SUMMARY
Toll-like receptors (TLRs) activate distinct, yet overlapping sets of signaling molecules, leading to inflammatory responses to pathogens. Toll-IL-1R (TIR) domains, present in all TLRs and TLR adapters, mediate protein interactions downstream of activated TLRs. A peptide library derived from TLR2 TIR was screened for inhibition of TLR2 signaling. Cell-permeable peptides derived from the D helix and the segment immediately N terminal to TLR2 TIR domain potently inhibited TLR2-mediated cytokine production. The D helix peptide, 2R9, also potently inhibited TLR4, TLR7, and TLR9, but not TLR3 or TNF-α signaling. Cell imaging, co-immunoprecipitation, and in vitro studies demonstrated that 2R9 preferentially targets TIRAP. 2R9 diminished systemic cytokine responses elicited in vivo by synthetic TLR2 and TLR7 agonists; it inhibited activation of macrophages infected with influenza strain A/PR/8/34 (PR8) and significantly improved survival of PR8-infected mice. Thus, 2R9 represents a TLR-targeting agent that blocks protein interactions downstream of activated TLRs.
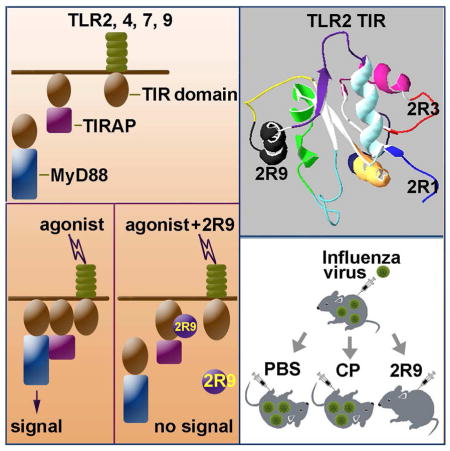
Graphical abstract
INTRODUCTION
Toll-like receptors (TLR) recognize exogenous microbial and endogenous damage-associated molecules to activate inflammatory responses critical for host recovery from infection or sterile tissue injury (Chen and Nunez, 2010; Kawai and Akira, 2011). Ligand interaction with TLR ectodomains induces dimerization of cytoplasmic Toll-Interleukin-1R (TIR) domains of two receptor molecules (Jin and Lee, 2008). This creates composite binding sites, to which adapter proteins are recruited through TIR domains present in each TLR adapter, leading to activation of several signaling cascades (Gay et al., 2014). In mammals, TIR domains are present in all TLRs, IL-1R family members and the adapters that transduce signals from these receptors (Gay and Keith, 1991; Medzhitov et al., 1997; O’Neill and Bowie, 2007). TIR domains are protein interaction domains that mediate transient interactions of signaling proteins. TIR domains tend to interact with other TIR domains; nevertheless, no common TIR:TIR binding motif has been identified (Pawson and Nash, 2003; Toshchakov and Vogel, 2007). TLRs and the proteins that transduce TLR signals are important therapeutic targets because excessive or prolonged TLR activation underlies many chronic inflammatory diseases and may be lethal (Brandes et al., 2013; Kawai and Akira, 2011; O’Neill et al., 2009).
TLR2 is activated by many ligands specific for Gram-positive bacteria, mycobacteria, or fungi (Means et al., 1999; Takeuchi et al., 1999; Underhill et al., 1999; Werts et al., 2001). TLR2 functions as a heterodimer with TLR1 or TLR6 and activates the MyD88-dependent signaling leading to activation of NF-κB and production of proinflammatory cytokines (Ozinsky et al., 2000). TLR2 utilizes two TIR domain-containing adapters: MyD88 and TIRAP, also called Mal. MyD88 is an adapter common to all human TIR-containing receptors, except TLR3, while TIRAP participates in fewer pathways (Kawai and Akira, 2010; Medzhitov et al., 1998). Early experiments demonstrated that cells obtained from TIRAP-deficient mice are hyporesponsive to TLR4 and TLR2 ligands, while capable of mounting a potent response to TLR3, TLR5, TLR7, and TLR9 agonists (Horng et al., 2002; Yamamoto et al., 2002). Later studies elaborated that the responsiveness of TIRAP-deficient cells to TLR2 agonists could be partly restored by prolonged exposure to a pathogen or increased agonist concentration (Cole et al., 2010; Kenny et al., 2009). Another study has found that, analogously to its role in TLR2 signaling, TIRAP facilitates TLR9 signaling because the TLR9 response is severely diminished by a targeted mutation of TIRAP gene in some cell types, as is the response to several viral pathogens (Bonham et al., 2014).
Although the general mechanism by which TLR activation induces formation of cytoplasmic signaling complexes has been determined, the specific location of sites that mediate the interactions of TIR-containing proteins in a functional signaling complex, as well as the composition and stoichiometry of components in the immediate receptor complexes, is still debated ((Gay et al., 2014) for a recent review; (Piao et al., 2013a) and (Enokizono et al., 2013) for an example of debate). Previously, we screened several libraries of peptides derived from putative TIR:TIR interaction sites of TLR4 and TIR-containing TLR4 adapters and identified several as potent TLR inhibitors that competitively block TIR:TIR interactions required for signal transduction (Couture et al., 2012; Piao et al., 2013a; Piao et al., 2013b; Toshchakov et al., 2011).
The present study extends our prior work by identification of a TLR2-derived peptide, 2R9, which is a potent, multispecific TLR inhibitor that inhibits cytokine activation elicited by TLR2, TLR4, TLR7, and TLR9 agonists. A cell-based FRET assay and co-immunoprecipitation studies demonstrated that 2R9 primarily targets TIRAP; however, a weak binding of 2R9 to TLR4 and TLR7 was also detected. Analysis of 2R9/TIRAP interaction using recombinant TIRAP has revealed that 2R9 binds TIRAP with nanomolar affinity due to a fast association rate coupled with slow dissociation of the complex. 2R9 binds albumin with micromolar affinity. 2R9 potently inhibits cytokine production by cultured macrophages infected with the mouse-adapted influenza virus strain A/PR/8/34 (PR8) and protects mice from lethal PR8 challenge.
RESULTS
Screening of TLR2 peptide library
TLR2 activation leads to a direct interaction of TLR2 TIR domain with the TIR domain of TLR1 or TLR6, followed by recruitment of TIRAP and MyD88, to the dimer, through TIR domains of these adapters. Therefore, TLR2 TIR participates in several TIR:TIR interactions, each of which might be competitively blocked by a peptide derived from a TLR2 TIR interface, leading to diminished signaling (Toshchakov and Vogel, 2007). To identify TLR2 inhibitors, we designed a library of cell-permeable TLR2-derived peptides and tested individual peptides for the ability to block TLR2-mediated signaling in primary mouse macrophages. All decoy peptide sequences were synthesized fused to the translocating segment of Antennapedia homeodomain (RQIKIWFQNRRMKWKK) at the N terminus (Derossi et al., 1994). Eleven peptides, named 2R1 – 2R11, each of which represents a separate patch of TLR2 TIR surface, were examined in the primary screening (Figure S1).
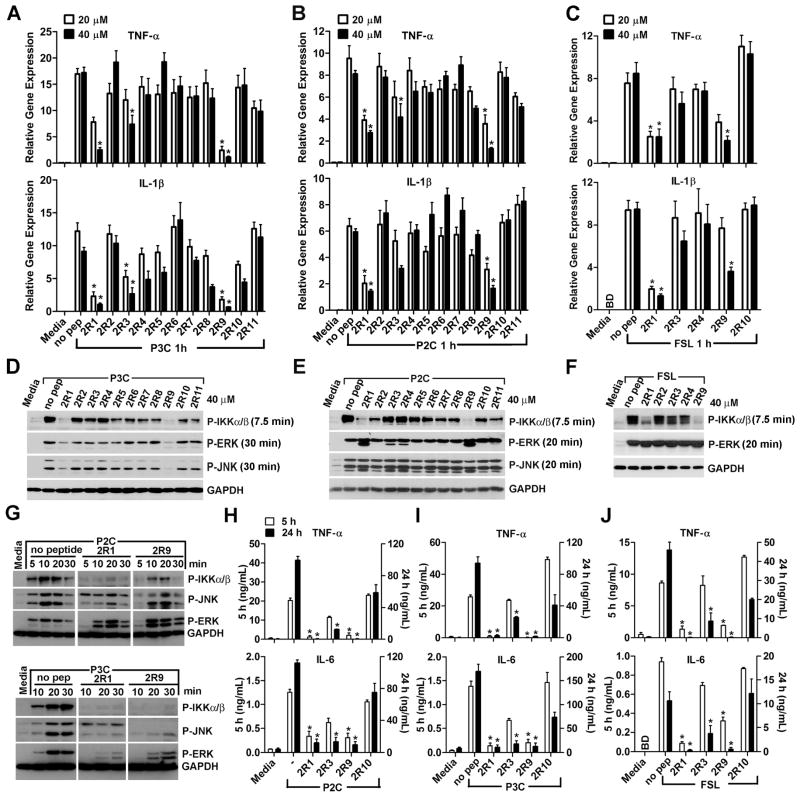
We first tested if the peptides inhibit cytokine mRNA transcription induced by S-[2,3- bis(palmitoyloxy)-(2-RS)-propyl]-N-palmitoyl-(R)-Cys-Ser-Lys4-OH (P3C), a synthetic lipopeptide that activates the TLR2/TLR1 heterodimer (Ozinsky et al., 2000), and by S-[2,3-bis- (palmitoyloxy)-(2-RS)-propyl]-[R]-Cys-Ser-Lys4-OH (P2C) or Fibroblast-Stimulating Lipopeptide-1 (FSL-1), two lipopeptides recognized by the TLR2/TLR6 heterodimer (Takeuchi et al., 2001). Two peptides, 2R1 and 2R9, potently inhibited TNF-α and IL-1β mRNA expression 1 h after macrophage stimulation with P3C, while the third peptide, 2R3, inhibited suboptimally (Figure 1A). 2R1 and 2R9 also potently inhibited mRNA induced by TLR2/TLR6 agonists, P2C or FSL-1 (Figure 1B–C). Although 2R4 did not inhibit TLR2 significantly (Figure 1A–F), we confirmed our previous data (Toshchakov et al., 2007) that 2BB, a peptide that represents the extended BB loop of TLR2 and includes all 2R4 residues, inhibits the P3C-induced cytokine production in macrophages (Figure S2).
