Abstract
Targeted imaging and antimicrobial photodynamic inactivation (PDI) are emerging methods for detecting and eradicating pathogenic microorganisms. This study describes two structurally related optical probes that are conjugates of a zinc(II)-dipicolylamine targeting unit and a BODIPY chromophore. One probe is a microbial targeted fluorescent imaging agent, mSeek, and the other is an oxygen photosensitizing analogue, mDestroy. The conjugates exhibited high fluorescence quantum yield and singlet oxygen production, respectively. Fluorescence imaging and detection studies examined four bacterial strains: E. coli, S. aureus, K. pneumonia, and B. thuringiensis vegetative cells and purified spores. The fluorescent probe, mSeek, is not phototoxic and enabled detection of all tested bacteria at concentrations of ~100 CFU/mL for B. thuringiensis spores, ~1000 CFU/mL for S. aureus and ~10,000 CFU/mL for E. coli. The photosensitizer analogue, mDestroy, inactivated 99–99.99% of bacterial samples and selectively killed bacterial cells in the presence of mammalian cells. However, mDestroy was ineffective against B. thuringiensis spores. Together, the results demonstrate a new two-probe strategy to optimize PDI of bacterial infection/contamination.
Keywords: photodynamic inactivation, bacteria, spores, BODIPY, singlet oxygen, fluorescence microscopy
Introduction
Pathogenic bacteria remain a serious threat to public health.1 Excessive antibiotic use has promoted natural selection of bacterial strains with an increasing number of resistance mechanisms.2 Photodynamic inactivation (PDI) is an attractive antimicrobial strategy that has been applied to blood sterilization, dental disinfection and surface decontamination.3–6 PDI employs a photosensitizer (PS) molecule to convert light energy into a cytotoxic event. In short, the photosensitizer excited state undergoes efficient intersystem crossing to produce an excited triplet state that either produces unstable radicals (type I photoprocess); or alternatively transfers energy to neighboring molecular oxygen to produce reactive singlet oxygen (type II photoprocess). In both cases, the reactive species induce cell death either by cleavage of biomolecules or membrane disruption. As an antibiotic therapy, this approach kills drug-resistant bacteria with no apparent induction of resistance.7, 8 An attractive secondary feature is the destruction of associated bacterial endotoxins, like lipopolysaccharide.9
Bacterial PDI has been explored primarily with organic dyes such as xanthenes, pthalocyanines, porphrins or chlorins that have little bacterial selectivity.10 Clinical studies have explored 5-aminolevulinic acid for skin infections and methylene blue for oral disinfection.10, 11 An emerging new approach is targeted PDI using PS conjugates with appended bacteria targeting units. Promising results have been obtained using the antimicrobial peptide (KLAKLAK)2 conjugated to eosin Y and the lipopolysaccharide neutralizing peptide YI13WF conjugated to protophophyrin IX.12, 13 Other studies have used larger bacteria targeting molecules such as antibodies, bacteriophages, polylysine, and polyethyleneimine.14–20 It is unlikely that a single targeted conjugate will be suitable for all types of applications. In some cases, highly selective targeting for a specific strain will be needed and in other cases, broad spectrum activity is desired. In many situations, such as surface decontamination, an important factor is cost effectiveness. Finally, there is a need to rapidly evaluate PDI efficacy, which means a complementary imaging method that can quantify the infection/contamination level before and/or after photodynamic treatment. Many of the literature PS are also weakly fluorescent, and thus have been used for imaging experiments. But the quality of the imaging data is compromised by the concomitant PS phototoxicity. We envision an alternative two-probe imaging and treatment strategy that employs a targeted non-phototoxic imaging probe for accurate fluorescence detection and a structurally related photosensitizing agent for PDI.
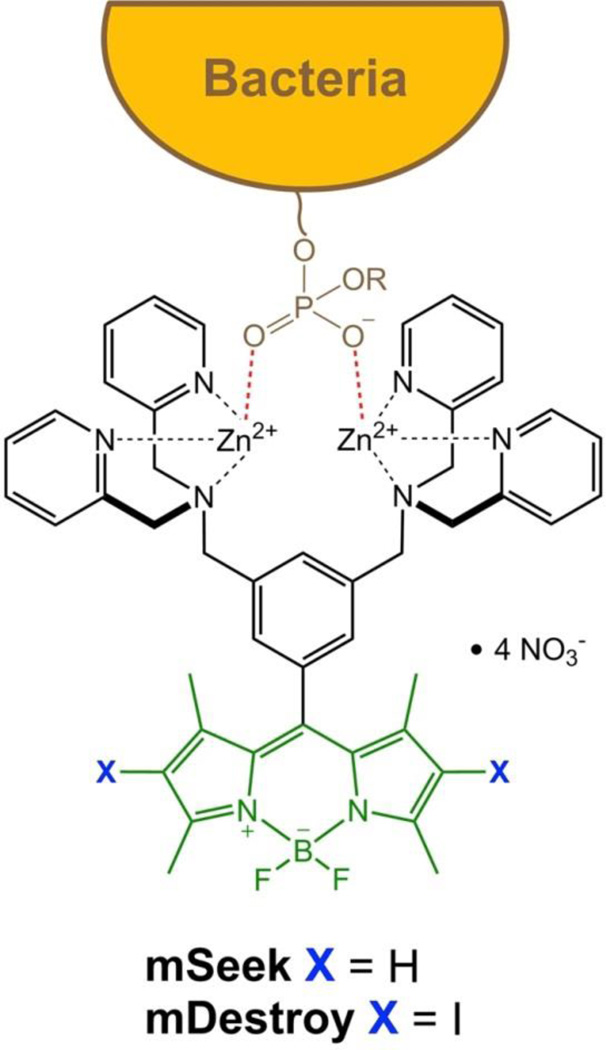
This report describes a novel pair of broad spectrum optical probes for all genera of bacteria. The common targeting unit in both probes is a zinc(II)-dipicolylamine (ZnDPA) coordination complex which can selectively associate with the anionic surfaces of bacteria, in preference to the near neutral membrane surfaces of healthy mammalian cells.21 ZnDPA complexes are known to bind the phosphorylated amphiphiles, phosphatidylglycerol, cardiolipin, lipoteichoic acid and lipopolysaccharide, which are ubiquitous in the bacterial envelope.22 Fluorescent and nuclear labeled ZnDPA conjugates have enabled microscopic imaging of cultured bacterial cells and meso-scale imaging of infection in living animal models.23–26 Additionally, some ZnDPA structures are bactericidal, apparently through the disruption of the bacterial membrane.27
With an eye on production scalability, a crucial factor for eventual large scale implementation, we chose boron dipyrromethane (BODIPY) as the photoactive chromophore.28–31 The structural factors that create highly fluorescent BODIPY dyes, or alternatively potent BODIPY photosensitizers, are well established.32 Building on this literature knowledge we designed two microbial targeted ZnDPA-BODIPY conjugates: a fluorescent imaging agent, mSeek, and a photosensitizing analogue, mDestroy (Scheme 1). In addition to probe synthesis and characterization we report the fluorescence detection sensitivity and phototoxicity against multiple strains of bacterial cells and spores.
Scheme 1.
Bacteria cell surface targeting by a ZnDPA-BODIPY fluorophore (mSeek) or photosensitizer (mDestroy). The prefix “m” designates “microbial”.
Experimental Section
Materials
All chemicals were reagent grade and used as purchased. Reactions were monitored by TLC analysis using 250 µm glass backed silica gel plates and compounds were visualized using a UV lamp. Luria-Bertani (LB) broth, LB agar and nutrient broth were purchased from BD Medical Supplies. ATCC liquid medium 3 was purchased from American Type Culture Collection (ATCC).
