Abstract
There are multiple platforms available for whole-exome enrichment and sequencing (WES). This protocol is based on the Agilent SureSelect Human All Exon platform, which targets ~50 Mb of the human exonic regions. The SureSelect system uses ~120-base RNA probes to capture known coding DNA sequences (CDS) from the NCBI Consensus CDS Database as well as other major RNA coding sequence databases, such as Sanger miRBase. The protocol can be performed at the benchside without the need for automation, and the resulting library can be used for targeted next-generation sequencing on an Illumina HiSeq 2000 sequencer.
MATERIALS
It is essential that you consult the appropriate Material Safety Data Sheets and your institution’s Environmental Health and Safety Office for proper handling of equipment and hazardous materials used in this protocol.
Reagents
Agarose gels, precast (E-Gel EX, 2% [Life Technologies G4020–02; 20 gels])
Agencourt AMPure XP magnetic bead-based purification system (Beckman Coulter A63881; 60 mL)
Bioanalyzer reagents (Agilent)
DNA 1000 Kit (5067-1504)
High-Sensitivity DNA Kit (5067-4626)
Buffer EB (QIAGEN 19086)
Dynabeads MyOne Streptavidin T1 (Life Technologies 65602; 10 mL)
Ethanol, molecular biology grade (Sigma-Aldrich E7023)
Prepare a solution of 70% ethanol.
Genomic DNA, human (high-quality, nondegraded, with an average fragment size of 40 kb or greater and an A260/A280 of 1.8–2.0)
Herculase II Fusion DNA Polymerase (Agilent 600679; 400 reactions)
Nuclease-free water (UltraPure Distilled Water [Life Technologies 10977-015] or equivalent)
SureSelectXT Human All Exon V5 + UTRs (Agilent 5190-6213; 16 reactions)
TE Buffer (Life Technologies 12090-015)
Equipment
2100 Bioanalyzer (Agilent)
Adhesive film (MicroAmp Clear [Life Technologies 4306311] or equivalent)
AFA microTUBES with snap caps (Covaris 520045)
DNA LoBind tubes, 1.5-mL (Eppendorf 022431021 or equivalent)
Heat blocks at 37°C and 65°C
Magnetic stand (Dynal DynaMag-2 [Life Technologies 12321D] or equivalent)
Microcentrifuge (Fisher Scientific accuSpin Micro 17 or equivalent)
Mini LabRoller (Labnet H5500 or equivalent)
Multichannel pipette, 12-channel (Rainin Pipet-Lite L12-20 or equivalent) (optional; see Step 31)
PCR plates, 96-well (Agilent 410088 or equivalent)
PCR tubes, 0.2-mL (sterile)
Pipette tips (sterile, nuclease-free with aerosol barrier)
Thermal cycler (DNA Engine Tetrad 2 [BioRad PTC-0240G])
Ultra-sonicator (Covaris S-series Single Tube Sample Preparation System, Model S2)
Set up the ultrasonicator following the manufacturer’s instructions. Make sure the tank is filled with fresh deionized H2O chilled to 2°C–5°C and degassed for at least 30 min before use.