Figure 1. Peptides 2R1 and 2R9 inhibit TLR2-mediated macrophage activation.
(A–C) Effect of TLR2 decoy peptides on cytokine mRNA expression. Mouse peritoneal macrophages were treated with 20 or 40 μM of indicated peptides for 30 min prior to stimulation with P3C (500 ng/ml) (A), P2C (50 ng/ml) (B), or FSL-1 (50 ng/ml) (C). Cytokine mRNA was measured 1 h after cell stimulation and is normalized to the expression of HPRT.
(D–G) IKKα/β and MAPK phosphorylation was measured by western blot in lysates of macrophages stimulated with a TLR2 agonist for indicated duration.
(H–J) TNF-α and IL-6 contents in macrophage supernatants 5 (open columns, scaled by left Y axes) or 24 h (black columns, right Y axes) after P3C (I), P2C (H), or FSL-1 (J) stimulation of mock- or peptide-treated macrophages. Peptides (40 μM) were added 30 minutes before TLR2 stimulation. BD, below detection limit. Data in each panel represent at least three independent experiments. See also Figures S1-3.
In accordance with strong effects of 2R1 and 2R9 on cytokine expression, both peptides potently inhibited phosphorylation of IKK-α/β, kinases of the NF-κB activation pathway, induced by all TLR2 agonists (Figure 1D–G). The inhibitory effect on MAPK activation, however, was limited to TLR2/1 signaling: 2R1 and 2R9 inhibited ERK and JNK induced by P3C, a TLR2/TLR1 agonist, but failed to inhibit activation of MAPKs induced by P2C or FSL-1, thus suggesting a distinct mechanism of MAPK activation via TLR2/1 and TLR2/6 (Figure 1D–G). 2R1 and 2R9 potently inhibited TNF-α and IL-6 secretion measured in macrophage supernatants 5 or 24 h after stimulation by all TLR2 agonists, indicating that peptides exert a long-lasting inhibitory effect (Figure 1H–J). To determine effect of peptides on cell viability, we conducted 3-[4,5-dimethylthiazol-2-yl]-2,5 diphenyltetrazolium bromide (MTT) incorporation assay. Macrophage viability was consistently greater than 80% for all peptides, at all conditions, and in particular, 2R1 and 2R9 exerted a minimal effect on cell viability that was comparable to the effect on cell viability caused by P3C stimulation alone (Figure S3). The negative effect of TLR2 activation on cell viability, particularly by P3C, has long been reported and is attributed to pro-apoptotic properties of TLR2 agonists (Aliprantis et al., 1999).
We examined if inhibitory peptides derived from mouse TLR2 inhibit human TLR2 and found that effects of both peptides are highly similar in human and mouse macrophages (Figure S4). Thus, we identified two TLR2 inhibitory peptides that block TLR2-mediated responses in vitro by 80–90% in human and mouse cells, with minimal effects on cell viability.
Specificity of signaling inhibition by TLR2-derived peptides
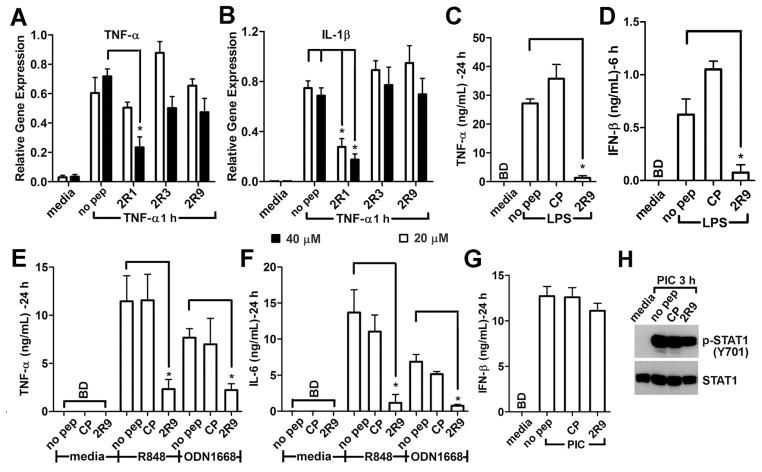
TLR2-derived peptides that inhibited TLR2 were next examined for inhibition of signaling induced by TNF-α, TLR4, TLR7, TLR9, and TLR3. 2R1, but not 2R9, inhibited TNF-α and IL-1β mRNA expression induced by TNF-α (Figure 2A, B), suggesting that 2R1 may target a protein that does not have a TIR domain. Peptides 2R9 and 2R1, but not Control Peptide (CP), also potently decreased TLR4-induced TNF-α and IL-1β expression (data not shown) and secretion (Figure 2C, D). R848, an imidazoquinoline compound that activates TLR7 (Diebold et al., 2004; Heil et al., 2004; Lund et al., 2004), and ODN1668, a class B CpG oligonucleotide that activates murine TLR9, induced robust production of TNF-α and IL-6 by cultured murine macrophages (Figure 2E, F). Treatment of cells with 2R9 at 20 μM, the lower effective dose used in Figure 1, decreased TLR7 and TLR9-activated cytokine production by 75 – 80% (Figure 2E, F). 2R9 did not affect the IFN-β secretion and STAT-1 activation induced by the TLR3 agonist, poly(I:C) (Figure 2G–H). Thus, the inhibitory effects of 2R1 are non-specific in contrast to 2R9 that does not block TNF-α- or TLR3-mediated signaling, but blocks TLR2, TLR4, TLR7, and TLR9.
Figure 2. Specificity of TLR inhibition by TLR2-derived peptides.
(A–B) Effect of TLR2 peptides on TNF-induced cytokine expression. Mouse peritoneal macrophages were treated with 40 μM (black bars) or 20 μM (open bars) of indicated peptides for 30 min prior to stimulation with TNF-α (10 ng/ml). Cells were lysed 1 h after stimulation.
(C–G) Effect of peptides on cytokine secretion induced in primary macrophages by LPS (100 ng/ml) (C and D), R848 (2.85 μM) and ODN1668 (1 μM) (E, F), or poly (I:C) (PIC) (50 μg/ml) (G).
(H) Macrophage extracts were analyzed for STAT1-Y701 phosphorylation by western blot.
CP, Control Peptide. See also Figure S4.
2R9 binds TIRAP and prevents adapter recruitment to TLR2 signaling complex
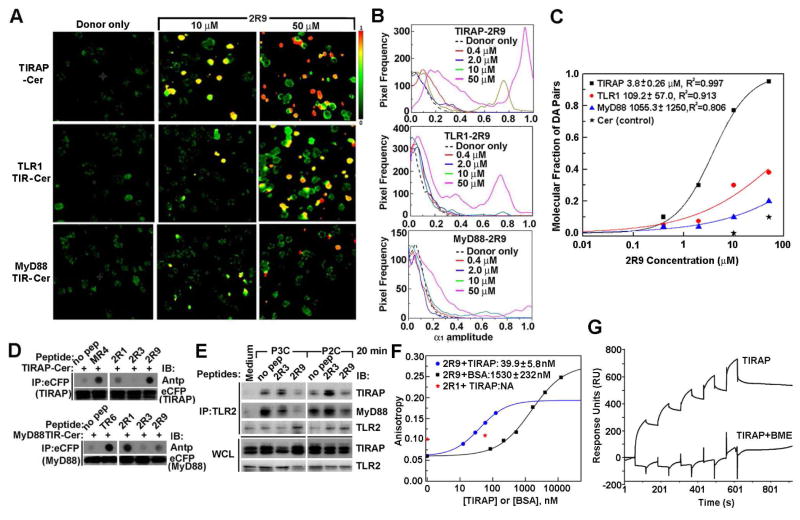
The specificity profile exhibited by 2R9 could either be a consequence of targeting several distinct TIR domains or result from targeting one TIR-containing adapter, common to several TLRs. To identify TIR domains targeted by 2R9, we used two approaches. The first is that a panel of fluorescently labeled TIR-containing proteins, each a candidate binding partner for 2R9, is expressed in a host cell line, and the resulting cell lines are treated with a decoy peptide labeled with a fluorescent dye that can serve as a FRET acceptor for the TIR label. If a peptide binds the TIR domain, it results in FRET and an associated decrease in donor fluorescence lifetime. Cerulean- (Cer-) labeled TIR domains and Bodipy-TMR-X- (BTX-) labeled decoy peptides were used as a FRET pair to study interactions of decoy peptides with TIR domains (Toshchakov et al., 2011). We recently developed a Fluorescence Lifetime IMaging- (FLIM-) based approach that enables quantitative comparisons of TIR-peptide binding affinities directly in cells (Szmacinski et al., 2014). This approach was used to analyze new FLIM images. The second approach that was developed to analyze TIR-peptide binding is the dot blot immunoprecipitation assay, in which labeled TIR domains are expressed in cells and then immunoprecipitated from cell lysates spiked with decoy peptides. In this approach, the level of peptide-TIR binding is estimated by quantifying peptides in immunoprecipitates (Piao et al., 2013a).
