Abstract
To date, genes identified and transcriptionally regulated by the AirSR TCS have been involved in energy production and cellular homeostasis of the staphylococcal cell. It is well accepted that the state of cellular metabolism impacts the expression of virulence factors in Staphylococcus aureus. For this reason, we conducted experiments to determine if the AirSR TCS contributes to the pathogenesis of S. aureus using an antisense RNA interference technology, an inducible overexpression system, and gene deletions. Depletion of AirSR by antisense RNA expression or deletion of the genes, results in significant decrease in bacterial survival in human blood. Conversely, overexpression of AirR significantly promotes survival of S. aureus in blood. AirR promotes the secretion of virulence factors that inhibits opsonin-based phagocytosis. This enhanced survival is partially linked to the transcriptional regulation of the sspABC operon, encoding V8 protease (SspA), staphopain B (SspB) and staphostatin B (SspC). SspA and SspB are known virulence factors which proteolytically digest opsonins and inhibit killing of S. aureus by professional phagocytes. This is the first evidence linking the AirSR TCS to pathogenesis of S. aureus.
Keywords: S. aureus, V8 protease, staphopain B, transcriptional regulation, AirSR (YhcSR)
Introduction
Staphylococcus aureus accounts for approximately 20% of bloodstream infections in the U.S. (Wisplinghoff et al., 2004). The bacteria gain access to the bloodstream commonly from the result of puncture wounds of the skin (Saravolatz et al., 1982; Control Centers for Disease Control and Prevention [CDC], 2003; Begier et al., 2004), surgical site infections, or insertion of central venous lines and catheters (Maki et al., 1997; Wisplinghoff et al., 2004). Once S. aureus enters the bloodstream, the bacteria have the ability to enter almost any site of the human body (Gordon and Lowy, 2008). S. aureus bloodstream infections often lead to septic shock and endocarditis (Lowy, 1998). Bacteremia was responsible for 75% of invasive S. aureus infections, which were identified by the Active Bacterial Core Surveillance program, a nationwide observation program of federal and state health officials. Septic shock and endocarditis accounted for an additional 10% of invasive infections (Klevens et al., 2007).
The pathogenicity of S. aureus partially relies on the coordinately regulated expression of virulence factors that allow the bacterium to evade the host immune system and/or promote survival during infection. Similar to other bacterial pathogens (Crosa, 1997; Cotter and DiRita, 2000; Ollinger et al., 2008; Tomaras et al., 2008; Hammerstrom et al., 2011; Ouyang et al., 2011), S. aureus has evolved a series of regulatory effectors (Crosa, 1997; Howell et al., 2003; Torres et al., 2007; Zheng et al., 2007; Montgomery et al., 2010) which allow the organism to sense and to adapt to changing environmental stimuli and survive within a particular niche by modulating specific cellular responses and virulence gene expression. Sixteen two-component systems are encoded in the core S. aureus genome, with many of them influencing the expression of virulence factors (Novick et al., 1993; Brunskill and Bayles, 1996; Giraudo et al., 1999; Fournier et al., 2001; Kuroda et al., 2003; Liang et al., 2005; Toledo-Arana et al., 2005; Meehl et al., 2007; Watkins et al., 2011, 2013). Some of these TCSs link cellular metabolism and virulence factor expression to the availability of extracellular nutrients, such as KdpDE and HssRS systems that sense extracellular K+ and heme, respectively (Torres et al., 2007; Xue et al., 2011). Analysis of AirSR to date has shown the two-component system to be a sensor of oxygen (Sun et al., 2011) that modulates the expression of pathways responsible for dissimilatory nitrate reduction (Yan et al., 2011), cellular osmotic balance (Yan et al., 2009) and alternative sugar catabolism pathways (Yan et al., 2012). Moreover, the AirSR TCS is important for aerobic and anaerobic growth of S. aureus (Sun et al., 2005; Hall and Ji, 2013).
The mechanisms by which S. aureus survives and subverts the vertebrate immune system have been studied for many decades. S. aureus produces various immune suppression factors, including V8 protease (sspA, serine endopeptidase), staphopain B (sspB, cysteine endopeptidase) and staphostatin B (sspC, inhibitor of Staphopain B). These proteases have been linked to a wide variety of innate immune system suppression pathways by their ability to degrade complement components (Jusko et al., 2013), induce vascular leakage and promote extracellular matrix structural damage (Imamura et al., 2005; Ohbayashi et al., 2011). In addition, the proteases inhibit neutrophil chemotaxis and induce apoptosis of neutrophils or engulfment of neutrophils by macrophages (Smagur et al., 2009a,b). The circulating neutrophils and monocytes are key innate cellular components to combat infection by S. aureus (Kapral and Shayegani, 1959; Mandell, 1974; Fournier and Philpott, 2005; DeLeo et al., 2009; Rigby and DeLeo, 2012; Spaan et al., 2013).
