Abstract
The Pim-1 kinase regulates cell survival, proliferation and differentiation and is overexpressed frequently in many malignancies including leukemia and skin cancer. In this study, we used kinase profiling analysis to demonstrate that 2′-hydroxycinnamicadelhyde (2′-HCA), a compound found in cinnamon, specifically inhibits Pim-1. Co-crystallography studies determined the hydrogen bonding pattern between 2′-HCA and Pim-1. Notably, 2′-HCA binding altered the apo kinase structure in a manner that shielded the ligand from solvent, thereby acting as a gatekeeper loop. Biologically, 2′-HCA inhibited the growth of human erythroleukemia or squamous epidermoid carcinoma cells by inducing apoptosis. The compound was also effective as a chemopreventive agent against EGF-mediated neoplastic transformation. Lastly, 2′-HCA potently suppressed the growth of mouse xenografts representing human leukemia or skin cancer. Overall, our results offered preclinical proof of concept for 2′-HCA as a potent anticancer principle arising from direct targeting of the Pim-1 kinase.
Keywords: 2′-hydroxycinnamicadelhyde, (2′-HCA), co-crystallography, xenograft, chemoprevention, human erythroleukemia
Introduction
Natural compounds found in food and Traditional Asian Medicines (TAM) represent a diverse and promising resource for treating chronic human diseases such as cancer, cardiovascular disease, diabetes, and Alzheimer’s disease (1). Although strong clinical evidence is lacking for many claimed health benefits, a long history of human consumption lends some credibility to their efficacy and safety. Research using modern biotechnology methods combining supercomputer technology, protein crystallography and biological assays (2) has shown that natural phytochemicals have a number of advantages compared to synthetic compounds (3). An important foundation for their wider use involves the elucidation of their direct molecular targets and mechanisms of action, as well as confirmation of their pharmacodynamic properties (2).
Cinnamon is one of the oldest and most popular spices that has been used by different cultures around the world and is popular as a flavoring agent and a preservative for beverages, bakery products and candies (4). The product is harvested from the dry bark and twigs of Cinnamomum spp and has been used in traditional Chinese medicine for treating dyspepsia, gastritis, blood circulation disturbances, inflammatory diseases and cancer in Korea, China and India (5). A number of recent studies have shown several potential health benefits of cinnamon for diabetes, neuropathy, cardiovascular disease and cancer (6). Cinnamon extracts have been shown to inhibit proliferation of lymphoma, melanoma, hepatocellular cancer, cervix cancer and colon cancer cells in vitro and melanoma in vivo (4,7-9). These extracts also reportedly suppress angiogenesis by targeting VEGFR2 (10,11).
2′-Hydroxycinnamicaldehyde (2′-HCA, Fig. 1Aa) is a major component of the essential oil of cinnamon and is present at levels of approximately 0.01-0.8 mg/g in commercial cinnamon powder (4,12). It is regarded as a major bioactive component of cinnamon (13) and reportedly inhibits proliferation of several human cancer cell lines including breast, colon, leukemia, ovarian, and lung tumor cells (14-18). 2′-HCA also attenuates the in vivo xenograft growth of colon HCT116 cancer cells and allograft growth of oral RK3E-ras-Fluc cancer cells (16,18). However, its direct molecular target(s) and mechanism(s) of action have not been clearly elucidated.
Figure 1.
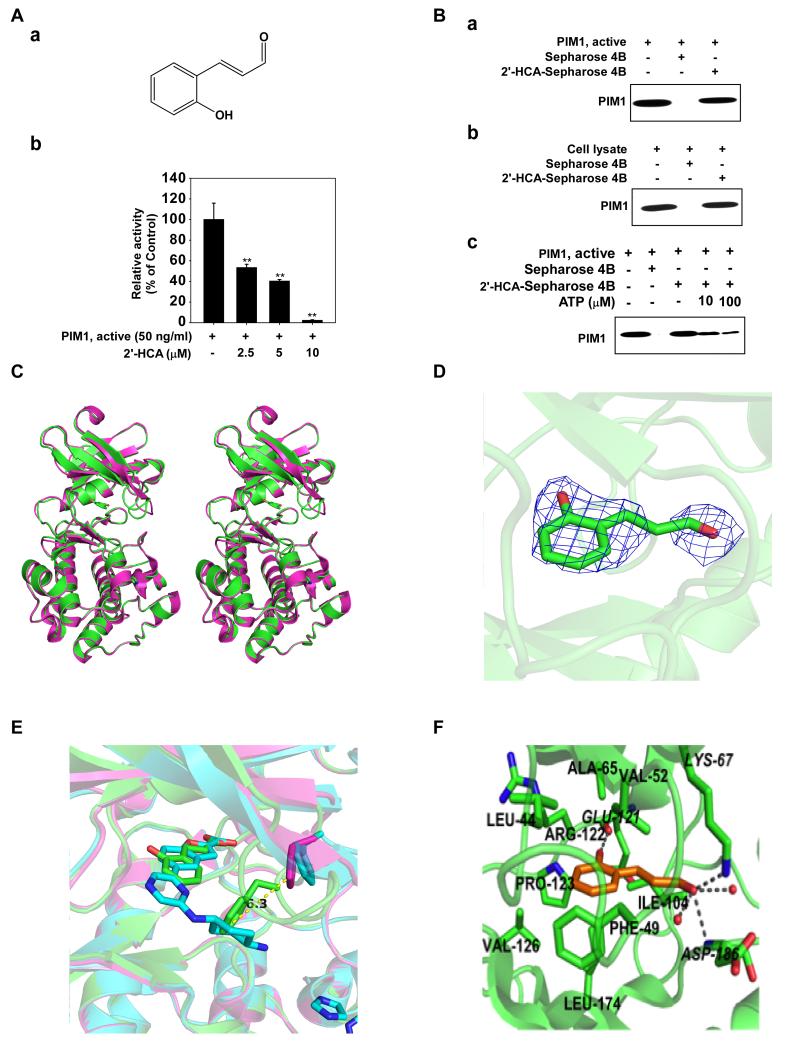
2′-Hydroxycinnamicaldehye (2′-HCA) inhibits Pim-1 activity. Aa, chemical structure of 2′-HCA. Ab, 2′-HCA inhibits Pim-1 activity. Active Pim-1 (50 ng) was mixed with 2′-HCA (0, 2.5, 5, or 10 μM) and then incubated with [γ-32P] ATP. The radioactive incorporation was determined using a scintillation counter. Ba, 2′-HCA binds Pim-1 directly in an ATP-competitive manner. Pim-1 and 2′-HCA binding was confirmed by immunoblotting with an antibody against Pim-1: lane 1 (input control), Pim-1 protein standard; lane 2 (negative control), Sepharose 4B beads were used for an immunoprecipitation assay; lane 3, Pim-1 was immunoprecipitated using 2′-HCA-Sepharose 4B beads. Bb, 2′-HCA binds Pim-1 ex vivo. Binding of 2′-HCA to Pim-1 in HEL cells was confirmed by immunoblotting with an antibody against Pim-1: lane 1 (input control), HEL cell lysate; lane 2 (negative control), HEL cell lysates were precipitated with Sepharose 4B beads; lane 3, HEL cell lysates were precipitated using 2′-HCA-Sepharose 4B beads. Bc, Active Pim-1 (0.2 μg) was incubated with ATP at the indicated concentrations (0, 10, or 100 μM) together with 100 μl 2′-HCA -Sepharose 4B beads or Sepharose 4B beads (negative control) added in reaction buffer to a final volume of 500 μl. The immunoprecipitated proteins were detected by immunoblotting with an antibody against Pim-1. Lane 1, negative control, showing that Pim-1 does not bind to Sepharose 4B beads alone; lane 2: positive control, showing that Pim-1 binds with 2′-HCA-Sepharose 4B beads. Crystal structure of Pim-1 in complex with 2′-HCA. C, Stereographic representation of apo Pim-1 alone (magenta) and in complex with 2′-HCA (green). D, 2Fo–Fc omit map of 2′-HCA electron density contoured at 1 σ. E, F, Molecular interactions between 2′-HCA and Pim-1. Residues involved in hydrophobic interactions are indicated in bold, and those involved in hydrogen bonding are in italics. All hydrogen bonds are marked as grey broken lines. Enlarged view of the glycine-rich loop showing the Phe49 residue that extends into the active site to interact with 2′-HCA. The glycine-rich loop in the structure of Pim-1 in complex with benzofuran-2-carboxylic acid, shown in blue, also adopts the same conformation as in the apo structure.