Chemical Synthesis
Alcohol 1
Synthesized by a literature procedure with comparable yield.33, 34
Aldehyde 2
Activated manganese dioxide (164 mg, 1.89 mmol) was added to a solution of 1 (100 mg, 0.188 mmol) in THF. The mixture was stirred at room temperature for 18 h and the reaction was followed by TLC. Extra portions (164 mg, 1.89 mmol) of MnO2 were added and the reaction stirred for another 18 h until complete disappearance of the starting material. The mixture was filtered through celite, then the solvent was removed by evaporation to reveal an oil which was purified by column chromatography to yield 2 as a light yellow oil (CH2Cl2: MeOH : NH4OH = 100 : 10 :1). Yield 90 %. 1H NMR (400 MHz, CDCl3) δ (ppm) 3.76 (s, 4H), 3.81 (s, 8H), 7.13 (m, 4H), 7.55 (d, J = 7.79 Hz, 4H), 7.62 (m, 4H), 7.74 (s, 1H), 7.79 (s, 2H), 8.51 (m, 4H), 9.99 (s, 1H). 13C NMR (100 MHz, CDCl3) δ (ppm) 192.4, 158.4, 148.9, 137.2, 136.9, 135.9, 129.5, 123.6, 122.6, 59.8, 58.1. ESI-MS: Found 529.2701, Calcd. C33H33N6O 529.2710 [M+H]+.
BODIPY 3a
Aldehyde 2 (134 mg, 0.25 mmol) and 2,4-dimethyl pyrrole (48 mg, 52.3 µL, 0.51 mmol) were dissolved in 10 mL anhydrous CH2Cl2 under argon atmosphere. TFA (140 µL) was added and the solution was stirred at room temperature for 24 h. A solution of tetrachlorobenzoquinone (66 mg, 0.27 mmol) in 10 mL CH2Cl2 was added, and stirring was continued for 30 minutes followed by the addition of Et3N (0.69 mL) and BF3OEt2 (0.69 mL). After stirring for 30 minutes the reaction mixture was washed three times with water, dried over Na2SO4 and the solvent was removed under vacuum. The residue was purified on silica gel column chromatography to yield pure 3a (CHCl3:MeOH = 95:5, v/v)35 Yield: 30%. 1H NMR (400 MHz, CDCl3) δ (ppm) 1.20 (s, 6H), 2.56 (s, 6H), 3.71 (s, 4H), 3.78 (s, 8H), 5.90 (s, 2H), 7.14 (m, 4H), 7.29 (s, 2H), 7.52 (m, 5H), 7.62 (m, 4H), 8.50 (m, 4H). 13C NMR (100 MHz, CDCl3) δ (ppm) 159.6, 149.3, 140.9, 136.7, 130.6, 127.3, 122.8, 122.3, 60.2, 58.5, 14.8, 14.6. ESI-MS: Found 747.3928, Calcd. C45H46BF2N8 747.3909 [M+H]+.
Iodo-BODIPY 3b
A solution of HIO3 (0.08 mmol, 3.52 mg) in a minimum amount of water was added dropwise to a solution of 3a (0.02 mmol, 15 mg) and I2 (0.1 mmol, 25 mg) in EtOH (2 mL). The mixture was heated to 60 °C for 20 min, then cooled to room temperature and extracted with sodium thiosulfate dissolved water and CH2Cl2. The organic phase was dried over Na2SO4 and concentrated under vacuum. The crude product was purified by silica gel column chromatography (CHCl3:MeOH 95:5, v/v) give pure 3b. Yield 40%. 1H NMR (400 MHz, CDCl3) δ (ppm) 1.18 (s, 6H), 2.65 (s, 6H), 3.72 (s, 4H), 3.78 (s, 8H), 7.15 (m, 4H), 7.28 (s, 1H), 7.51 (m, 5H), 7.56 (s, 1H), 7.63 (m, 4H), 8.51 (m, 4H). 13C NMR (100 MHz, CDCl3) δ (ppm) 156.8, 148.8, 140.5, 137.0, 131.2, 127.3, 123.1, 122.5, 59.4, 58.0, 17.0, 16.0. ESI-MS: Found 999.1868, Calcd. C45H44BF2N8I2 999.1842 [M+H]+.
BODIPY 4
Synthesized by a literature procedure with comparable yield.36 1H NMR (400 MHz, CDCl3) δ (ppm) 1.37 (s, 6H), 2.57 (s, 6H), 6.00 (s, 2H), 7.45 (d, 2H), 8.24 (d, 2H). ESI-MS: Found 369.1592, Calcd. C20H20BF2N2O2 369.1584 [M+H]+.
mSeek and mDestroy
Separate methanolic solutions of 3a (6.70 µmol) or 3b (6.70 µmol) were mixed with a aqueous solution of Zn(NO3)2 (14.1 µmol) and stirred for 45 minutes at room temperature. The solvent was removed and the residue lyophilized to afford mSeek or mDestroy in quantitative yield.
Singlet Oxygen Generation
The rates of singlet oxygen (1O2) generation due to photosensitization of ZnDPA conjugates and control dyes were determined using the 1O2 trap 1,3-diphenylisobenzofuran (DPBF). DPBF (λabs = 415 nm) readily reacts with 1O2 to form a bleached cycloadduct. Twenty molar equivalents of DPBF were added to separate solutions of each dye in acetonitrile (5.0 µM) and absorption spectra were acquired at various time points while the samples were irradiated with green light (150 W Xenon lamp with long pass filter >495 nm) at a fluence of 50 mW/cm2 (25 °C).
Mammalian Cell Toxicity
Cell viability was measured using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) cell viability assay. The number of living cells is directly correlated to the amount of reduced MTT which is monitored by absorbance at 570 nm. Only active reductase enzymes in viable cells can perform the reduction reaction. CHO-K1 (Chinese hamster ovary) cells that were purchased from American Type Culture Collection (ATCC), spread into 96-microwell plates, and grown to confluency of 85% in RPMI or F-12K media supplemented with 10% fetal bovine serum, and 1% streptavidin L-glutamate at 37 °C and 5% CO2. The Vybrant MTT cell proliferation Assay Kit (Invitrogen, Eugene, USA) was performed according to the manufacture’s protocol. The cells were treated with either mSeek or mDestroy (0−20 µM) and incubated for 24 h at 37 °C. The medium was replaced with 100 µL of F-12K media containing MTT (1.2 mM) and incubated at 37 °C and 5% CO2 for an additional 4 hours. An SDS-HCl detergent solution was added and the absorbance of each well was read at 570 nm and normalized to wells containing cells but no added probe (N = 5).
Bacterial Strains and Spore Preparation
Escherichia coli DH5α was a gift from Dr. Holly Goodson at the University of Notre Dame Department of Chemistry and Biochemistry. Klebsiella pneumoniae subsp. pneumoniae (ATCC 33495) and Staphylococcus aureus NRS 11 were gifts from Dr. Shahriar Mobashery at the University of Notre Dame Department of Chemistry and Biochemistry. Bacillus thuringiensis strain NRS 1124 (ATCC 19268) was purchased from ATCC. E. coli and S. aureus were grown in LB broth, K. pneumoniae in nutrient broth and B. thuringiensis in ATCC liquid medium 3. Stocks were made for each strain and used to streak sterile agar plates. Colonies from plates were used to inoculate overnight cultures, which were grown aerobically at 37 °C, 200 rev/min. Fresh cultures were inoculated the next day in a 1:1000 dilution of overnight culture and used for experiments after growth to mid log phase (OD600 ~0.4−0.6). B. thuringiensis spores were prepared using a sporulation media and a previously described literature procedure.37 Refined soybean oil was added as an antifoaming agent to a final concentration of ~1%. Water was deionized and vacuum filtered through a Millipore water system. Sporulation was performed in 500 mL of broth in a 1 L glass flask and incubated in a shaker-incubator at 30 °C and 300 rev/minutes for 7 days. Cultures were filtered through two layers of sterile cheesecloth to remove highly aggregated spores and then pelleted at 16,000 g for 10 min. Spore pellets were re-suspended in sterile water with 1% Tween-80 and dispensed into Eppendorf tubes and frozen at −60 °C. Aliquots were removed to determine spore viability as determined by counting colony forming units (CFU). Samples were either heated at 80 °C for 30 minutes to determine spore viability or warmed to room temperature to determine the total CFU of the sample (spores + vegetative cells). Samples were serially diluted in ATCC liquid medium 3 and dilutions were used to inoculate blood agar plates (ATCC) and incubated for 20 hours.