In this study, we found that the overproduction of AirR resulted in enhanced survival of S. aureus in human blood and inhibited opsonin-mediated phagocytosis. We identified that AirSR activates expression of the sspABC protease operon. Analysis of an sspAB mutant revealed the proteases are only one of many, as yet unidentified, proteins that contribute to AirSR-mediated survival in blood and inhibition of opsonophagocytic clearance of the bacteria.
Materials and Methods
Bacterial Strains, Plasmids, and Growth Media
The bacterial strains and plasmids used in this study are listed in Tables 1 and 2. The S. aureus cells were cultured in trypticase soy broth (TSB) at 37°C with shaking. Escherichia coli strains were grown in Luria-Bertani (LB) broth. Transformants containing recombinant plasmids were selected on LB agar containing ampicillin (100 μg/ml), kanamycin (50 μg/ml), or erythromycin (300 μg/ml) for E. coli, and trypticase soy agar (TSA) containing chloramphenicol (10 μg/ml), tetracycline (5 μg/ml), and/or erythromycin (5 μg/ml) for S. aureus. All overnight cultures grew to similar OD600nm values.
Table 1.
Bacterial strains.
| Strain | Description | Reference |
|---|---|---|
| DC10B | Dam- Escherichia coli which allows for direct electroporation of purified plasmid DNA into wild-type Staphylococcus aureus | Monk et al. (2012) |
| BL21(DE3) | E. coli used for protein expression; IPTG inducible; Cmr and Kanr | Invitrogen |
| RN4220 | Laboratory S. aureus strain (RsbU-) | Kreiswirth et al. (1983) |
| WCUH29 | Encapsulated human clinical HA-MRSA isolate | NCIMB40771 |
| AH1263 | USA300 CA-MRSA Erms (LAC*) | (White et al., 2014) |
| AH2084 | AH1263ΔairSR | White et al. (2014) |
| JE2 | Plasmid cured derivative of LAC strain, community-acquired MRSA isolate | Fey et al. (2013) |
| MW2 | USA300 Community acquired MRSA isolate | |
| MRSA923 | Community-acquired MRSA isolate | |
| JSAS909 | WCUH29 with pYJY909; Ermr | Sun et al. (2005) |
| WAirR | WCUH29 with pAirR; Ermr | This Study |
| JE2/pYH4 | JE2 with pYH4; Ermr | This Study |
| JE2/pAirR | JE2 with pAirR; Ermr | This Study |
| MRSA923/pYH4 | MRSA923 with pYH4; Ermr | This Study |
| MRSA923/pAirR | MRSA923 with pAirR; Ermr | This Study |
| MW2/pYH4 | MW2 with pYH4; Ermr | This Study |
| MW2/pAirR | MW2 with pAirR; Ermr | This Study |
| NE1787 | JE2 with bursa aurealis Tn in srtA | Fey et al. (2013) |
| NE1787/pYH4 | NE1787 with pYH4; Ermr | |
| NE1787/pAirR | NE1787 with pAirR; Ermr | |
| WSspB | WCUH29 with pSspB; Ermr | This Study |
| Control Pssp-lux | WCUH29/pYH4 with Pssp-lux; Ermr | This Study |
| WAirR/Pssp-lux | WAirR with Pssp-lux; Ermr | This Study |
| AH1263/Pssp-lux | AH1263/with Pssp-lux; Ermr | This Study |
| AH2084/Pssp-lux | AH2084/with Pssp-lux; Ermr | This Study |
| ΔSspAB | WCUH29 with in-frame deletion of sspAB | This Study |
| ΔSspAB/pYH4 | WCUH29ΔsspAB with pYH4; Ermr | This Study |
| ΔSspAB/pAirR | WCUH29ΔsspAB with pAirR; Ermr | This Study |
| ΔSspAB/pSspABC | WCUH29ΔsspAB with pSspABC; Ermr | This Study |
Table 2.
