI] [PubMed] [Google Scholar]
- 66.Stemmann O, Zou H, Gerber SA, Gygi SP, Kirschner MW. Dual inhibition of sister chromatid separation at metaphase. Cell. 2001;107:715–726. doi: 10.1016/s0092-8674(01)00603-1. [DOI] [PubMed] [Google Scholar]
- 67.Boos D, Kuffer C, Lenobel R, Körner R, Stemmann O. Phosphorylation-dependent binding of cyclin B1 to a Cdc6-like domain of human separase. J Biol Chem. 2008;283:816–823. doi: 10.1074/jbc.M706748200. [DOI] [PubMed] [Google Scholar]
- 68.Hauf S, Waizenegger IC, Peters JM. Cohesin cleavage by separase required for anaphase and cytokinesis in human cells. Science. 2001;293:1320–1323. doi: 10.1126/science.1061376. [DOI] [PubMed] [Google Scholar]
- 69.Uhlmann F, Wernic D, Poupart MA, Koonin EV, Nasmyth K. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell. 2000;103:375–386. doi: 10.1016/s0092-8674(00)00130-6. [DOI] [PubMed] [Google Scholar]
- 70.Unal E, Heidinger-Pauli JM, Koshland D. DNA double-strand breaks trigger genome-wide sister-chromatid cohesion through Eco1 (Ctf7). Science. 2007;317:245–248. doi: 10.1126/science.1140637. [DOI] [PubMed] [Google Scholar]
- 71.Ström L, Karlsson C, Lindroos HB, Wedahl S, Katou Y, Shirahige K, Sjögren C. Postreplicative formation of cohesion is required for repair and induced by a single DNA break. Science. 2007;317:242–245. doi: 10.1126/science.1140649. [DOI] [PubMed] [Google Scholar]
- 72.Sjögren C, Nasmyth K. Sister chromatid cohesion is required for postreplicative double-strand break repair in Saccharomyces cerevisiae. Curr Biol. 2001;11:991–995. doi: 10.1016/s0960-9822(01)00271-8. [DOI] [PubMed] [Google Scholar]
- 73.Ström L, Lindroos HB, Shirahige K, Sjögren C. Postreplicative recruitment of cohesin to double-strand breaks is required for DNA repair. Mol Cell. 2004;16:1003–1015. doi: 10.1016/j.molcel.2004.11.026. [DOI] [PubMed] [Google Scholar]
- 74.Unal E, Arbel-Eden A, Sattler U, Shroff R, Lichten M, Haber JE, Koshland D. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol Cell. 2004;16:991–1002. doi: 10.1016/j.molcel.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 75.Game JC, Birrell GW, Brown JA, Shibata T, Baccari C, Chu AM, Williamson MS, Brown JM. Use of a genome-wide approach to identify new genes that control resistance of Saccharomyces cerevisiae to ionizing radiation. Radiat Res. 2003;160:14–24. doi: 10.1667/rr3019. [DOI] [PubMed] [Google Scholar]
- 76.Nagao K, Adachi Y, Yanagida M. Separase-mediated cleavage of cohesin at interphase is required for DNA repair. Nature. 2004;430:1044–1048. doi: 10.1038/nature02803. [DOI] [PubMed] [Google Scholar]
- 77.Kim B-J, Li Y, Zhang J, Xi Y, Li Y, Yang T, Jung SY, Pan X, Chen R, Li W, et al. Genome-wide reinforcement of cohesin binding at preexisting cohesin sites in response to ionizing radiation in human cells. J Biol Chem. 2010;285:22784–22792. doi: 10.1074/jbc.M110.134577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.van der Lelij P, Godthelp BC, van Zon W, Van Gosliga D, Oostra AB, Steltenpool J, de Groot J, Scheper RJ, Wolthuis RM, Waisfisz Q, et al. The cellular phenotype of Roberts Syndrome fibroblasts as revealed by ectopic expression of ESCO2. PLoS One. 2009;4:e6936. doi: 10.1371/journal.pone.0006936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Caron P, Aymard F, Iacovoni JS, Briois S, Canitrot Y, Bugler B, Massip L, Losada A, Legube G. Cohesin protects genes against γH2AX induced by DNA double-strand breaks. PLoS Genet. 