Abstract
A number of Helicobacter species may confound experimental data because of their association with disease progressing in various kinds of laboratory animals. Screening of Helicobacter species is particularly desirable, because they are prevalent in commercial and research animal facilities. The aim of the present study was to compare three diagnostic methods [e.g. Helicobacter stool antigen kit (HpSA), polymerase chain reaction (PCR) and rapid urease test (RUT)] for the identification of Helicobacter spp. in stools or gastric biopsy specimens collected from eight dogs suffering from gastritis. The gastroscopic biopsy specimens were tested using RUT and PCR, while stool specimens were evaluated using both HpSA and PCR. DNAs from the gastric biopsies and stool specimens were analyzed by both a consensus PCR that amplified the RNA polymerase beta-subunit-coding gene (rpoB) of Helicobacter spp. and a species-specific PCR to amplify the urease B gene of Helicobacter heilmannii, Helicobacter pylori, and Helicobacter felis. Helicobacter spp. were detected in 62.5% of the dogs, while H. heilmannii and H. felis were identified in 37.5 and 25% of the dogs, respectively. The HpSA did not efficiently detect Helicobacter spp. in the stool samples compared to the RUT and PCR assays, both of which successfully detected Helicobacter spp. in the two sample types. Finally, we recommend that consensus PCR with stool specimens could be used before the species-specific PCR for identifying Helicobacter species in laboratory dogs.
Keywords: Helicobacter, stool antigen kit, stool, PCR, dog
Helicobacter spp. are gram-negative, microaerobic, motile, fusiform, spiral-shaped bacteria; they may have a curved to spiral rod morphology and move using flagella that vary in number and location among different species [1,2]. Most of the species are associated with either hepatic or gastric infections [2]. At least 24 Helicobacter spp. have been reported to date and it is likely that several more await discovery [1]. Helicobacter spp. colonize the gastrointestinal tract of humans and several animal species, such as cats, dogs, ferrets, pigs, cheetahs, and monkeys [3]. In humans, Helicobacter pylori is a major cause of chronic diffuse superficial gastritis and peptic ulcers. It is also considered a cofactor in the development of gastric malignancies.
A number of Helicobacter species may confound experimental data because of their association with disease progressing in various kinds of laboratory animals [4,5,6]. Thus, Helicobacter infection of laboratory animals may influence the results of research, and it is necessary to clarify the current status of Helicobacter contamination in laboratory animal colonies. Screening of Helicobacter species in laboratory animals is particularly desirable, because they are prevalent in commercial and research animal facilities [7,8,9]. The main gastric Helicobacter spp. in dogs are Helicobacter heilmannii and Helicobacter felis [3]. To date, H. heilmannii has not been reliably cultured in vitro [10]. However, both H. heilmannii and H. felis can be identified by electron microscopy or by analysis of their 16S rRNA and urease gene sequences [11,12].
Several methods have been used to diagnose H. pylori infection. There is an increasing interest in non-invasive tests, as they do not require endoscopic assessment [13]. The 13C-urea breath test (UBT) is the most recommended non-invasive test for detecting H. pylori infection and has a high sensitivity and specificity [14]. However, the UBT cannot be applied to animals due to its high cost and the requirement for expensive analytical instruments [15]. For this reason, many researchers have used polymerase chain reaction (PCR) assays to monitor infection in animal stools [16,17]. However, PCR assays can be time-consuming and expensive [18]. In addition, DNA extraction and amplification from stool samples can be difficult [19]. Recently, several commercial companies have developed H. pylori stool antigen (HpSA) test kits. HpSA tests are non-invasive diagnostic assays for the detection of H. pylori infection in human stool samples [20,21,22]. Presently, there is little information on the usefulness of the HpSA test in detecting other Helicobacter spp. In this study, Helicobacter spp. infection was detected in dogs with gastric disease by PCR and RUT from stool samples or gastric biopsy specimens. In addition, the ability of the HpSA assay to detect Helicobacter spp. in the stools or gastric biopsies of the infected dogs was compared to that of PCR and RUT.