Fluorescence Microscopy
Bacterial strains cultured overnight were harvested and washed twice with sterile HEPES buffer (pH 7.4). The washed cells were resuspended in HEPES at an OD600 of 0.7 and 100 µL aliquots were treated with 5 µM of mSeek. After incubation for 10 minutes in the dark (20 °C), the cells were washed with sterile HEPES buffer by centrifugation (8,500 g, 1 min) to remove unbound probe and a drop of the dispersed suspension was placed on a glass slide followed by a glass coverslip. Imaging was performed on an epifluorescence microscope (Nikon Eclipse TE2000-U epifluorescence) using bright field and the following fluorescence filter sets: UV-2E/C (ex = 360 ± 20 nm, em = 460 nm ± 25 nm), GFP (ex = 470 ± 20 nm, em = 425 ± 25 nm), and Cy3 (ex = 540 ± 20 nm, em = 640 ± 30 nm). Images were captured with Nikon NIS Elements Software and analysed using Image J 1.45s.
Flow Cytometry
Bacterial samples were prepared as described above. Then 100 µL aliquots containing 108 CFU were treated with 10 µM of mSeek or control dye 4. After incubation for 20 minutes in the dark at 37 °C, the cells were washed as described. The samples were injected into a flow cytometer (Beckman FC-500) equipped with a Biosense flow cell and a 6 W argon ion laser. The excitation laser was tuned for 480-nm emission (500 mW) and emission light was measured using a 530-nm-long pass absorbance filter. Histograms represent a total of 50,000 to 100,000 events each. Fluorescence and count data were normalized to "polychromatic" calibration beads.
Detection Sensitivity Studies
The bacterial detection limit of mSeek was determined using an imaging station containing a charged coupled device (CCD) camera. Aliquots of bacteria (106 CFU) were treated with mSeek (30 µM, 10 min) and washed with HEPES buffer. Serial dilutions of treated bacteria were aliquoted into a multiwell plate and imaged using an IVIS Imaging Station. Following imaging, a region-of-interest analysis of each well was conducted using ImageJ 1.45s software to determine the fluorescence intensity produced from each dilution of treated bacteria. The CCD background cutoff was defined as the fluorescence detection error when no fluorophore was present.
Photodynamic Inactivation of Bacterial Cells
Bacteria were grown and washed as described above. A bacteria suspension sample at OD600 ~0.5 in sterile HEPES buffer was diluted to a final concentration 106 CFU/mL. Aliquots of mDestroy or mSeek stock solution (1 mM) were added to the bacteria and the suspension incubated in the dark for 20 minutes at room temperature. The sample was then washed three times with sterile HEPES buffer and resuspended in a final volume of 2 mL in a 4 mL fluorescence quartz cuvette. The cuvette was irradiated with light from a 150 W Xenon lamp filtered through a long pass 495 nm filter and set at a distance of 15 cm from the light source. The fluence was measured to be 50 mW/cm2 and the samples were stirred at 200 rpm. The samples were illuminated for 60 minutes with continuous O2 bubbling or kept in the dark. Aliquots (100 µL) were removed and diluted into 900 µL of the appropriate sterile bacteria media. Further 10-fold serial dilutions gave a dilution range of 101−105. From each diluted sample, a 50 µL aliquot was removed, spread on an agar plate, and incubated overnight at 37 °C. For B. Thuringiensis spores, no additional treatment to enhance spore germination was used. Colonies were counted the next morning and the survival fraction was plotted on a log scale. Plates containing untreated bacteria, non-irradiated bacteria treated with mDestroy and bacteria treated with mSeek were included as negative controls.
Selective Staining/Inactivation of Bacteria over Mammalian Cells
Fluorescence microscopy was used to demonstrate the selective staining and inactivation of bacterial cells over healthy mammalian cells. Separate samples of Jurkat cells (105) were mixed with an aliquot of E. Coli DH5α (106 CFU) in a 1.5 mL Eppendorf tube containing HEPES buffer. For demonstration of selective staining, mSeek (5 µM) was added to the cell mixture, which was incubated for 15 minutes and then imaged using epifluorescence microscope. Fluorescence images were captured using the GFP filter set. For demonstration of selective inactivation, the same Jurkat/E. coli cell mixture was incubated with mDestroy (10 µM) for 10 minutes, then transferred to a quartz cuvette and irradiated for 45 minutes with green light at a fluence of 50 mW/cm2 using the lamp setup described above. Following irradiation, the sample was incubated with propidium iodide (5 µM) for 15 min. A 10 µL aliquot of the mixture was added to glass slides and covered with a glass coverslip. Fluorescence micrographs were captured using the Cy3 filter set.
Results
Synthesis and Characterization
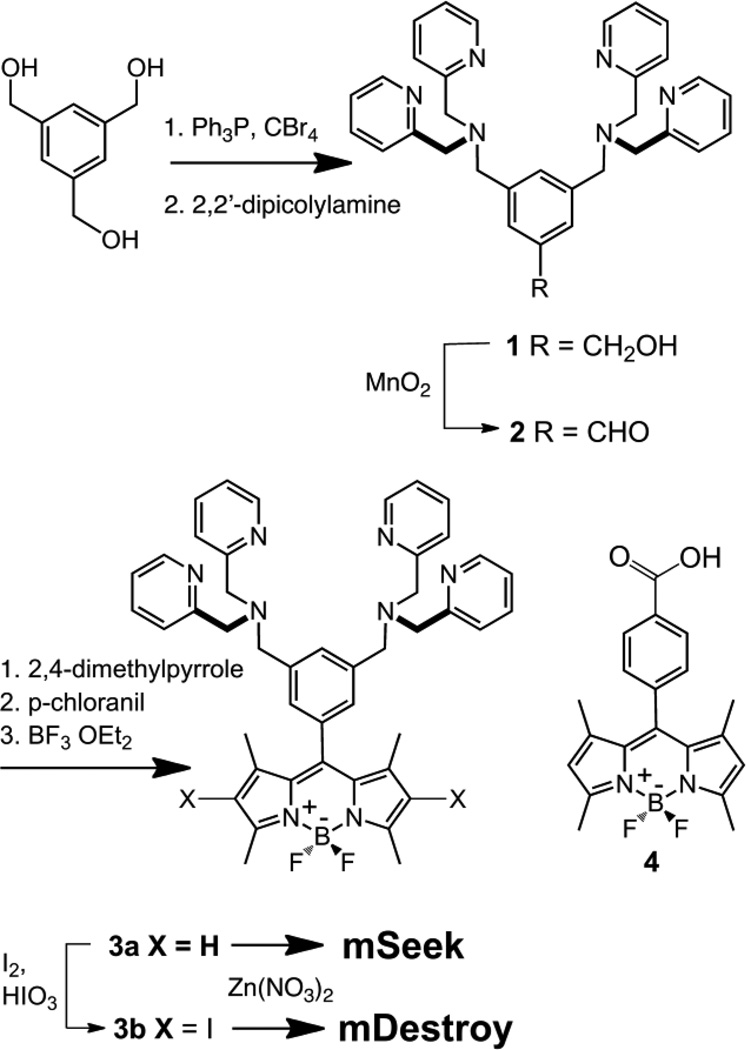
The synthetic pathways to make mSeek and mDestroy are described in Scheme 2. The ZnDPA conjugated BODIPY dye 3a was synthesized in five steps. The known alcohol 1,34 was oxidized by active manganese dioxide to afford aldehyde 2, which was converted, in a one-pot multistep process, to BODIPY dye 3a in 30% overall yield. Halogenation of 3a using I2/HIO3 gave the 2,6-diiodo derivative 3b. The dyes were complexed with Zn(NO3)2 to give the two desired probes. The control dye 4, without a ZnDPA targeting unit, was also prepared.