Plasmids.
| Plasmids | Description | Reference |
|---|---|---|
| pCY1006 | Shuttle vector carrying agr promoter-gfp-lux reporter, derives from pSB2019; Cmr, Ampr | Liang et al. (2006) |
| pETairR | pET24b based for production of AirR in E. coli BL21(DE3) | Yan et al. (2012) |
| pSAS909 | pYH3 with airSR antisense downstream of TetR promoter; Ampr, Ermr | Sun et al. (2005) |
| pYH4 | pYH3 with Ampr removed; Ermr | Huang et al. (2004) |
| pAirR | airR cloned downstream of pYH4 TetR promoter for overproduction; Ermr | This Study |
| pSspB | sspB cloned downstream of pYH4 TetR promoter for overproduction; Ermr | This Study |
| Pssp-lux | sspABC promoter cloned upstream of promoterless luxABCDE; derived from pCY1006; Cmr, Ampr | This Study |
| pJB38-sspAB | pJB38 with in-frame sspAB upstream/downstream deletion region | White et al. (2014) |
| pSspABC | sspABC operon cloned downstream of pYH4 TetR promoter for overproduction; Ermr | This Study |
Blood Survival Assay
Strains were cultured in TSB with appropriate antibiotics. Inducer anhydrotetracycline (ATc) was added when indicated to overnight cultures. Following 18 h of culturing, the bacteria were washed twice in sterile PBS and suspended to an OD of 0.14 using a Behring photometer in PBS. Fresh venous human whole blood was collected using heparin containing Vacutainer tubes (BD) from outwardly healthy adult donors. The blood was then immediately used in the assay. Approximately 5 × 106 CFU in 50 μl of PBS were added to 450 μl of blood per microcentrifuge tube with appropriate antibiotics and ATc, where indicated. Microcentrifuge tubes were capped and placed in a rotisserie incubator and incubated at 37°C with end-over-end mixing. At indicated time points a 20 μl sample was removed from each sample, serially diluted, and plated on TSA to determine the surviving CFU count for each sample. The percentage of surviving bacteria was calculated as CFUtimepoint/CFUinitialinput*100. Blood collection was approved by the University of Minnesota Institutional Review Board.
Gene Deletion
Deletion of sspAB was carried out following the pKOR1 allelic exchange protocol as described (Bae and Schneewind, 2006) and primers in listed in Table 3. Plasmid pJB38 is a modified version of pKOR1 (Bose et al., 2013) and pJB38-sspAB was kindly provided by Alex Horswill (Mootz et al., 2013). All deletions were confirmed by diagnostic PCR.
Table 3.
Oligonucleotide sequencesa.
| Primers | Sequence |
|---|---|
| ssp Pro For EcoRI | 5′-TAGCGAATTCGATATGTTGAACT GGACGTCGTGAAC-3′ |
| ssp Pro Rev XmaI | 5′-TTCCCGGGCTAAAAACCTCCAA AAAATTTATTTACAAGTTAAATA TAACAC-3′ |
| sspB-For | 5′-AGGAGGTTTAAACTATGAATAGTTCATA TAAATCTAGAGTATTCA ATATTATAAGC-3′ |
| sspB-Rev-AscI | 5′-TTGGCGCGCCTTAGTAACC TATCATTGAACCATACCAG-3′ |
| sspABC-For | 5′-AGGAGGTTTAAACTATGAAAGGT AAATTTTTAAAAGTTAGTTC TTTATTCG-3′ |
| sspABC-Rev-AscI | 5′-TT GGCGCGCCTTATACTAAGCG CTCATAAACGATTGG-3′ |
| AirROE-for | 5′-AAACTATGGAACAAAGG ACGCGAC-3′ |
| AirROE-rev | 5′-TTGGCGCGCCCTATTTTA TAGGAATTGTGAATTG-3′ |
| pJB38-ssp For | 5′-GAATATGATATTAAGTC ACTTGCGTCG-3′ |
| pJB38-ssp Rev | 5′-GCTTATGAAATGGATGTT TTAAAAGAAGGTATG-3′ |
| luxArev | 5′-GCT CCA GTA ACC ATA CGG TAT C- 3′ |
| TetRfor | 5′-CAA TAC AAT GTA GGC TGC -3′ |
| TTrev | 5′-CTC AGG AGA GCG TTC AC -3′ |
aPurchased from IDT DNA, Coralville, IA.