Materials and Methods
Study animals
Eight dogs displaying symptoms of gastritis (e.g. vomiting) were evaluated in this study. None of the animals had been treated with antibiotics during the four-week period prior to examination. The age of the dogs ranged from six months to eight years (mean 3 years). Five of the dogs were male and three were female. The time interval from the last vomiting episode and the sampling was recorded. All studies were performed in accordance with the Guide for Animal Experimentation by Wonkwang University and approved by the Institutional Animal Care and Use Committee of Wonkwang University (Approval No. WKU11-125.) All efforts were made to minimize pain or discomfort of animals used.
Gastroscopic biopsy and stool specimens
Fasting dogs were anesthetized with diazepam (0.2 mg/kg of body weight) and ketamine (3 to 5 mg/kg given until effective); the dogs were intubated, and anesthesia was maintained with halothane-oxygen. During gastroscopy, the macroscopic appearances of the mucosa were recorded and biopsy samples were taken and then analyzed by the RUT and PCR. Endoscope and biopsy forceps were disinfected with 4% Sekusept Plus solution (Henkel, Muttenz, Switzerland) for 30min and thoroughly flushed with tap water prior to use.
Stool samples were collected in sterile screw-capped containers and stored at room temperature. Half of the sample was used for the HpSA and the other half for PCR analysis. The sample for PCR analysis was processed within 24 h of being collected. Each collection swab was resuspended in 2 mL of 0.1 M phosphate buffered saline (PBS) buffer and vortexed. The PBS suspension was then used to extract genomic DNA for PCR analysis.
Rapid urease test with gastroscopic biopsy specimens
The gastroscopic biopsy specimens were minced and tested for urease activity using a CLO Helicobacter-detection kit (Asan Pharm Co., Ltd., Seoul, Korea). The specimens were incubated at 35℃ for 24 h prior to reading the assay. Negative and positive reactions were indicated as bright yellow and dark red color changes, respectively.
HpSA kit
Stool samples were evaluated using the commercially available SD Bioline HpSA kit following the manufacturer's instructions. Samples (250 mg) were incubated at room temperature for 30 min in the diluents provided in the kit and then 100 µL was transferred to the assay device. The test results were read after 15 min. One red line indicated negative and double red line indicated Helicobacter-positive result.
DNA extraction from gastric biopsies and stool specimens
Gastroscopic biopsy tissues were homogenized and resuspended in PBS for DNA extraction. Briefly, genomic DNA was extracted using an AccuPrep Genomic DNA Extraction kit (Bioneer Corp., Daejeon, Korea) according to the manufacturer's instructions. DNA was eluted in Tris-EDTA buffer (pH 8.0) and was stored at -20℃ until required. Aliquots (x µL) were used for PCR amplification.
Genomic DNA from the stool samples of the dogs was extracted using the AccuPrep Stool DNA Extraction Kit (Bioneer, Korea) according to the manufacturer's instructions. AccuPrep Stool DNA Extraction Kit is designed for the rapid extraction of DNA from fresh or frozen stools containing PCR inhibitors. The kit uses a glass filter fixed in a column tube that can efficiently bind DNA in the presence of chaotropic salts. Using the spin-column method, contaminants and enzyme inhibitors such as heparin, bilirubin bile salts, and porphyrin are eliminated, and after the washing steps, which remove proteins and salt, high-purity DNA is finally eluted using a low-concentration elution buffer, which is ready for use in a variety of applications [18]. It yields between 2 and 5 µg of DNA from 100 mg stool [18].