Scheme 2.
Synthesis of mSeek and mDestroy.
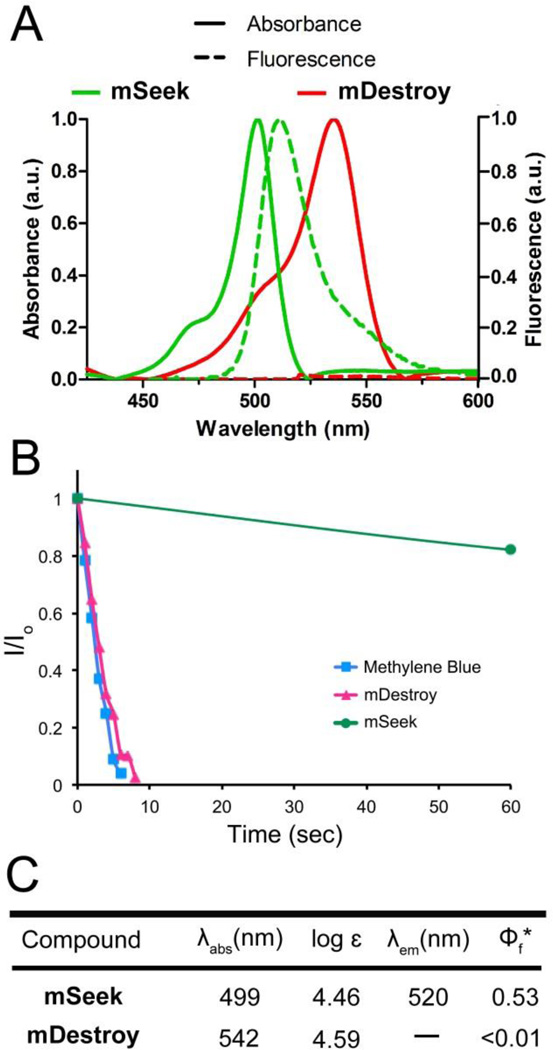
As expected, mSeek and control dye 4 emit strong green fluorescence while mDestroy is essentially non-fluorescent (Figure 1A, 1C). The oxygen photosensitization capability was determined by standard chemical trapping experiments with 1,3-diphenylisobenzofuran (DPBF) as a reactive trap for photogenerated 1O2.38 The experiments compared mSeek, mDestroy and the well-known photosensitizer methylene blue. In each case, an acetonitrile solution of dye (5.0 µM) and excess DPBF (100 µM) was irradiated with green light and monitored periodically by absorption spectroscopy. The photogenerated 1O2 reacted rapidly with the DPBF to produce a colorless product. The rate of 1O2 production was measured by the decrease in DPBF absorption at 415 nm (Figure S1). As expected, mSeek was found to be a very weak oxygen photosensitizer, whereas mDestroy generated 1O2 at the same rate as methylene blue (Figure 1B) which has a reported singlet oxygen quantum yield of 0.57.39 Additional tests of probe photostability found that there was little degradation after 45 minutes of constant bench-top irradiation of green light (Figure S2).40 Mammalian cell viability assays using Chinese hamster ovary (CHO) cells indicated that both probes were non-toxic in the absence of light (Figure S3).
Figure 1.
[A] Absorption and fluorescence emission profiles of mSeek (ex. 470 nm) and mDestroy (ex. 500 nm) in PBS, pH 7.4. [B] Normalized rates of singlet oxygen production measured by the ratio of DPBF absorption after reacting with singlet oxygen (I) and the initial DPBF absorption (Io). Lines are provided only to guide the reader’s eye. [C] Spectral properties. *Φf represents quantum yield of fluorescence with compared to fluorescein (0.92, in aqueous solution).66 Values are averages of three separate experiments.
Bacterial Imaging Using mSeek
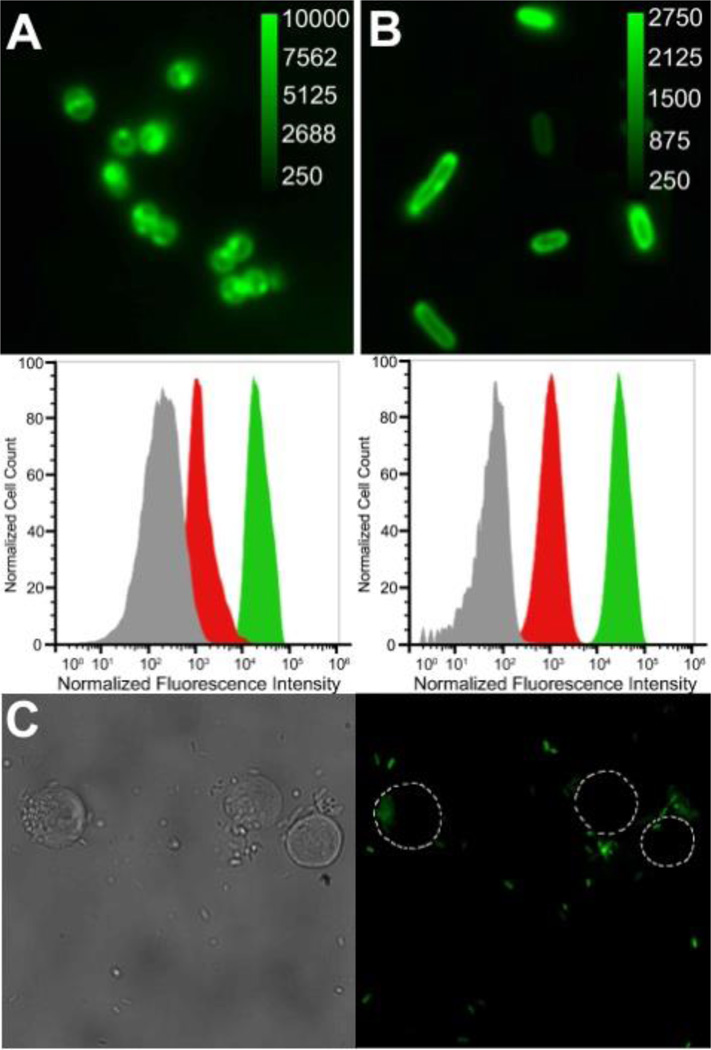
The bacterial targeting ability of fluorescent mSeek was assessed by fluorescence microscopy and flow cytometry using cultures of vegetative S. aureus, E. coli, B. thuringiensis and K. pneumoniae, and B. thuringiensis spores. Separate samples of bacteria (108 CFU) were treated with mSeek (10 µM) in HEPES buffer. After incubation for 10 min, each sample was rinsed twice with fresh buffer and then subjected to fluorescence microscopy. In agreement with previous studies,25 strong fluorescence staining of all bacteria was observed with most of the fluorescence emission localized to the bacterial envelope. Shown in Figure 2A-B are typical micrographs obtained with the Gram-positive S. aureus and Gram-negative E. coil using a green fluorescence set. In relative terms, the stained Gram-negative bacteria were slightly less fluorescent indicating some protection by the outer membrane. The requirement for a ZnDPA targeting ligand was demonstrated by comparing the bacterial staining by mSeek with the staining by control dye 4. Flow cytometry histograms of S. aureus and E. coli cells treated with mSeek showed ~10 times higher fluorescence than cells treated with 4 (Figure 2). Similar results were obtained when K. pneumoniae bacteria or B. thuringiensis spores were treated with the two probes (Figure S4). Additional cell microscopy experiments demonstrated the selectivity of mSeek for bacterial cells over healthy mammalian cells. A suspension of E. coli (106 CFU/mL) was mixed with Jurkat cells, a non-adherent lymphocyte mammalian cell line, followed by addition of mSeek (5 µM) and the mixture was incubated for 15 minutes. In Figure 2C is a typical fluorescent micrograph showing selective mSeek staining of the bacterial cells with no obvious fluorescence detected on the surface of Jurkat cells.