Vacuum concentrator (Thermo Scientific Savant SPD111V or equivalent)
Vortex (Fisher Scientific 02-215-370 or equivalent)
METHOD
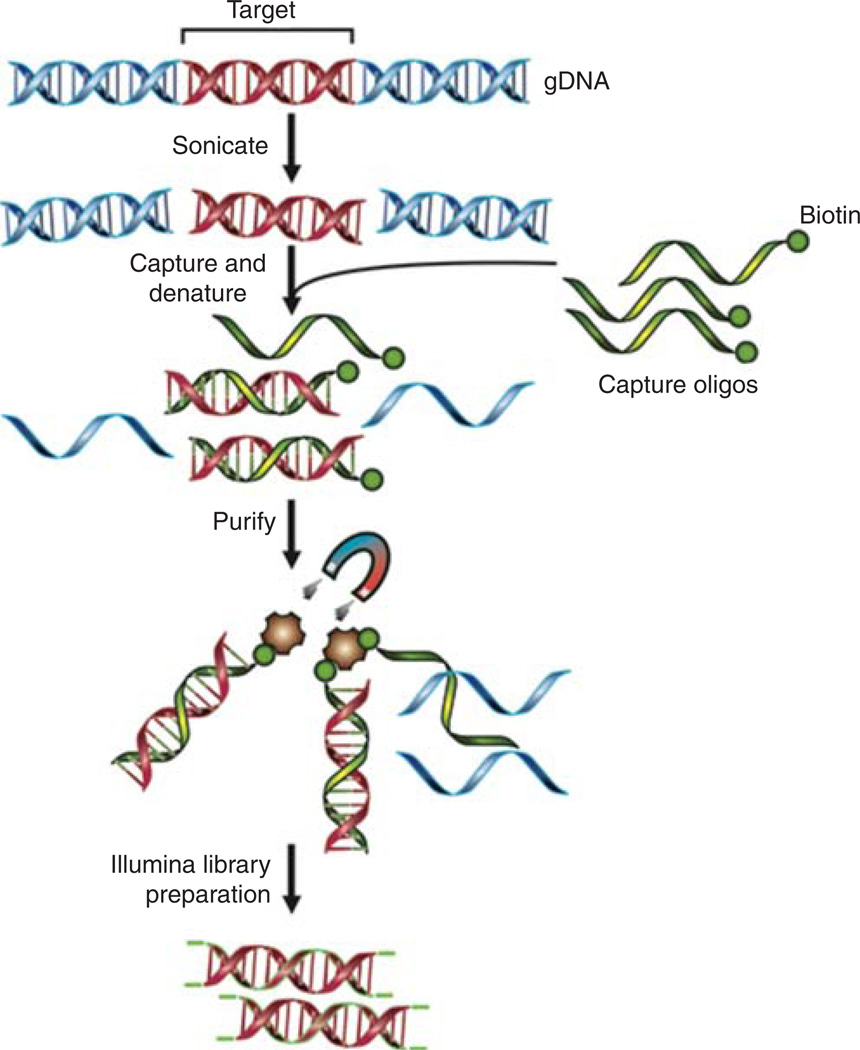
The general scheme of DNA preparation for hybridization-based whole-exome capture and sequencing is diagrammed in Figure 1. The following protocol is based on the original method provided by Agilent (SureSelectXT Target Enrichment System for Illumina Paired-End Sequencing Library, version 1.3.1), with minor modifications to streamline the process based on our experience. A detailed performance evaluation of this method as well as a comparison to alternative platforms can be found in our previous publication (Clark et al. 2011); see Related Information.
FIGURE 1.
Common scheme for hybridization-based whole-exome enrichment methods. In summary, genomic DNA is fragmented, denatured, and hybridized with capture oligos during library preparation for high-throughput sequencing. The captured sequences are then enriched with streptavidin-conjugated paramagnetic beads and further amplified before being subjected to Illumina sequencing. Diagrammed DNA sequences in red, on-target regions; sequences in blue, off-target regions; single-stranded oligos in green, capture probes labeled with biotin; brown particles, streptavidin-conjugated paramagnetic beads.
Fragmenting Genomic DNA
-
1
In a 1.5-mL LoBind tube, dilute 3 µg of human genomic DNA in TE buffer to a total volume of 120 µL. Transfer the diluted DNA to an AFA microTUBE.
-
2
Load the microTUBE into the tube holder of the ultrasonicator and shear the DNA using the following settings: mode, frequency sweeping; duty cycle, 10%; intensity, 5; cycles per burst, 200; duration, 60 sec × 6 cycles; temperature, 4°C–7°C.
-
3Transfer the sheared DNA to a fresh 1.5-mL LoBind tube. Check the shearing by running 2 µL of the sample on a 2% E-Gel EX gel.The average DNA size should be ~150–200 bp.It is safe to stop here and store the sample at −20°C.
Purifying DNA with Agencourt AMPure XP Magnetic Beads
-
4
Equilibrate the Agencourt AMPure XP reagent for at least 30 min to room temperature and mix well by vortexing.
-
5
Add 216 µL of homogenous AMPure XP reagent to the sheared DNA sample (~120 µL) and mix well by vortexing. Incubate the mixture in the Mini LabRoller for 5 min at room temperature.
-
6
Place the tube in the magnetic stand. Wait for the solution to clear (~2 min) and carefully discard the cleared solution without touching the beads.
-
7
Wash the beads twice with 500 µL of 70% ethanol for 1 min per wash.
-
8
Dry the beads for 5 min in a heat block at 37°C.
-
9
Add 50 µL of nuclease-free water to the beads and mix thoroughly by vortexing. Incubate the mixture on the Mini LabRoller for 5 min at room temperature.