Consensus polymerase chain reaction to detect Helicobacter genus
A set of primers HF (5'-ACTTTAAACGCATGAA GATAT-3') and HR (5'-ATATTTTGACCTTCTGGGGT-3') was used to amplify the Helicobacter rpoB gene (458 bp) [18]. The template DNA (50 ng) and 20 pmol of each primer were added to a PCR tube (Maxime PCR PreMix; iNtRON Biotechnology, Korea) containing 1 U of Taq DNA polymerase, 250 µM each deoxynucleoside triphosphate, 50 mM Tris-HCl (pH 8.3), 40 mM KCl, and 1.5 mM MgCl2. The volume was adjusted with distilled water to 20 µL. The reaction conditions were 5 min at 95℃ followed by 40 cycles of 30 s at 94℃, 30 s at 52℃, 45 s at 72℃ with a final 5 min extension at 72℃. The PCR products were electrophoresed on a 1.2% (wt/vol) agarose gel. Positive samples were further analyzed using a species-specific PCR and DNA sequencing to identify individual Helicobacter spp.
Multiplex species-specific PCR
For the multiplex species-specific PCR, primers were designed based on the urease B gene sequence of each Helicobacter spp. Table 1 shows the sequences of the primers, the EMBL accession numbers of the urease B gene, and the expected amplicon lengths [17]. The template DNA (50 ng) and 20 pmol of each primer were added to a PCR tube containing 1 U of Taq DNA polymerase, 250 µM of each deoxynucleoside triphosphate, 50 mM Tris-HCl (pH 8.3), 40 mM KCl, and 1.5 mM MgCl2. The thermocycling parameters were as follows: one cycle of 94℃ for 3 min, 57℃ for 2 min, and 72℃ for 3 min followed by 31 cycles of 94℃ for 30 s, 57℃ for 30 s, and 72℃ for 1 min and a final extension at 72℃ for 5 min. PCR products were analyzed by electrophoresis on a 1.2% agarose gel. The sizes of the expected amplicons were 580, 1,707, and 1,150 bp for H. heilmannii, H. pylori, and H. felis, respectively.
Table 1. Primers used for the amplification of the urease B gene of H. heilmannii, H. pylori, and H. felis [17].
| Species | Primer and sequence | EMBL accession no. | Product size (bp) |
|---|---|---|---|
| H. heilmannii | F, 5'-GGGCGATAAAGTGCGCTTG-3' | L25079 | 580 |
| R, 5'-CTGGTCAATGAGAGCAGG-3' | |||
| H. pylori | F, 5'-GGAATTCCAGATCTATGAAAAAGATTAGCAGAAAAG-3' | M60398 | 1,707 |
| R, 5'-GGAATTCGTCGACCTAGAAAATGCTAAAGAGTTG-3' | |||
| H. felis | F, 5'-ATGAAACTAACGCCTAAAGAACTAG-3' | X69080 | 1,150 |
| R, 5'-GGAGAGATAAAGTGAATATGCGT-3' |
F, forward; R, reverse.
Antibiotic therapy and treatment evaluation
Following a positive diagnosis of Helicobacter spp. infection, the dogs underwent a triple therapy with clarithromycin (0.24 mg/head), metronidazole (0.72 mg/head), and omeprazole (0.021 mg/head). The treatment was administered once a day for two weeks. For the evaluation of the therapy, stool specimens were collected from the dogs and genomic DNA was extracted as previously described. A multiplex Helicobacter speciesspecific PCR to amplify the urease B gene was performed. The dogs were observed for a further three months following the antibiotic therapy.
Results
RUT and HpSA test
Table 2 shows the result of the RUT. Biopsies from five (62.5%) of the dogs gave positive results. Stool specimens containing less water were collected. Table 2 shows the results of the HpSA test. Two (25%) dogs were deemed positive using this kit.