Figure 2.
Epifluorescence microscopy of S. aureus NRS11 [A] and E. coli DH5α [B] after incubation with mSeek (10 µM). Images viewed at 1500× magnification with pixel intensity scale bar in arbitrary units. The corresponding flow cytometry histograms show cell counts vs. normalized fluorescence intensity for S. aureus NRS11 and E. coli DH5α after no treatment (grey), control dye 4 (red) and mSeek (green); n = 3 for both cell lines. [C] Representative brightfield (left) and fluorescence micrograph (right) of a mixture of Jurkat and E. Coli DH5α cells that had been treated with mSeek.
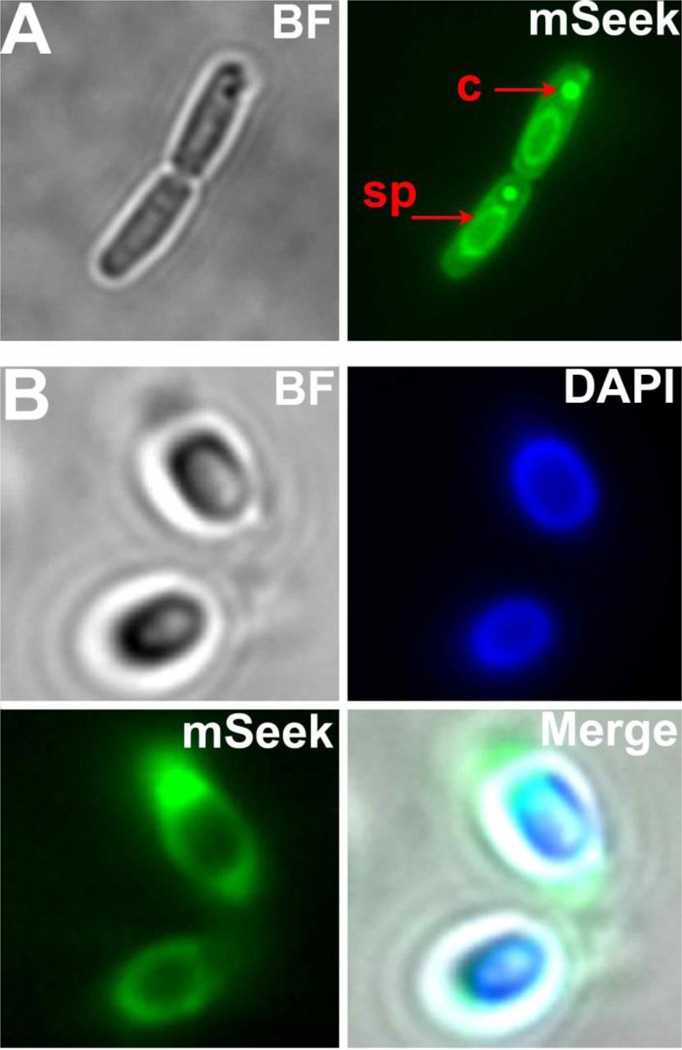
Further fluorescence imaging studies revealed that mSeek efficiently stains bacillus cells and spores. Fluorescence microscopy of sporulating B. thuringiensis cells treated with mSeek showed fluorescence throughout the cytoplasm with strong staining of the developing spore and the protein endotoxin crystals (Figure 3A).41 Micrographs of purified spores that had been incubated with mSeek and the known exosporium stain DAPI (20 µM, 2 hours) show colocalized staining of the exosporium (Figure 3B).42
Figure 3.
Epifluorescence microscopy of B. thuringiensis during sporulation [A] and purified spores [B] stained with mSeek (5 µM, 15 minutes) and DAPI (20 µM, 2 hours) followed by fluorescence microscopy using green and blue emission channels. sp = developing spore , c = protein endotoxin crystal.
The bacterial cell detection limit of mSeek was determined by culture imaging experiments using a CCD camera (Figure 4A). An aliquot containing 106 bacterial cells was treated with mSeek, washed with HEPES buffer and serially diluted in a multiwell plate. Fluorescent images of the wells (Figure 4B) were subjected to region-of-interest analysis which compared pixel intensity to the background signal of an untreated well (Figure 4C). Detectable signals were observed for the following numbers of mSeek-stained bacteria: B. thuringiensis spores at ~100 CFU/mL, S. aureus at ~1000 CFU/mL, and E. coli at ~10,000 CFU/mL (Figure 4D).
Figure 4.
[A] CCD camera setup for sensitivity detection. [B] Representative fluorescent image of a multiwell plate containing mSeek-stained B. thuringiensis spores diluted from 106 – 0 CFU/mL. [C] Normalized fluorescence intensity of various bacterial strains that had been treated with mSeek. [D] Expanded region of panel [C] showing CCD background level. Fluorescence intensities are an average of nine measurements and error bars represent mean ± standard error.
Bacterial PDI Using mDestroy
The effectiveness of mDestroy for bacterial PDI was evaluated using Gram-positive and Gram-negative bacteria strains as well as bacillus spores. Separate aliquots of bacteria (106) were treated with various concentrations of the probe and exposed to green light (50 mW/cm2) irradiation for one hour (Figure S5). After light treatment, the sample was serially diluted and streaked onto agar plates followed by overnight incubation. Various control experiments used mSeek and control dye 4, and also tested the presence and absence of light. Bacteria exposed to light alone or mDestroy without irradiation did not produce a bactericidal effect (Figure S6). Similarly, light treatment of cells treated with mSeek had no significant effect on bacteria viability (Figure S7). The photodynamic activity of mDestroy (10 µM) against vegetative bacteria was substantial with 99−99.99% of the bacteria killed (Figure 5). Greater phototoxicity was observed with the Gram-positive bacteria strains compared to the Gram-negative strains. However, there was no phototoxic effect against the B. thuringiensis spores.
Figure 5.
Fractions of killed bacterial cells treated with increasing concentrations of mDestroy and one hour of green light irradiation (50 mW/cm2). Data points are fit to linear regression.
The selectivity of the photodynamic effect was tested by incubating mDestroy (10 µM) with a mixture of E. coli bacteria and Jurkat cells for 15 minutes followed by green light irradiation of the mixture for 45 minutes. The cell mixture was then treated with propidium iodide (5 µM), a membrane impermeant dye that is excluded from viable cells and internalized within compromised cells. Fluorescence microscopy showed that only the E. coli cells were stained with propidium iodide indicating that they were selectively photoinactivated (Figure 6). A colony-forming assay after serial dilutions of the sample confirmed that more than 99% of the E. coli bacteria were killed.
Figure 6.
Representative merge of brightfield and fluorescence micrographs of Jurkat cells and E. Coli DH5α after treatment with mDestroy and 45 minutes of green light irradiation followed by staining with propidium iodide.
Discussion
Although PDI of bacteria has been known for over a century, its use in the clinic is limited.43, 44 Most studies have focused on neutral or cationic PS that generate high amounts of singlet oxygen. Photodynamic activity typically correlates with PS lipophilicity which imparts a propensity to associate with biological membranes.45, 46 However, lipophilic PS do not typically discriminate bacterial cells over mammalian cells. Cationic PS are more selective for bacteria and Gram-positive bacteria are usually more sensitive to PDI than Gram-negative strains.47–49 The fluorescence imaging data with mSeek and the PDI data with mDestroy demonstrate that the ZnDPA targeting unit has high selectivity for bacterial cells over healthy mammalian cells. The slightly weaker staining and inactivation of Gram-negative bacteria is probably due to the protective second, outer membrane.50 Most likely the ZnDPA units must compete with the Ca2+ and Mg2+ cations that bridge the lipopolysaccharides in the membrane.