Proviral insertion in murine lymphomas-1 (Pim-1) is a proto-oncogene involved in pivotal cellular processes (19). Pim-1 is overexpressed in various tumors and has been linked to a poor prognosis (20). Its deregulation also interferes with apoptosis and cell cycle-related pathways to promote neoplastic transformation in many forms of malignancy. Over-expression of Pim-1 in mice leads to a greater susceptibility to spontaneous tumor formation, while increasing susceptibility to radiation- and chemically-induced tumorigenesis (21). Accordingly, interest in developing small molecule inhibitors of Pim-1 has been growing (21).
We sought to further investigate the underlying mechanism of the anti-tumorigenic effects of 2′-HCA and its relationship with Pim-1. In this study, we observed that Pim-1 is a direct molecular target of 2′-HCA and revealed direct molecular binding using X-ray co-crystallography. The significance of these findings was further explored using a number of in vitro and in vivo approaches.
Materials and Methods
Reagents
2′-HCA was purchased from Tokyo Chemical Industry (Tokyo, Japan). Active Pim-1 and the Pim-1 substrate peptide (KRRRLASLR) were purchased from SignalChem (Richmond, Canada). Antibodies specific for detecting total Bad, phosphorylated Bad (Ser112), total ERKs, phosphorylated ERKs (Thr202/Tyr204), total RSK, phosphorylated RSK (Thr356/Ser360), PARP, caspase-3, and caspase-9 were purchased from Cell Signaling Technology (Beverly, MA). Antibodies against total Pim-1, PARP, Bad, p27KIP1, p21CIP1 and β-actin were purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Cell culture
All cell lines were purchased from the American Type Culture Collection and were cytogenetically tested and authenticated before being frozen. Each vial of frozen cells was thawed and maintained in culture for a maximum of 8 weeks. Enough frozen vials were available for each cell line to ensure that all cell-based experiments were conducted on cells that had been tested and in culture for 8 weeks or less. The human erythroleukemia (HEL) cell line was cultured in RPMI-1640 supplemented with 10% fetal bovine serum (FBS, Atlanta Biologicals, Flowery Branch, GA) and 1% antibiotic-antimycotic (Thermo Fisher Scientific Inc., Waltham, MA). HaCaT skin keratinocytes and A431 human epidermoid carcinoma cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM, Thermo Fisher Scientific, Inc.) supplemented with 10% FBS and 1% antibiotic-antimycotic. JB6 P+ mouse skin epidermal cells were cultured in minimum essential medium (MEM) supplemented with 5% FBS and 1% antibiotic-antimycotic.
Direct and cell-based pull-down assays
An active Pim-1 protein (200 ng) or a HEL cellular supernatant fraction (500 μg) was incubated with 2′-HCA Sepharose 4B beads or Sepharose 4B beads alone as a control (100 μL, 50% slurry) in reaction buffer (50 mM Tris, pH 7.5, 5 mM EDTA, 150 mM NaCl, 1 mM DTT, 0.01% Nonidet P-40, 2 μg/mL BSA, 0.02 mM PMSF, and 1× protease inhibitor mixture). After incubation with gentle rocking overnight at 4°C, the beads were washed 5× with reaction buffer (50 mM Tris, pH 7.5, 5 mM EDTA, 150 mM NaCl, 1 mM DTT, 0.01% Nonidet P-40, and 0.02 mM PMSF), and proteins bound to the beads were analyzed by Western blotting.
ATP and 2′-HCA competition assay
Recombinant Pim-1 (0.2 μg) was incubated with 100 μL of 2′-HCA-Sepharose 4B beads or 100 μL of Sepharose 4B beads in reaction buffer (see above) for 12 h at 4°C. ATP was added at either 10 or 100 μM to a final volume of 500 μL, followed by incubation for 30 min. The samples were washed and proteins were detected by Western blotting.
In vitro kinase assays
The kinase assays were performed in accordance with instructions provided by EMD Millipore (Billerica, MA). Briefly, the reaction was carried out in the presence of 10 μCi of [γ-32P] ATP with each compound in 40 μl of reaction buffer containing 20 mM N-2-hydroxyethylpiperazine-N-2-ethanesulfonic acid (pH 7.4), 10 mM MgCl2, 10 mM MnCl2 and 1 mM dithiothreitol (DTT). Diluted [γ-32P] ATP solution (10 μL) was incubated at 30°C for 10 min in assay buffer (described above) and substrate peptide and then 15-μL aliquots were transferred onto p81 paper and washed 3× with 0.75% phosphoric acid (5 min per wash) and once with acetone (5 min). The incorporation of radioactivity was determined using a scintillation counter (Beckman Coulter, Indianapolis, IN)
Crystallization and data collection
Co-crystallography of Pim-1 and 2′-HCA was performed by Drs. Derek Logan and Björn Walse at SARomics Biostructures AB (Lund, Sweden). Diffraction quality crystals were grown in MRC Maxi plates (Molecular Dimensions, Suffolk, UK) containing 150 nl Pim-1 at 10 mg/ml in 25 mM Tris-HCl (pH 7.5), 250 mM NaCl, and 5 mM DTT mixed with a 50 nl reservoir consisting of 0.1 M imidazole (pH 6.3), and 1.2–1.4 M sodium acetate. Crystals grew to their full size of about 0.05 × 0.05 × 0.4 mm over the course of 1 week at 4°C. A stock solution of 150 mM 2′-HCA in 50% DMSO was prepared. Suitable crystals were transferred from the crystallization drops into a 3 μl drop of soaking solution consisting of 0.1 M imidazole pH 6.3, 1.4 M sodium acetate, 10 mM 2′-HCA and 3.3% DMSO. The drop was sealed over reservoir solution and incubated for 48 h. A soaked crystal measuring 0.04 × 0.04 × 0.2 mm was flash-frozen in cryo solution (0.1 M imidazole pH 6.3, 1.4 M sodium acetate, 10 mM 2′-HCA, 3.3% DMSO, 20% glycerol) and data were collected at 100 K. The data were collected to 2.5 Å at beamline I911-3 of the MAX-II synchrotron in Lund, Sweden using a 165 mm CCD detector from MarResearch (Norderstedt, Germany). These data were processed with XDS, scaled in XSCALE and converted to the MTZ format with XDSCONV (22) (see Supplemental Table 1 for details).
Structure determination and refinement
The structure of Pim-1 in complex with benzofuran-2-carboxylic acid (PDB code 3R04) with the ligand removed was used as a search model for molecular replacement using the Phaser program in the CCP4 program suite (23,24). Five percent of the reflections were used for calculation of the free R-value (see values in parentheses in Supplemental Table 1). Iterative rebuilding of the model and addition of solvent molecules was conducted using Coot (24). The model was finally improved by restrained refinement using REFMAC5 (25). The density for the 2′-HCA ligand was not completely visible at a contour level of 1.0 σ, so the ligand placement was aided by the hydrogen-bonding pattern between the ligand and the neighboring water molecules and side chains. The refinement statistics have been summarized in Supplemental Table 1.
Lentiviral infection
The lentiviral expression vectors, including Gipz-shPim-1 (TATACACAGTCAGCAATGC, RNAi core, University of Minnesota), and packaging vectors, including pMD2.0G and psPAX, were purchased from Addgene, Inc. (Cambridge, MA). To prepare shPim-1 viral particles, each viral vector and the packaging vectors (pMD2.0G and psPAX) were transfected using jetPEI into HEK293T cells following the manufacturer’s instructions. The transfection medium was changed at 4 h after transfection and the cells were then cultured for 36 h. The viral particles were harvested by filtration using a 0.45-mm syringe filter, then combined with 8 μg/mL polybrene (EMD Millipore) and infected into 60% confluent HEL, A431 or HaCaT cells overnight. The cell culture medium was replaced with fresh complete growth medium for 24 h before the cells were selected using puromycin (2 μg/mL; Sigma, St. Louis, MO) over 36 h. The selected cells were then used for further experiments.
Immunoblot analysis
Cell lysates were prepared with radio-immunoprecipitation assay (RIPA) buffer (50 mM Tris–HCl pH 7.4, 1% NP-40, 0.25% sodium deoxycholate, 0.1% sodium dodecyl sulfate, 150 mM NaCl, 1 mM ethylenediaminetetraacetic acid, and 1× protease inhibitor tablet). Equal loading of protein was confirmed using a bicinchoninic acid assay (Pierce, Rockford, IL). Proteins were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluoride (PVDF) membranes (EMD Millipore). Membranes were blocked with 5% non-fat dry milk for 1 h at room temperature (RT) and incubated with the appropriate primary antibodies overnight at 4°C. After washing with phosphate-buffered saline containing 0.1% Tween 20, the membrane was incubated with a horseradish peroxidase (HRP)-conjugated secondary antibody at a 1:5000 dilution and the signal was detected with a chemiluminescence reagent (GE Healthcare, Piscataway, NJ).