Upon TLR2 activation, TLR2 TIR dimerizes with the TIR domain of TLR1 or TLR6 and the dimer recruits TIRAP and MyD88 through TIR domains of these adapters. Therefore, TLR1, MyD88, and TIRAP TIR domains were selected as the primary candidate targets of TLR2-derived decoy peptides, and mammalian expression vectors that encode these TIR domains fused at the C terminus with Cer were generated. Figures 3A–C present results of analysis of Cer fluorescence lifetime in images of HeLa cells transfected with Cer-fused, TIR-containing proteins and incubated in the presence of different concentrations of BTX-labeled 2R9 (BTX-2R9) for 1 h. Images were analyzed using the bi-exponential fluorescence decay model with the fixed components’ lifetimes of 2.96 ns (free donor) and 0.9 ns (donor-acceptor pair), determined as described previously (Szmacinski et al., 2014). Figure 3A demonstrates cell images in pseudocolor based on the fractional amplitude of the short lifetime component, α1; this parameter reflects the portion of acceptor-bound donor molecules (Szmacinski et al., 2014). Incubation of TIRAP-Cer-expressing cells with BTX-2R9 caused a significant, dose-dependent increase of α1, reflecting the higher molecular fraction of donor-acceptor pairs. This finding indicates a direct interaction between TIRAP and 2R9. The selective quenching of TIRAP-Cer fluorescence by BTX-2R9 is clearly visible in the α1 images of cells treated with 10 or 50 μM of 2R9 as shown by pseudocolor (Figure 3A) and by the appearance of distinct peaks at high α1 values in histograms of Figure 3B. Formation of donor-acceptor pairs in the presence of 2R9 was considerably less for the TLR1-expressing cells and less yet for the MyD88-expressing cells (Figure 3A–C). Importantly, Cer not fused with a TIR domain was not quenched to a comparable degree even by the highest concentration of BTX-2R9 used (Figure 3C), indicating that 2R9 indeed targets TIRAP in the TIRAP-Cer fusion protein.
Figure 3. 2R9 binding specificity.
(A) The fractional amplitude of the short lifetime component in FLIM images of cells transfected with TIRAP-Cer (upper row), TLR1-Cer (middle row), and MyD88 TIR-Cer (bottom row) and treated with 10 or 50 μM of BTX-2R9.
(B) Distribution of fractional amplitude of the short lifetime component in FLIM images of BTX-2R9-treated HeLa cells that express TIRAP-Cer, TLR1-Cer, or MyD88-Cer.
(C) Concentration dependence of the apparent molecular fraction of acceptor-bound TIR domains for TIRAP-Cer-, TLR1-Cer-, or MyD88-Cer-expressing HeLa cells. The molecular fractions were calculated from characteristic amplitudes of lifetime components in images of cells incubated with different acceptor concentrations.
(D) Peptide-TIR co-immunoprecipitation. Lysates of HEK293T cells that express indicated fusion proteins were incubated with 20 μM of indicated peptides for 1 h and immunoprecipitated with anti-eCFP Ab. Peptide contents in the immunoprecipitates were measured by the dot blot assay using Ab to Antennapedia translocating sequence (Antp). Peptides TR6, MR4, and 4BB were used as a positive binding control for MyD88, TIRAP, and TLR4 respectively. See also Figure S5.
(E) 2R9 decreases co-immunoprecipitation of activated TLR2 with TLR2 adapters. Primary peritoneal macrophages were treated with 40 μM of indicated peptide for 30 min prior to treatment with a TLR2 agonist. 500 μg of cell lysates were immunoprecipitated by anti-TLR2 Ab, and the immune complex was assessed using anti-TIRAP or anti-MyD88 Ab. Data represent 3 individual experiments.
(F) FP analysis of 2R9 protein binding. 2R9 binds TIRAP with high affinity. BTX-labeled peptides were dissolved in PBS at 50 nM and titrated with recombinant mouse TIRAP or bovine serum albumin (BSA).
(G) 2R9/TIRAP SPR single cycle sensorgrams. Mouse TIRAP at concentrations of 21, 43, 86, 178, and 344 μM was injected to flow cells with immobilized 2R9 or control Antennapedia peptide in the presence or absence of BME.
FLIM images of cells incubated at varied concentrations of 2R9 were used to estimate the apparent TIR-2R9 binding affinities in cellular milieu. The titration plots (Figure 3C) were constructed assuming that amplitude α1 corresponds to the molecular fraction of donor-acceptor pairs as α1= [DA]/([D]+[DA]). At each acceptor concentration, the α1 values in Figure 3C correspond to the predominant pixel frequency derived from several images using histograms such as that in Figure 3B. Fitting the experimental points with logistic function revealed that mid-points are at ~3.8, 109.2, and 1055.3 μM for TIRAP, TLR1, and MyD88, respectively. The >10-fold higher affinity of 2R9 binding to TIRAP compared to binding to TLR1 or MyD88 suggests that 2R9 selectively binds TIRAP. The apparent affinity of 2R9/TIRAP binding in cells is in the low micromolar range and thus corresponds to the effective 2R9 dose for signaling inhibition and suggests that 2R9/TIRAP binding may account for TLR inhibition by this peptide.
Co-immunoprecipitation studies confirmed the robust 2R9/TIRAP binding and indicated that 2R9 binds TIRAP more strongly than other TLR2-derived inhibitory peptides (Figure 3D). The binding of 2R9 to MyD88 TIR domain, however, was much weaker compared to that of 2R1 or TR6, a TIRAP C helix peptide, the strong binding of which to the MyD88 TIR domain was previously reported (Couture et al., 2012; Piao et al., 2013a). 2R9 binding to TLR1 and TLR6 was weaker than that for two other TLR2 peptides (Figure S5A–B); however, we noted considerable binding of 2R9 to TLR4 (Figure S5C). 2R9/TLR4 binding was only slightly less than that of 4BB, a TLR4-derived peptide that strongly binds TLR4 (Szmacinski et al., 2014; Toshchakov et al., 2011). Interestingly, regions of TIR surface represented by 2R1 and 2R3 peptides are juxtaposed (Figure S1B) and both peptides preferentially interact with TLR2 co-receptors, TLR1 and TLR6. These observations may indicate the TLR2 TIR interface that mediates interaction with co-receptors.
Dot blot analysis suggested a weak interaction of 2R9 with TLR4 TIR domain (Figure S5C); therefore, we sought to evaluate binding of 2R9 to TIR domains of other TLRs inhibited by 2R9 and generated expression vectors encoding Cer-labeled TLR4, TLR7, and TLR9 TIR domains. Analysis of 2R9 binding to these proteins was complicated by significant differences in cellular abundance of these proteins after ectopic expression in HeLa or HEK293T cells. Particularly, the TLR7-TIR-Cer abundance 2 days post-transfection was not sufficient for FLIM. Apparently, cells quickly eliminated the TLR7 TIR encoding vector because shortening of time between transfection and treatment of cells with 2R9-BTX from two to one day resulted in better Cer expression and fluorescent signals sufficient for FLIM (Figure S5D–F). Analysis of several independently obtained images suggested that there was detectable binding of 2R9 to TLR7 and TLR4 (Figure S5D, E), which was ~10 times weaker than 2R9-TIRAP binding (Figure S5F). We detected no binding between 2R9 and TLR9 TIR (Figure S5D–G).
In accordance with the strong effect of 2R9 on TLR2 signaling, 2R9 decreased agonist-induced recruitment of both TIRAP and MyD88 to TLR2 in co-immunoprecipitation assay (Figure 3E). Thus, our data suggest that 2R9 suppresses TLR signaling primarily through direct binding to TIRAP and a consequent indirect effect on MyD88 recruitment; however, 2R9 may also affect TLR4 and TLR7 signaling through weak binding to TIR domains of these receptors.
Affinity and kinetics of 2R9/TIRAP binding in a binary system
Having identified TIRAP as the main target of the 2R9 peptide, we next studied this interaction in vitro using recombinant mouse TIRAP. Two approaches were used. The first is the fluorescence polarization assay (FP). In this assay, a fluorescently labeled ligand, BTX-2R9, is incubated with TIRAP and the interaction between the two detected based on increased polarization of ligand fluorescence due to decreased rotational mobility of the protein-bound ligand. Figure 3F demonstrates that BTX-2R9 fluorescence polarization (shown as fluorescence anisotropy, r = 3P/(3-P)) indeed increases dose-dependently in the presence of recombinant TIRAP. The TIRAP-caused increase in anisotropy clearly indicates direct 2R9/TIRAP binding in solution. Fitting the observed dependence of BTX-2R9 anisotropy on TIRAP concentration to the four parameter logistic function suggested the apparent KD of 2R9/TIRAP interaction is ~40 nM. Incubation of BTX-labeled 2R1 in the presence of TIRAP did not change fluorescence anisotropy, indicating that 2R9/TIRAP interaction is specific (Figure 3F). Interestingly, the FP experiments detected binding of 2R9 to bovine albumin (BSA), with apparent KD in the low micromolar range (Figure 3F).
