Abstract
Background
Epidermal growth factor receptor (EGFR) mutation is the key predictor of EGFR tyrosine kinase inhibitors (TKIs) efficacy in non-small cell lung cancer (NSCLC). We conducted this study to verify the feasibility of EGFR mutation analysis in cytological specimens and investigate the responsiveness to gefitinib treatment in patients carrying EGFR mutations.
Methods
A total of 210 cytological specimens were collected for EGFR mutation detection by both direct sequencing and amplification refractory mutation system (ARMS). We analyzed EGFR mutation status by both methods and evaluated the responsiveness to gefitinib treatment in patients harboring EGFR mutations by overall response rate (ORR), disease control rate (DCR) and progression free survival (PFS).
Results
Of all patients, EGFR mutation rate was 28.6% (60/210) by direct sequencing and 45.2% (95/210) by ARMS (P<0.001) respectively. Among the EGFR wild type patients tested by direct sequencing, 26.7% of them were positive by ARMS. For the 72 EGFR mutation positive patients treated with gefitinib, the ORR, DCR and median PFS were 69.4%, 90.2% and 9.3 months respectively. The patients whose EGFR mutation status was negative by direct sequencing but positive by ARMS had lower ORR (48.0% vs. 80.9%, P=0.004) and shorter median PFS (7.4 vs. 10.5 months, P=0.009) as compared with that of EGFR mutation positive patients by both detection methods.
Conclusions
Our study verified the feasibility of EGFR analysis in cytological specimens in advanced NSCLC. ARMS is more sensitive than direct sequencing in EGFR mutation detection. EGFR Mutation status tested on cytological samples is applicable for predicting the response to gefitinib. Abundance of EGFR mutations might have an influence on TKIs efficacy.
Keywords: Non-small cell lung cancer (NSCLC), epidermal growth factor receptor (EGFR) mutation, cytological specimen, amplification refractory mutation system (ARMS), gefitinib
Introduction
Non-small cell lung cancer (NSCLC) accounts for over 80% of lung cancers which is the most leading cause of cancer death in the whole world (1). About 85% patients are initially diagnosed with NSCLC at late stage and lost the chance of operation. The median survival time of advanced NSCLC is only about 8 to 10 months with systemic chemotherapy. The situation had not improved for decades until epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs) were applied to EGFR mutation patients. EGFR mutation is the key predictor of TKI efficacy in NSCLC (2,3). Overall response rate (ORR) is over 70% in patients with EGFR activating mutations, and the overall survival (OS) time is prolonged to around 30 months in patients treated with TKIs (4). However patients of EGFR wild type cannot benefit from TKIs treatment (2). Therefore, detection of EGFR mutation status is very important in treatment decision making in advanced NSCLC.
Direct sequencing is a classic way to detect greater than 10% EGFR mutation frequency, while amplification refractory mutation system (ARMS) is more sensitive and be able to detect mutation frequency as low as 1% (5). So ARMS has been more accepted as the standard EGFR mutation detection method now. The samples for EGFR mutation testing are generally histological specimens, which are used as criteria in all randomized clinical trials all over the world. But the practical situation is that sufficient tissues are often difficult to obtain for further molecular detection in many advanced stage patients. However, cytological specimens such as sputum, pleural effusion, or lavage are easier to get for most NSCLC patients. Therefore, clinicians pay close attention to the feasibility of EGFR mutation testing on cytological specimens. Some investigators have reported their research on the detection of EGFR mutations on body cytological specimens by either direct sequencing or ARMS way (6-8). We conducted this study to further verify the feasibility of EGFR mutation detection in cytological specimens in a larger NSCLC population, by both direct sequencing and ARMS, and to investigate the practical significance of cytology molecular testing in predicting response to TKIs treatment.