Cloning, Expression, and Purification of AirR-His Tagged Fusion Protein in Escherichia coli
The purification of AirR-6x His was carried out as described using the previously constructed pETairR plasmid (Yan et al., 2012). The only modification to the protocol was the use of Pro-LyseTM Bacterial Lysis Buffer (Lamda Biotech) to lyse the E. coli.
SDS-PAGE Analysis of Exported Proteins, Mass Spectrometry Peptide-Protein Identification, and Immunoblotting
The culture supernatants were collected from the overnight cultures of S. aureus strains grown in TSB medium with 5 μg/ml of erythromycin and 250 ng/ml inducer ATc. Bacterial cells were pelleted by centrifugation at 3900 × g for 20 min. The culture supernatants were then passed through a 0.2 μm syringe filter to remove bacterial cells. The exported proteins were precipitated from an equal volume of supernatant using ethanol as described (Ji et al., 1999). The exported protein profiles were detected by 12% SDS-PAGE and Coomassie Blue staining. Prominent overproduced protein bands were cut from the gel and in-gel digested (Shevchenko et al., 1996). Samples were submitted to the University of Minnesota Mass Spectrometry Core for mass spectrometry. Immunoblotting for SspB was conducted as described previously (Liang et al., 2006) using a SspB antibody kindly provided by Alex Horswill (Mootz et al., 2013) and an alkaline phosphatase conjugated anti-chicken secondary antibody (Sigma). Overnight cultures grew to similar OD600nm values and equal volume of precipitated protein from each culture supernatant was loaded rather than equal protein concentration so that differences in protein concentration could be observed.
Zymography Analysis
Induced cultures were grown in TSB with appropriate antibiotics and 250 ng/ml of inducer ATc overnight at 37°C with shaking. The following day the bacterial cells were pelleted and the TSB culture supernatant was filter sterilized with a 0.2 μm syringe filter. Twenty five milliliters of each culture supernatant, along with sterile TSB as a vehicle control, were concentrated 50-fold using a Millipore Centrifugal Protein Concentrator with a 10 kD nominal molecular weight limit. Proteins were resolved using 12% SDS-PAGE, gelatin was added to a final concentration of 0.1% (v/v) for zymography analysis. An equal volume of each concentrated culture supernatant sample was mixed with protein solubilization buffer (5X, 50% glycerol, 10% (w/v) SDS, and 0.5 M Tris-HCl, pH 6.8) and incubated at room temperature for 30 min.
Each sample was loaded and resolved in the gelatin SDS-PAGE. After electrophoresis, the gel was placed in a plastic wash container and washed with 1X SDS removal buffer (2.5% Triton X-100, 5 mM MgCl2, 25 mM Tris-Cl, pH 7.5) for 60 min at room temperature. The SDS removal buffer was replaced after 30 min with fresh removal buffer and then rinsed gently with deionized (DI) water. Development buffer (0.1% Triton X-100, 5 mM MgCl2, 25 mM Tris-Cl, and pH 7.5) was added until it covered the gel and the container was incubated at 37°C overnight. After the development period, the development buffer was removed and the gel was rinsed gently with DI water. Stain buffer (50% DI water, 35% MetOH, 15% Glacial Acetic Acid, 0.25% Coomassie Blue R-250) was added to cover the gel in the container. The gel was incubated until it was no longer visible in the stain buffer. Stain buffer was removed and the gel was rinsed gently with DI water. Fixing buffer (2% Glacial Acetic Acid, 98% DI water) was added to the container until it covered the gel and incubated at room temperature for 24 h.
Analysis of Transcriptional Regulation Using a Promoter-luxABCDE Reporter Fusion System
The upstream ssp promoter region was PCR amplified with primers ssp Pro For/ssp Pro Rev listed in Table 3, digested with EcoRI and XmaI (NEB) and replaced the agr promoter fragment in pCY1006. The re-constructed ssp-lux promoter reporter was confirmed by diagnostic PCR. Plasmids were purified from E. coli DC10B and electroporated into the S. aureus strains as indicated in Table 1. Bioluminescence intensity and optical density of the cultures were measured at different times of the experiment in duplicate. The Relative Light Units (RLU) were calculated by dividing the average bioluminescence reading by the average OD600nm reading (lum/OD600nm) at each time point. The experiment was repeated three times with separate colonies of each strain.