Table 2. Comparison of the results from three diagnostic assays used to identify Helicobacter spp. in dogs.
| Type | Assay | Result | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | Positive percent | ||
| Non-invasive | Stool Ag Kit | ● | ○ | ○ | ● | ○ | ○ | ○ | ○ | 25.0 %, CIa 7.1-59.0 |
| PCRb of stool specimen | ● | ● | ○ | ● | ● | ● | ○ | ○ | 62.5 %, CI 30.5-86.3 | |
| Invasive | RUTb) | ● | ● | ○ | ● | ● | ● | ○ | ○ | 62.5 %, CI 30.5-86.3 |
| PCR of gastric specimen | ● | ● | ○ | ● | ● | ● | ○ | ○ | 62.5 %, CI 30.5-86.3 | |
○, negative; •, positive.
a Incidence percentage (95% confidential interval) was calculated with MiniTab statistical software.
bIt was detected by polymerase chain reaction using consensus primers.
Consensus PCR of gastric biopsy and stool samples
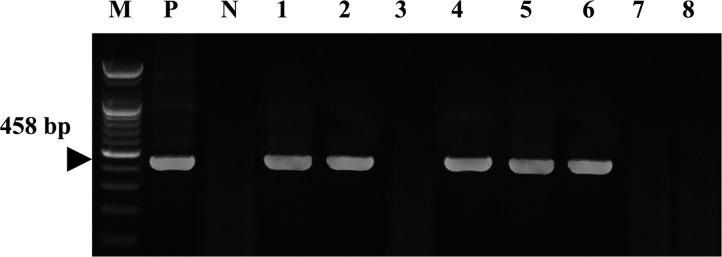
A consensus PCR to amplify the rpoB gene was used to detect Helicobacter spp. in both gastric biopsies and stool samples. The detection limit of the consensus PCR was 0.1 pg of Helicobacter DNA [18]. Five dogs (62.5%) were positive using the consensus PCR using gastric biopsies (Figure 1). Table 2 shows that the consensus PCR detect successfully with both gastric biopsies and stool samples.
Figure 1. Amplification of Helicobacter rpoB DNAs from consensus PCR with gastroscopic biopsy specimens was identified on a 1.2% agarose gel electrophoresis. M: Size marker, P: Positive control (H. pylori), N: Distilled water, Lane 1~8: Gastroscopic biopsy specimen of dog No. 1~8.
Species-specific PCR using gastric biopsies and stool specimens
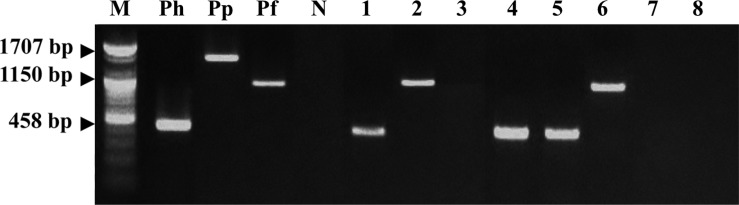
A multiplex species-specific PCR on gastroscopic biopsy and stool specimens was conducted with the primers for the urease B gene. No amplification products corresponding to H. pylori were detected, but some specimens tested were positive for H. felis or H. heilmannii (Figure 2). Table 3 shows the results of the multiplex species-specific PCR which reveal that three (37.5%) and two (25%) of the eight dogs were infected with H. heilmannii and H. felis, respectively. All of the samples that were positive by the consensus PCR were confirmed using the species-specific PCR.
Figure 2. Results of the multiplex species-specific PCR to amplify urease B gene for H. heilmannii, H. pylori and H. felis. M: Size marker, Ph: Positive control (H. heilmannii), Pp: Positive control (H. pylori), Pf: Positive control (H. felis), N: Distilled water, Lane 1~8: Gastroscopic biopsy specimen of dog No. 1~8.