Materials and methods
Patients
From Jan 2011 to Apr 2014, cytology samples were collected from a total of 210 advanced NSCLC patients who were unavailable for histology EGFR mutation test in Beijing Hospital, which included 174 pleural effusion, 17 sputum, 14 bronchoscopy lavages, and 5 fine-needle aspiration. The informed consent was obtained from each patient for EGFR mutation test. All patients’ clinical features were recorded including age, gender, pathological type, and treatment outcomes. Patients treated with gefitinib (Iressa, AstraZeneca) were followed and efficacy was assessed every 6 to 8 weeks with computed tomography (CT) scans for the ORR and disease control rate (DCR) based on the Response Evaluation Criteria in Solid Tumor (RECIST 1.1). Progression-free survival was assessed when patients achieved progression disease, or death from any cause.
Samples preparation
Pleural effusion specimens were collected in an amount of 20 to 500 mL. Sputum specimens were continuously collected in an iced box in 12 to 24 hours. After hematoxylin and eosin (H&E) staining sections were firstly made, all samples were sent to the pathologist to evaluate if tumor cells could be found and enough for further EGFR mutation testing. All samples were centrifuged at 1,500 revolutions per minute for 5 minutes. Sputum samples should be mixed with 10% dithiothreitol and shaken for 5 minutes before centrifugation. Next, formalin fixed paraffin-embedded cell blocks were prepared. Further immunocytochemical were performed on the sections made from the blocks for final pathological diagnose of the patients.
EGFR mutation detection
All molecular tests were performed on the cell blocks by experienced pathologists in Beijing Hospital. In general, the samples with >100 tumor cells could undergo EGFR mutation testing. While for those with less than 100 tumor cells, micro dissection might be needed to enrich the tumor cells. DNA extractions were undergone according to the protocol of ADx kit (Amoy diagnostics, Xiamen, China). Both direct sequencing and ARMS were performed on the same DNA extraction in all samples. Exons 18, 19, 20, and 21 of the EGFR gene were amplified by polymerase chain reaction (PCR). The primers were designed based on the report of Lynch et al. (3). The PCR mixture was bought from American Applied Biosystems Incorporation. The final products were cleared and sequenced with internal primers using ABI PRISM 3730 DNA Analyser (Applied Biosystems, Foster City, CA, USA). The patients’ DNA was also tested by ARMS by using ADx EGFR Mutations Detection Kit (Amoy diagnostics, Xiamen, China) and covered the 29 EGFR mutations from exon 18 to 21. PCR experiments were carried out with the instrument ABI 7500 Fast (Applied Biosystems Inc., CA, USA). The CT values used to determine whether a sample was positive or negative were based on extensive validation.
Statistical analysis
EGFR mutation status and ORR between different groups were compared using χ2 tests. Progression free survival under gefitinib treatment was analyzed by the Kaplan-Meier method and was compared between different groups by the log-rank test. The value of P≤0.05 was considered statistically significant. All statistical analyses were performed using IBM SPSS Statistics 18 software package (SPSS INC., Chicago, IL, USA).
Results
Characteristics of patients and EGFR mutation rates
Among the 210 patients with NSCLC, 118 males (56.2%) and 92 females (43.8%) were at stage IIIB or IV. The mean age was 68 years ranging from 33 to 91 years (Table 1). There were 203 cases (96.7%) of adenocarcinoma and 7 cases (3.3%) of squamous cell carcinoma.
Table 1. Patient characteristics and EGFR mutation status.
| Characteristics | Total, n (%) | EGFR mutations, n (%) |
||
|---|---|---|---|---|
| Sequencing | ARMS | P | ||
| No. of patients | 210 | 60 (28.6) | 95 (45.2) | <0.001 (χ2=12.526) |
| Age (years) | ||||
| Mean | 68 | |||
| Range | 33-91 | |||
| Gender | ||||
| Male | 118 (56.2) | 28 (23.7)* | 48 (40.7)** | |
| Female | 92 (43.8) | 32 (34.8)* | 47 (51.1)** | |
| Pathological type | <0.001 (χ2=12.784) | |||
| Adenocarcinoma | 203 (96.7) | 60 (29.6) | 95 (46.8) | |
| Squamous cell carcinoma | 7 (3.3) | 0 (0) | 0 (0) | |
*, P=0.079 (χ2=3.095); **, P=0.133 (χ2=2.261). EGFR, epidermal growth factor receptor; ARMS, amplification refractory mutation system.