Construction of Overproduction Plasmids
Gene ORFs were obtained by PCR using Q5 high-fidelity polymerase (NEB) with the primers (AirROE-for/AirROE-rev; sspB-For/sspB-Rev-AscI; sspABC-For/sspABC-Rev-AscI) in Table 3. Purified PCR fragments were digested with AscI. The pYH4 plasmid carrying the TetR regulated, ATc inducible promoter was digested with PmeI and AscI. Digested PCR fragments were ligated into the digested pYH4 plasmid with T4 DNA ligase (Promega) and confirmed by diagnostic PCR using the Tetfor/TTrev primer pair listed in Table 3 that are specific for regions upstream and downstream of the MCS in pYH4.
HL-60 Opsonophagocytic Killing Assay
To determine the effect of S. aureus strains exported proteins on the activity of serum antibodies and complement, induced cultures were grown in TSB with appropriate antibiotics and 250 ng/ml of inducer ATc overnight at 37°C with shaking. The following day the bacterial cells were pelleted and the TSB culture supernatant was filter sterilized with a 0.2 μm syringe filter. Twenty five milliliters of each culture supernatant, along with sterile TSB as a vehicle control, were concentrated 50-fold using a Millipore Centrifugal Protein Concentrator with a 10 kD nominal molecular weight limit. Before the HL-60 phagocytic assay, each concentrated culture supernatant and TSB was mixed 1:1 with the human serum or rabbit complement, incubated at 37°C for 30 min, and then placed on ice. In the assay, 40 and 20 μl of the serum mixture and complement mixture, respectively, were added to each well. Additional buffer was added to the complement mixture wells so all wells were of equal 100 μl volume.
HL-60 pluripotent cells were differentiated to granulocytic cells and cultured for 5 days as described (Kim et al., 2003). The basic assay consisted of 1,000 CFUs of S. aureus WCUH29 placed in duplicate of a 96 well microtiter plate. Pre-treated human serum and complement from 3 to 4 week of white rabbits (Life Technologies), respectively, were added to each well. Lastly, 4 × 105 differentiated HL-60 granulocytes were added to each well to initiate the assay. The plates were incubated for 60 min at 37°C and with a CO2 concentration of 5%. Each well was mixed gently and 10 μl of sample from each well was drop plated in triplicate on TSA plates to determine surviving CFU. The percent survival was calculated as the number of surviving CFU/number of input CFU multiplied by 100, (CFUf/CFUi*100). The experiment was repeated at least 4 times.
Statistical Analysis
Statistical data analysis was performed in Microsoft Excel for Mac 2011 using unpaired Student’s t-tests with a alpha level ≤ 0.05. Significant differences are noted by the addition of the p-value over the data being compared.
Results
AirSR Contributes to the Survival of S. aureus in Human Blood
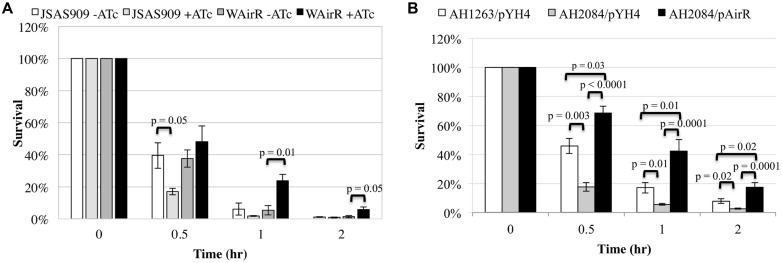
The AirSR TCS is essential for growth in S. aureus WCUH29 (Sun et al., 2005), and it is important to validate the in vivo essentiality of any gene as some genes found to be essential in vitro may not be essential in vivo (Gandotra et al., 2007; Brinster et al., 2009). Survival of the airS antisense RNA strain (JSAS909) in human blood was examined as an initial step to determine the importance of airSR for survival the human host. An equal number of colony forming units (CFUs) per strain were inoculated into a defined volume of freshly isolated venous blood and depletion of AirSR by induction of airSR antisense RNA with ATc (Sun et al., 2005) resulted in a significantly decreased percentage of ATc induced JSAS909 CFUs surviving in the first half hour of incubation in human blood compared to the non-induced inoculum (Figure 1A, 18% vs. 40%). After 1 h, fewer ATc induced JSAS909 CFUs survived compared to the non-induced JSAS909, but was not statistically different. After 2 h of incubation, a similar percentage of CFUs survived for both strains (Figure 1A). Uninduced (–ATc) and induced (+ATc) empty plasmid control strains survived equally well (data not shown).