Table 3. The percentage of dogs positive for Helicobacter spp.
| Gender | n | Detection % (95% CIa) | |||
|---|---|---|---|---|---|
| bHelicobacter spp. | Speciesc | ||||
| H. heilmannii | H. felis | H. pylori | |||
| Male | 5 | 50.0 (21.5-78.4) | 37.5 (13.6-69.4) | 12.5 (2.2-47.0) | 0 (0-32.4) |
| Female | 3 | 12.5 (2.2-47.0) | 0 (0-32.4) | 12.5 (2.2-47.0) | 0 (0-32.4) |
| Total | 8 | 62.5 CI 30.5-86.3 | 37.5 (13.6-69.4) | 25.0, CIa 7.1-59.0 | 0 (0-32.4) |
a95% confidential interval was calculated using the MINITAB program.
bDetected by PCR using consensus primers.
cDetected by PCR using species-specific primers
Discussion
In laboratory animals, a lot of Helicobacter species have identified and revealed highly prevalent in animal facilities [7,8,9]. Therefore, the monitoring and control of Helicobacter species has been required for better experimental results in laboratory animals. Dogs can be infected with different Helicobacter spp., and coinfections are also know to occur [3,23]. The presence of Helicobacter spp. in the stools of the dogs meant that they were harboring the bacterium. Therefore, the detection rate of Helicobacter species is very important. This study reports a prevalence of fecal Helicobacter spp. in five (62.5%) of eight pet dogs studied. In addition, three (37.5%) and two (25%) of the dogs were infected with H. heilmannii and H. felis, respectively. A high prevalence of Helicobacter spp. has previously been reported in pet dogs and cats with gastritis and so our present data confirms these findings [18,24].
The current gold standard for diagnosing H. pylori infection is endoscopic biopsy of the gastric tissue to be analyzed by RUT, histology, and culture. However, these invasive procedures have major disadvantages, which include discomfort to the patient, the use of anesthesia, and possible ethical issues [25]. In contrast, noninvasive tests are easy to perform and do not produce any significant discomfort for the patient. Such non-invasive tests for H. pylori include serological antibody testing, the UBT, and the HpSA test [25].
The HpSA test and PCR of stool specimens for diagnosing Helicobacter spp. infection offers a useful non-invasive method without having to sacrifice animals. In this study, the HpSA did not efficiently detect Helicobacter spp. in stool samples. On the other hand, the RUT and PCR assay using gastric and stool samples did detect successfully Helicobacter spp. It must be considered, however, that the poor performance of the HpSA may be due to the degradation of the Helicobacter spp. as it passes through the intestine. In addition, excretion of Helicobacter spp. from the intestine of the animal may vary with time. Furthermore, use of Nacetylcysteine-like mucolytic agents may decrease the accuracy of the diagnosis [24]. Cut off titer, though difficult to decide but crucial to reach the conclusion by using antigen detection technique [26]. Several limitations with PCR analysis have also been reported for Helicobacter spp. [18,27]. Stool specimens are easy to obtain and are consequently of high interest for the development of direct methods for the detection of Helicobacter spp. [18,23]. PCR has been successfully used to detect bacteria in stools even though stool samples remain the most difficult specimens from which DNA can be extracted and amplified [19]. This difficulty is due to the presence of DNA polymerase inhibitors like complex polysaccharides in the stools [19]. In this study, the spincolumn method was used, effectively eliminating contaminants and enzyme inhibitors such as heparin, bilirubin bile salts, and porphyrin. As a result, between 2 and 5 µg of high-purity DNA was obtained.
In conclusion, we have shown that the PCR amplification of stool samples is a useful method for detecting Helicobacter spp. infection in laboratory dogs. This assay may be useful as a screening test for infection and could be used to address questions relevant to pathogenesis and therapy [28].We recommend that the consensus PCR for the urease B gene be used on samples first, followed by the species-specific multiplex assay.