2012;8:e1002460. doi: 10.1371/journal.pgen.1002460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sansam CL, Pezza RJ. Connecting by breaking and repairing: mechanisms of strand exchange in meiotic recombination. FEBS J. 2015 doi: 10.1111/febs.13317. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kurdzo EL, Dawson DS. Centromere pairing – tethering partner chromosomes in meiosis I. FEBS J. 2015 doi: 10.1111/febs.13280. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gorbsky GJ. The spindle checkpoint and chromosome segregation in meiosis. FEBS J. 2014 doi: 10.1111/febs.13166. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jones KT, Lane SIR. Molecular causes of aneuploidy in mammalian eggs. Development. 2013;140:3719–3730. doi: 10.1242/dev.090589. [DOI] [PubMed] [Google Scholar]
- 84.Wolstenholme J, Angell RR. Maternal age and trisomy–a unifying mechanism of formation. Chromosoma. 2000;109:435–438. doi: 10.1007/s004120000088. [DOI] [PubMed] [Google Scholar]
- 85.Chiang T, Duncan FE, Schindler K, Schultz RM, Lampson MA. Evidence that weakened centromere cohesion is a leading cause of age-related aneuploidy in oocytes. Curr Biol. 2010;20:1522–1528. doi: 10.1016/j.cub.2010.06.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jeffreys C, Burrage P, Bickel S. A model system for increased meiotic nondisjunction in older oocytes. Curr Biol. 2003;13:498–503. doi: 10.1016/s0960-9822(03)00134-9. [DOI] [PubMed] [Google Scholar]
- 87.Herrán Y, Gutiérrez-Caballero C, Sánchez-Martín M, Hernández T, Viera A, Barbero JL, de Álava E, de Rooij DG, Suja JÁ, Llano E, et al. The cohesin subunit RAD21L functions in meiotic synapsis and exhibits sexual dimorphism in fertility. EMBO J. 2011;30:3091–3105. doi: 10.1038/emboj.2011.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hodges C, Revenkova E, Jessberger R, Hassold T, Hunt P. SMC1beta-deficient female mice provide evidence that cohesins are a missing link in age-related nondisjunction. Nat Genet. 2005;37:1351–1355. doi: 10.1038/ng1672. [DOI] [PubMed] [Google Scholar]
- 89.Weng KA, Jeffreys CA, Bickel SE. Rejuvenation of meiotic cohesion in oocytes during prophase I is required for Chiasma maintenance and accurate chromosome segregation. PLoS Genet. 2014;10:e1004607. doi: 10.1371/journal.pgen.1004607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Daum JR, Potapova TA, Sivakumar S, Daniel JJ, Flynn JN, Rankin S, Gorbsky GJ. Cohesion fatigue induces chromatid separation in cells delayed at metaphase. Curr Biol. 2011;21:1018–1024. doi: 10.1016/j.cub.2011.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Stevens D, Gassmann R, Oegema K, Desai A. Uncoordinated loss of chromatid cohesion is a common outcome of extended metaphase arrest. PLoS One. 2011;6:e22969. doi: 10.1371/journal.pone.0022969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ponticelli AS, Smith GR. Meiotic recombination-deficient mutants of Schizosaccharomyces pombe. Genetics. 1989;123:45–54. doi: 10.1093/genetics/123.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Klein F, Mahr P, Galova M, Buonomo SB, Michaelis C, Nairz K, Nasmyth K. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell. 1999;98:91–103. doi: 10.1016/S0092-8674(00)80609-1. [DOI] [PubMed] [Google Scholar]
- 94.Xu H, Beasley MD, Warren WD, van der Horst GTJ, McKay MJ. Absence of mouse REC8 cohesin promotes synapsis of sister chromatids in meiosis. Dev Cell. 2005;8:949–961. doi: 10.1016/j.devcel.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 95.Bannister LA, Reinholdt LG, Munroe RJ, Schimenti JC. Positional cloning and characterization of mouse mei8, a disrupted allelle of the meiotic cohesin Rec8. Genesis. 2004;40:184–194. doi: 10.1002/gene.20085. [DOI] [PubMed] [Google Scholar]