Overall, 60 (28.6%) of the samples were identified with EGFR mutations on exon 18, 19, 20 or 21 by direct sequencing, and 95 (45.2%) of specimens were EGFR mutation positive by ARMS (P<0.001). For the 118 male and 92 female patients, EGFR mutation rate was 23.7% and 34.8% (P=0.079) respectively by direct sequencing; and was 40.7% and 51.1% (P=0.133) respectively by ARMS. All patients carrying EGFR mutations were of adenocarcinoma pathology type, and the EGFR mutation rate was 29.6% by sequencing and 46.8% by ARMS in adenocarcinoma (P<0.001).
EGFR mutation status
Further analysis of EGFR mutation status (Table 2) showed that in-frame deletions in exon 19 and L858R mutation in exon 21 were the most common activating mutations, with mutation rate of 50.0% and 43.3% respectively by direct sequencing, and similarly with mutation rate of 47.3% and 41.1% respectively by ARMS. Some other minor mutations such as exon 20 insertion, S768I mutation, G719X in exon 18, and L861Q in exon 21, etc. were also found by both detection ways. Among the 150 EGFR wild type patients tested by direct sequencing, 35 (23.3%) cases were EGFR mutation positive by ARMS. And ARMS seemed to be able to identify more minor mutations (11.6% vs. 6.7%). One treatment-naïve patient was detected as both exon 19 deletion and T790M mutation positive by ARMS.
Table 2. Comparison of EGFR mutation status in sequencing and ARMS detection way.
| EGFR mutation type | Sequencing, n (%) | ARMS, n (%) | S-A+, n (%) |
|---|---|---|---|
| Total | 60 | 95 | 35 |
| 19 deletion | 30 (50.0) | 45 (47.3) | 15 (42.9) |
| 21 L858R | 26 (43.3) | 39 (41.1) | 13 (37.1) |
| Minor mutations | 4 (6.7) | 11 (11.6) | 7 (20.0) |
| 20 ins 1 | 20 ins 1 | 20 ins 0 | |
| S768I 1 | S768I 4 | S768I 3 | |
| L861Q 1 | L861Q 4 | L861Q 3 | |
| G719X 1 | G719X 1 | G719X 0 | |
| T790M + 19 del 1 (treatment-naive) | 790M + 19 del 1 (treatment-naive) |
EGFR, epidermal growth factor receptor; ARMS, amplification refractory mutation system; S-A+, sequencing negative but ARMS positive; 20 ins, insertion mutations in exon 20.
Treatment outcomes
Among the patients carrying activating mutations, 72 patients were treated with single agent of gefitinib and followed for their treatment outcomes (Table 3). The other mutated patients were treated with other EGFR-TKI dugs, or refused to get TKI treatment. The ORR was 69.4% with 7 patients (9.7%) achieving complete response (CR) and 43 patients (59.7%) achieving partial response (PR). Seven patients experienced disease progression, so DCR was 90.2%. Median PFS of these patients was 9.3 months. Among these 72 patients, 25 patients’ EGFR mutation status were sequencing negative but ARMS positive. So we performed further analysis on this group of patients and also on those patients positive in both ways. We found that in sequencing negative but ARMS positive group, 12 patients (48.0%) partially responsed to gefitinib treatment, and 9 patients were with stable disease. So ORR in this group was 48.0%, which was lower than that of both sequencing and ARMS positive group (80.9%, P=0.004), and DCR were 84.0% vs. 93.6%, P=0.190. The median PFS in the patients positive in both sequencing and ARMS was 10.0 and 7.4 months in sequencing negative but ARMS positive patients (P=0.009) (Figure 1).
Table 3. Clinical efficacy of gefitinib in EGFR mutated patients detected by different way.
| Response | A+ (%) | S+A+ (%) | S-A+ (%) | P |
|---|---|---|---|---|
| Total | 72 | 47 | 25 | |
| CR | 7 (9.7) | 7 (14.9) | 0 (0) | |
| PR | 43 (59.7) | 31 (66.0) | 12 (48.0) | |
| ORR | 50 (69.4) | 38 (80.9) | 12 (48.0) | 0.004 (χ2=8.300) |
| DCR | 65 (90.2) | 44 (93.6) | 21 (84.0) | 0.190 (χ2=1.720) |
EGFR, epidermal growth factor receptor; A+, ARMS positive; S+A+, sequencing positive and ARMS positive; S-A+, sequencing negative but ARMS positive; CR, complete response; PR, partial response; ORR, overall response rate; DCR, disease control rate; ARMS, amplification refractory mutation system.