Cell proliferation assays
Cells were seeded (1 × 103 cells per well) in 96-well plates and incubated for 24 h, before treatment with the indicated doses of each compound. After incubation for 1, 2, or 3 days, 20 μL of CellTiter96 AQueous One Solution (Promega, Madison, WI) were added and the cells were incubated for 1 h at 37°C in a 5% CO2 incubator. Absorbance was measured at 492 nm.
Anchorage-independent cell growth
Cells (8 × 103 per well) suspended in Basal Medium Eagle (BME) supplemented with 10% FBS and 1% antibiotics were added to 0.3% agar with varying doses of each compound in the top layer over a base layer of 0.6% agar. The cultures were maintained at 37°C in a 5% CO2 incubator for 3 weeks before colonies were counted under a microscope using the Image-Pro Plus Software (vs.4) program (Media Cybernetics, Rockville, MD).
Annexin V staining
Apoptosis was assessed using the annexin V-FITC apoptosis detection kit (MBL International Corp., Watertown, MA) following the manufacturer’s instructions. Cells were treated and then harvested and washed with phosphate-buffered saline and incubated for 5 min at RT with annexin V-FITC plus propidium iodide (PI). Cells were analyzed using a FACSCalibur flow cytometer (BD Biosciences, San Jose, CA).
Xenograft mouse model
The experimental protocol was approved by the Animal Care and Use Committee of Seoul National University (SNU-130904-1). Male or female mice (6-week-old, BALB/c-nu) implanted with HEL or A431 cells, respectively, were used for the xenograft assay. A431 (1 × 106) or HEL (2 × 107) cells, suspended in 100 uL of serum-free media containing 50% Matrigel (BD Biosciences), were implanted subcutaneously in both dorsal flanks of mice. Cells were allowed to form tumors, and once the tumors reached a size of 50 mm3, the mice were randomly assigned into groups (8 mice/group). In previous studies by others, mice were treated with 2′-HCA with doses ranging from 10 to 50 mg/kg/day by intraperitoneal (IP) injection. Based on this information, we performed a pilot study to determine the optimal dosage of 2′-HCA treatment, and chose to treat with 2′-HCA at 5 and 20 mg/kg as optimal dosages to show a dose dependency of its tumor inhibition effect. 2′-HCA suspended in vehicle (10% DMSO in PBS) was administered at a dose of 5 (2′-HCA_5) or 20 (2′-HCA_20) mg/kg body weight intraperitoneally 5 days a week. Tumor volume was measured every week using calipers and calculated according to a standard formula: V = (L × H × W) π/6. Tumor mass was measured after excision on the final day of the experiment and tumor tissues were preserved for further analysis. Note that no study specific study has focused on the pharmacokinetics of 2′-HCA. But one study (26) used cinnamaldehyde and observed blood levels of over 1 μM in the blood. Thus the concentrations used in this study seem reasonable based on the literature.
Immunostaining analysis
The immunostaining assay was conducted on tumor samples that were frozen using tissue-freezing medium immediately after dissection and stored at −70°C. Cryostat sections were cut into fragments of 10-mm thickness and fixed with cold acetone for 10 min at RT and left to dry. Specimens were incubated in 5% goat serum in PBS containing 0.3% Triton X-100 (PBS-T) for 1 h at RT in order to permeabilize them and block any non-specific antibody binding. Primary antibodies were incubated at 4°C overnight, and secondary antibodies for 2 h at RT. Fluorescent images were obtained by confocal microscopy and signal-positive area densities were measured by analysis of pixel-based fluorescence intensities using the ImageJ software program (NIH).
Statistical analysis
All quantitative results are expressed as mean values ± S.D. or S.E. Statistically significant differences were determined using the Student t test or by one-way ANOVA. Values of p < 0.05 were considered to be statistically significant.
Results
Pim-1 is a direct molecular target of 2′-HCA
To elucidate the molecular target of 2′-HCA, we screened 77 cancer-related kinases using KinaseProfiler provided by EMD Millipore. Results of the screening with 20 μM 2′-HCA indicated that only the activity of Pim-1 was decreased by more than 80% (Supplemental Table 2). The activity was reduced in a concentration-dependent manner with an IC50 of approximately 3 μM (Fig. 1Ab). To identify the mechanism by which 2′-HCA modulates Pim-1 activity, we determined whether 2′-HCA could bind directly to Pim-1. Pull-down assay results revealed that 2′-HCA physically binds to the active Pim-1 protein (Fig. 1Ba, lane 3), but not to unconjugated Sepharose 4B beads (Fig. 1Ba, lane 2). The input lane (Fig. 1Ba, lane 1) showing the loading of 20 ng of the active Pim-1 protein as a marker, verified that the detected band represents the Pim-1 protein. We also observed binding of 2′-HCA to Pim-1 in HEL cells (Fig. 1Bb). Next, to examine the mode of 2′-HCA binding to Pim-1, we performed ATP competitive-binding assays. ATP was found to compete with 2′-HCA for Pim-1 binding (Fig. 1Bc), suggesting that 2′-HCA binds to the Pim-1 ATP-binding pocket.
Crystal structure of Pim-1 in complex with 2′-HCA
We determined the crystal structure of Pim-1 in complex with 2′-HCA to a resolution of 2.5 Å. The structure includes amino acids 33 to 305, the 2′-HCA ligand, an imidazole molecule and 52 water molecules. The final R-factor for the model was 0.175 (Rfree = 0.219). The overall structure is very similar to the apo structure except for some loop regions that are shifted by a few Å (Fig. 1C). The most significant difference is the relocation of the glycine-rich loop (Leu44–Val51) to block the entrance to the binding site. A 2Fo–Fc omit map of 2′-HCA electron density contoured at 1 σ (Fig. 1D). Analysis of the glycine-rich loop showed that the Phe49 residue extends into the active site to interact with 2′-HCA, while the same loop in the structure of Pim-1 in complex with benzofuran-2-carboxylic acid, was also shown to adopt the same conformation as in the apo structure (Fig. 1E, F).
2′-HCA inhibits anchorage-dependent and -independent cell growth of HEL and A431 cells
To identify an accurate cell model to determine the anticancer effects of 2′-HCA, we first used the Human Protein Atlas database to search for cell lines expressing the highest Pim-1-levels (27). A human erythroleukemia cell line, HEL, was identified in the database as having the highest Pim-1 expression. A431 cells overexpressing EGFR have been shown to grow in athymic nude mice, which represent an excellent model for studying EGF-mediated skin cancers. We therefore measured anchorage-independent and -dependent cell growth of HEL and A431 cells by soft agar and MTS assays, respectively. 2′-HCA treatment (2.5-10 μM) was found to suppress both anchorage-independent (Fig. 2A) and -dependent (Fig. 2B) growth of HEL (Fig. 2Aa-d, Ba) and A431 cells (Fig. 2Ae-h, Bb).
Figure 2.