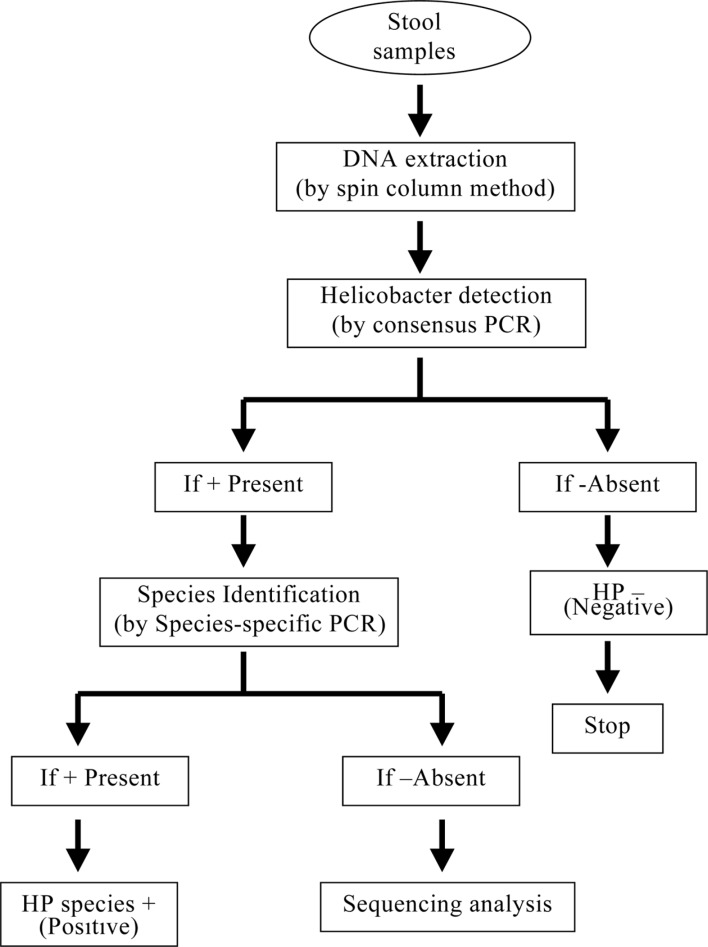
Previously, the consensus PCR has been used to successfully detect Helicobacter spp. This consensus PCR was recommended for diagnosing Helicobacter spp. in dogs. In this study, the consensus PCR was able to successfully detect Helicobacter spp. in the stool specimens of dogs. We suggested that the application of preceding consensus PCR before the species-specific PCRs as shown in Figure 3 might be the most effective strategy for the identification of Helicobacter species in laboratory dogs.
Figure 3. Scheme of detection of Helicobacter species in the canine stools.
Acknowledgments
This study was supported by a Wonkwang University research grant in 2015. We wish to thank Gi-Wook Oh, (Center for Animal Resources Development, Wonkwang University) for technical support.
Footnotes
Conflict of interests: The authors declare that there is no financial conflict of interests to publish these results.
References
- 1.Vandamme P, Falsen E, Pot B, Kersters K, De Ley J. Identification of Campylobacter cinaedi isolated from blood and feces of children and adult females. J Clin Microbiol. 1990;28(5):1016–1020. doi: 10.1128/jcm.28.5.1016-1020.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fox JG. The non-H pylori helicobacters: their expanding role in gastrointestinal and systemic diseases. Gut. 2002;50(2):273–283. doi: 10.1136/gut.50.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim O. Helicobacter-An emerging new zoonotic pathogen. In: Lorenzo-Morales J, editor. Zoonosis. 1st ed. Rijeka, Croatia: InTec; 2012. pp. 89–100. [Google Scholar]
- 4.Ward JM, Fox JG, Anver MR, Haines DC, George CV, Collins MJ, Gorelick PL, Nagashima K, Gonda MA, Gilden RV, Tully JG, Russell RJ, Benveniste RE, Paster BJ, Dewhirst FE, Donovan JC, Anderson LM, Rice JM. Chronic active hepatitis and associated liver tumors in mice caused by a persistent bacterial infection with a novel Helicobacter species. J Natl Cancer Inst. 1994;86(16):1222–1227. doi: 10.1093/jnci/86.16.1222. [DOI] [PubMed] [Google Scholar]
- 5.Eaton KA, Dewhirst FE, Paster BJ, Tzellas N, Coleman BE, Paola J, Sherding R. Prevalence and varieties of Helicobacter species in dogs from random sources and pet dogs: animal and public health implications. J Clin Microbiol. 1996;34(12):3165–3170. doi: 10.1128/jcm.34.12.3165-3170.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chin EY, Dangler CA, Fox JG, Schauer DB. Helicobacter hepaticus infection triggers inflammatory bowel disease in T cell receptor alpha beta mutant mice. Comp Med. 2000;50(6):586–594. [PubMed] [Google Scholar]