The study also evaluated staining and inactivation of bacterial spores, dormant structures that must undergo germination before regaining metabolic activity.51 The B. thuringiensis spores used in this study are genetically and structurally similar to highly toxic Bacillus anthracis spores which have been formulated for bioterrorism.52 The intense and rapid staining of B. thuringiensis spores by mSeek is attributed to the strong affinity of the ZnDPA targeting unit for the high number of phosphate and carboxylate groups in the exosporium.53 As shown in Figure 4A, mSeek appears to cross the vegetative portion of the B. thuringiensis bacterium to associate with developing spores and crystal toxin proteins. The plasma membrane is permeable during late stage sporulation prior to spore expulsion which likely permits mSeek uptake.54 The crystal toxin contains high fractions of anionic aspartate and glutamate residues that attract the cationic ZnDPA unit.55 The strong spore staining provided a low detection sensitivity of ~100 CFU/ml using the CCD experimental setup. Spores appear to possess more anionic surfaces charges compared to vegetative bacteria strains which could explain the enhanced accumulation.56, 57 Despite the strong targeting, the B. thuringiensis spores were much more resistant to mDestroy PDI than the tested vegetative strains. The literature on spore resistance to PDI is mixed with reports citing various degrees of sporicidal effect.58–60 Complete spore inactivation requires damage to the bacterial DNA or disruption of the inner membrane which is vital for the spore to undergo vegetative cell transition.61 The DNA and inner membrane within the spore core are surrounded by layers of lipids and proteinaceous architecture that inhibit PS penetration. It is likely that the mDestroy molecules do not penetrate deep enough to deliver 1O2 to the core components. This hypothesis is supported by the fluorescence micrographs of spores stained with mSeek which showed essentially no core penetration (Figure 3). It is worth noting that without the valuable imaging information provided by mSeek we would have little idea why the PDI experiments with mDestroy was ineffective. One idea for future efforts to enhance PDI using mDestroy is to pretreat the spores with protease enzymes or germinates that degrade the exosporium. Enzymes such as lysozyme and Spore-cortex-lytic enzyme B are known to breakdown the spore coat.62 Germinates such as L-alanine and inosine initiate exosporium shedding and conversion to vegetative cells which could improve PS penetration.63
From a practical perspective, the green wavelengths of the BODIPY chromophores in mSeek and mDestroy are appropriate for superficial applications such as surface detection and decontamination. It is possible that these ZnDPA targeted probes can be applied to other microbial species, since an anionic membrane is a common biomarker for multiple disease-causing pathogens.64, 65 Deep tissue therapeutic applications may require modified probe structures with red-shifted chromophores that enable optical imaging and PDI using longer wavelength light that penetrates further through biological matrices.
Conclusions
Two ZnDPA-BODIPY conjugates were prepared in good yield and shown to selectively target bacteria over mammalian cells. Fluorescence studies proved that the highly fluorescent and non-phototoxic mSeek stains multiple strains of bacteria, enabling sensitive fluorescence microscopy and detection using a CCD camera. The bacteria were inactivated effectively using the complementary photosensitizer, mDestroy. As a synergistic combination, the two probes offer a new strategy for optimized detection and PDI of bacterial infection/contamination.
Supplementary Material
Acknowledgements
We are grateful for funding support from the Defense Threat Reduction Agency (HDTRA1-13-1-0016 to B.D.S.) and the NIH (R01GM059078 to B.D.S. and T32GM075762 to D.R.R.). We thank M. Leevy and S. Chapman in the Notre Dame Integrated Imaging Facility (NDIIF) for technical assistance with the imaging systems.
Abbreviations
- PDI
photodynamic inactivation
- PS
photosensitizer
- 1O2
singlet oxygen
- ZnDPA
Zinc(II)-dipicolylamine
Footnotes
Electronic supplementary information available.
References
- 1.Spellberg B, Guidos R, Gilbert D, Bradley J, Boucher HW, Scheld WM, Bartlett JG, Edwards J., Jr The epidemic of antibiotic-resistant infections: a call to action for the medical community from the Infectious Diseases Society of America. Clin.lnfect. Dis. 2008;46:155–164. doi: 10.1086/524891. [DOI] [PubMed] [Google Scholar]
- 2.Sydnor ER, Perl TM. Hospital epidemiology and infection control in acute-care settings. Clin. Microbiol. Rev. 2011;24:141–173. doi: 10.1128/CMR.00027-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sperandio FF, Huang YY, Hamblin MR. Antimicrobial photodynamic therapy to kill Gram-negative bacteria. Recent Pat. Antiinfect. Drug Discov. 2013;8:108–120. doi: 10.2174/1574891x113089990012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wardlaw JL, Sullivan TJ, Lux CN, Austin FW. Photodynamic therapy against common bacteria causing wound and skin infections. Vet. J. 2012;192:374–377. doi: 10.1016/j.tvjl.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 5.Konopka K, Goslinski T. Photodynamic therapy in dentistry. J. Dent. Res. 2007;86:694–707. doi: 10.1177/154405910708600803. [DOI] [PubMed] [Google Scholar]
- 6.Noimark S, Dunnill CW, Parkin IP. Shining light on materials-a self-sterilising revolution. Adv. Drug Deliv. Rev. 2013;65:570–580. doi: 10.1016/j.addr.2012.07.003. [DOI] [PubMed] [Google Scholar]
- 7.Maisch T, Hackbarth S, Regensburger J, Felgentrager A, Baumler W, Landthaler M, Roder B. Photodynamic inactivation of multi-resistant bacteria (PIB) - a new approach to treat superficial infections in the 21st century. J. Dtsch. Dermatol. Ges. 2011;9:360–366. doi: 10.1111/j.1610-0387.2010.07577.x. [DOI] [PubMed] [Google Scholar]
- 8.Lauro FM, Pretto P, Covolo L, Jori G, Bertoloni G. Photoinactivation of bacterial strains involved in periodontal diseases sensitized by porphycene-polylysine conjugates. Photochem. Photobiol. Sci. 2002;1:468–470. doi: 10.1039/b200977c. [DOI] [PubMed] [Google Scholar]
- 9.Komerik N, Wilson M, Poole S. The effect of photodynamic action on two virulence factors of gram-negative bacteria. Photochem. Photobiol. 2000;72:676–680. doi: 10.1562/0031-8655(2000)072<0676:teopao>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 10.Dai T, Huang YY, Hamblin MR. Photodynamic therapy for localized infections--state of the art. Photodiagnosis Photodyn. Ther. 2009;6:170–188. doi: 10.1016/j.pdpdt.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lim Z, Cheng JL, Lim TW, Teo EG, Wong J, George S, Kishen A. Light activated disinfection: an alternative endodontic disinfection strategy. Aust. Dent. J. 2009;54:108–114. doi: 10.1111/j.1834-7819.2009.01102.x. [DOI] [PubMed] [Google Scholar]