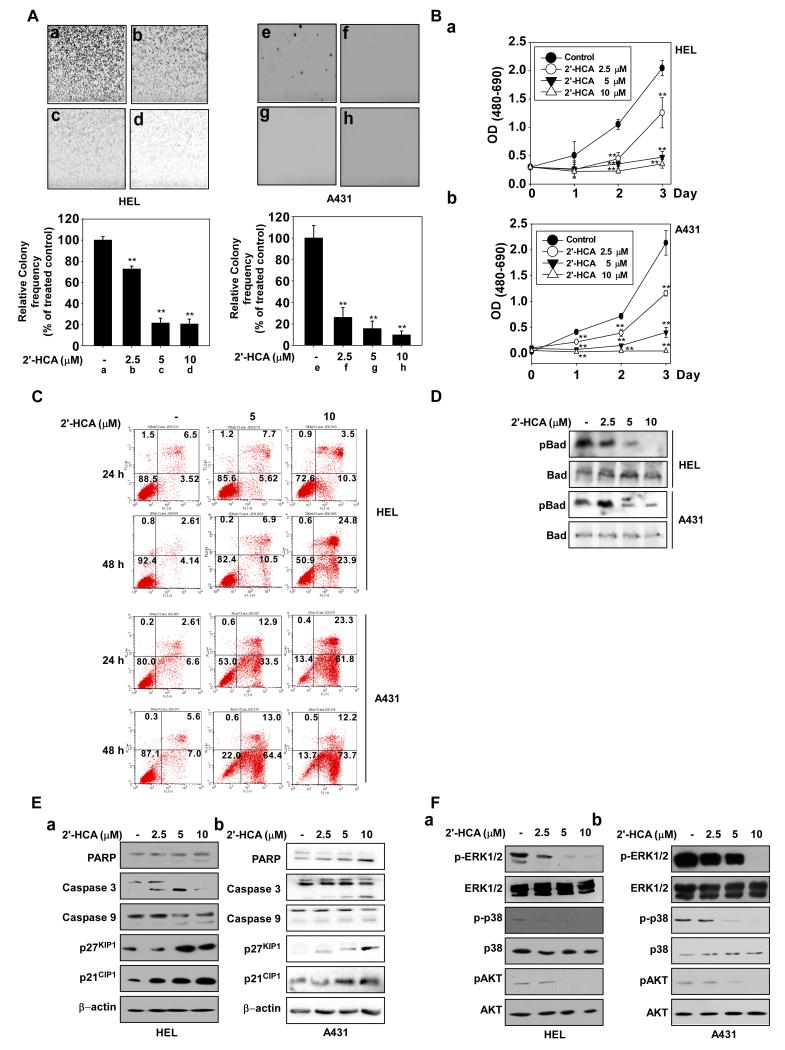
2′-HCA inhibits both anchorage-dependent and -independent HEL and A431 cell growth. A, 2′-HCA inhibits anchorage-independent growth of both HEL (a-d) and A431 (e-h) cells. A soft agar assay was performed with 2′-HCA (0, 2.5, 5, or 10 μM) and the number of colonies was counted under a microscope with the aid of the Image-Pro Plus software program (vs. 6.2). Results are represented as mean values ± S.E. (n = 3). The asterisks (*, **) indicate a significant difference (p < 0.05, p < 0.01) compared with the untreated control group. B, 2′-HCA inhibits the anchorage-dependent growth of both HEL (a) and A431 (b) cells. Cell growth was evaluated by the MTS assay with 2′-HCA (0, 2.5, 5, or 10 μM). The asterisks (*, **) indicate a significant difference of (p < 0.05, p < 0.01) compared with the untreated control group. C, 2′-HCA induces apoptosis in both HEL (upper panels) and A431 (lower panels) cells. Apoptosis was analyzed by annexin V/PI staining. 2′-HCA induces apoptosis in HEL and A431 cells in a dose-dependent manner. D, 2′-HCA regulates Pim-1 downstream signaling. 2′-HCA inhibits Bad phosphorylation in HEL (upper panel) and A431 (3rd panel) cells. E, 2′-HCA regulates apoptosis-related signaling pathways in HEL (a) and A431 (b) cells. F,2′-HCA inhibits ERK1/2, p38 and AKT phosphorylation in HEL (a) and A431 (b) cells. Cells were treated with 2′-HCA (0, 2.5, 5, or 10 μM) for 48 h and harvested. Immunoblotting was conducted using specific antibodies.
2′-HCA induces apoptosis and inhibits Bad phosphorylation
Pim-1 plays a pivotal role in resistance to apoptosis and cell cycle progression, as well as hypoxic responses (28). Pim-1 inactivates the pro-apoptotic Bad protein by phosphorylating its Ser112 residue (29). HEL and A431 cells were treated with 2′-HCA (2.5-10 μM) for various time periods and apoptosis was assessed by Annexin V staining and flow cytometry. 2′-HCA treatment induced substantial apoptosis in both HEL and A431 cell lines (Fig. 2C). We also observed that 2′-HCA (2.5-10 μM) inhibits phosphorylation of Bad at Ser112, which is a direct phosphorylation target of Pim-1 (Fig. 2D). Analysis of apoptosis-related proteins revealed that PARP, caspase 9, and caspase 3 were activated by 2′-HCA, in association with increased expression of p27KIP and p21CIP (Fig. 2Ea, b). Previous studies have shown that Pim-1 regulates the mitogen activated protein kinase and AKT pathways (30-33). We observed that 2′-HCA (2.5-10 μM) inhibits the phosphorylation of ERKs, p38 and AKT in HEL (Fig. 2Fa) and A431 cells (Fig. 2Fb).
2′-HCA inhibits EGF-induced transformation of HaCaT and JB6 cells
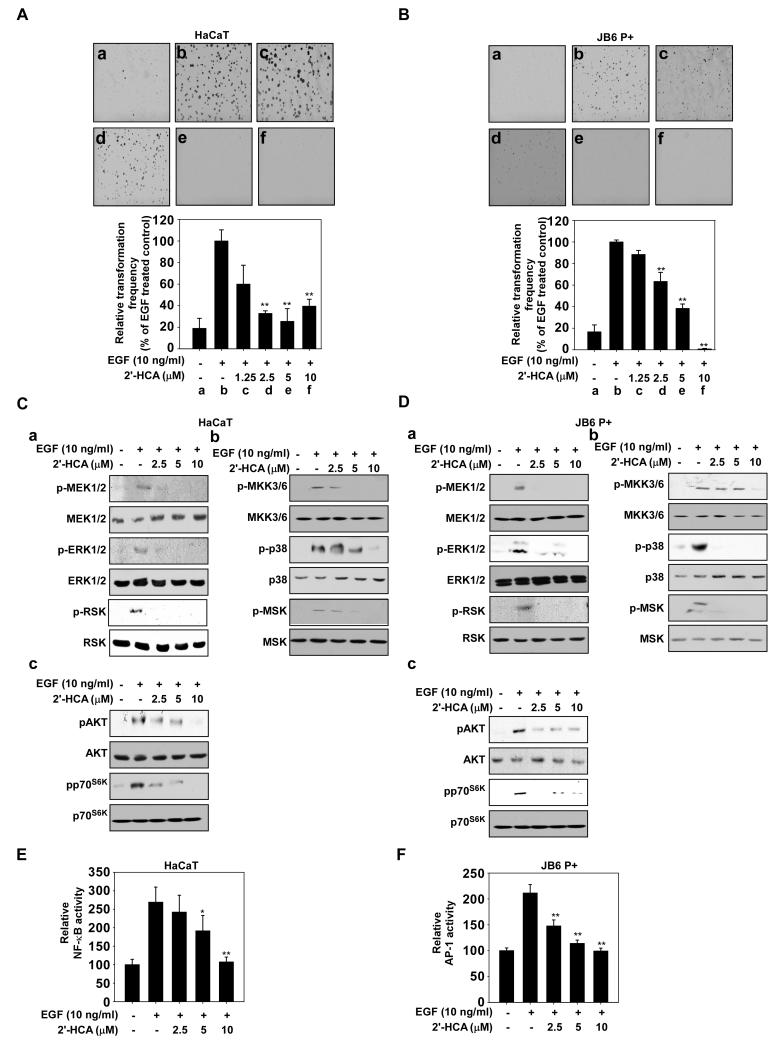
The epidermal growth factor (EGF)-induced cell transformation models with JB6 P+ or HaCaT cells are some of the best models for studying cancer prevention. JB6 P+ and HaCaT cells are normal skin cell lines and treatment of these cells with EGF can transform them to cancerous cells in soft agar. Anchorage-independent cell growth in soft agar is a well-accepted marker for transformation. Thus we used these neoplastic cell transformation models to measure the chemopreventive effects of 2′-HCA in JB6 P+ and HaCaT cells. Quantification of cell colonies revealed that EGF-induced neoplastic transformation was markedly inhibited in HaCaT (Fig. 3Aa-f) and JB6 P+ (Fig. 3Ba-f) cells treated with 5 μM 2′-HCA. Similar to HEL and A431 cells, 2′-HCA inhibited EGF-induced signaling in HaCaT (Fig. 3Ca-c) and JB6 P+ (Fig. 3Da-c) cells. NF-κB and AP-1 are well-known transcription factors activated by EGF and play pivotal roles in neoplastic transformation (34). A luciferase reporter gene assay with stable NF-κB and AP-1 luciferase-transfected HaCaT or JB6 P+ cells showed that 2′-HCA treatment (2.5-10 μM) inhibited both NF-κB (Fig. 3E) and AP-1 (Fig. 3F) transactivation.
Figure 3.