Figure 1.
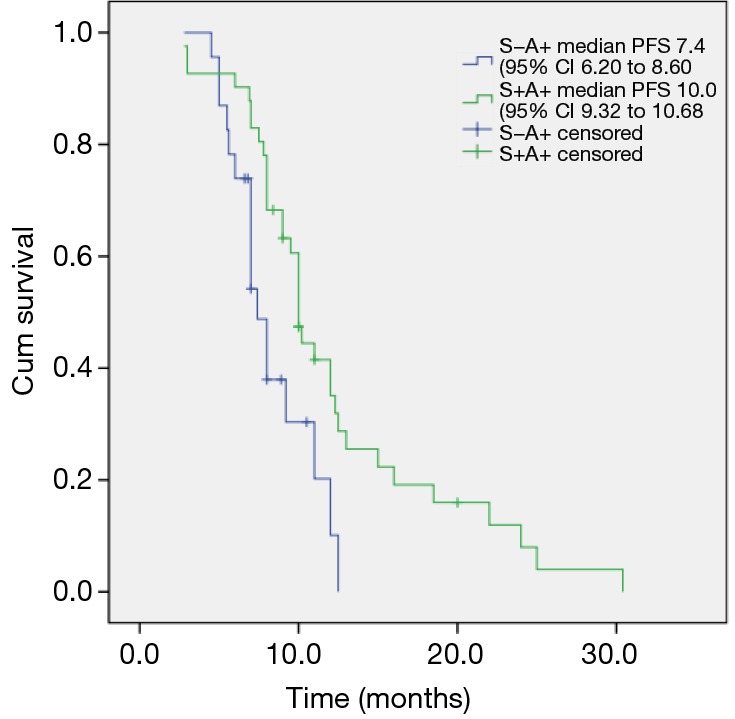
Progression-free survival (PFS) of patients carrying EGFR mutations treated with gefitinib in different mutation groups. S-A+, sequencing negative but ARMS positive; S+A+, sequencing positive and ARMS positive; log-rank, P=0.009; ARMS, Scorpion amplification refractory mutation system.
Treatment outcome in minor mutations patients
There were seven patients with less frequent EGFR mutations treated with gefitinib. One of them was with G719X mutation and achieved PR with PFS of 7.1 months. All the three L861Q mutation patients were detected sequencing negative but ARMS positive. One achieved stable disease with PFS of 3.3 months, the other two achieved PR with PFS of 8.2 and 6.0 months. Among the three S768I mutated patients, two achieved stable disease with PFS of 6.2 and 4.3 months respectively, and the third one experienced disease progression after initial gefitinib treatment.
Mutations after gefitinib resistance
Among all the patients treated with gefitinib, 26 patients were able to be reevaluated of EGFR mutation by cytological specimens after experiencing disease progression. Twenty of them were tested by pleural effusion and six by sputum samples. T790M mutations were detected in 11 (42.3%) of the patients in addition to their previous mutations. The rest 15 patients remained their previous mutations without T790M mutation.
Discussion
Detection of EGFR mutation status is critical for EGFR-TKIs treatment. Therefore, how to acquire suitable samples for further molecular test is a key issue to oncologists. In this study we showed that cytological samples were reliable for EGFR mutation testing in advanced NSCLC, and could predict treatment benefit of EGFR-TKIs.