- 12.Johnson GA, Muthukrishnan N, Pellois JP. Photoinactivation of Gram positive and Gram negative bacteria with the antimicrobial peptide (KLAKLAK)(2) conjugated to the hydrophilic photosensitizer eosin Y. Bioconjugate Chem. 2013;24:114–123. doi: 10.1021/bc3005254. [DOI] [PubMed] [Google Scholar]
- 13.Liu F, Soh Yan Ni A, Lim Y, Mohanram H, Bhattacharjya S, Xing B. Lipopolysaccharide neutralizing peptide-porphyrin conjugates for effective photoinactivation and intracellular imaging of Gram-negative bacteria strains. Bioconjugate Chem. 2012;23:1639–1647. doi: 10.1021/bc300203d. [DOI] [PubMed] [Google Scholar]
- 14.Bhatti M, MacRobert A, Henderson B, Shepherd P, Cridland J, Wilson M. Antibody-targeted lethal photosensitization of Porphyromonas gingivalis. Antimicrob. Agents Chemother. 2000;44:2615–2618. doi: 10.1128/aac.44.10.2615-2618.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Embleton ML, Nair SP, Cookson BD, Wilson M. Selective lethal photosensitization of methicillin-resistant Staphylococcus aureus using an IgG-tin(IV) chlorin e6 conjugate. J. Antimicrob. Chemother. 2002;50:857–864. doi: 10.1093/jac/dkf209. [DOI] [PubMed] [Google Scholar]
- 16.Embleton ML, Nair SP, Cookson BD, Wilson M. Antibody-directed photodynamic therapy of methicillin resistant Staphylococcus aureus. Microb. Drug Resist. 2004;10:92–97. doi: 10.1089/1076629041310000. [DOI] [PubMed] [Google Scholar]
- 17.Embleton ML, Nair SP, Heywood W, Menon DC, Cookson BD, Wilson M. Development of a novel targeting system for lethal photosensitization of antibiotic-resistant strains of Staphylococcus aureus. Antimicrob. Agents Chemother. 2005;49:3690–3696. doi: 10.1128/AAC.49.9.3690-3696.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soukos NS, Ximenez-Fyvie LA, Hamblin MR, Socransky SS, Hasan T. Targeted antimicrobial photochemotherapy. Antimicrob. Agents Chemother. 1998;42:2595–2601. doi: 10.1128/aac.42.10.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tegos GP, Anbe M, Yang C, Demidova TN, Satti M, Mroz P, Janjua S, Gad F, Hamblin MR. Protease-stable polycationic photosensitizer conjugates between polyethyleneimine and chlorin(e6) for broad-spectrum antimicrobial photoinactivation. Antimicrob. Agents Chemother. 2006;50:1402–1410. doi: 10.1128/AAC.50.4.1402-1410.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu CL, Yang QO, Liu LB, Lv FT, Li SY, Yang GQ, Wang S. Multifunctional cationic poly(p-phenylene vinylene) polyelectrolytes for selective recognition, imaging, and killing of bacteria over mammalian cells. Adv. Mater. 2011;23:4805–4810. doi: 10.1002/adma.201102850. [DOI] [PubMed] [Google Scholar]
- 21.O'Neil EJ, Smith BD. Anion recognition using dimetallic coordination complexes. Coord. Chem. Rev. 2006;250:3068–3080. [Google Scholar]
- 22.Hancock RE. Alterations in outer membrane permeability. Annu. Rev. Microbiol. 1984;38:237–264. doi: 10.1146/annurev.mi.38.100184.001321. [DOI] [PubMed] [Google Scholar]
- 23.Leevy WM, Johnson JR, Lakshmi C, Morris J, Marquez M, Smith BD. Selective recognition of bacterial membranes by zinc(II)-coordination complexes. Chem. Commun. 2006:1595–1597. doi: 10.1039/b517519d. [DOI] [PubMed] [Google Scholar]
- 24.Rice DR, Plaunt AJ, Turkyilmaz S, Smith M, Wang Y, Rusckowski M, Smith BD. Evaluation of 111[In]-labeled zinc-dipicolylamine tracers for SPECT imaging of bacterial infection. Mol. Imaging Biol. 2014:1–10. doi: 10.1007/s11307-014-0758-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.White AG, Fu N, Leevy WM, Lee JJ, Blasco MA, Smith BD. Optical imaging of bacterial infection in living mice using deep-red fluorescent squaraine rotaxane probes. Bioconjugate Chem. 2010;21:1297–1304. doi: 10.1021/bc1000998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.White AG, Gray BD, Pak KY, Smith BD. Deep-red fluorescent imaging probe for bacteria. Bioorg. Med. Chem. Lett. 2012;22:2833–2836. doi: 10.1016/j.bmcl.2012.02.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DiVittorio KM, Leevy WM, O'Neil EJ, Johnson JR, Vakulenko S, Morris JD, Rosek KD, Serazin N, Hilkert S, Hurley S, Marquez M, Smith BD. Zinc(II) coordination complexes as membrane-active fluorescent probes and antibiotics. ChemBioChem. 2008;9:286–293. doi: 10.1002/cbic.200700489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yogo T, Urano Y, Ishitsuka Y, Maniwa F, Nagano T. Highly efficient and photostable photosensitizer based on BODIPY chromophore. J. Am. Chem. Soc. 2005;127:12162–12163. doi: 10.1021/ja0528533. [DOI] [PubMed] [Google Scholar]
- 29.Wood TE, Thompson A. Advances in the chemistry of dipyrrins and their complexes. Chem. Rev. 2007;107:1831–1861. doi: 10.1021/cr050052c. [DOI] [PubMed] [Google Scholar]
- 30.Jang HG, Park M, Wishnok JS, Tannenbaum SR, Wogan GN. Hydroxyl-specific fluorescence labeling of ABP-deoxyguanosine, PhIP-deoxyguanosine, and AFB1-formamidopyrimidine with BODIPY-FL. Anal. Biochem. 2006;359:151–160. doi: 10.1016/j.ab.2006.08.030. [DOI] [PubMed] [Google Scholar]
- 31.Bonardi L, Kanaan H, Camerel F, Jolinat P, Retailleau P, Ziessel R. Fine-tuning of yellow or red photo- and electroluminescence of functional difluoro-boradiazaindacene films. Adv. Funct. Mater. 2008;18:401–413. [Google Scholar]
- 32.Awuah SG, Polreis J, Biradar V, You Y. Singlet oxygen generation by novel NIR BODIPY dyes. Org. Lett. 2011;13:3884–3887. doi: 10.1021/ol2014076. [DOI] [PubMed] [Google Scholar]
- 33.Diez-Barra E, Garcia-Martinez JC, Merino S, del Rey R, Rodriguez-Lopez J, Sanchez-Verdu P, Tejeda J. Synthesis, characterization, and optical response of dipolar and non-dipolar poly(phenylenevinylene) dendrimers. J. Org. Chem. 2001;66:5664–5670. doi: 10.1021/jo015764r. [DOI] [PubMed] [Google Scholar]
- 34.Yamaguchi S, Yoshimura I, Kohira T, Tamaru S, Hamachi I. Cooperation between artificial receptors and supramolecular hydrogels for sensing and discriminating phosphate derivatives. J. Am. Chem. Soc. 2005;127:11835–11841. doi: 10.1021/ja052838y. [DOI] [PubMed] [Google Scholar]
- 35.Ozlem S, Akkaya EU. Thinking outside the silicon box: molecular and logic as an additional layer of selectivity in singlet oxygen generation for photodynamic therapy. J. Am. Chem. Soc. 2009;131:48–49. doi: 10.1021/ja808389t. [DOI] [PubMed] [Google Scholar]
- 36.Nepomnyashchii AB, Pistner AJ, Bard AJ, Rosenthal J. Synthesis, photophysics, electrochemistry and electrogenerated chemiluminescence of PEG-modified BODIPY dyes in organic and aqueous solutions. J.Phys. Chem. C. 2013;117:5599–5609. doi: 10.1021/jp312166w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buhr TL, McPherson DC, Gutting BW. Analysis of broth-cultured Bacillus atrophaeus and Bacillus cereus spores. J. Appl. Microbiol. 2008;105:1604–1613. doi: 10.1111/j.1365-2672.2008.03899.x. [DOI] [PubMed] [Google Scholar]
- 38.Peck EM, Collins CG, Smith BD. Thiosquaraine rotaxanes: synthesis, dynamic structure, and oxygen photosensitization. Org. Lett. 2013;15:2762–2765. doi: 10.1021/ol401097f. [DOI] [PubMed] [Google Scholar]