- 7.Shames B, Fox JG, Dewhirst F, Yan L, Shen Z, Taylor NS. Identification of widespread Helicobacter hepaticus infection in feces in commercial mouse colonies by culture and PCR assay. J Clin Microbiol. 1995;33(11):2968–2972. doi: 10.1128/jcm.33.11.2968-2972.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goto K, Ohashi H, Takakura A, Itoh T. Current status of Helicobacter contamination of laboratory mice, rats, gerbils, and house musk shrews in Japan. Curr Microbiol. 2000;41(3):161–166. doi: 10.1007/s002840010111. [DOI] [PubMed] [Google Scholar]
- 9.Whary MT, Cline JH, King AE, Hewes KM, Chojnacky D, Salvarrey A, Fox JG. Monitoring sentinel mice for Helicobacter hepaticus, H rodentium, and H bilis infection by use of polymerase chain reaction analysis and serologic testing. Comp Med. 2000;50(4):436–443. [PubMed] [Google Scholar]
- 10.Andersen LP, Norgaard A, Holck S, Blom J, Elsborg L. Isolation of a "Helicobacter heilmanii"-like organism from the human stomach. Eur J Clin Microbiol Infect Dis. 1996;15(1):95–96. doi: 10.1007/BF01586196. [DOI] [PubMed] [Google Scholar]
- 11.Solnick JV, O'Rourke J, Lee A, Paster BJ, Dewhirst FE, Tompkins LS. An uncultured gastric spiral organism is a newly identified Helicobacter in humans. J Infect Dis. 1993;168(2):379–385. doi: 10.1093/infdis/168.2.379. [DOI] [PubMed] [Google Scholar]
- 12.Solnick JV, O'Rourke J, Lee A, Tompkins LS. Molecular analysis of urease genes from a newly identified uncultured species of Helicobacter. Infect Immun. 1994;62(5):1631–1638. doi: 10.1128/iai.62.5.1631-1638.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mégraud F. The most important diagnostic modalities for Helicobacter pylori, now and in the future. Eur J Gastroenterol Hepatol. 2012;9(Suppl 1):S13–S15. [PubMed] [Google Scholar]
- 14.Leal YA, Flores LL, Fuentes-Pananá EM, Cedillo-Rivera R, Torres J. 13C-urea breath test for the diagnosis of Helicobacter pylori infection in children: a systematic review and metaanalysis. Helicobacter. 2011;16(4):327–337. doi: 10.1111/j.1523-5378.2011.00863.x. [DOI] [PubMed] [Google Scholar]
- 15.Nyan DC, Welch AR, Dubois A, Coleman WG., Jr Development of a noninvasive method for detecting and monitoring the time course of Helicobacter pylori infection. Infect Immun. 2004;72(9):5358–5364. doi: 10.1128/IAI.72.9.5358-5364.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Santos AM, Lopes T, Oleastro M, Chaves P, Cordeiro R, Ferreira M, Pereira T, Machado J, Guerreiro AS. Role of 13C-urea breath test in experimental model of Helicobacter pylori infection in mice. Helicobacter. 2011;16(4):320–326. doi: 10.1111/j.1523-5378.2011.00847.x. [DOI] [PubMed] [Google Scholar]
- 17.Neiger R, Dieterich C, Burnens A, Waldvogel A, Corthésy-Theulaz I, Halter F, Lauterburg B, Schmassmann A. Detection and prevalence of Helicobacter infection in pet cats. J Clin Microbiol. 1998;36(3):634–637. doi: 10.1128/jcm.36.3.634-637.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim S, Cho S, Kim O. Detection and identification of secreting Helicobacter species from cats. Lab Anim Res. 2006;22(3):243–247. [Google Scholar]
