INTRODUCTION
Chemotherapy induced peripheral neuropathy (CIPN) is a dose limiting toxicity of paclitaxel, termed here paclitaxel induced peripheral neuropathy (PIPN). PIPN affects a minority of patients and cannot be predicted from clinical patient characteristics. Therefore, a phamacogenomic basis has been proposed for PIPN (1).
Genome wide association studies (GWAS) provide an unbiased approach for the identification of novel genetic loci associated with complex traits (2). Schneider et al. performed the first PIPN GWAS in 2011, (3) finding that the risk of PIPN was linked to two single nucleotide variants (SNV) in two genes that had previously not been known to be associated with this condition or with CIPN due to any other drug. The SNV rs2296308 in RWDD3 was reported to be associated with PIPN with a HR=1.5 and a significance level of p=8.5 × 10−8. The rs1829 SNV in TECTA was reported to be associated with PIPN with a HR= 2.1 and a significance level of p=3.2×10−7. Both SNV thereby passed the threshold set by the authors for genome-wide significance thereby representing important novel PIPN gene candidates.
Bergmann et al. investigated rs2296308 and rs1829 in a Scandinavian ovarian cancer patient cohort (4). Their study failed to corroborate the report by Schneider et al., thereby calling that GWAS result into question. The original GWAS consisted of a very large cohort, 2204 patients, while the validation attempt by Bergmann et al. was limited to a smaller cohort of 241 patients. Both studies used similar phenotyping and analyses relying on Common Toxicity Criteria Adverse Effects (CTCAE) reporting of toxicity and time-to-grade 2 or worse scores as the primary endpoint. These conflicting results thereby created uncertainty in the field regarding the role of RWWD3 and TECTA.
We recently reported a new PIPN cohort, N08C1, for which improved phenotyping was available using serial assessments of PIPN symptoms with the European Organization for Research and Treatment of Cancer (EORTC) Quality of Life Questionnaire (QLQ) CIPN-20 (CIPN20) (5). Here, we report testing of the association of rs2296308 in RWWD3 and rs1829 in TECTA with PIPN in N08C1.
METHODS
Patients
The patient cohort N08C1 was recently reported (6). In brief, patients were exposed to paclitaxel chemotherapy and evaluated by repeat assessments with the CIPN20 instrument. An “extreme phenotyping” approach was used to select cases and controls. Extreme phenotyping improves the power of genetic studies and has been discussed and used and discussed by numerous authors before (7, 8). In N08C1, cases were selected as those patients that had significantly worse progression of PIPN symptoms over time and controls were selected as those patients that had significantly less progression of PIPN symptoms over time. Patients with equivocal progression of PIPN symptoms were excluded from the study. Informed consent was obtained from patients for CIPN assessment and collection of blood for genetic testing.
Genotyping of rs2296308 in RWDD3
The SNV rs2296308 was genotyped by the approach described previously (6). In brief, a short read sequencing approach was employed using Illumina reagents (True Seq and target gene enrichment kit) and sequenced on Illumina HiSeq2000 platform. Genotype calling from short reads was performed with the bioinformatics analysis pipeline Genome Analysis Toolkit (GATK) [McKenna 2011] and Variant Quality Score Recalibration (VQSR) with analysis parameters set as described before (6).
Genotyping of rs1829 in TECTA
The SNV rs1829 was located in the intronic region and therefore was not captured by the above capture sequencing approach. Therefore, we used traditional Taqman PCR to genotype rs1829. Genomic DNA (gDNA) for PCR genotyping was available for 114 patients. gDNA was isolated from peripheral blood leukocytes using commercially available kits. PCR amplification of gDNA encompassing the loci of interest was achieved to generate DNA amplicons. TaqMan probes were designed specifically to assay the locus. Analysis of the fluroscence signal from the Taqman PCR reaction and allele discrimination was executed with the SDS 2.0 software (Applied Biosystems).
RESULTS
73 cases and 46 controls were identified with the extreme phenotyping approach forming the cohort as previously described (6). All 119 patients were successfully genotyped at rs2296308, which is located in exon 3 of the RWDD3 gene that codes for a non-synonymous SNV. The average sequencing depth across this locus was 138 fold exceeding the sequencing depth requirements that are typically set at 20 to 100 fold. For rs1829, PCR genotyping succeeded in 113 patients out of the 114 with available gDNA (the PCR reaction failed in one patient) providing results on 71 cases and 42 controls, on which the subsequent analysis was based.