FIGURE 1.
The AirSR TCS is important for survival in human blood. (A) Percent survival of the induced Staphylococcus aureus airSR antisense strain (JSAS909, 500 ng/ml ATc) and AirR overproduction strain (WAirR, 250 ng/ml ATc) in human blood during induction. (B) Percent survival of the wildtype LAC* (AH1263/pYH4), LAC*ΔairSR (AH2084/pYH4), and LAC*ΔairSR /AirR overproduction strain (AH2084/pAirR) in human blood during with 250 ng/ml of inducer ATc. Data represents the mean and SEM of at least three experiments.
The induction of airSR antisense RNA, results in a delayed growth phenotype in WCUH29 (Sun et al., 2005). To eliminate impact of growth factors on bacterial survival in human blood, we determined if the overproduction of the AirR response regulator could promote survival in blood using an inducible overproduction strain (WAirR). ATc induced overproduction of AirR promoted survival of S. aureus WCUH29 over the course of the 3-h experiment compared to non-induced WAirR (Figure 1A). By hour two of the experiment, a significantly greater percentage of the initial inoculum of the induced WAirR strain survived in the blood compared to the uninduced WAirR strain (Figure 1A, 24% vs. 5%) and the increased survival of induced WAirR continued into hour three of the experiment.
To determine if the enhanced survival of S. aureus during AirR overproduction was applicable to other genetic backgrounds of S. aureus, we examined the effect of AirR overproduction in community-acquired methicillin resistant S. aureus (CA-MRSA) strains, MW2, MRSA923, and JE2. Similar to the results found with WCUH29, ATc induced overproduction of AirR greatly increased the percentage of CFUs that survived in human blood for all strains (Supplementary Figure S1).
Since the first publication on the identification and essentiality of airSR in strain WCUH29 (Sun et al., 2005), there have been several other research articles published investigating various aspects of the biological function of AirSR and the essentiality of airSR in other S. aureus strains has been disputed in these articles (Sun et al., 2011; White et al., 2014). This difference in essentiality may be due to distinct genetic differences between the strains of S. aureus used in each study. Most recently, the clean deletion of airSR was reported in S. aureus AH1263, a derivative of LAC (White et al., 2014). Following this publication, the pJB38-airSR deletion plasmid was introduced into both WCUH29 and S. aureus JE2, respectively. Approximately 100 colonies of WCUH29 and JE2 were screened for deletion of airSR using diagnostic colony PCR. Deletion of airSR was not detected in strain WCUH29 but was readily detected in the JE2 strain (data not shown), indicating the essentiality of airSR is strain dependent.
To determine if the airSR null deletion in AH1263 impacted bacterial survival in human blood, we conducted blood survival assays. Similar to our results with strain JSAS909, deletion of airSR significantly impaired the ability of AH1263/pYH4 to survive in whole blood (Figure 1B). The decreased survival of the AH2084/pYH4 strain was more than complemented by introduction and ATc induction of the AirR overproduction plasmid, with AH2084/pAirR having significantly enhanced survival relative to the AH1263/pYH4 and AH2084/pYH4 (Figure 1B). All three strains were assayed as group, thus the empty pYH4 control strain was introduced into the AH1263 and AH2084 to control for potential effects caused by the use of erythromycin and inducer ATc during the blood survival assay.
AirR-Mediated Secreted Factors are Important for Enhanced Survival and Inhibited Opsonophagocytic Killing of S. aureus WCUH29
Staphylococcus aureus produces numerous LPXTG cell-surface linked MSCRAMMs and exported virulence factors involved in inhibition of complement and antibody mediated phagocytosis that enhance survival in blood and tissues (Zecconi and Scali, 2013; Foster et al., 2014). Since the JE2 strain showed similar enhanced survival to WCUH29, we utilized the srtA JE2 bursa aurelis Tn mutant, NE1787, to determine which surface factor(s) are involved in the enhanced survival in blood mediated by AirR. Sortase A is a transpeptidase responsible for proper LPXTG-MSCRAMM attachment to the cell surface. The AirR overproduction plasmid (pAirR) was electroporated into NE1787. We found Tn mutagenesis of srtA had no influence on AirR enhanced bacterial survival in human blood (data not shown), indicating SrtA processed MSCRAMMs are not responsible for the AirR-mediated enhanced survival in blood.