- 19.Monteiro L, Bonnemaison D, Vekris A, Petry KG, Bonnet J, Vidal R, Cabrita J, Mégraud F. Complex polysaccharides as PCR inhibitors in feces: Helicobacter pylori model. J Clin Microbiol. 1997;35(4):995–998. doi: 10.1128/jcm.35.4.995-998.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moon DI, Shin EH, Oh HG, Oh JS, Hong S, Chung Y, Kim O. Usefulness of a Helicobacter pylori stool antigen test for diagnosing H. pylori infected C57BL/6 mice. Lab Anim Res. 2013;29(1):27–32. doi: 10.5625/lar.2013.29.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shimoyama T. Stool antigen tests for the management of Helicobacter pylori infection. World J Gastroenterol. 2013;19(45):8188–8191. doi: 10.3748/wjg.v19.i45.8188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patel SK, Pratap CB, Jain AK, Gulati AK, Nath G. Diagnosis of Helicobacter pylori: what should be the gold standard? World J Gastroenterol. 2014;20(36):12847–12859. doi: 10.3748/wjg.v20.i36.12847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee H, Park Y, Kim O. Prevalence of Helicobacter Species in Feces of Dogs. Lab Anim Res. 2007;23(3):339–344. [Google Scholar]
- 24.Demirtürk L, Yazgan Y, Tarçin O, Ozel M, Diler M, Oncül O, Yildirim S. Does N-acetyl cystein affect the sensitivity and specificity of Helicobacter pylori stool antigen test? Helicobacter. 2003;8(2):120–123. doi: 10.1046/j.1523-5378.2003.00132.x. [DOI] [PubMed] [Google Scholar]
- 25.Hoshina S, Kahn SM, Jiang W, Green PH, Neu HC, Chin N, Morotomi M, LoGerfo P, Weinstein IB. Direct detection and amplification of Helicobacter pylori ribosomal 16S gene segments from gastric endoscopic biopsies. Diagn Microbiol Infect Dis. 1990;13(6):473–479. doi: 10.1016/0732-8893(90)90079-b. [DOI] [PubMed] [Google Scholar]
- 26.Malfertheiner P, Megraud F, O'Morain CA, Atherton J, Axon AT, Bazzoli F, Gensini GF, Gisbert JP, Graham DY, Rokkas T, El-Omar EM, Kuipers EJ European Helicobacter Study Group. Management of Helicobacter pylori infection--the Maastricht IV/Florence Consensus Report. Gut. 2012;61(5):646–664. doi: 10.1136/gutjnl-2012-302084. [DOI] [PubMed] [Google Scholar]
- 27.Rautelin H, Lehours P, Mégraud F. Diagnosis of Helicobacter pylori infection. Helicobacter. 2003;8(Suppl 1):13–20. doi: 10.1046/j.1523-5378.2003.00168.x. [DOI] [PubMed] [Google Scholar]
- 28.Shinozaki JK, Sellon RK, Cantor GH, Besser TE, Mealey KL, Vaden SL. Fecal polymerase chain reaction with 16S ribosomal RNA primers can detect the presence of gastrointestinal Helicobacter in dogs. J Vet Intern Med. 2002;16(4):426–432. doi: 10.1892/0891-6640(2002)016<0426:fpcrwr>2.3.co;2. [DOI] [PubMed] [Google Scholar]