2′-HCA inhibits epidermal growth factor (EGF)-induced neoplastic transformation of HaCaT and JB6 P+ cells. 2′-HCA inhibits EGF-induced neoplastic transformation of (A) HaCaT and (B) JB6 P+ cells. The effect of 2′-HCA on EGF-induced cell transformation compared with untreated control cells (a); and cells treated with EGF alone (b); EGF and 1.25 μM 2′-HCA (c); EGF and 2.5 μM 2′-HCA (d); or EGF and 5 μM 2′-HCA (e); or EGF and 10 μM 2′-HCA (f). The colonies were counted under a microscope with the aid of the Image-Pro Plus software program (vs. 6.2). Results are represented as mean values ± S.E. (n = 3). The asterisks (*, **) indicate a significant difference (p < 0.05, p < 0.01) compared with the EGF-treated group. 2′-HCA inhibits ERK1/2, p38 and AKT signaling in (C) HaCaT and (D) JB6 P+ cells. HaCaT or JB6 P+ cells were treated with 2′-HCA (0, 2.5, 5, or 10 μM) for 1 h before treatment with EGF (10 ng/mL) and then harvested after 15 min. Immunoblot analysis was conducted as described in Materials and Methods. 2′-HCA suppresses EGF-induced (E) NF-κB and (F) AP-1 transactivation. JB6 P+ cells, which were stably transfected with NF-κB or AP-1 luciferase reporter plasmids, were pretreated with 2′-HCA (0, 2.5, 5, or 10 μM) for 1 h before being exposed to EGF (10 ng/ml) and harvested 6 h later. Relative luciferase activities were determined and data are presented as mean values ± S.D. The asterisks (*, **) indicate a significant (* p < 0.05, ** p < 0.01) inhibition of luciferase activity by 2′-HCA compared to the group treated with EGF alone.
Knockdown of Pim-1 mimics the inhibitory effects of 2′-HCA treatment
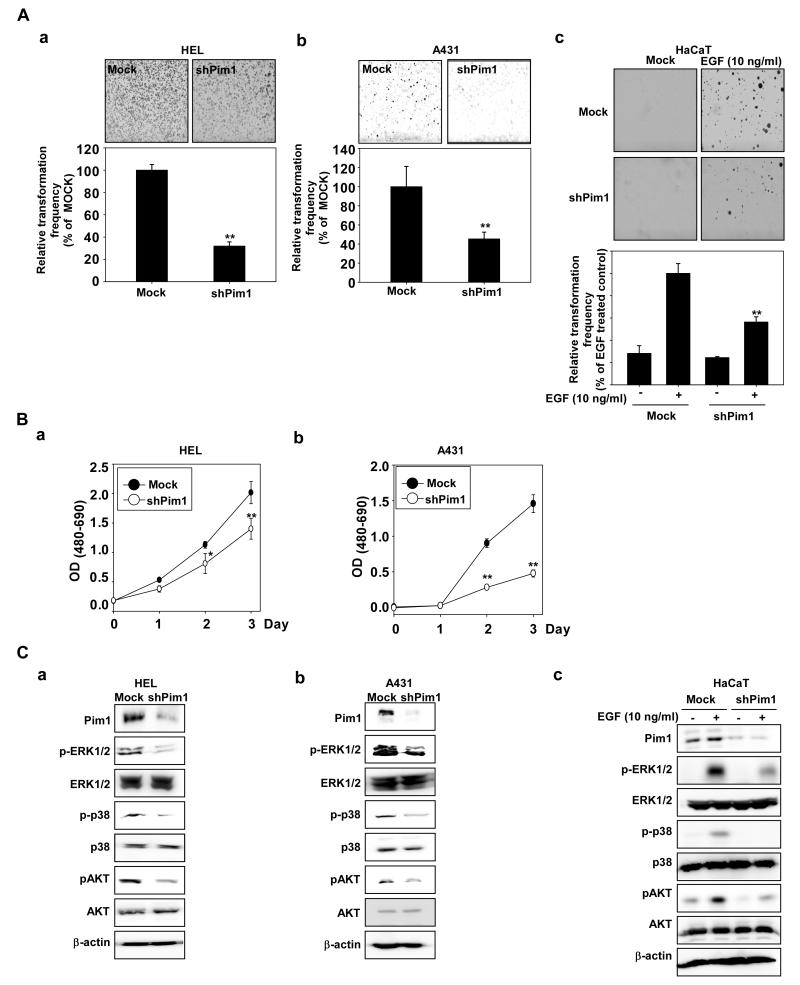
We next investigated the effects of knocking down Pim-1 expression on anchorage-dependent and -independent growth of HEL and A431 cells, and on EGF-induced cell transformation of the human HaCaT cells. Anchorage-independent cell growth of HEL and A431 cells silenced with shPim-1 was slower than that of mock-transfected cells (Fig. 4Aa, b). EGF-induced transformation in HaCaT cells was also decreased by knockdown of Pim-1 (Fig. 4Ac). Treatment with 2′-HCA also decreased anchorage-dependent growth of HEL (Fig. 4Ba) and A431 (Fig. 4Bb) cells. Notably, compared with HEL and A431 cells expressing green fluorescent protein–shRNA (shMock), HEL and A431 cells expressing shPim-1 exhibited a substantially reduced abundance of endogenous Pim-1 (Fig. 4C). Knockdown of Pim-1 inhibited phosphorylation of ERKs, p38, and AKT (Fig. 4Ca,b), as well as EGF-induced ERKs, p38, and AKT phosphorylation (Fig. 4Cc).
Figure 4.
Knockdown of Pim-1 inhibits growth of HEL and A431 cells and EGF-induced neoplastic growth of HaCaT cells. A, Knockdown of Pim-1 decreases soft agar growth of (a) HEL, (b) A431, and (c) HaCaT cells. A soft agar assay was performed using Mock or shPim-1 transfected HEL, A431, or HaCaT cells and the number of colonies was counted under a microscope with the aid of the Image-Pro Plus software program (vs. 6.2). B, Knockdown of Pim-1 inhibits anchorage-dependent growth of (a) HEL and (b) A431 cells. Growth was evaluated by MTS assay using Mock- or shPim-1-transfected HEL or A431 cells. The asterisks (*, **) indicate a significant (* p < 0.05, ** p < 0.01) difference between Mock- and shPim-1-transfected cells. C, 2′-HCA inhibits ERK1/2, p38 and AKT phosphorylation in (a) HEL, (b) A431 and (c) EGF-induced HaCaT cells. HEL or A431 cells were seeded and incubated for 48 h before the proteins were recovered. Mock- or shPim-1-transfected HaCaT cells were treated with EGF (10 ng/mL) and harvested after 15 min. Immunoblotting was conducted using specific antibodies.