The 2R9/TIRAP interaction was studied further using the surface plasmon resonance (SPR) single cycle kinetics analysis. The 2R9 peptide was immobilized on the surface of the flow cell using amine coupling. As a control, the Antennapedia peptide that contained the cell-permeating sequence of 2R9, but not its decoy sequence, was immobilized in the control flow cell. The SPR analysis confirmed TIRAP/2R9 interaction (Figure 3G). The addition of reducing agent β-mercaptoethanol (BME) to the sample buffer abolished TIRAP binding to 2R9, indicating that TIRAP tertiary structure is important for this interaction (Figure 3G). A notable feature of the sensorgrams is that TIRAP binding to 2R9 has a fairly fast association rate compared to apparently slow dissociation.
Thus, both FP and SPR analyses suggest that 2R9 specifically binds TIRAP with high affinity. The FP experiments demonstrated 2R9 also binds albumin with low affinity.
TLR7 signaling is diminished in TIRAP-deficient cells
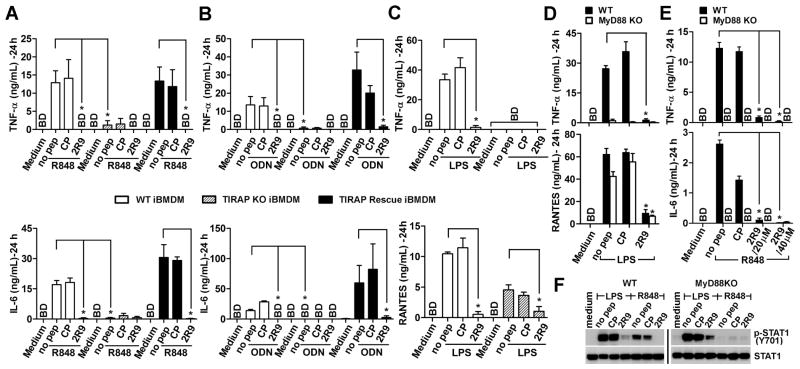
We discovered that 2R9 inhibits TLR2, TLR4, TLR7, and TLR9, but not TLR3 (Figure 2). This specificity pattern may suggest that 2R9 targets MyD88 because MyD88 is involved in each blocked pathway, whereas this adapter is dispensable for TLR3 signaling (Kawai and Akira, 2010). However, our binding studies demonstrated that 2R9 binds TIRAP, not MyD88 (Figure 3). The available literature only partly supports the notion that TIRAP is involved in the pathways inhibited by 2R9. TIRAP has long been known to be important for TLR4 and TLR2 signaling (Horng et al., 2002; Yamamoto et al., 2002). These studies, however, suggested that TIRAP is not involved in all MyD88-dependent TLR signaling because TIRAP-deficient mice and cells from TIRAP-deficient mice remained capable of responding through some TLRs, including TLR9 and TLR7. However, a more recent study specified that, although the response to TLR9 agonists can occur in the absence of TIRAP, TIRAP facilitates the MyD88-dependent response mediated by endosomal TLRs, especially in cells that are less endocytic, such as immortalized BMDM (iBMDM) (Bonham et al., 2014). This study also showed that TLR9 signaling and the response to viruses that activate TLR9 and TLR7 is diminished in TIRAP-deficient cells (Bonham et al., 2014). However, the authors did not study whether TLR7 signaling is TIRAP-dependent. We, therefore, studied the effects of 2R9 on TLR-driven activation of wild-type (WT) iBMDM, TIRAP-deficient iBMDMs (TIRAP KO iBMDM), and TIRAP KO iBMDM retrovirally transfected with TIRAP (TIRAP rescue iBMDM).
Figure 4A confirms findings of Bonham et al. that TLR9-induced secretion of TNF-α and IL-6 in iBMDM is TIRAP-dependent because TIRAP KO iBMDMs do not respond to ODN1668 (Figure 4A). This figure also demonstrates that 2R9 potently blocks TLR9-induced production of TNF-α and IL-6 in WT iBMDMs and in TIRAP rescue iBMDMs (Figure 4A). Figure 4B expands these findings to TLR7-mediated activation of iBMDMs.
Figure 4. TIRAP is required for full TLR7- and TLR9-mediated inflammatory response in iBMDM.
(A–C) Wild-type (WT) (open columns), TIRAP-deficient (TIRAP KO) (dashed columns), or TIRAP KO iBMDMs retrovirally transfected with TIRAP (TIRAP Rescue iBMDM) (black columns) were pretreated with a 20 μM peptide and stimulated with (A) ODN1668 (1 μM), (B) R848 (2.85 μM), or (C) LPS (100 ng/ml). TNF-α, IL-6, and RANTES were measured in 24 h supernatants.
(D, E) Cytokine secretion by thioglycollate-elicited peritoneal macrophages from WT (black columns) or MyD88-deficient (MyD88KO) mice (open columns) pretreated with 20 μM peptide and stimulated with LPS (100 ng/ml) (D) or R848 (2.85 μM) (E). Cytokine levels were measured in 24 h supernatants.
(F) STAT-1 phosphorylation in peritoneal macrophages from WT or MyD88KO mice. Cells were stimulated as described in legend for panel D and E and lysed 5 h after stimulation.
Unlike TLR2, TLR7, or TLR9, TLR4 activates two signaling pathways resulting in activation of a subset of genes in MyD88- and TIRAP-independent manner (Kawai and Akira, 2010). Surprisingly, the TLR4-driven activation of a MyD88- and TIRAP-independent cytokine, RANTES, was sensitive to 2R9 in iBMDM (Figure 4C) and in peritoneal macrophages (Figure 4D), albeit less sensitive than the genes that are completely MyD88- and TIRAP-dependent (Figure 4E). Confirming the latter observation, the MyD88-independent activation of STAT1, shown previously to be the consequence of LPS-induced IFN-β acting back on the macrophages through the IFNAR (Toshchakov et al., 2002), was also diminished in the presence of 2R9, albeit less potently than in WT macrophages (Figure 4E). These findings likely indicate that direct binding of 2R9 to TLR4 demonstrated by the dot blot assays and FLIM (Figure S5C–F) constitutes an additional mechanism of TLR4 inhibition by this peptide. Our data also confirm that unlike TLR4-mediated activation of STAT1, TLR7-mediated activation of STAT1 and TNF-α is completely MyD88-dependent (Figure 4E, F).
Thus, TIRAP is important for endosomal TLR signaling through TLR7 and TLR9, and, therefore, targeting of TIRAP may account for the observed pattern of TLR inhibition by 2R9. Yet, binding to additional TIR domains may also be a part of 2R9 functionality as it is likely the case for 2R9/TLR4 and 2R9/TLR7 binding. It is noteworthy that we previously observed that some highly inhibitory TLR-targeting decoy peptides demonstrate similar, ‘multispecific’ binding behavior. One peptide that binds several TIR domains is TF5, a TRIF-derived TLR4 inhibitor that binds TRAM strongly and TLR4 less strongly, while it does not bind MyD88 or TIRAP TIRs (Piao et al., 2013a).
2R9 blocks TLR2- and TLR7-mediated systemic cytokine response in mice
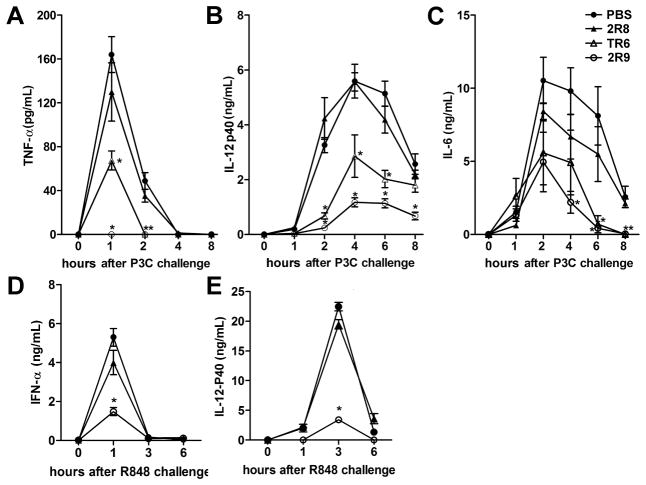
We next examined whether 2R9 is effective in vivo. C57BL/6J mice were pretreated i.p. with 2R9, or 2R8, a cell-permeating peptide that does not inhibit TLRs, or TR6, a TIRAP-derived peptide that inhibits TLR4 and TLR2 (Couture et al., 2012), and challenged with P3C (3.3 nmol/g). Peptides were administered at a dose of 10 nmol/g of mouse weight 1 h before P3C. TNF-α. IL-12 p40, and IL-6 were measured in plasma samples collected immediately before and after stimulation. 2R9 pretreatment decreased the P3C-induced systemic TNF-α levels below the limit of detection (Figure 5A). IL-12 p40 was also significantly decreased (Figure 5B). Although the 2R9-mediated IL-6 decrease was not statistically significant at the peak time point, i.e., 2 h after administration of P3C, the tendency was there and the P3C-induced IL-6 augmentation was significantly lower at 4, 6, and 8 h post-stimulation in the 2R9-treated group (Figure 5C). TR6, a TIRAP-derived peptide that binds MyD88 (Couture et al., 2012; Piao et al., 2013a), also decreased the P3C-induced cytokine levels; however, the effect of TR6 was less than that of 2R9 (Figure 5A–C).