-
10Place the tube back into the magnetic stand and wait for the solution to clear (~2 min). Transfer the supernatant containing the eluted DNA to a fresh 1.5-mL LoBind tube.It is safe to stop here and store the sample at −20°C.
Preparing the Adaptor-Ligated DNA Library
Repairing DNA Ends
-
11For each sample of sheared DNA, prepare the following reaction in a 0.2-mL PCR tube using the reagents provided in the SureSelectXT Human All Exon Kit.
Reagent Amount to add DNA sample from Step 10 ~50 µL 10× End Repair Buffer 10 µL dNTP Mix 1.6 µL Nuclease-free water to 94.8 µL T4 DNA Polymerase 1 µL Klenow DNA Polymerase 2 µL T4 Polynucleotide Kinase 2.2 µL Total volume 100 µL -
12
Incubate the reaction for 30 min at 20°C in a thermal cycler without the heated lid.
-
13
Purify the end-repaired DNA as described in Steps 4–10, with the following modifications: Add 180 µL of homogenous AMPure XP reagent to the reaction, and elute the purified DNA in 32 µL of nuclease-free water.
Adding an Adenine Base to End-Repaired Fragments
-
14For each sample, prepare the following reaction in a 0.2-mL PCR tube using the reagents provided in the SureSelectXT Human All Exon Kit.
Reagent Amount to add DNA sample from Step 13 ~32 µL 10× Klenow Polymerase Buffer 5 µL Nuclease-free water to 46 µL dATP 1 uL Exo(-) Klenow 3 µL Total volume 50 µL -
15
Incubate the reaction for 30 min at 37°C in a thermal cycler.
-
16
Purify the DNA as described in Steps 4–10, with the following modifications: add 90 µL of homogenous AMPure XP reagent to the reaction, and elute the purified DNA in 28.5 µL of nuclease-free water.
Ligating the Indexing-Specific Paired-End Adaptor
-
17For each sample, prepare the following reaction in a 0.2-mL PCR tube using the reagents provided in the SureSelectXT Human All Exon Kit.
Reagent Amount to add DNA sample from Step 16 ~28.5 uL 5× T4 DNA ligase buffer 10 µL Nuclease-free water to 38.5 µL SureSelect Adaptor Oligo Mix 10 µL T4 DNA ligase 1.5 µL Total volume 50 µL -
18
Incubate the reaction for 15 min at 20°C in a thermal cycler without the heated lid.
-
19
Purify the ligated DNA as described in Steps 4–10, with the following modifications: add 90 µL of homogenous AMPure XP reagent to the reaction, and elute the purified DNA in 32 µL of nuclease-free water.
Amplifying the Adaptor-Ligated Library
-
20For each sample, prepare the following reaction in a 0.2-mL PCR tube using the reagents provided in the SureSelectXT Human All Exon Kit and Herculase II Fusion DNA polymerase.
Reagent Amount to add Indexing adaptor-ligated library DNA from Step 19 15 µL Nuclease-free water 21 µL SureSelect primer 1.25 µL SureSelect ILM indexing pre capture PCR reverse primer 1.25 µL 5× Herculase II reaction buffer 10 µL 100 mM dNTP mix 0.5 µL Herculase II Fusion DNA Polymerase 1 µL Total volume 50 µL -
21
Place the tube in a thermal cycler and amplify the library DNA using the following program: 2 min at 98°C, 6 cycles of (30 sec at 98°C, 30 sec at 65°C, 1 min at 72°C), 10 min at 72°C.
-
22
Purify the DNA as described in Steps 4–10 with the following modifications: add 90 µL of homogenous AMPure XP reagent to the reaction, and elute the purified DNA in 30 µL of nuclease-free water.
-
23Check the quality of the amplified preenrichment library by running 1 µL on a DNA 1000 chip using a 2100 Bioanalyzer following the manufacturer’s instructions. Record the measured concentration of the library.The amplified library should have a peak size of ~350 bp.It is safe to stop here and store the sample at −20°C.
Hybridizing the Library
-
24Aliquot 500 ng of the amplified library DNA into a 1.5-mL LoBind tube. Perforate the lid of the tube with a 21-gauge needle. Centrifuge the tube in a vacuum concentrator at ≤45°C to reduce the total volume to 3.4 µL (147 ng/µL).It is also safe to completely lyophilize the DNA and reconstitute in 3.4 µL nuclease-free water.