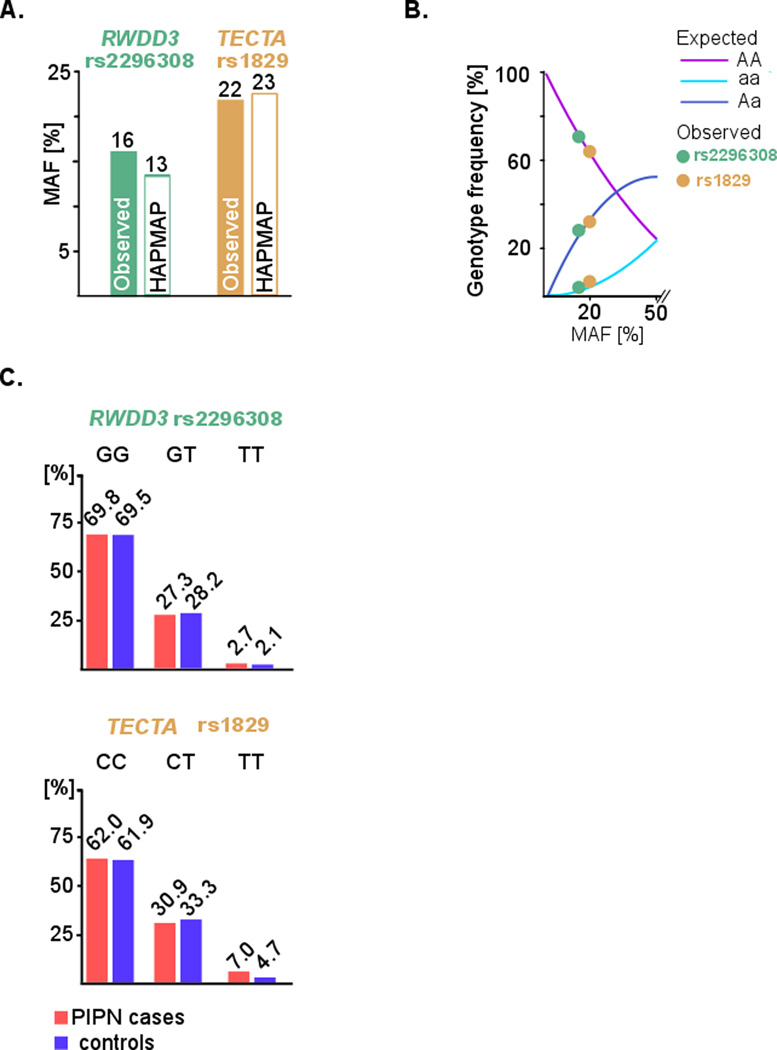
The minor allele frequency (MAF) detected by genetic testing in the present study was 16% for rs2296308 and 22% for rs1829. Thereby, the observed MAF was similar to the MAF reported for a large reference dataset, the dbSNP141 database using HAPMAP-CEU as the reference population. Fig 1 panel A shows this comparison between the observed and the reference MAF. The genotype frequencies detected in the present study were also in agreement with expectations of Hardy Weinberg equilibrium as depicted in panel B of Fig 1. These data suggest that the N08C1 genotyping results were faithful without dropout of an allele and free from any possible inconsistencies.
Figure 1. RWDD3 rs2296308 and TECTA rs1829 quality control metrics for the N08C1 cohort and testing for an association with paclitaxel induced peripheral neuropathy (PIPN).
A. Minor allele frequency (MAF). The MAF of rs2296308 and rs1829 observed in N08C1 was similar to the MAF of the public reference dataset HAPMAP (reference population = CEU).
B. Hardy-Weinberg equilibrium (HWE) testing. The observed distribution of genotypes for rs2296308 and rs1829 were consistent with expectations for SNV that are not under strong natural selection thereby further supporting that the assay employed in the study was not affected by allele drop-out. No departure from the HWE was noted.
C. Association testing. Genotype frequencies for SNV in PIPN cases and controls identified by extreme phenotyping are shown. For rs1829 in the data is presented on 71 cases and 42 controls.
The minor risk allele ‘T’ for rs2296308 in the gene RWDD3 was found in 22 patients among 73 cases and 14 patients among 46 controls, which corresponded to similar fraction of 30.1% in cases and 30.4% controls respectively. 2 patients among cases and 1 among controls were homozygous for the minor allele.
The minor risk allele ‘T’ for rs1829 in the gene TECTA was found in 27 patients among 71 cases and 16 patients among 42 controls, which corresponded to fraction of 38% in CIPN cases and 38% in controls. 5 patients among cases and 2 patients among controls were homozygous for the minor allele. Fig. 1 panel C shows the genotype distribution for the two SNV in PIPN cases and controls.
Allelic variation can be associated with a phenotype through different mechanisms involving either one or both alleles. To accommodate all possibilities of association with PIPN testable with the available data, we calculated an odds ratio (OR) and significance values for an association of each SNV with PIPN for the dominant and the additive model. Results of the association testing are shown in Table 1. Under the dominant model, where only one risk allele is needed to express the disease phenotype, rs2296308 had an OR=0.98 and p= 0.59. rs1829 had an OR=0.99 and p=0.58. An OR that is close to 1 is found, when no genetic association exists. Therefore, the OR results of 0.98 and 0.99 with non-significant p-values demonstrate that there was no effect of these two SNV on PIPN. The results for the additive were similarly negative. Few patients were homozygous for the minor allele rendering tests for a recessive inheritance model ineffective. Thus, no genetic association was observed in the present study for either SNV under any of the possible inheritance models.