- 96.Llano E, Herran Y, Garcia-Tunon I, Gutierrez-Caballero C, deAlava E, Barbero JL, Schimenti J, deRooij DG, Sanchez-Martin M, Pendas AM. Meiotic cohesin complexes are essential for the formation of the axial element in mice. J Cell Biol. 2012;197:877–885. doi: 10.1083/jcb.201201100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tachibana-Konwalski K, Godwin J, van der Weyden L, Champion L, Kudo NR, Adams DJ, Nasmyth K. Rec8-containing cohesin maintains bivalents without turnover during the growing phase of mouse oocytes. Genes Dev. 2010;24:2505–2516. doi: 10.1101/gad.605910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lee J, Hirano T. RAD21L, a novel cohesin subunit implicated in linking homologous chromosomes in mammalian meiosis. J Cell Biol. 2011;192:263–276. doi: 10.1083/jcb.201008005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ishiguro K-I, Kim J, Fujiyama-Nakamura S, Kato S, Watanabe Y. A new meiosis-specific cohesin complex implicated in the cohesin code for homologous pairing. EMBO Rep. 2011;12:267–275. doi: 10.1038/embor.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ishiguro KI, Kim J, Shibuya H, Hernandez-Hernandez A, Suzuki A, Fukagawa T, Shioi G, Kiyonari H, Li XC, Schimenti J, et al. Meiosis-specific cohesin mediates homolog recognition in mouse spermatocytes. Genes Dev. 2014;28:594–607. doi: 10.1101/gad.237313.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gutiérrez-Caballero C, Herrán Y, Sánchez-Martín M, Suja JÁ, Barbero JL, Llano E, Pendás AM. Identification and molecular characterization of the mammalian α-kleisin RAD21L. Cell Cycle. 2011;10:1477–1487. doi: 10.4161/cc.10.9.15515. [DOI] [PubMed] [Google Scholar]
- 102.Severson AF, Ling L, van Zuylen V, Meyer BJ. The axial element protein HTP-3 promotes cohesin loading and meiotic axis assembly in C. elegans to implement the meiotic program of chromosome segregation. Genes Dev. 2009;23:1763–1778. doi: 10.1101/gad.1808809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pasierbek P, Jantsch M, Melcher M, Schleiffer A, Schweizer D, Loidl J. A Caenorhabditis elegans cohesion protein with functions in meiotic chromosome pairing and disjunction. Genes Dev. 2001;15:1349–1360. doi: 10.1101/gad.192701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Severson AF, Meyer BJ. Divergent kleisin subunits of cohesin specify mechanisms to tether and release meiotic chromosomes. Elife. 2014;3:e03467. doi: 10.7554/eLife.03467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Revenkova E, Eijpe M, Heyting C, Gross B, Jessberger R. Novel meiosis-specific isoform of mammalian SMC1. Mol Cell Biol. 2001;21:6984–6998. doi: 10.1128/MCB.21.20.6984-6998.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Revenkova E, Eijpe M, Heyting C, Hodges CA, Hunt PA, Liebe B, Scherthan H, Jessberger R. Cohesin SMC1β is required for meiotic chromosome dynamics, sister chromatid cohesion and DNA recombination. Nat Cell Biol. 2004;6:555–562. doi: 10.1038/ncb1135. [DOI] [PubMed] [Google Scholar]