-
25Prepare the Hybridization Buffer using the reagents provided in the SureSelectXT Human All Exon Kit as follows (per sample). If precipitation is visible in the component tubes, warm to 65°C and incubate until all precipitation dissolves before buffer preparation.
Reagent Amount to add SureSelect Hyb #1 25 µL SureSelect Hyb #2 1 µL SureSelect Hyb #3 10 µL SureSelect Hyb #4 13 µL Total volume 49 µL -
26Prepare the Block Mix using the reagents provided in the SureSelectXT Human All Exon Kit as follows (per sample).
Reagent Amount to add SureSelect Indexing Block #1 2.5 µL SureSelect Block #2 2.5 µL SureSelect Indexing Block #3 0.6 µL Total volume 5.6 µL -
27Dilute the SureSelect RNase Block by adding 2 µL of nuclease-free water to 1 µL of SureSelect RNase Block for each hybridization reaction. Prepare the Capture Library Mix using the reagents provided in the SureSelectXT Human All Exon Kit as follows (per sample) and keep on ice.
Reagent Amount to add SureSelect Human All Exon 50M Library 5 µL Diluted SureSelect RNase Block 2 µL Total volume 7 µL -
28Mix 3.4 µL of amplified library DNA (Step 24) with 5.6 µL of Block Mix (Step 26). Load the mixture into one well in row B of a fresh 96-well PCR plate. Seal the plate with adhesive film, denature the mixture in a thermal cycler for 5 min at 95°C, and hold the temperature at 65°C.To minimize evaporation, it is recommended to avoid placing samples in the outermost wells (Columns 1 and 12) of the 96-well plate.
-
29
Into the corresponding well of the same column in row A, load 40 µL of Hybridization Buffer (Step 25). Seal the plate and incubate for 5 min at 65°C.
-
30
Into the corresponding well of the same column in row C, load 7 µL of Capture Library Mix (Step 27). Seal the plate and incubate for 2 min at 65°C.
-
31Transfer the following reagents, in order, to row C without changing pipette tips. Use a multichannel pipette if working with more than one sample.
- Transfer 13 µL of hybridization buffer from row A to the corresponding well in row C.
- Transfer the contents of row B to the corresponding well in row C.
- Pipette up and down 10 times to mix.
-
32Seal the plate securely with two new adhesive films to prevent excessive evaporation: Cut the first film slightly smaller so the size is just enough to seal all the wells, and then apply the second film to seal the whole plate. Incubate the sealed plate in a thermal cycler for 24 h at 65°C with the heated lid set to 105°C.We apply the adhesive seal while the plate is inside the PCR machine to avoid possible air pressure differences inside and outside the wells.
Performing Hybrid Capture Selection with Dynabeads
MyOne Streptavidin T1
-
33
Aliquot and prewarm SureSelect Wash 2 (1.65 mL/sample including 10% extra) in a heat block at 65°C.
-
34
Resuspend the Dynabeads MyOne Streptavidin T1 in the storage vial by vortexing. For each hybridization, aliquot 50 µL of resuspended beads to a fresh 1.5-mL LoBind tube.
-
35
Wash the beads three times with 200 µL of SureSelect Binding Buffer per wash. For each wash, vortex for 5 sec, separate the beads with a magnetic stand, and remove the supernatant when clear. Resuspend the beads in 200 µL of SureSelect Binding Buffer.
-
36
Add the hybridization mixture from Step 32 directly to the resuspended beads while maintaining the PCR plate at 65°C. Mix well by pipetting up and down 10 times. Incubate the mixture on the Mini LabRoller for 30 min at room temperature.
-
37
Briefly centrifuge the tube to collect the mixture. Separate the beads from the supernatant on the magnetic stand. Discard the supernatant and resuspend the beads in 500 µL of SureSelect Wash 1. Incubate the tube for 15 min at room temperature.
-
38Briefly centrifuge the tube to collect the mixture. Separate the beads from the supernatant on the magnetic stand. Discard the supernatant and wash the beads three times as follows.
- Resuspend the beads with 500 µL of SureSelect Wash 2 (prewarmed to 65°C) and vortex for 5 sec.
- Incubate the sample for 10 min at 65°C.
- Briefly spin to collect the mixture. Separate the beads from the supernatant on the magnetic stand and discard the supernatant.