- 39.Cakmak Y, Kolemen S, Duman S, Dede Y, Dolen Y, Kilic B, Kostereli Z, Yildirim LT, Dogan AL, Guc D, Akkaya EU. Designing excited states: theory-guided access to efficient photosensitizers for photodynamic action. Angew. Chem. Int. Ed. 2011;50:11937–11941. doi: 10.1002/anie.201105736. [DOI] [PubMed] [Google Scholar]
- 40.Dulin D, Le Gall A, Perronet K, Soler N, Fourmy D, Yoshizawa S, Bouyer P, Westbrook N. Reduced photobleaching of BODIPY-FL. Physcs Proc. 2010;3:1563–1567. [Google Scholar]
- 41.Swiecicka I, Bideshi DK, Federici BA. Novel isolate of Bacillus thuringiensis subsp. thuringiensis that produces a quasicuboidal crystal of Cry1Ab21 toxic to larvae of Trichoplusia ni. Appl. Environ. Microbiol. 2008;74:923–930. doi: 10.1128/AEM.01955-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Magge A, Setlow B, Cowan AE, Setlow P. Analysis of dye binding by and membrane potential in spores of Bacillus species. J. Appl. Microbiol. 2009;106:814–824. doi: 10.1111/j.1365-2672.2008.04048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moan J, Peng Q. An outline of the hundred-year history of PDT. Anticancer Res. 2003;23:3591–3600. [PubMed] [Google Scholar]
- 44.Hamblin MR, Hasan T. Photodynamic therapy: a new antimicrobial approach to infectious disease? Photochem. Photobiol. Sci. 2004;3:436–450. doi: 10.1039/b311900a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Valenzeno DP, Pooler JP. Cell-membrane photo-mondification - relative effects of halogenated fluorosciens for photohemolysis. Photochem. Photobiol. 1982;35:343–350. doi: 10.1111/j.1751-1097.1982.tb02572.x. [DOI] [PubMed] [Google Scholar]
- 46.Pooler JP. Photooxidation of cell-membranes using eosin derivatives that locate in lipid or protein to study the role of diffusible intermediates. Photochem. Photobiol. 1989;50:55–68. doi: 10.1111/j.1751-1097.1989.tb04129.x. [DOI] [PubMed] [Google Scholar]
- 47.Usacheva MN, Teichert MC, Biel MA. Comparison of the methylene blue and toluidine blue photobactericidal efficacy against Gram-positive and Gram-negative microorganisms. Lasers Surg. Med. 2001;29:165–173. doi: 10.1002/lsm.1105. [DOI] [PubMed] [Google Scholar]
- 48.Maisch T, Bosl C, Szeimies RM, Lehn N, Abels C. Photodynamic effects of novel XF porphyrin derivatives on prokaryotic and eukaryotic cells. Antimicrob. Agents Chemother. 2005;49:1542–1552. doi: 10.1128/AAC.49.4.1542-1552.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nitzan Y, Gutterman M, Malik Z, Ehrenberg B. Inactivation of Gram-negative bacteria by photosensitized porphyrins. Photochem. Photobiol. 1992;55:89–96. doi: 10.1111/j.1751-1097.1992.tb04213.x. [DOI] [PubMed] [Google Scholar]
- 50.Xu ZQ, Flavin MT, Flavin J. Combating multidrug-resistant Gram-negative bacterial infections. Expert Opin. Investig. Drugs. 2014;23:163–182. doi: 10.1517/13543784.2014.848853. [DOI] [PubMed] [Google Scholar]
- 51.Leggett MJ, McDonnell G, Denyer SP, Setlow P, Maillard JY. Bacterial spore structures and their protective role in biocide resistance. J. Appl. Microbiol. 2012;113:485–498. doi: 10.1111/j.1365-2672.2012.05336.x. [DOI] [PubMed] [Google Scholar]
- 52.Tufts JA, Calfee MW, Lee SD, Ryan SP. Bacillus thuringiensis as a surrogate for Bacillus anthracis in aerosol research. World. J. Microbiol. Biotechnol. 2014;30:1453–1461. doi: 10.1007/s11274-013-1576-x. [DOI] [PubMed] [Google Scholar]
- 53.Ghosal S, Leighton TJ, Wheeler KE, Hutcheon ID, Weber PK. Spatially resolved characterization of water and ion incorporation in Bacillus spores. Appl. Environ. Microbiol. 2010;76:3275–3282. doi: 10.1128/AEM.02485-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Errington J. Bacillus subtilis sporulation: regulation of gene expression and control of morphogenesis. Microbiol. Rev. 1993;57:1–33. doi: 10.1128/mr.57.1.1-33.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Senthil Kumar B, Ralte Z, Passari AK, Mishra VK, Chutia BM, Singh BP, Guruswami G, Nachimuthu SK. Characterization of Bacillus thuringiensis Cry1 class proteins in relation to their insecticidal action. Interdiscip. Sci. 2013;5:127–135. doi: 10.1007/s12539-013-0160-2. [DOI] [PubMed] [Google Scholar]
- 56.Pesce G, Rusciano G, Sasso A, Isticato R, Sirec T, Ricca E. Surface charge and hydrodynamic coefficient measurements of Bacillus subtilis spore by optical tweezers. Colloid. Surface. B. 2014;116:568–575. doi: 10.1016/j.colsurfb.2014.01.039. [DOI] [PubMed] [Google Scholar]
- 57.Terada A, Yuasa A, Kushimoto T, Tsuneda S, Katakai A, Tamada M. Bacterial adhesion to and viability on positively charged polymer surfaces. Microbiology. 2006;152:3575–3583. doi: 10.1099/mic.0.28881-0. [DOI] [PubMed] [Google Scholar]
- 58.Schafer M, Schmitz C, Facius R, Horneck G, Milow B, Funken KH, Ortner J. Systematic study of parameters influencing the action of rose bengal with visible light on bacterial cells: comparison between the biological effect and singlet-oxygen production. Photochem. Photobiol. 2000;71:514–523. doi: 10.1562/0031-8655(2000)071<0514:ssopit>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 59.Demidova TN, Hamblin MR. Photodynamic inactivation of Bacillus spores, mediated by phenothiazinium dyes. Appl. Environ. Microbiol. 2005;71:6918–6925. doi: 10.1128/AEM.71.11.6918-6925.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Oliveira A, Almeida A, Carvalho CM, Tome JP, Faustino MA, Neves MG, Tome AC, Cavaleiro JA, Cunha A. Porphyrin derivatives as photosensitizers for the inactivation of Bacillus cereus endospores. J. Appl. Microbiol. 2009;106:1986–1995. doi: 10.1111/j.1365-2672.2009.04168.x. [DOI] [PubMed] [Google Scholar]
- 61.Setlow P. Spores of Bacillus subtilis: their resistance to and killing by radiation, heat and chemicals. J. Appl. Microbiol. 2006;101:514–525. doi: 10.1111/j.1365-2672.2005.02736.x. [DOI] [PubMed] [Google Scholar]
- 62.Chirakkal H, O'Rourke M, Atrih A, Foster SJ, Moir A. Analysis of spore cortex lytic enzymes and related proteins in Bacillus subtilis endospore germination. Microbiology. 2002;148:2383–2392. doi: 10.1099/00221287-148-8-2383. [DOI] [PubMed] [Google Scholar]
- 63.Pinzon-Arango PA, Nagarajan R, Camesano TA. Effects of L-alanine and inosine germinants on the elasticity of Bacillus anthracis spores. Langmuir. 2010;26:6535–6541. doi: 10.1021/la904071y. [DOI] [PubMed] [Google Scholar]
- 64.Moller-Tank S, Maury W. Phosphatidylserine receptors: enhancers of enveloped virus entry and infection. Virology. 2014;468–470:565–580. doi: 10.1016/j.virol.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wanderley JL, Thorpe PE, Barcinski MA, Soong L. Phosphatidylserine exposure on the surface of Leishmania amazonensis amastigotes modulates in vivo infection and dendritic cell function. Parasite Immunol. 2013;35:109–119. doi: 10.1111/pim.12019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Magde D, Wong R, Seybold PG. Fluorescence quantum yields and their relation to lifetimes of rhodamine 6G and fluorescein in nine solvents: improved absolute standards for quantum yields. Photochem. Photobiol. 2002;75:327–334. doi: 10.1562/0031-8655(2002)075<0327:fqyatr>2.0.co;2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.