Pim-1 knockdown elicits resistance to 2′-HCA
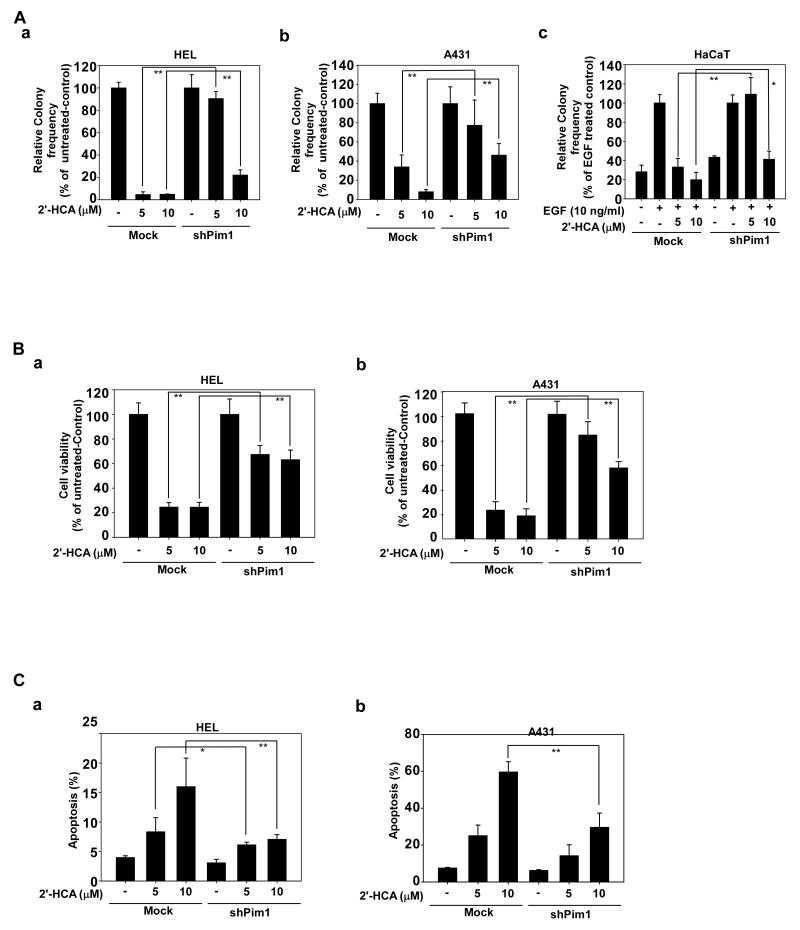
To confirm the specificity of 2′-HCA, we compared its effects on mock- and shPim-1-transfected cells. Because growth rates are altered by knockdown of Pim-1, we designated the proliferation rate of 2′-HCA untreated cells as 100% to compare the mock and shPim-1 HEL and A431 groups. For HaCaT cells, the EGF-only group was set as the 100% comparator. The results indicated that cells expressing shPim-1 were resistant to the inhibitory effects of 2′-HCA against anchorage-independent cell growth in HEL (Fig. 5Aa), A413 (Fig. 5Ab) cells and EGF-induced transformation of HaCaT cells (Fig. 5Ac), compared to cells expressing shMock (Fig. 5A). We also examined the effect of 2′-HCA on anchorage-dependent cell growth between mock and shPim-1 HEL and A413 cells. The results showed that the inhibitory effect of 2′-HCA (5-10 μM) on shPim-1 cells was much less than its effect on shMock-transfected HEL (Fig. 5Ba) or A413 cells (Fig. 5Bb). In addition, 2′-HCA (5-10 μM) induced apoptosis to a greater extent in shMock cells compared with shPim-1 cells (Fig. 5Ca, b). These findings suggest that the anticancer effects of 2′-HCA are at least partially dependent on Pim-1 protein expression.
Figure 5.
Pim-1 knockdown cells are resistant to the effects of 2′-HCA. A, Knockdown of Pim-1 causes resistance to the inhibitory effects of 2′-HCA against anchorage-independent growth of A431 or HEL cells or EGF-induced neoplastic growth of HaCaT cells. A soft agar assay was performed with Mock- or shPim1-transfected (a) HEL, (b) A431, or (c) EGF-induced HaCaT cells treated with 2′-HCA (0, 2.5, 5, or 10 μM). The number of colonies was counted under a microscope with the aid of the Image-Pro Plus software program (vs. 6.2). The untreated control was calculated as 10% of Mock- or shPim-1-transfected HEL and A431 cells and the EGF-treated group was calculated as 100% of Mock- or shPim-1-transfected HaCaT cells. B, Growth was evaluated by MTS assay using Mock- or shPim1-transfected (a) HEL and (b) A431 cells treated with 2′-HCA (0, 2.5, 5, or 10 μM) for 48 h. C, Apoptosis was analyzed by annexin V/PI staining of Mock- or shPim1-transfected (a) HEL and (b) A431 cells treated with (0, 2.5, 5, or 10 μM) 2′-HCA in for 24 h. The asterisks (*, **) indicate a significant difference (p < 0.05, p < 0.01) between Mock- or shPim-1-transfected HEL and A431 cells.
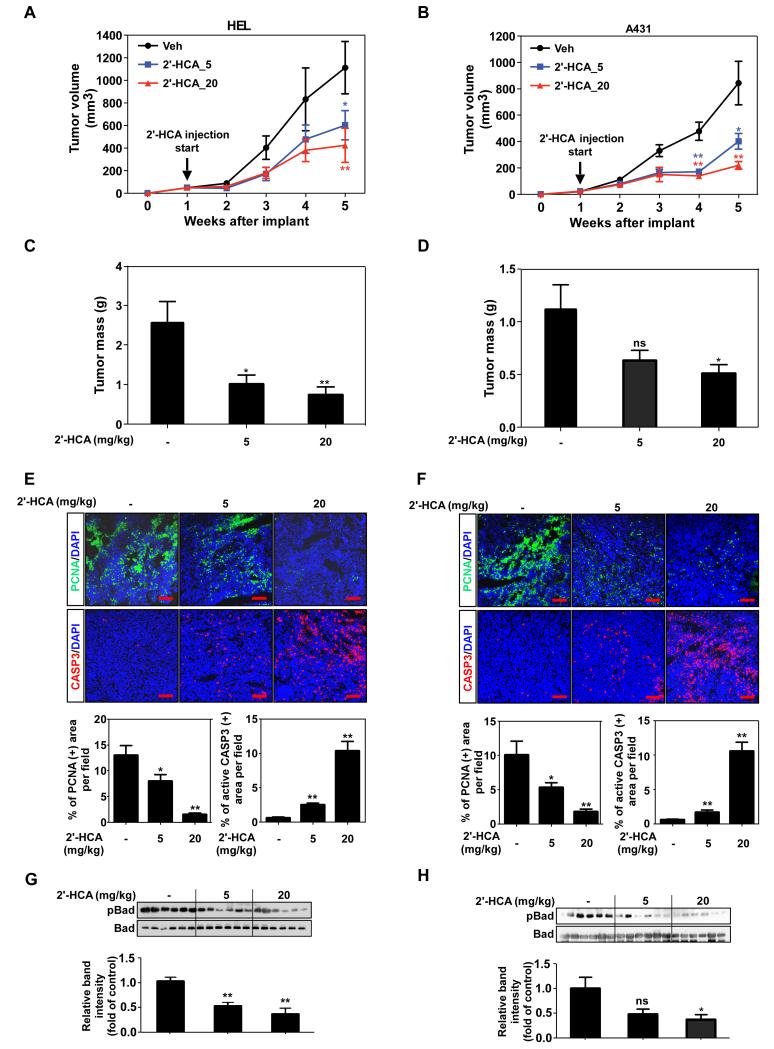
2′-HCA inhibits leukemia and skin cancer tumor growth in a xenograft mouse model
To investigate the anti-cancer effects of 2′-HCA in vivo, HEL or A431 cells were subcutaneously transplanted into nude mice, followed by treatment with 2′-HCA (5 or 20 mg/kg). Mice treated with 2′-HCA exhibited significantly suppressed HEL and A431-derived tumor growth (Fig. 6 A, B) that occurred in a dose-dependent manner (Fig. 6 C, D). Immuno-staining data demonstrated that the levels of PCNA, a cell proliferation marker, were lower in the 2′-HCA-treated HEL and A431 tumor groups compared to the vehicle-treated groups (Fig. 6E, F). In addition, the levels of cleaved caspase 3, a cellular marker of apoptosis, were higher in the 2′-HCA treated tumors compared to the vehicle-treated group (Fig. 6 E, F). Immunoblot data showed that both the HEL (Fig. 6G) and A431 (Fig. 6H) groups treated with 2′-HCA had tumors exhibiting lower phosphorylation levels of Bad, which is a direct downstream substrate of Pim-1. No significant body weight loss or conspicuous physical changes were observed in mice treated with 2′-HCA, suggesting that the compound was not overtly toxic to the animals (Supplementary Fig. 1A-C).
Figure 6.
2′-HCA inhibits HEL and A431 tumor growth in a mouse xenograft model. Mice implanted with (A) HEL or (B) A431 cells were administered 2′-HCA or the vehicle control. Tumors were removed at 5 weeks after implantation of cells. HEL and A431 tumor volume (A, B) and tumor mass (C, D) were determined as described in Materials and Methods (n = 16). Representative images and quantification of PCNA (green) and cleaved caspase 3 (red) staining of (E) HEL and (F) A431 tumors (n = 12). Scale bars = 100 μM. Data are represented as mean values ± S.E. Immunoblotting and quantification of phosphorylated and basal levels of Bad in (G) HEL and (H) A431 tumor lysates (n = 6). Data are represented as mean values ± S.E. The asterisks (*, **) indicate a significant (p < 0.05, p < 0.01) difference between the vehicle-treated and 2′-HCA-treated groups.