Figure 5. Effect of inhibitory peptides on TLR2- and TLR7-elicited cytokine activation in mice.
C57BL/6J female mice were mock-treated with PBS or treated i.p. with 10 nmol/g of a peptide 1 h before i.p. administration of P3C (3.3 nmol/g) (A–C) or R-848 (2.5 nmol/g) (D–E). Plasma samples were obtained from 6 (A–C) or 4 (D–E) animals. See also Figure S6.
The effect of 2R8 and 2R9 on TLR7-induced systemic cytokines was also studied. 2R9, but not the inert peptide, significantly decreased cytokine induction by R848 (Figure 5D–E). These data establish 2R9 as a potent inhibitor of TLR2 and TLR7 in vivo.
2R9 suppresses PR8-induced macrophage activation and protects mice from lethal PR8 challenge
A broad TLR inhibitory specificity of a candidate therapeutic, such as that demonstrated by 2R9, may provide a distinct advantage clinically because such a compound may be expected to modulate TLR response to pathogens, which elicit inflammatory responses through multiple TLRs. Influenza virus is sensed by several innate receptors, including TLR7, TLR8, TLR10, and also by RIG-I, although TLR8 and TLR10 are not functional genes in mice (Diebold et al., 2004; Heil et al., 2004; Iwasaki and Pillai, 2014; Kawai and Akira, 2011; Lee et al., 2014; Lund et al., 2004). Secondary TLR4 activation by endogenous ligands associated with tissue damage also plays a significant role in influenza pathogenesis by promoting injurious inflammation (Shirey et al., 2013). Therefore, we hypothesized that 2R9 can modulate the innate immune response to influenza. Cultured mouse peritoneal macrophages were challenged in vitro with the mouse-adapted H1N1 influenza PR8 strain at a multiplicity of infection (MOI) = 1 for 2 h, then the virus was removed, and cells washed with PBS and mock-treated or treated with 2R9. Cytokine levels were measured in supernatants 24 h later. 2R9, but not a control peptide, potently inhibited PR8-induced secretion of TNF-α, IL-6, and IFN-β (Figure 6A–C).
Figure 6. 2R9 suppresses PR8-induced cytokine secretion by macrophages and improves survival in a mouse model of influenza.
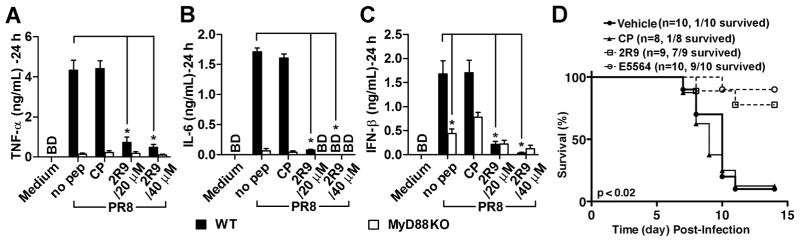
(A–C) Primary peritoneal macrophages obtained from C57BL/6J WT (black columns) or MyD88-deficient (open columns) female mice were kept in cell culture overnight and challenged with PR8 at 1 MOI for 2 hours. Cytokine content was measured in supernatants 24 hours post infection.
(D) Survival of mice infected with PR8. C57BL/6J female mice received intranasal inoculate of PR8 (~7500 TCID50; ~ LD90), on day “0”. Starting from day 2, the mice received 2R9, CP, Eritoran (E5564), or vehicle once daily for the course of 5 successive days. Peptides were administered i.p. at the dose 200 nmol. Eritoran was administered i.v. at the dose 200 μg (Shirey et al., 2013).
MyD88-deficient macrophages infected with PR8 induced TNF-α and IL-6 poorly (Figure 6A, B), indicating a critical contribution of TLRs to viral induction of these genes. In contrast, the PR8-induced production of IFN-β was less dependent on MyD88 (Figure 6C), reflecting an important contribution of RIG-I-like receptors (RLRs) to viral induction of type I IFNs reported for a variety of cell types (Kato et al., 2005). Accordingly, the MyD88-independent IFN-β induction was only slightly suppressed by 2R9 (Figure 6C). The residual sensitivity of IFN-β expression to 2R9 reflects complexity of immune response to live pathogens and also might be explained by indirect secondary activation of TLR4 by endogenous ligands produced in response to influenza virus (Imai et al., 2008; Shirey et al., 2013).
We next examined if 2R9 would improve survival of mice infected with an LD90 of PR8. Groups of PR8-infected C57BL/6J mice received a control cell-permeating peptide, or 2R9 (200 nmol/mouse), or vehicle i.p. once daily for 5 days, starting 48 h after PR8 infection. In the 2R9-treated group, 78% of mice survived, whereas only 12.5% and 10% survived in the CP and vehicle control groups, respectively (p < 0.02 for both control groups; Mantel-Cox test) (Figure 6D).
DISCUSSION
TLRs are important therapeutic targets because excessive or prolonged TLR activation leads to inflammation that can cause disease or lethality (O’Neill et al., 2009). Due to a more significant effort expended in the past on the development of TLR antagonists that antagonize the agonist binding sites of TLR ectodomains, the currently available pool of TLR inhibitors that block interactions of cytoplasmic proteins is less advanced (Connolly and O’Neill, 2012; Wang et al., 2013). Although targeting the transient protein interactions of signaling proteins is challenging (Smith and Gestwicki, 2012), this approach may be advantageous because some intracellular signaling molecules participate in several pathways or participate in a pathway in a stoichiometrically higher quantity. Thus, broader specificity and/or higher potency of signaling inhibition might be anticipated as a result of targeting a cytoplasmic signaling protein compared to agonist antagonism.
To develop TLR inhibitors that act intracellularly by blocking the protein interactions downstream of activated TLRs, we designed and screened a library of peptides derived from putative interaction sites of the TLR2 TIR domain, for inhibition of TLR2. The TLR2 library is the second library derived from a receptor TIR domain that we have examined. Originally, TLR4 peptides were designed analogously and screened for TLR4 inhibition (Toshchakov et al., 2011). The screening of the TLR2 library revealed two peptides that potently inhibit TLR2: 2R1 corresponds to the region that connects the TIR domain to transmembrane region, while 2R9 represents the D helix of the TIR. Regions represented by these inhibitory peptides are located at opposite sides of TIR domain (Figure 7A). Two other TLR2 peptides weakly suppressed TLR2, i.e., the AB loop peptide, 2R3, and the BB loop peptide (2BB), previously shown to inhibit TLR2 and TLR4 moderately (Toshchakov et al., 2007).
Figure 7. Position of regions represented by TLR2 peptides.
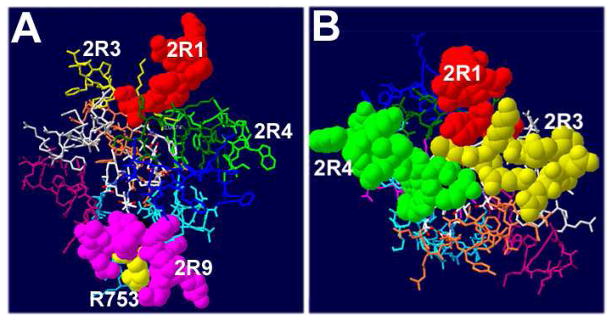
(A) D helix is located on the side opposite to 2R1. Polymorphic R753 is shown in yellow. Leu752 (black) has surface exposure less than 2%.
(B) Presumable dimerization interface of TLR2.
Interestingly, peptides derived from structurally homologous regions of TLR4 also inhibited signaling mediated by parent receptor (Toshchakov et al., 2011). The sequence similarity of TLR4 and TLR2 inhibitory peptides from structurally homologous regions is, however, rather limited. Thus, regions 1 in TLR4 and TLR2 have only 4 of 14 amino acids conserved, whereas regions 9 are completely dissimilar and have only one amino acid positioned similarly, which is the central leucine, a residue that is completely buried in the TLR2 TIR (Figure 7B). Peptides 3 and 4 (BB) are ~50% conserved in TLR2 and TLR4.
We previously reported that TLR4 peptides 4R1, 4R3, and 4BB inhibit TLR4, bind TLR4 TIR domain, and together form a cluster on the TIR surface (Toshchakov et al., 2011). We proposed that this cluster forms the TLR4 dimerization interface. Interestingly, peptides from structurally homologous regions of TLR2, i.e., 2R1, 2R3 and 2BB, also inhibit cognate receptor, albeit 2R3 and 2BB are less potent. 2R1 binds TLR2 co-receptors TLR1 and TLR6 (Figure S5A–B). 2R1, however, inhibited not only TLR2, but multiple signaling pathways, i.e., it suppressed all TLRs tested and also TNF-α-induced signaling. This finding might suggest that 2R1 binds several proteins with similar affinity. Co-immunoprecipitation studies confirmed a certain lack of specificity of 2R1 interaction with TIR domains because significant binding of this peptide to MyD88 and both TLR2 co-receptors was observed (Figures 3D and S5A–B). Interestingly, 4R1, the 2R1 homolog from TLR4, unlike other TLR4 inhibitory peptides, also demonstrated ‘multispecific’ binding in a cell-based FRET assay and bound both TLR4 and TLR2 TIRs (Toshchakov et al., 2011). The BB loop is the most conserved surface-exposed region of TIR domains (Slack et al., 2000); e.g., BB loops of murine TLR4 and TLR2 are ~50% conserved. Therefore, peptides derived from BB loop of different proteins might be predicted to demonstrate similar properties. Along this line, TLR4 and TLR2 BB loop peptides were found to cross-react as each peptide suppresses both TLR4 and TLR2 signaling (Toshchakov et al., 2007).