Most of the samples in this study were pleural effusion which is more common in advanced adenocarcinoma. Sputum, bronchoscopy lavage, and fine-needle aspirates could also be applicable to extract DNA for further EGFR testing so long as there were enough tumor cells in samples. Evaluations of the cell amount by H&E staining slides were important before DNA extraction for all cytological samples. Usually more than 100 cells were needed for a successful test of EGFR mutation, which were also reported in previous study (7). The result of EGFR mutation was obtained from one patient in this study with only 20 mL of pleural effusion which contained enough tumor cells. Although sputum samples did not account for a large proportion (8.1%) in this study, it did show the feasibility in EGFR mutation detection so long as sputum containing enough tumor cells could be collected. Cough is a common symptom in advanced NSCLC patients. Therefore, sputum could be a good option for EGFR mutation testing in patients whose histological samples were difficult to obtain.
EGFR mutation rate is approximately 30-40% in Asian population as compared with 10-20% in non-Asian patients (9-11). In this study, EGFR mutation rate was 45.2% by ARMS in the whole group. The high mutation rate may be attributed to the overwhelming majority of adenocarcinoma patients, in which pleural effusion appeared more common. EGFR mutations were not detected in squamous cell carcinoma patients. The EGFR mutation rate of adenocarcinoma was 46.8% in our study, which was similar as the prospective molecular epidemiology study of EGFR mutations in Asian patients with advanced adenocarcinoma (PIONEER) (12), finding that overall EGFR mutation frequency was 51.4%.
Direct sequencing was used as a classic way for EGFR mutation testing (3,13). Since ARMS detection method was found to be more sensitive and the EGFR mutation status is a predictor of TKIs efficacy (5,14), ARMS was recommended as a standard testing method nowadays. In our study, we carried out EGFR mutation test in both ways on all samples, and detected more frequent EGFR mutations by ARMS than that by direct sequencing. We found about 26.7% of sequencing negative samples were turned out to be positive by ARMS. The phase II study of comparing pemetrexed with gefitinib as the second-line treatment in wild-type EGFR nonsquamous NSCLC patients (CTONG 0806) reported of 29.6% ARMS positive rate in sequencing negative samples (15) and some other studies also showed similar results (5,16). All these showed that ARMS was a more sensitive detection way than direct sequencing, rare mutated tumor cells in samples therefore could be more probably detected by ARMS than by direct sequencing.
The most common EGFR mutations are in-frame deletions in exon 19 (19 del) and L858R point mutation in exon 21, which account for approximately 90% of all EGFR mutations (3,17).We also detected around 90% of these two mutations, and the mutation rate of 19 del was a little higher than that of L858R mutation. Some minor EGFR mutations could also be found in exon 18-21 less frequently (18), such as S768I, L861Q, etc. In our study, we detected a higher rate of minor mutations by ARMS than by direct sequencing. We speculated that the amounts of minor mutations might be less abundant so that they could be better detected by more sensitive way.
The study of gefitinib in previous untreated advanced adenocarcinoma patients in East Asia (IPASS) (2), using ARMS to test EGFR mutation and showed mutation patients could achieve good gefitinib treatment results with ORR 71.2% and median PFS 9.5 months. In our study, we also followed the EGFR mutated patients treated with gefitinib and found that ORR was 69.4%, DCR was 90.7% and median PFS was 9.3 months. This result was quite similar as that in IPASS, in which histological samples were used to test EGFR mutations. Some other reported studies testing on cytological samples also showed good clinical response to TKIs treatment in EGFR mutation positive patients (6,8). Therefore, cytological samples which were easier to obtain in advanced NSCLC patients were feasible for EGFR mutation testing and predictable for clinical TKIs treatment.
For the group of patients who were sequencing negative but ARMS positive, we found the ORR was 48.0% which was much lower than that of patients positive in both detection ways. The median PFS was also shorter. From another study, Zhou et al. (19) showed similar result as ours that in a group of 18 patients who were sequencing negative but ARMS positive, the ORR was 44.4%, which was lower than both positive patients but higher than the wild type patients when treated with gefitinib, so was the median PFS and OS. We proposed that the relative abundance of EGFR mutation might have influence on the clinical benefits from TKIs. However, the patient number of the study was limited, and there were not many studies reported on this issue until now. We would further follow on more patients on this group and also observe their overall survival time. We hypothesized that along with the development of EGFR mutation detecting technology, quantifying the abundance of EGFR mutations might be more important and predictable for TKIs clinical benefits. More investigations need to be explored on it.