- 107.Biswas U, Wetzker C, Lange J, Christodoulou EG, Seifert M, Beyer A, Jessberger R. Meiotic cohesin SMC1β provides prophase I centromeric cohesion and is required for multiple synapsis-associated functions. PLoS Genet. 2013;9:e1003985. doi: 10.1371/journal.pgen.1003985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Winters T, McNicoll F, Jessberger R. Meiotic cohesin STAG3 is required for chromosome axis formation and sister chromatid cohesion. EMBO J. 2014;33:1256–1270. doi: 10.1002/embj.201387330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fukuda T, Fukuda N, Agostinho A, Hernández-Hernández A, Kouznetsova A, Höög C. STAG3-mediated stabilization of REC8 cohesin complexes promotes chromosome synapsis during meiosis. EMBO J. 2014;33:1243–1255. doi: 10.1002/embj.201387329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hopkins J, Hwang G, Jacob J, Sapp N, Bedigian R, Oka K, Overbeek P, Murray S, Jordan PW. Meiosis-specific cohesin component, Stag3 is essential for maintaining centromere chromatid cohesion, and required for DNA repair and synapsis between homologous chromosomes. PLoS Genet. 2014;10:e1004413. doi: 10.1371/journal.pgen.1004413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kitajima TS, Yokobayashi S, Yamamoto M, Watanabe Y. Distinct cohesin complexes organize meiotic chromosome domains. Science. 2003;300:1152–1155. doi: 10.1126/science.1083634. [DOI] [PubMed] [Google Scholar]
- 112.Hunt PA, Hassold TJ. Sex matters in meiosis. Science. 2002;296:2181–2183. doi: 10.1126/science.1071907. [DOI] [PubMed] [Google Scholar]
- 113.Turner JMA. Meiotic sex chromosome inactivation. Development. 2007;134:1823–1831. doi: 10.1242/dev.000018. [DOI] [PubMed] [Google Scholar]
- 114.Ding D-Q, Sakurai N, Katou Y, Itoh T, Shirahige K, Haraguchi T, Hiraoka Y. Meiotic cohesins modulate chromosome compaction during meiotic prophase in fission yeast. J Cell Biol. 2006;174:499–508. doi: 10.1083/jcb.200605074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sherwood R, Takahashi TS, Jallepalli PV. Sister acts: coordinating DNA replication and cohesion establishment. Genes Dev. 2010;24:2723–2731. doi: 10.1101/gad.1976710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Watrin E, Peters J-M. Cohesin and DNA damage repair. Exp Cell Res. 2006;312:2687–2693. doi: 10.1016/j.yexcr.2006.06.024. [DOI] [PubMed] [Google Scholar]
- 117.Takahashi TS, Basu A, Bermudez V, Hurwitz J, Walter JC. Cdc7-Drf1 kinase links chromosome cohesion to the initiation of DNA replication in Xenopus egg extracts. Genes & Dev. 2008;22:1894–1905. doi: 10.1101/gad.1683308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gillespie PJ, Hirano T. Scc2 couples replication licensing to sister chromatid cohesion in Xenopus egg extracts. Curr Biol. 2004;14:1598–1603. doi: 10.1016/j.cub.2004.07.053. [DOI] [PubMed] [Google Scholar]
- 119.Takahashi TS, Yiu P, Chou MF, Gygi S, Walter JC. Recruitment of Xenopus Scc2 and cohesin to chromatin requires the pre-replication complex. Nat Cell Biol. 2004;6:991–996. doi: 10.1038/ncb1177. [DOI] [PubMed] [Google Scholar]
- 120.Lopez-Serra L, Kelly G, Patel H, Stewart A, Uhlmann F. The Scc2-Scc4 complex acts in sister chromatid cohesion and transcriptional regulation by maintaining nucleosome-free regions. Nat Genet. 2014;46:1147–1151. doi: 10.1038/ng.3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lin W, Jin H, Liu X, Hampton K, Yu H-G. Scc2 regulates gene expression by recruiting cohesin to the chromosome as a transcriptional activator during yeast meiosis. Mol Biol Cell. 2011;22:1985–1996. doi: 10.1091/mbc.E10-06-0545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gause M, Webber HA, Misulovin Z, Haller G, Rollins RA, Eissenberg JC, Bickel SE, Dorsett D. Functional links between Drosophila Nipped-B and cohesin in somatic and meiotic cells. Chromosoma. 2008;117:51–66. doi: 10.1007/s00412-007-0125-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kuleszewicz K, Fu X, Kudo NR. Cohesin loading factor Nipbl localizes to chromosome axes during mammalian meiotic prophase. Cell Division. 