New and published findings suggest that TLR4 and TLR2 peptides from regions 1, 3, and 4 interact with TIR domains of corresponding dimerization partners; yet each peptide may have additional binding partners among TIR domains. Less specific binding to the dimerization interface compared to the region that mediates the recruitment of a specific protein from cytosol may be anticipated because dimerization of receptor TIR domains is, in a sense, a ‘forced’ interaction driven by the agonist-induced conformational change in the ectodomains. In contrast, the adapter recruitment site has to be specific to function properly because binding of unrelated proteins to this site would slow down the formation of receptor complexes. Although the extent to which these speculations are applicable to peptides derived from TIR:TIR interfaces is not certain at this time, the available study suggests that the bound conformation of protein-derived peptides is often highly similar to the conformation of the peptides in folded proteins (Vanhee et al., 2009).
Based on observation that the D helix peptide from TLR4 inhibits TLR4, but does not bind TLR4 TIR, we proposed that TLR4 D helix mediates adapter recruitment (Toshchakov et al., 2011). Our current study finds that, analogously to TLR4, the TLR2 D helix is an adapter recruitment site too; we further specify that this site binds TIRAP. Another interesting analogy is that for both TLR4 and TLR2, one TIR of the dimer seems to interact with the other TIR via a large surface formed by AB and BB loops with an important contribution of the region that is immediately N-terminal to the TIR domain (Figure 7B). We previously proposed that, in the case of TLR4, this region interacts with E helix of the other TIR of the dimer (Toshchakov et al., 2011). At this time, it is not known whether the mode of interaction of TLR2 co-receptors with TLR2 is similar.
Available mutagenesis data support the importance of TLR2 BB loop and the D helix for TLR2 function (Gautam et al., 2006; Xiong et al., 2012). A rare SNP allele (rs5743708), that results in R753Q replacement in the middle of D helix of TLR2 TIR (Figure 7A), is associated with higher susceptibility to infections caused by several bacterial and viral pathogens (Lorenz et al., 2000; Ogus et al., 2004; Schroder et al., 2005).
Studies of 2R9 binding to putative components of the primary TLR2 signaling complex indicated this peptide specifically binds TIRAP (Figure 3). FLIM measurements suggested that the apparent 2R9/TIRAP binding affinity measured directly in cells is >10 times higher than that for the 2R9/TLR1 or 2R9/MyD88 pairs (Figure 3C). The FP experiments using recombinant TIRAP confirmed the high binding affinity with KD of ~40 nM for the 2R9/TIRAP pair. Although the FP studies also detected binding of 2R9 to BSA (Figure 3F), 2R9 binds albumin with the affinity that is ~40 times lower than TIRAP (Figure 3F). Albumin is the most abundant serum protein that has several multifaceted binding sites capable of binding a range of small and medium size molecules (Ghuman et al., 2005). Binding to albumin significantly improves circulatory half-life of protein- and peptide-based drugs (Kratz, 2008; Sleep et al., 2013). Improving the pharmaceutical properties of drug candidates through engineering of molecules with enhanced binding to albumin has become an established strategy in drug development (Larsen et al., 2001; Sleep et al., 2013). The observed ratio of affinities of 2R9 binding to TIRAP versus albumin together with the high 2R9 dose required for TLR inhibitory effect in cells and in vivo suggest that a significant portion of total 2R9 in the system is likely bound to albumin under conditions of our in vitro and in vivo experiments and is thus protected from proteolysis. Although additional studies are required to elucidate if reversible binding to albumin is critical for the observed in vivo efficacy of 2R9, it is likely so. The observed 2R9/TIRAP binding affinity is very high compared to previously reported interaction of TIRAP with the viral Bcl-like protein A46 (apparent KD of 1.5 μM) (Oda et al., 2011). This study, however, failed to measure TIRAP binding with the TLR4-inhibitory peptide VIPER that is derived from A46 (Lysakova-Devine et al., 2010; Oda et al., 2011). A separate SPR study of IL-17RA binding to the CC loop decoy peptide from SEFIR domain of NF-κB activator 1 reported a very high KD value of 117 μM (Liu et al., 2011).
2R9 blocked TLR2, TLR4, TLR7, and TLR9, but not TLR3 or TNF-α-mediated signaling. Such a specificity profile agrees with the notion that this peptide binds TIRAP and precludes TIRAP interactions with TLR TIR domains. Although TIRAP-dependency of TLR7 signaling was not reported previously, our data suggest that, analogously to TLR9-mediated response, the TLR7-mediated activation of iBMDM strongly depends on TIRAP (Figure 4A and 4B). In accordance with these data, the TIRAP-targeting peptide, 2R9, potently inhibited TLR7 in cells and in vivo (Figures 4 and 5). The possibility that, in addition to TIRAP, 2R9 binds other TIR domain proteins, so that the observed functionality of this peptide is due to targeting of several TIR domains, cannot be excluded at this time. Particularly, the binding of 2R9 to TLR4 (Figure S5C–F) likely accounts for inhibition of LPS-induced cytokines in MyD88- and TIRAP-deficient cells (Figure 4C, D) and, also, increases the potency of TLR4 inhibition by 2R9 in WT cells. A similar mechanism of targeting several TIR domains may also play a role in inhibition of TLR7 by 2R9 because our data suggest that, in addition to strong binding to TIRAP, 2R9 directly binds the TIR of TLR7, albeit with apparent TLR7 binding affinity that is ~10 times less. Whether or not this secondary binding to TLR7 TIR is important for 2R9 activity in vivo remains to be elucidated.
2R9 also potently suppressed systemic cytokine activation induced in live animals by a synthetic TLR2 agonist. 2R9 blocked TLR2-mediated cytokine production more potently than TR6, a TIRAP-derived peptide that inhibits TLR4 and TLR2 and binds MyD88 (Couture et al., 2012; Piao et al., 2013a). The in vivo efficacy of 2R9 is confirmed by its effect on TLR7-induced systemic cytokine levels (Figure 5D–F).
Influenza virus, a single-stranded RNA virus, is sensed by a number of pattern recognition receptors, although TLR7 appears to play the primary role in mice (Diebold et al., 2004; Kawai and Akira, 2011; Lund et al., 2004). Accordingly, 2R9 that potently blocks TLR7 significantly inhibited the MyD88-dependent cytokine production by PR8-infected macrophages in cell culture experiments (Figure 6A–C). 2R9 less potently affected the PR8-induced IFN-β because of significant contribution of RLRs to induction of this gene.
TLR4 and TLR2 activation by endogenous, damage-associated molecules secondary to acute lung injury, caused by a pathogen or chemically induced, significantly contributes to severity of influenza (Imai et al., 2008; Shirey et al., 2013). The potent TLR4 antagonist, Eritoran, protected mice from PR8 infection; however, TLR2−/− mice could not be protected with Eritoran, indicating complexity of TLR roles in influenza and Eritoran-mediated protection. Therefore, a TLR antagonist that is multispecific and blocks a set of TLRs, including TLR2 and TLR4, may have distinct advantages for treatment of systemic inflammation in infectious disease, compared to a monospecific TLR antagonist. Our data demonstrate that 2R9, a cell-permeating decoy peptide, that inhibits TLR7, TLR4, and TLR2, indeed provided robust protection against PR8-induced lethality in mice, equivalently to Eritoran (Figure 6D). The broad specificity of TLR inhibition by 2R9 also suggests this peptide might be effective for modulation of inflammation caused by bacterial/viral co-infections.
Excessive inflammatory response elicited by lethal pathogens is the prevailing mechanistic cause of lethality in acute infections. Nonetheless, therapeutic interventions to reduce inflammatory response to pathogens, so that the injurious component is mitigated, yet the remaining response is adequate to activate anti-microbial defenses, are not developed sufficiently, despite significant efforts. Our study has resulted in the identification of a peptide that blocks several TLRs. We have demonstrated that this D helix peptide, 2R9, predominantly targets TIRAP, is effective in vivo, and protects mice from deadly PR8 infection. Importantly, potency and biological stability of the inhibitor is amenable to improvement in a number of rational approaches, including peptidomimicry, to develop more effective therapeutics. Although the screening of peptide library derived from an interaction domain of TLR2 has resulted in identification of a potent TLR inhibitor in this study; further studies will be necessary to determine if this decoy peptide approach is generally applicable to targeting the intracellular signaling events downstream of activated receptors and search for signaling inhibitors and therapeutic leads.
EXPERIMENTAL PROCEDURES
Animals, cells, and cell culture
C57BL/6J WT and MyD88-deficient mice were obtained from the Jackson Laboratory (Bar Harbor, Maine). Harvesting, culturing, and stimulation of peritoneal macrophages were described previously (Toshchakov et al., 2005). THP-1 cells were differentiated by incubation in the presence of 10 nM PMA in 10% FCS RPMI-1640 for 3 days. TLR agonists, P3C, P2C, FSL-1, R848, ODN1668, and high molecular weight poly (I:C) were purchased from InvivoGen (San Diego, CA). Escherichia coli K235 LPS was phenol-purified. Mouse rTNF was purchased from Biolegend, Inc.