-
39
Resuspend the beads with 50 µL of SureSelect Elution Buffer by vortexing for 5 sec. Incubate the samples for 10 min at room temperature.
-
40
During the incubation in Step 39, aliquot 50 µL of SureSelect Neutralization Buffer to a fresh 1.5-mL LoBind tube.
-
41
When the incubation is completed, briefly centrifuge the sample to collect the mixture. Separate the beads from the supernatant on the magnetic stand. Transfer the supernatant to the tube containing the SureSelect Neutralization Buffer (Step 40). Mix well by pipetting up and down 10 times.
-
42Purify the sample as described in Steps 4–10, with the following modifications: add 180 µL of homogenous AMPure XP reagent to the reaction, and elute the purified DNA in 30 µL of nuclease-free water.It is safe to stop here and store the sample at −20°C.
Adding the Index by Posthybridization Amplification
-
43Select the desired index for each library. Prepare the amplification reaction in a 0.2-mL PCR tube using the reagents provided in the SureSelectXT Human All Exon Kit and Herculase II Fusion DNA Polymerase as follows, using one of the 16 provided primers according to the desired index.
Reagent Amount to add Captured DNA from Step 42 14 µL Nuclease-free water 22.5 µL 5× Herculase II reaction buffer 10 µL 100 mM dNTP mix 0.5 µL Herculase II Fusion DNA Polymerase 1 µL SureSelect ILM indexing post capture forward PCR primer 1 µL PCR primer (Index 1–Index 16) 1 µL Total volume 50 µL -
44
Place the tubes in a thermal cycler and amplify the DNA using the following program: 2 min at 98°C, 12 cycles of (30 sec at 98°C, 30 sec at 57°C, 1 min at 72°C), 10 min at 72°C.
-
45
Purify the final indexed library as described in Steps 4–10, with the following modifications: Add 90 µL of homogenous AMPure XP reagent to the reaction, and elute the purified DNA in 30 µL of QIAGEN buffer EB.
-
46Check the quality of the final library as follows.
- Run 1 µL on a high-sensitivity DNA chip using the Agilent 2100 Bioanalyzer following the manufacturer’s instructions.The amplified library should have a peak size of ~350 bp.
- Record the measured concentration of the library for reference when pooling multiple libraries.We recommend pooling libraries based on their molar concentration as determined by the Bioanalyzer.The library is now ready for paired-end sequencing on an Illumina HiSeq 2000 sequencer. Alternatively, the library can be placed at −80°C for long-term storage.
RELATED INFORMATION
Multiple platforms are available for WES, particularly for use with Illumina next-generation sequencers. In our previous publication (Clark et al. 2011), we compared the performance of three of the most popular hybridization-based whole exome capture and sequencing platforms for the human exome, including the Agilent SureSelect Human All Exon system described here; protocols for the two other platforms are provided in Whole-Exome Enrichment with the Roche Nimblegen SeqCap EZ Exome Library SR Platform (Chen et al. 2015a) and Whole-Exome Enrichment with the Illumina TruSeq Exome Enrichment Platform (Chen et al. 2015b). In summary, the performance of each platform was generally on par with the others, with specific, but minor, pros and cons. Researchers would be wise to choose among the platforms based on their specific experimental needs (e.g., genomic regions of interest, coverage vs. depth, etc.). The size of the targeted regions for each platform is flexible: By designing custom probes with Agilent, Roche, or Illumina, each protocol is suitable for enrichment of a lower number of custom targets for Illumina sequencing.
ACKNOWLEDGMENTS
This publication is supported by the National Institute of Diabetes and Digestive and Kidney Diseases of the National Institutes of Health under Award Number 3U54DK10255602S2.
REFERENCES
- Chen R, Im H, Snyder M. Whole-exome enrichment with the Roche Nimblegen SeqCap EZ Exome Library SR platform. Cold Spring Harb Protoc. 2015a doi: 10.1101/pdb.prot084855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Im H, Snyder M. Whole-exome enrichment with the Illumina TruSeq Exome Enrichment platform. Cold Spring Harb Protoc. 2015b doi: 10.1101/pdb.prot084863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MJ, Chen R, Lam HY, Karczewski KJ, Euskirchen G, Butte AJ, Snyder M. Performance comparison of exome DNA sequencing technologies. Nat Biotechnol. 2011;29:908–914. doi: 10.1038/nbt.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]