2013;8:12–19. doi: 10.1186/1747-1028-8-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lightfoot J, Testori S, Barroso C, Martinez-Perez E. Loading of meiotic cohesin by SCC-2 is required for early processing of DSBs and for the DNA damage checkpoint. Curr Biol. 2011;21:1421–1430. doi: 10.1016/j.cub.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 125.Qiao H, Lohmiller LD, Anderson LK. Cohesin proteins load sequentially during prophase I in tomato primary microsporocytes. Chromosome Res. 2011;19:193–207. doi: 10.1007/s10577-010-9184-1. [DOI] [PubMed] [Google Scholar]
- 126.Valdeolmillos AM, Viera A, Page J, Prieto I, Santos JL, Parra MT, Heck MMS, Martínez-A C, Barbero JL, Suja JA, et al. Sequential loading of cohesin subunits during the first meiotic prophase of grasshoppers. PLoS Genet. 2007;3:e28. doi: 10.1371/journal.pgen.0030028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kawauchi S, Calof AL, Santos R, Lopez-Burks ME, Young CM, Hoang MP, Chua A, Lao T, Lechner MS, Daniel JA, et al. Multiple organ system defects and transcriptional dysregulation in the Nipbl (+/−) mouse, a model of Cornelia de Lange Syndrome. PLoS Genet. 2009;5:e1000650. doi: 10.1371/journal.pgen.1000650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Moldovan G-L, Pfander B, Jentsch S. PCNA controls establishment of sister chromatid cohesion during S phase. Mol Cell. 2006;23:723–732. doi: 10.1016/j.molcel.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 129.Evans EB, Hogarth C, Evanoff RM, Mitchell D, Small C, Griswold MD. Localization and regulation of murine Esco2 during male and female meiosis. Biol Reprod. 2012;87:1–8. doi: 10.1095/biolreprod.112.099978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Mahadevaiah SK, Turner JM, Baudat F, Rogakou EP, de Boer P, Blanco-Rodríguez J, Jasin M, Keeney S, Bonner WM, Burgoyne PS. Recombinational DNA double-strand breaks in mice precede synapsis. Nat Genet. 2001;27:271–276. doi: 10.1038/85830. [DOI] [PubMed] [Google Scholar]
- 131.Lyons NA, Morgan DO. Cdk1-dependent destruction of Eco1 prevents cohesion establishment after S phase. Mol Cell. 2011;42:378–389. doi: 10.1016/j.molcel.2011.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lu L-Y, Xiong Y, Kuang H, Korakavi G, Yu X. Regulation of the DNA damage response on male meiotic sex chromosomes. Nat Commun. 2013;4:2105. doi: 10.1038/ncomms3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kagami A, Sakuno T, Yamagishi Y, Ishiguro T, Tsukahara T, Shirahige K, Tanaka K, Watanabe Y. Acetylation regulates monopolar attachment at multiple levels during meiosis I in fission yeast. EMBO Rep. 2011;12:1189–1195. doi: 10.1038/embor.2011.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sakuno T, Tada K, Watanabe Y. Kinetochore geometry defined by cohesion within the centromere. Nature. 2009;458:852–858. doi: 10.1038/nature07876. [DOI] [PubMed] [Google Scholar]
- 135.Whelan G, Kreidl E, Wutz G, Egner A, Peters J-M, Eichele G. Cohesin acetyltransferase Esco2 is a cell viability factor and is required for cohesion in pericentric heterochromatin. EMBO J. 2011;31:71–82. doi: 10.1038/emboj.2011.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Mönnich M, Kuriger Z, Print CG, Horsfield JA. A Zebrafish Model of Roberts Syndrome reveals that Esco2 depletion interferes with development by disrupting the cell cycle. PLoS One. 2011;6:e20051. doi: 10.1371/journal.pone.0020051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Goh ES-Y, Li C, Horsburgh S, Kasai Y, Kolomietz E, Morel CF. The Roberts syndrome/SC phocomelia spectrum–a case report of an adult with review of the literature. Am J Med Genet. 2010;152A:472–478. doi: 10.1002/ajmg.a.33261. [DOI] [PubMed] [Google Scholar]