PR8 Infection
The mouse-adapted H1N1 A/PR/8/34 strain of influenza virus (PR8) was obtained from ATCC (Manassas, VA) and propagated as previously described (Shirey et al., 2013). Peritoneal macrophages were infected at MOI=1. After a 2 h incubation in the presence of PR8, cells were washed with PBS and incubated in RPMI supplemented with 2% FCS, without or with a decoy peptide. C57BL/6J female mice were infected with 7500 TCID50 (LD90) of PR8 intranasally. Two days post-inoculation, mice received a 5 day course of 2R9 or 2R8 as a single i.p. administration daily at the dose of 200 nmol. Eritoran (200 μg/mouse), administered i.v. daily for 5 days starting on day 2, was included as a positive control (Shirey et al., 2013). Survival was monitored for 14 days. All animal experiments were conducted with approval from the UMB IACUC.
Supplementary Material
Acknowledgments
This work was supported by NIH grants AI-082299 (VYT), AI-018797 (SNV), and GM-107986, EB-006521 and RR-26370 (JRL). Genetically engineered iBMDM lines were a kind gift of Dr. Jonathan Kagan (Boston Children’s Hospital and Harvard Medical School).
Footnotes
AUTHOR CONTRIBUTIONS
WP designed and performed experiments and wrote the paper. LWR screened peptide library. WL measured IFN-β. WP, KAS, and SNV proposed testing the efficacy of 2R9 against PR8 and carried out studies of antiviral protection by 2R9. HS and JRL designed and implemented the FLIM-based approach to analysis of peptide-TIR binding in cells and also conducted FP experiments. GAS and EJS studied biophysics of 2R9/TIRAP interaction by SPR. VYT designed the study and peptides, analyzed data and wrote the paper.
Supplemental Information includes Extended Experimental Procedures and six figures.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aliprantis AO, Yang RB, Mark MR, Suggett S, Devaux B, Radolf JD, Klimpel GR, Godowski P, Zychlinsky A. Cell activation and apoptosis by bacterial lipoproteins through toll-like receptor-2. Science. 1999;285:736–739. doi: 10.1126/science.285.5428.736. [DOI] [PubMed] [Google Scholar]
- Bonham KS, Orzalli MH, Hayashi K, Wolf AI, Glanemann C, Weninger W, Iwasaki A, Knipe DM, Kagan JC. A promiscuous lipid-binding protein diversifies the subcellular sites of toll-like receptor signal transduction. Cell. 2014;156:705–716. doi: 10.1016/j.cell.2014.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandes M, Klauschen F, Kuchen S, Germain RN. A systems analysis identifies a feedforward inflammatory circuit leading to lethal influenza infection. Cell. 2013;154:197–212. doi: 10.1016/j.cell.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GY, Nunez G. Sterile inflammation: sensing and reacting to damage. Nature reviews Immunology. 2010;10:826–837. doi: 10.1038/nri2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole LE, Laird MH, Seekatz A, Santiago A, Jiang Z, Barry E, Shirey KA, Fitzgerald KA, Vogel SN. Phagosomal retention of Francisella tularensis results in TIRAP/Mal-independent TLR2 signaling. Journal of leukocyte biology. 2010;87:275–281. doi: 10.1189/jlb.0909619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly DJ, O’Neill LA. New developments in Toll-like receptor targeted therapeutics. Current opinion in pharmacology. 2012;12:510–518. doi: 10.1016/j.coph.2012.06.002. [DOI] [PubMed] [Google Scholar]
- Couture LA, Piao W, Ru LW, Vogel SN, Toshchakov VY. Targeting Toll-like receptor (TLR) signaling by Toll/interleukin-1 receptor (TIR) domain-containing adapter protein/MyD88 adapter-like (TIRAP/Mal)-derived decoy peptides. J Biol Chem. 2012;287:24641–24648. doi: 10.1074/jbc.M112.360925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derossi D, Joliot AH, Chassaing G, Prochiantz A. The third helix of the Antennapedia homeodomain translocates through biological membranes. J Biol Chem. 1994;269:10444–10450. [PubMed] [Google Scholar]
- Diebold SS, Kaisho T, Hemmi H, Akira S, Reis e Sousa C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303:1529–1531. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- Enokizono Y, Kumeta H, Funami K, Horiuchi M, Sarmiento J, Yamashita K, Standley DM, Matsumoto M, Seya T, Inagaki F. Structures and interface mapping of the TIR domain-containing adaptor molecules involved in interferon signaling. Proc Natl Acad Sci U S A. 2013;110:19908–19913. doi: 10.1073/pnas.1222811110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautam JK, Ashish Comeau LD, Krueger JK, Smith MF., Jr Structural and functional evidence for the role of the TLR2 DD loop in TLR1/TLR2 heterodimerization and signaling. J Biol Chem. 2006;281:30132–30142. doi: 10.1074/jbc.M602057200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay NJ, Keith FJ. Drosophila Toll and IL-1 receptor. Nature. 1991;351:355–356. doi: 10.1038/351355b0. [DOI] [PubMed] [Google Scholar]
- Gay NJ, Symmons MF, Gangloff M, Bryant CE. Assembly and localization of Toll-like receptor signaling complexes. Nature Immunology. 2014;2014:546–558. doi: 10.1038/nri3713. [DOI] [PubMed] [Google Scholar]
- Ghuman J, Zunszain PA, Petitpas I, Bhattacharya AA, Otagiri M, Curry S. Structural basis of the drug-binding specificity of human serum albumin. J Mol Biol. 2005;353:38–52. doi: 10.1016/j.jmb.2005.07.075. [DOI] [PubMed] [Google Scholar]
- Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, Lipford G, Wagner H, Bauer S. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303:1526–1529. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- Horng T, Barton GM, Flavell RA, Medzhitov R. The adaptor molecule TIRAP provides signalling specificity for Toll-like receptors. Nature. 2002;420:329–333. doi: 10.1038/nature01180. [DOI] [PubMed] [Google Scholar]
- Imai Y, Kuba K, Neely GG, Yaghubian-Malhami R, Perkmann T, van Loo G, Ermolaeva M, Veldhuizen R, Leung YHC, Wang HL, et al. Identification of oxidative stress and toll-like receptor 4 signaling as a key pathway of acute lung injury. Cell. 2008;133:235–249. doi: 10.1016/j.cell.2008.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki A, Pillai PS. Innate immunity to influenza virus infection. Nature reviews Immunology. 2014;14:315–328. doi: 10.1038/nri3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin MS, Lee JO. Structures of the toll-like receptor family and its ligand complexes. Immunity. 2008;29:182–191. doi: 10.1016/j.immuni.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Kato H, Sato S, Yoneyama M, Yamamoto M, Uematsu S, Matsui K, Tsujimura T, Takeda K, Fujita T, Takeuchi O, et al. Cell type-specific involvement of RIG-I in antiviral response. Immunity. 2005;23:19–28. doi: 10.1016/j.immuni.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34:637–650. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- Kenny EF, Talbot S, Gong M, Golenbock DT, Bryant CE, O’Neill LA. MyD88 adaptor-like is not essential for TLR2 signaling and inhibits signaling by TLR3. J Immunol. 2009;183:3642–3651. doi: 10.4049/jimmunol.0901140. [DOI] [PubMed] [Google Scholar]
- Kratz F. Albumin as a drug carrier: design of prodrugs, drug conjugates and nanoparticles. Journal of controlled release : official journal of the Controlled Release Society. 2008;132:171–183. doi: 10.1016/j.jconrel.2008.05.010. [DOI] [PubMed] [Google Scholar]
- Larsen PJ, Fledelius C, Knudsen LB, Tang-Christensen M. Systemic administration of the long-acting GLP-1 derivative NN2211 induces lasting and reversible weight loss in both normal and obese rats. Diabetes. 2001;50:2530–2539. doi: 10.2337/diabetes.50.11.2530. [DOI] [PubMed] [Google Scholar]
- Lee SM, Kok KH, Jaume M, Cheung TK, Yip TF, Lai JC, Guan Y, Webster RG, Jin DY, Peiris JS. Toll-like receptor 10 is involved in induction of innate immune responses to influenza virus infection. Proc Natl Acad Sci U S A. 2014;111:3793–3798. doi: 10.1073/pnas.1324266111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Swaidani S, Qian W, Kang Z, Sun P, Han Y, Wang C, Gulen MF, Yin W, Zhang C, et al. A CC’ loop decoy peptide blocks the interaction between Act1 and IL-17RA to attenuate IL-17- and IL-25-induced inflammation. Science signaling. 2011;4:ra72. doi: 10.1126/scisignal.2001843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz E, Mira JP, Cornish KL, Arbour NC, Schwartz DA. A novel polymorphism in the toll-like receptor 2 gene and its potential association with staphylococcal infection. Infect Immun. 2000;68:6398–6401. doi: 10.1128/iai.68.11.6398-6401.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund JM, Alexopoulou L, Sato A, Karow M, Adams NC, Gale NW, Iwasaki A, Flavell RA. Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc Natl Acad Sci U S A. 2004;101:5598–5603. doi: 10.1073/pnas.0400937101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysakova-Devine T, Keogh B, Harrington B, Nagpal K, Halle A, Golenbock DT, Monie T, Bowie AG. Viral inhibitory peptide of TLR4, a peptide derived from vaccinia protein A46, specifically inhibits TLR4 by directly targeting MyD88 adaptor-like and TRIF-related adaptor molecule. J Immunol. 2010;185:4261–4271. doi: 10.4049/jimmunol.1002013. [DOI] [PubMed] [Google Scholar]