In our study, we also detected some uncommon mutations, and ARMS seemed to be of a more sensitive way to detect the mutations. When these patients were treated with gefitinib, most of them achieved stable diseases with lower ORR and shorter PFS when compared as those with 19 del or 21 exon L858R mutations. Although uncommon EGFR mutations account for only a small proportion in NSCLC patients, TKIs efficacy in these patients are attracting more and more attention. The study reported on the World Conference of Lung Cancer (WCLC) in 2013 showed that afatinib could achieve comparable treatment results in patients carrying some uncommon mutations with that observed in patients with activating mutations (20). Several other studies showed that gefitinib and erlotinib are active in patients with uncommon mutations, such as G719X/L861Q/S768I mutations, but less effective than in those with common mutations (21-23).
Resistance usually occurred after 9-10 months of TKIs treatment. Mechanisms has been investigated and found that T790M mutation accounted for about 50% of resistance cases (24). We also detected EGFR mutations in 26 TKI treatment resistant patients using their cytological samples. We identified a rate of 42.3% of T790M mutations in addition to their former activating mutations. The rest patients remained their former mutations without T790M mutation. It indicated that using cytological samples might be helpful in re-evaluating EGFR mutation status to exploring the resistance mechanism. This could provide clinicians more information for making further treatment decisions. On the 2014 ASCO Conference, some new TKIs drugs such as AZD9291 and CO1686 showed promising results in treating TKIs resistant patients with T790M mutation with ORR of about 60% (25,26). With the development of molecular investigation, retesting EGFR mutations when diseases progressed after TKI treatment was of more and more importance. Since using cytological specimen was feasible in EGFR mutation testing, it would also provide more choices for re-evaluation of molecular status in resistance.
In conclusion, our study proved that cytological specimens were feasible in testing EGFR mutations and predicting TKIs treatment outcome. Using ARMS way could detect more patients with mutations as compared to direct sequencing. Abundance of EGFR mutations might have an influence on the clinical treatment efficacy.
Acknowledgements
Disclosure: The authors declare no conflict of interest.
References
- 1.Jemal A, Siegel R, Xu J, et al. Cancer statistics, 2010. CA Cancer J Clin 2010;60:277-300. [DOI] [PubMed] [Google Scholar]
- 2.Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009;361:947-57. [DOI] [PubMed] [Google Scholar]
- 3.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004;350:2129-39. [DOI] [PubMed] [Google Scholar]
- 4.Mitsudomi T, Morita S, Yatabe Y, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol 2010;11:121-8. [DOI] [PubMed] [Google Scholar]
- 5.Ellison G, Donald E, McWalter G, et al. A comparison of ARMS and DNA sequencing for mutation analysis in clinical biopsy samples. J Exp Clin Cancer Res 2010;29:132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han X, Zhang Z, Wu D, et al. Suitability of surgical tumor tissues, biopsy, or cytology samples for epidermal growth factor receptor mutation testing in non-small cell lung carcinoma based on chinese population. Transl Oncol 2014;7:795-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai G, Wong R, Chhieng D, et al. Identification of EGFR mutation, KRAS mutation, and ALK gene rearrangement in cytological specimens of primary and metastatic lung adenocarcinoma. Cancer Cytopathol 2013;121:500-7. [DOI] [PubMed] [Google Scholar]
- 8.Malapelle U, Bellevicine C, De Luca C, et al. EGFR mutations detected on cytology samples by a centralized laboratory reliably predict response to gefitinib in non-small cell lung carcinoma patients. Cancer Cytopathol 2013;121:552-60. [DOI] [PubMed] [Google Scholar]
- 9.Tokumo M, Toyooka S, Kiura K, et al. The relationship between epidermal growth factor receptor mutations and clinicopathologic features in non-small cell lung cancers. Clin Cancer Res 2005;11:1167-73. [PubMed] [Google Scholar]