Discussion
The traditional approach to elucidating the beneficial effects of phytochemicals begins with the screening of their effects, followed by the identification of the molecular targets and the overall mechanism of action. Many potential obstacles can arise at each step, which causes significant hurdles for researchers. In this study, we took an opposite approach and first identified the molecular target of a natural compound found in cinnamon. At this time no one else has studied or identified a direct target for this cinnamon compound. By screening a library of protein kinases, we discovered that Pim-1 is a target of 2′-HCA. Using structural biology methods, we then demonstrated the physical binding mode between Pim-1 and 2′-HCA. We further identified the specific disease subtypes relevant for the effects of 2′-HCA on Pim-1. Pim-1 is highly expressed in leukemia cells and 2′-HCA exhibits inhibitory effects on these cells in vitro and in vivo. Based on the xenograft tumor volume and mass data of both HEL and A431 tumors, treatment with 2′-HCA resulted in dose-dependent tumor growth inhibitory effects. In HEL tumor data, 2′-HCA treatment significantly decreased tumor volume and mass in a dose-dependent manner with the higher dose seeming more effective. Moreover in the A431 tumor data, only the tumors in the 20 mg/kg treated group were significantly decreased. These results demonstrated that 2′-HCA treatment inhibited tumor growth in a dose-dependent manner. Additionally, 2′-HCA was also found to have chemopreventive effects in both leukemia and skin cancer cell lines, stemming from Pim-1 inhibition.
Pim-1 is a kinase that is primarily involved in transcriptional activation and cellular signal transduction pathways related to cell cycle progression and apoptosis (28). Pim-1 overexpression has been observed in many types of cancer, including leukemia, squamous cell carcinoma, prostate cancer, gastric carcinoma, and bladder cancer (35), and it phosphorylates the anti-apoptosis protein, Bad, to inhibit apoptosis (29). We also measured the phosphorylation levels of Bad, which is representative of Pim1 activity and 2′-HCA treatment inhibited the phosphorylation of Bad in a dose-dependent manner in both HEL and A431 tumors. Myc, p21cip and p27kip are also substrates of Pim-1 (33,36,37). Many studies have shown that the inhibition of Pim-1 using pharmacological inhibitors, shRNA and Pim-1-specific antibodies regulates signal transduction through the MAPK and AKT pathways, inducing apoptosis (30,31,38). Because of its importance in leukemia, drugs targeting the Pim-1 kinase, such as AZD1208, are being tested in Phase 1 clinical trials. In this study, using 3 types of experimental models, we demonstrated that inhibition of Pim-1 by 2′-HCA creates a similar pattern to that for inhibition of Pim-1 by shRNA. Anchorage-dependent and -independent cell growth were suppressed in the presence of either 2′-HCA and shPim-1.
The specificity of small molecules for their designated cellular targets is a major point of concern in drug development. In this study, we screened 77 protein kinases, and found that 2′-HCA inhibited Pim-1 by at least 80%, an effect that was not observed for any other kinase. 2′-HCA treatment or expression of shPim-1 resulted in similar effects on signal transduction. Additionally, we showed that 2′-HCA resistance arises in Pim-1 knockdown cells. These data support the notion that 2′-HCA is a specific inhibitor of Pim-1. To determine how 2′-HCA specifically targets Pim-1, we used structural biology approaches. Pim-1 is functionally and structurally different from other classes of kinases (39), and has a unique hinge region sequence as well as novel hinge architecture (40). Our co-crystallography data revealed that 2′-HCA binds at the unique hinge region of Pim-1. This underscores the possibility of developing an effective therapeutic agent with high specificity for Pim-1 (39).
In the crystallography results, the density for the 2′-HCA ligand was not completely visible at a contour level of 1.0 σ, so the ligand placement was aided by the hydrogen-bonding pattern between the ligand and the neighboring water molecules and side chains. The observation that a ligand this small is not completely visible in an active site that was meant for a much larger ligand is not surprising. The molecule appears to be a relatively weak binder, without many steric constraints to exclude alternative orientations. However, 2′-HCA overlaps with and is coplanar with the aromatic ring system of benzofuran-2-carboxylic acid in PDB entry 3R04 (Fig. 1E). The 2-hydroxy group also makes hydrogen bond contacts with Lys67 and the backbone amino group of Asp168. The interactions at the innermost part of the binding pocket are conserved. An interesting feature of the interaction of 2′-HCA is the recruitment of Phe49 that moves 6.3 Å away from its position in the apo structure to shield the ligand from the exterior solvent. This movement completely remodels the glycine-rich loop such that it blocks the entrance to the binding pocket acting as a gatekeeper loop.
Elucidation of the direct molecular targets of new potential medicines is the first step towards personalized therapeutic approaches (41). Our study has highlighted the feasibility of such approaches, with the possibility of 2′-HCA-derived therapies having potential application for the treatment of patients with high Pim-1 activity. The further elucidation of targeted agents like 2′-HCA combined with genetic analysis of each patient will help usher in an accelerated era of personalized medicine.
Supplementary Material
Acknowledgments
Financial Support
Co-crystallography between Pim-1 and 2′-HCA was performed by Drs. Derek Logan and Björn Walse at SARomics Biostructures AB (Lund, Sweden). This work was supported by the National Leap Research Program (No. 2010-0029233) through the National Research Foundation of Korea funded by the Ministry of Science, ICT and Future Planning (MSIP) of Korea. This work was also supported by The Hormel Foundation and National Institutes of Health grants CA172457, CA166011, R37CA081064 and CA027502.
Footnotes
Conflict of Interest: The authors declare no conflict of interest.
References
- 1.Grayson M. Traditional Asian medicine. Nature. 2011;480(7378):S81–S81. doi: 10.1038/480S81a. [DOI] [PubMed] [Google Scholar]
- 2.Lee KW, Bode AM, Dong Z. Molecular targets of phytochemicals for cancer prevention. Nature reviews Cancer. 2011;11(3):211–8. doi: 10.1038/nrc3017. [DOI] [PubMed] [Google Scholar]
- 3.Cheung F. Modern TCM: Enter the clinic. Nature. 2011;480(7378):S94–S95. doi: 10.1038/480S94a. [DOI] [PubMed] [Google Scholar]
- 4.Choi J, Lee KT, Ka H, Jung WT, Jung HJ, Park HJ. Constituents of the essential oil of the Cinnamomum cassia stem bark and the biological properties. Archives of pharmacal research. 2001;24(5):418–23. doi: 10.1007/BF02975187. [DOI] [PubMed] [Google Scholar]
- 5.Qin B, Panickar KS, Anderson RA. Cinnamon: potential role in the prevention of insulin resistance, metabolic syndrome, and type 2 diabetes. Journal of diabetes science and technology. 2010;4(3):685–93. doi: 10.1177/193229681000400324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vangalapati M, Sree Satya N, Surya Prakash DV, Avanigadda S. A review on pharmacological activities and clinical effects of Cinnamon species. Research Journal of Pharmaceutical, Biological and Chemical Sciences. 2012;3(1):653–63. [Google Scholar]
- 7.Koppikar SJ, Choudhari AS, Suryavanshi SA, Kumari S, Chattopadhyay S, Kaul-Ghanekar R. Aqueous cinnamon extract (ACE-c) from the bark of Cinnamomum cassia causes apoptosis in human cervical cancer cell line (SiHa) through loss of mitochondrial membrane potential. BMC cancer. 2010;10:210. doi: 10.1186/1471-2407-10-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kwon HK, Hwang JS, So JS, Lee CG, Sahoo A, Ryu JH, et al. Cinnamon extract induces tumor cell death through inhibition of NFkappaB and AP1. BMC cancer. 2010;10:392. doi: 10.1186/1471-2407-10-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schoene NW, Kelly MA, Polansky MM, Anderson RA. Water-soluble polymeric polyphenols from cinnamon inhibit proliferation and alter cell cycle distribution patterns of hematologic tumor cell lines. Cancer letters. 2005;230(1):134–40. doi: 10.1016/j.canlet.2004.12.039. [DOI] [PubMed] [Google Scholar]