- Means TK, Lien E, Yoshimura A, Wang S, Golenbock DT, Fenton MJ. The CD14 ligands lipoarabinomannan and lipopolysaccharide differ in their requirement for Toll-like receptors. J Immunol. 1999;163:6748–6755. [PubMed] [Google Scholar]
- Medzhitov R, Preston-Hurlburt P, Janeway CA., Jr A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature. 1997;388:394–397. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- Medzhitov R, Preston-Hurlburt P, Kopp E, Stadlen A, Chen C, Ghosh S, Janeway CA., Jr MyD88 is an adaptor protein in the hToll/IL-1 receptor family signaling pathways. Molecular cell. 1998;2:253–258. doi: 10.1016/s1097-2765(00)80136-7. [DOI] [PubMed] [Google Scholar]
- O’Neill LA, Bowie AG. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nature reviews Immunology. 2007;7:353–364. doi: 10.1038/nri2079. [DOI] [PubMed] [Google Scholar]
- O’Neill LA, Bryant CE, Doyle SL. Therapeutic targeting of Toll-like receptors for infectious and inflammatory diseases and cancer. Pharmacological reviews. 2009;61:177–197. doi: 10.1124/pr.109.001073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda S, Franklin E, Khan AR. Poxvirus A46 protein binds to TIR domain-containing Mal/TIRAP via an alpha-helical sub-domain. Mol Immunol. 2011;48:2144–2150. doi: 10.1016/j.molimm.2011.07.014. [DOI] [PubMed] [Google Scholar]
- Ogus AC, Yoldas B, Ozdemir T, Uguz A, Olcen S, Keser I, Coskun M, Cilli A, Yegin O. The Arg753GLn polymorphism of the human toll-like receptor 2 gene in tuberculosis disease. The European respiratory journal. 2004;23:219–223. doi: 10.1183/09031936.03.00061703. [DOI] [PubMed] [Google Scholar]
- Ozinsky A, Underhill DM, Fontenot JD, Hajjar AM, Smith KD, Wilson CB, Schroeder L, Aderem A. The repertoire for pattern recognition of pathogens by the innate immune system is defined by cooperation between toll-like receptors. Proc Natl Acad Sci U S A. 2000;97:13766–13771. doi: 10.1073/pnas.250476497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawson T, Nash P. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;300:445–452. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- Piao W, Ru LW, Piepenbrink KH, Sundberg EJ, Vogel SN, Toshchakov VY. Recruitment of TLR adapter TRIF to TLR4 signaling complex is mediated by the second helical region of TRIF TIR domain. Proc Natl Acad Sci U S A. 2013a;110:19036–19041. doi: 10.1073/pnas.1313575110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao W, Vogel SN, Toshchakov VY. Inhibition of TLR4 signaling by TRAM-derived decoy peptides in vitro and in vivo. J Immunol. 2013b;190:2263–2272. doi: 10.4049/jimmunol.1202703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder NW, Diterich I, Zinke A, Eckert J, Draing C, von Baehr V, Hassler D, Priem S, Hahn K, Michelsen KS, et al. Heterozygous Arg753Gln polymorphism of human TLR-2 impairs immune activation by Borrelia burgdorferi and protects from late stage Lyme disease. J Immunol. 2005;175:2534–2540. doi: 10.4049/jimmunol.175.4.2534. [DOI] [PubMed] [Google Scholar]
- Shirey KA, Lai W, Scott AJ, Lipsky M, Mistry P, Pletneva LM, Karp CL, McAlees J, Gioannini TL, Weiss J, et al. The TLR4 antagonist Eritoran protects mice from lethal influenza infection. Nature. 2013;497:498–502. doi: 10.1038/nature12118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack JL, Schooley K, Bonnert TP, Mitcham JL, Qwarnstrom EE, Sims JE, Dower SK. Identification of two major sites in the type I interleukin-1 receptor cytoplasmic region responsible for coupling to pro-inflammatory signaling pathways. J Biol Chem. 2000;275:4670–4678. doi: 10.1074/jbc.275.7.4670. [DOI] [PubMed] [Google Scholar]
- Sleep D, Cameron J, Evans LR. Albumin as a versatile platform for drug half-life extension. Biochimica et biophysica acta. 2013;1830:5526–5534. doi: 10.1016/j.bbagen.2013.04.023. [DOI] [PubMed] [Google Scholar]
- Smith MC, Gestwicki JE. Features of protein-protein interactions that translate into potent inhibitors: topology, surface area and affinity. Expert reviews in molecular medicine. 2012;14:e16. doi: 10.1017/erm.2012.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szmacinski H, Toshchakov V, Lakowicz JR. Application of phasor plot and autofluorescence correction for study of heterogeneous cell population. Journal of biomedical optics. 2014;19:046017. doi: 10.1117/1.JBO.19.4.046017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O, Hoshino K, Kawai T, Sanjo H, Takada H, Ogawa T, Takeda K, Akira S. Differential roles of TLR2 and TLR4 in recognition of gram-negative and gram-positive bacterial cell wall components. Immunity. 1999;11:443–451. doi: 10.1016/s1074-7613(00)80119-3. [DOI] [PubMed] [Google Scholar]
- Takeuchi O, Kawai T, Muhlradt PF, Morr M, Radolf JD, Zychlinsky A, Takeda K, Akira S. Discrimination of bacterial lipoproteins by Toll-like receptor 6. International immunology. 2001;13:933–940. doi: 10.1093/intimm/13.7.933. [DOI] [PubMed] [Google Scholar]
- Toshchakov V, Jones BW, Perera PY, Thomas K, Cody MJ, Zhang S, Williams BR, Major J, Hamilton TA, Fenton MJ, et al. TLR4, but not TLR2, mediates IFN-beta-induced STAT1alpha/beta-dependent gene expression in macrophages. Nat Immunol. 2002;3:392–398. doi: 10.1038/ni774. [DOI] [PubMed] [Google Scholar]
- Toshchakov VU, Basu S, Fenton MJ, Vogel SN. Differential involvement of BB loops of toll-IL-1 resistance (TIR) domain-containing adapter proteins in TLR4- versus TLR2-mediated signal transduction. J Immunol. 2005;175:494–500. doi: 10.4049/jimmunol.175.1.494. [DOI] [PubMed] [Google Scholar]
- Toshchakov VY, Fenton MJ, Vogel SN. Cutting Edge: Differential inhibition of TLR signaling pathways by cell-permeable peptides representing BB loops of TLRs. J Immunol. 2007;178:2655–2660. doi: 10.4049/jimmunol.178.5.2655. [DOI] [PubMed] [Google Scholar]
- Toshchakov VY, Szmacinski H, Couture LA, Lakowicz JR, Vogel SN. Targeting TLR4 signaling by TLR4 Toll/IL-1 receptor domain-derived decoy peptides: identification of the TLR4 Toll/IL-1 receptor domain dimerization interface. J Immunol. 2011;186:4819–4827. doi: 10.4049/jimmunol.1002424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toshchakov VY, Vogel SN. Cell-penetrating TIR BB loop decoy peptides a novel class of TLR signaling inhibitors and a tool to study topology of TIR-TIR interactions. Expert opinion on biological therapy. 2007;7:1035–1050. doi: 10.1517/14712598.7.7.1035. [DOI] [PubMed] [Google Scholar]
- Underhill DM, Ozinsky A, Hajjar AM, Stevens A, Wilson CB, Bassetti M, Aderem A. The Toll-like receptor 2 is recruited to macrophage phagosomes and discriminates between pathogens. Nature. 1999;401:811–815. doi: 10.1038/44605. [DOI] [PubMed] [Google Scholar]
- Vanhee P, Stricher F, Baeten L, Verschueren E, Lenaerts T, Serrano L, Rousseau F, Schymkowitz J. Protein-peptide interactions adopt the same structural motifs as monomeric protein folds. Structure. 2009;17:1128–1136. doi: 10.1016/j.str.2009.06.013. [DOI] [PubMed] [Google Scholar]
- Wang X, Smith C, Yin H. Targeting Toll-like receptors with small molecule agents. Chemical Society reviews. 2013;42:4859–4866. doi: 10.1039/c3cs60039d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werts C, Tapping RI, Mathison JC, Chuang TH, Kravchenko V, Saint Girons I, Haake DA, Godowski PJ, Hayashi F, Ozinsky A, et al. Leptospiral lipopolysaccharide activates cells through a TLR2-dependent mechanism. Nat Immunol. 2001;2:346–352. doi: 10.1038/86354. [DOI] [PubMed] [Google Scholar]
- Xiong Y, Song C, Snyder GA, Sundberg EJ, Medvedev AE. R753Q polymorphism inhibits Toll-like receptor (TLR) 2 tyrosine phosphorylation, dimerization with TLR6, and recruitment of myeloid differentiation primary response protein 88. J Biol Chem. 2012;287:38327–38337. doi: 10.1074/jbc.M112.375493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Sato S, Hemmi H, Sanjo H, Uematsu S, Kaisho T, Hoshino K, Takeuchi O, Kobayashi M, Fujita T, et al. Essential role for TIRAP in activation of the signalling cascade shared by TLR2 and TLR4. Nature. 2002;420:324–329. doi: 10.1038/nature01182. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.