- 10.Yoshida K, Yatabe Y, Park JY, et al. Prospective validation for prediction of gefitinib sensitivity by epidermal growth factor receptor gene mutation in patients with non-small cell lung cancer. J Thorac Oncol 2007;2:22-8. [PubMed] [Google Scholar]
- 11.Kawaguchi T, Matsumura A, Fukai S, et al. Japanese ethnicity compared with Caucasian ethnicity and never-smoking status are independent favorable prognostic factors for overall survival in non-small cell lung cancer: a collaborative epidemiologic study of the National Hospital Organization Study Group for Lung Cancer (NHSGLC) in Japan and a Southern California Regional Cancer Registry databases. J Thorac Oncol 2010;5:1001-10. [DOI] [PubMed] [Google Scholar]
- 12.Shi Y, Au JS, Thongprasert S, et al. A prospective, molecular epidemiology study of EGFR mutations in Asian patients with advanced non-small-cell lung cancer of adenocarcinoma histology (PIONEER). J Thorac Oncol 2014;9:154-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paez JG, Jänne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 2004;304:1497-500. [DOI] [PubMed] [Google Scholar]
- 14.Kimura H, Fujiwara Y, Sone T, et al. High sensitivity detection of epidermal growth factor receptor mutations in the pleural effusion of non-small cell lung cancer patients. Cancer Sci 2006;97:642-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang JJ, Cheng Y, Zhao MF, et al. A phase II trial comparing pemetrexed with gefitinib as the second-line treatment of nonsquamous NSCLC patients with wild-type EGFR (CTONG0806). J Clin Oncol 2013;31;abstr 8042.
- 16.Liu Y, Liu B, Li XY, et al. A comparison of ARMS and direct sequencing for EGFR mutation analysis and tyrosine kinase inhibitors treatment prediction in body fluid samples of non-small-cell lung cancer patients. J Exp Clin Cancer Res 2011;30:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharma SV, Bell DW, Settleman J, et al. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer 2007;7:169-81. [DOI] [PubMed] [Google Scholar]
- 18.Chou TY, Chiu CH, Li LH, et al. Mutation in the tyrosine kinase domain of epidermal growth factor receptor is a predictive and prognostic factor for gefitinib treatment in patients with non-small cell lung cancer. Clin Cancer Res 2005;11:3750-7. [DOI] [PubMed] [Google Scholar]
- 19.Zhou Q, Zhang XC, Chen ZH, et al. Relative abundance of EGFR mutations predicts benefit from gefitinib treatment for advanced non-small-cell lung cancer. J Clin Oncol 2011;29:3316-21. [DOI] [PubMed] [Google Scholar]
- 20.Yang JC, Sequist LV, Geater S, et al. Activity of afatinib in uncommon epidermal growth factor receptor (EGFR) mutations: Findings from three trials of afatinib in EGFR mutation-positive lung cancer. J Thorac Oncol 2013;8:S141. [Google Scholar]
- 21.Wu JY, Yu CJ, Chang YC, et al. Effectiveness of tyrosine kinase inhibitors on "uncommon" epidermal growth factor receptor mutations of unknown clinical significance in non-small cell lung cancer. Clin Cancer Res 2011;17:3812-21. [DOI] [PubMed] [Google Scholar]
- 22.Watanabe S, Minegishi Y, Yoshizawa H, et al. Effectiveness of gefitinib against non-small-cell lung cancer with the uncommon EGFR mutations G719X and L861Q. J Thorac Oncol 2014;9:189-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chiu CH, Yang CT, Shih JY, et al. Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitor Treatment Response in Advanced Lung Adenocarcinomas with G719X/L861Q/S768I Mutations. J Thorac Oncol 2015;10:793-9. [DOI] [PubMed] [Google Scholar]
- 24.Sequist LV, Waltman BA, Dias-Santagata D, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med 2011;3:75ra26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sequist LV, Soria JC, Gadeel SM, et al. First-in-human evaluation of CO-1686, an irreversible, highly selective tyrosine kinase inhibitor of mutations of EGFR (activating and T790M). J Clin Oncol 2014;32:abstr 8010.
- 26.Janne PA, Ramalingam SS, Yang JC, et al. Clinical activity of the mutant-selective EGFR inhibitor AZD9291 in patients (pts) with EGFR inhibitor-resistant non-small cell lung cancer (NSCLC). J Clin Oncol 2014;32:abstr 8009.