- 10.Kwon HK, Jeon WK, Hwang JS, Lee CG, So JS, Park JA, et al. Cinnamon extract suppresses tumor progression by modulating angiogenesis and the effector function of CD8+ T cells. Cancer letters. 2009;278(2):174–82. doi: 10.1016/j.canlet.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 11.Lu J, Zhang K, Nam S, Anderson RA, Jove R, Wen W. Novel angiogenesis inhibitory activity in cinnamon extract blocks VEGFR2 kinase and downstream signaling. Carcinogenesis. 2010;31(3):481–8. doi: 10.1093/carcin/bgp292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kiridena W, Miller KG, Poole CF. Identification of 2-hydroxycinnamaldehyde in the cinnamons of commerce. Journal of Planar Chromatography - Modern TLC. 1995;8(3):177–83. [Google Scholar]
- 13.Hong SH, Kim J, Kim J-M, Lee S-Y, Shin D-S, Son K-H, et al. Apoptosis induction of 2′-hydroxycinnamaldehyde as a proteasome inhibitor is associated with ER stress and mitochondrial perturbation in cancer cells. Biochem Pharmacol. 2007;74(4):557–65. doi: 10.1016/j.bcp.2007.05.016. [DOI] [PubMed] [Google Scholar]
- 14.Lee CW, Lee SH, Lee JW, Ban JO, Lee SY, Yoo HS, et al. 2-hydroxycinnamaldehyde inhibits SW620 colon cancer cell growth through AP-1 inactivation. J Pharmacol Sci (Tokyo, Jpn) 2007;104(1):19–28. doi: 10.1254/jphs.fp0061204. [DOI] [PubMed] [Google Scholar]
- 15.Ismail IA, Kang HS, Lee H-J, Chang H, Yun J, Lee CW, et al. 2-Hydroxycinnamaldehyde inhibits the epithelial-mesenchymal transition in breast cancer cells. Breast Cancer Res Treat. 2013;137(3):697–708. doi: 10.1007/s10549-012-2388-7. [DOI] [PubMed] [Google Scholar]
- 16.Kim S-A, Sung Y-K, Kwon B-M, Yoon J-H, Lee H, Ahn S-G, et al. 2′-hydroxycinnamaldehyde shows antitumor activity against oral cancer in vitro and in vivo in a rat tumor model. Anticancer Res. 2010;30(2):489–94. [PubMed] [Google Scholar]
- 17.Hwang H, Jeon H, Ock J, Hong SH, Han Y-M, Kwon B-M, et al. 2′-Hydroxycinnamaldehyde targets low-density lipoprotein receptor-related protein-1 to inhibit lipopolysaccharide-induced microglial activation. J Neuroimmunol. 2011;230(1-2):52–64. doi: 10.1016/j.jneuroim.2010.08.021. [DOI] [PubMed] [Google Scholar]
- 18.Lee MA, Park HJ, Chung H-J, Kim WK, Lee SK. Antitumor Activity of 2-Hydroxycinnamaldehyde for Human Colon Cancer Cells through Suppression of β-Catenin Signaling. J Nat Prod. 2013;76(7):1278–84. doi: 10.1021/np400216m. [DOI] [PubMed] [Google Scholar]
- 19.Brault L, Gasser C, Bracher F, Huber K, Knapp S, Schwaller J. PIM serine/threonine kinases in the pathogenesis and therapy of hematologic malignancies and solid cancers. Haematologica. 2010;95(6):1004–15. doi: 10.3324/haematol.2009.017079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weirauch U, Beckmann N, Thomas M, Grunweller A, Huber K, Bracher F, et al. Functional role and therapeutic potential of the pim-1 kinase in colon carcinoma. Neoplasia (New York, NY) 2013;15(7):783–94. doi: 10.1593/neo.13172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pierce AC, Jacobs M, Stuver-Moody C. Docking study yields four novel inhibitors of the protooncogene Pim-1 kinase. Journal of medicinal chemistry. 2008;51(6):1972–5. doi: 10.1021/jm701248t. [DOI] [PubMed] [Google Scholar]
- 22.Kabsch W. XDS. Acta crystallographica Section D, Biological crystallography. 2010;66(Pt 2):125–32. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, et al. Overview of the CCP4 suite and current developments. Acta crystallographica Section D, Biological crystallography. 2011;67(Pt 4):235–42. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. Journal of applied crystallography. 2007;40(Pt 4):658–74. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta crystallographica Section D, Biological crystallography. 2011;67(Pt 4):355–67. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao H, Xie Y, Yang Q, Cao Y, Tu H, Cao W, et al. Pharmacokinetic study of cinnamaldehyde in rats by GC-MS after oral and intravenous administration. Journal of pharmaceutical and biomedical analysis. 2014;89:150–7. doi: 10.1016/j.jpba.2013.10.044. [DOI] [PubMed] [Google Scholar]
- 27.Uhlen M, Oksvold P, Fagerberg L, Lundberg E, Jonasson K, Forsberg M, et al. Towards a knowledge-based Human Protein Atlas. Nature biotechnology. 2010;28(12):1248–50. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- 28.Swords R, Kelly K, Carew J, Nawrocki S, Mahalingam D, Sarantopoulos J, et al. The Pim kinases: new targets for drug development. Current drug targets. 2011;12(14):2059–66. doi: 10.2174/138945011798829447. [DOI] [PubMed] [Google Scholar]
- 29.Aho TL, Sandholm J, Peltola KJ, Mankonen HP, Lilly M, Koskinen PJ. Pim-1 kinase promotes inactivation of the pro-apoptotic Bad protein by phosphorylating it on the Ser112 gatekeeper site. FEBS letters. 2004;571(1-3):43–9. doi: 10.1016/j.febslet.2004.06.050. [DOI] [PubMed] [Google Scholar]
- 30.Gu JJ, Wang Z, Reeves R, Magnuson NS. PIM1 phosphorylates and negatively regulates ASK1-mediated apoptosis. Oncogene. 2009;28(48):4261–71. doi: 10.1038/onc.2009.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu XF, Li J, Vandervalk S, Wang Z, Magnuson NS, Xing PX. PIM-1-specific mAb suppresses human and mouse tumor growth by decreasing PIM-1 levels, reducing Akt phosphorylation, and activating apoptosis. The Journal of clinical investigation. 2009;119(2):362–75. doi: 10.1172/JCI33216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li J, Loveland BE, Xing PX. Anti-Pim-1 mAb inhibits activation and proliferation of T lymphocytes and prolongs mouse skin allograft survival. Cellular immunology. 2011;272(1):87–93. doi: 10.1016/j.cellimm.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 33.Wang J, Anderson PD, Luo W, Gius D, Roh M, Abdulkadir SA. Pim1 kinase is required to maintain tumorigenicity in MYC-expressing prostate cancer cells. Oncogene. 2012;31(14):1794–803. doi: 10.1038/onc.2011.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bode AM, Dong Z. Signal transduction pathways in cancer development and as targets for cancer prevention. Progress in nucleic acid research and molecular biology. 2005;79:237–97. doi: 10.1016/S0079-6603(04)79005-4. [DOI] [PubMed] [Google Scholar]
- 35.Blanco-Aparicio C, Carnero A. Pim kinases in cancer: diagnostic, prognostic and treatment opportunities. Biochemical pharmacology. 2013;85(5):629–43. doi: 10.1016/j.bcp.2012.09.018. [DOI] [PubMed] [Google Scholar]
- 36.Morishita D, Katayama R, Sekimizu K, Tsuruo T, Fujita N. Pim kinases promote cell cycle progression by phosphorylating and down-regulating p27Kip1 at the transcriptional and posttranscriptional levels. Cancer research. 2008;68(13):5076–85. doi: 10.1158/0008-5472.CAN-08-0634. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Y, Wang Z, Magnuson NS. Pim-1 kinase-dependent phosphorylation of p21Cip1/WAF1 regulates its stability and cellular localization in H1299 cells. Molecular cancer research : MCR. 2007;5(9):909–22. doi: 10.1158/1541-7786.MCR-06-0388. [DOI] [PubMed] [Google Scholar]
- 38.Natarajan K, Xie Y, Burcu M, Linn DE, Qiu Y, Baer MR. Pim-1 kinase phosphorylates and stabilizes 130 kDa FLT3 and promotes aberrant STAT5 signaling in acute myeloid leukemia with FLT3 internal tandem duplication. PloS one. 2013;8(9):e74653. doi: 10.1371/journal.pone.0074653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ogawa N, Yuki H, Tanaka A. Insights from Pim1 structure for anti-cancer drug design. Expert opinion on drug discovery. 2012;7(12):1177–92. doi: 10.1517/17460441.2012.727394. [DOI] [PubMed] [Google Scholar]
- 40.Larid PW, Van der Lugt NMT, Clarke A, Domen J, Linders K, McWhir J, et al. In vivo analysis of Pim-1 deficiency. Nucleic Acids Research. 1993;21(20):4750–55. doi: 10.1093/nar/21.20.4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kang NJ, Shin SH, Lee HJ, Lee KW. Polyphenols as small molecular inhibitors of signaling cascades in carcinogenesis. Pharmacology & therapeutics. 2011;130(3):310–24. doi: 10.1016/j.pharmthera.2011.02.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.