Table 1.
| Gene/ SNV |
Inheritence model |
Odds-ratio (OR) | p-val |
|---|---|---|---|
|
RWDD3 rs2296308 |
Dominant | 0.98 (0.41–2.37) | 0.59 |
| Additive | 1.01 (0.47–2.16) | 0.56 | |
| Recessive | NA* | 0.66 | |
|
TECTA rs1829 |
Dominant | 0.99 (0.42–2.32) | 0.58 |
| Additive | 1.06 (0.53–2.15) | 0.49 | |
| Recessive | NA* | 0.48 |
NA*-Not applicable as counts are too few
Bergmann et al observed that the minor allele of rs1829 in the gene TECTA was possibly protective as indicated by a HR=0.21 and urged caution that this unexpected result should prompt further studies. The results from our study suggest a middle ground, specifically, that rs1829 is neither protective nor a risk allele bearing no association with PIPN.
DISCUSSION
Validation in independent cohorts is an important objective in studies involving the genetic basis of complex traits including in pharmacogenomics. Moreover, publication of negative results is critical for a field, because if negative data is suppressed while “positive” findings are reported, this systematic over-reporting of false positive results is an important source of bias termed publication bias (9). The contrasting reports by Schneider et al. and by Bergmann et al. created uncertainty, thereby leading us to readdress the question in a new clinical trial cohort.
N08C1 results did not support the original report by Schneider et al., but, instead mostly replicated the findings of Bergmann et al. In the case of TECTA, Schneider et al. found that the minor allele ‘T’ at the SNV rs1829 was a risk allele, while Bergmann et al. found the same minor allele to be protective. Bergmann et al. presented their observation with careful restraint suggesting that it would require replication. Our results suggest that rs1829 has no effect, because the OR=0.99 of was so close to 1.
In the case of RWWD3, Schneider et al. reported that the minor allele was a risk allele, while Bergmann et al. found no effect. Our findings support Bergmann’s result and conclusion.
Taken together, the result by Bergmann et al. and the present report suggest that the association of RWDD3 and TECTA with PIPN may have been a false positive signal possibly related to lack of correction for effect magnitude in the GWAS.
REFERENCES
- 1.Cavaletti G, Alberti P, Marmiroli P. Chemotherapy-induced peripheral neurotoxicity in the era of pharmacogenomics. The Lancet Oncology. 2011;12(12):1151–1161. doi: 10.1016/S1470-2045(11)70131-0. [DOI] [PubMed] [Google Scholar]
- 2.McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JP, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nature reviews Genetics. 2008;9(5):356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 3.Schneider BP, Li L, Miller K, Flockhart D, Radovich M, Hancock BA, et al. Genetic associations with taxane-induced neuropathy by a genome-wide association study (GWAS) in E5103. 2011 ASCO Annual Meeting Abstracts; 2011. http://meeting.ascopubs.org/cgi/content/short/29/15_suppl/1000 - otherarticles. [Google Scholar]
- 4.Bergmann TK, Vach W, Feddersen S, Eckhoff L, Green H, Herrstedt J, et al. GWAS-based association between RWDD3 and TECTA variants and paclitaxel induced neuropathy could not be confirmed in Scandinavian ovarian cancer patients. Acta Oncol. 2012;52(4):871–874. doi: 10.3109/0284186X.2012.707787. [DOI] [PubMed] [Google Scholar]
- 5.Postma TJ, Aaronson NK, Heimans JJ, Muller MJ, Hildebrand JG, Delattre JY, et al. The development of an EORTC quality of life questionnaire to assess chemotherapy-induced peripheral neuropathy: the QLQ-CIPN20. Eur J Cancer. 2005;41(8):1135–1139. doi: 10.1016/j.ejca.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 6.Beutler AS, Kulkarni AA, Kanwar R, Klein CJ, Therneau TM, Qin R, et al. Sequencing of Charcot-Marie-Tooth disease genes in a toxic polyneuropathy. Annals of neurology. 2014;76:727–737. doi: 10.1002/ana.24265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barnett IJ, Lee S, Lin X. Detecting rare variant effects using extreme phenotype sampling in sequencing association studies. Genetic epidemiology. 2013;37(2):142–151. doi: 10.1002/gepi.21699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li D, Lewinger JP, Gauderman WJ, Murcray CE, Conti D. Using extreme phenotype sampling to identify the rare causal variants of quantitative traits in association studies. Genetic epidemiology. 2011;35(8):790–799. doi: 10.1002/gepi.20628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ioannidis JP. Why most published research findings are false. PLoS medicine. 2005;2(8):e124. doi: 10.1371/journal.pmed.0020124. [DOI] [PMC free article] [PubMed] [Google Scholar]