- 138.Haarhuis JHI, Elbatsh AMO, van den Broek B, Camps D, Erkan H, Jalink K, Medema RH, Rowland BD. WAPL-mediated removal of cohesin protects against segregation errors and aneuploidy. Curr Biol. 2013;23:2071–2077. doi: 10.1016/j.cub.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 139.Kuroda M, Oikawa K, Ohbayashi T, Yoshida K, Yamada K, Mimura J, Matsuda Y, Fujii-Kuriyama Y, Mukai K. A dioxin sensitive gene, mammalian WAPL, is implicated in spermatogenesis. FEBS Lett. 2005;579:167–172. doi: 10.1016/j.febslet.2004.11.070. [DOI] [PubMed] [Google Scholar]
- 140.Zhang N, Kuznetsov S, Sharan S, Li K, Rao P, Pati D. A handcuff model for the cohesin complex. J Cell Biol. 2008;183:1019–1031. doi: 10.1083/jcb.200801157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Nishiyama T, Sykora MM, Huis In ‘t Veld PJ, Mechtler K, Peters J-M. Aurora B and Cdk1 mediate Wapl activation and release of acetylated cohesin from chromosomes by phosphorylating Sororin. Proc Natl Acad Sci USA. 2013;110:12404–13409. doi: 10.1073/pnas.1305020110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zhang Z, Ren Q, Yang H, Conrad MN, Guacci V, Kateneva A, Dresser ME. Budding yeast PDS5 plays an important role in meiosis and is required for sister chromatid cohesion. Mol Microbiol. 2005;56:670–680. doi: 10.1111/j.1365-2958.2005.04582.x. [DOI] [PubMed] [Google Scholar]
- 143.Jin H, Guacci V, Yu H-G. Pds5 is required for homologue pairing and inhibits synapsis of sister chromatids during yeast meiosis. J Cell Biol. 2009;186:713–725. doi: 10.1083/jcb.200810107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.van Heemst D, James F, Pöggeler S, Berteaux-Lecellier V, Zickler D. Spo76p is a conserved chromosome morphogenesis protein that links the mitotic and meiotic programs. Cell. 1999;98:261–271. doi: 10.1016/s0092-8674(00)81020-x. [DOI] [PubMed] [Google Scholar]
- 145.Zhang B, Chang J, Fu M, Huang J, Kashyap R, Salavaggione E, Jain S, Shashikant K, Deardorff MA, Uzielli MLG, et al. Dosage effects of cohesin regulatory factor PDS5 on mammalian development: implications for cohesinopathies. PLoS One. 2009;4:e5232. doi: 10.1371/journal.pone.0005232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Fukuda T, Höög C. The mouse cohesin-associated protein PDS5B is expressed in testicular cells and is associated with the meiotic chromosome axes. Genes (Basel) 2010;1:484–494. doi: 10.3390/genes1030484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Schöckel L, Möckel M, Mayer B, Boos D, Stemmann O. Cleavage of cohesin rings coordinates the separation of centrioles and chromatids. Nat Cell Biol. 2011;13:966–972. doi: 10.1038/ncb2280. [DOI] [PubMed] [Google Scholar]
- 148.Tsou M, Stearns T. Mechanism limiting centrosome duplication to once per cell cycle. Nature. 2006;442:947–951. doi: 10.1038/nature04985. [DOI] [PubMed] [Google Scholar]
- 149.Cabral G, Sans SS, Cowan CR, Dammermann A. Multiple mechanisms contribute to centriole separation in C. elegans. Curr Biol. 2013;23:1380–1387. doi: 10.1016/j.cub.2013.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Oliveira RA, Nasmyth K. Cohesin cleavage is insufficient for centriole disengagement in Drosophila. Curr Biol. 2013;23:R601–R603. doi: 10.1016/j.cub.2013.04.003. [DOI] [PubMed] [Google Scholar]