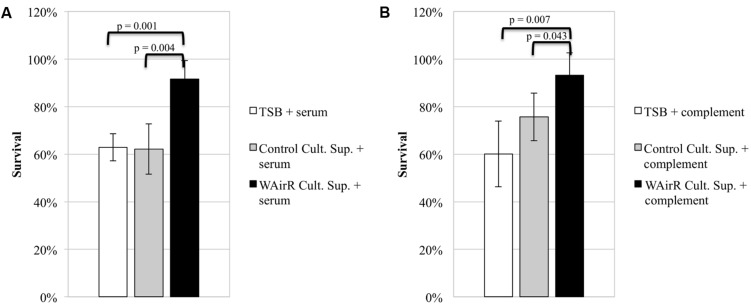
To investigate if exported proteins contribute to AirSR regulated anti-phagocytic mechanisms, we determined the effect of culture supernatants on bacterial anti-phagocytic capacity using a HL-60 opsonophagocytic killing assay (see Materials and Methods). If the induced WAirR gives rise to more anti-phagocytic virulence factors, a greater percentage of wild-type S. aureus WCUH29 CFUs would be expected to survive when the fractions are incubated with ATc induced WAirR culture supernatant compared to sterile concentrated TSB or concentrated ATc induced empty plasmid control supernatants. Indeed, significantly more wild-type S. aureus WCUH29 survived when the serum fraction (Figure 2A, 90% vs. 60%) or complement fraction of the assay (Figure 2B, 90% vs. 75%) was pre-incubated with concentrated induced WAirR culture supernatant compared to the induced control supernatant. As a control, concentrated TSB growth medium was included and did not impact the killing S. aureus relative to the induced control plasmid supernatant. These data suggest the AirSR two-component system contributes to S. aureus survival in human blood by promoting production of anti-opsonophagocytic virulence factors that inhibit serum- and complement-mediated mechanisms.
FIGURE 2.
AirSR regulated exported proteins inhibit complement and antibody mediated opsonophagocytic killing of S. aureus. Sterile TSB and culture supernatants of ATc (250 ng/ml) induced control (Control Cult. Sup) and AirR overproducing S. aureus (WAirR Cult. Sup) were concentrated 50-fold. The serum (A) and complement (B) components of the assay were pre-incubated 30 min with concentrated sterile TSB or individual culture supernatants before addition to the assay. Data represents the mean and standard error of four individual experiments.
Identification of Overproduced Exported Proteins Resulting from AirR Overproduction in S. aureus
To identify which exported protein(s) are overexpressed resulting from AirR overproduction, we prepared the exported proteins from cell-free culture supernatants of ATc induced pYH4 control and WAirR strains. The exported proteins were visually compared using SDS-PAGE, and protein bands that were obviously over-represented or bands that only appeared in the WAirR lane were processed for peptide identification by mass spectrometry (see Materials and Methods).
The subsequently identified proteins were cross-referenced with published studies to identify proteins that are involved in innate immune suppression via inhibition of the humoral and/or innate cellular response. Peptides from the cysteine endopeptidase, staphopain B (SspB) dominated one of the over-represented bands from ATc induced WAirR culture supernatants. More than 80% of the processed active form of SspB was identified by mass spectrometry (Supplementary Figure S2).
AirR Overproduction Results in Increased Functional Staphopain B Production
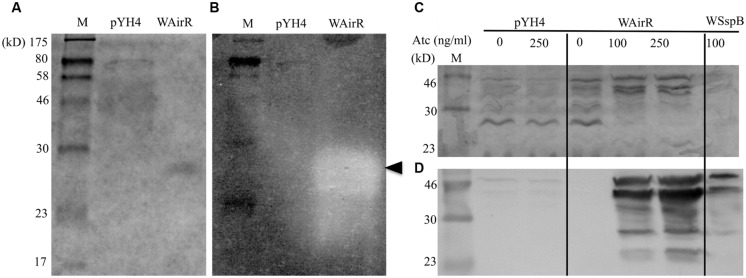
To examine if the overproduced SspB is functional, we conducted gelatin zymography assays using the cell-free culture supernatants, as SspB is able to degrade collagen (Ohbayashi et al., 2011). Coomassie Blue staining and gelatin zymography analyzed was used to analyze an equal volume of concentrated culture supernatant from each induced strain. A single band in the induced WAirR lane and the disappearance of other proteins relative to the pYH4 control lane was detected by Coomassie Blue (Figure 3A). Gelatin zymography analysis of the same samples revealed very little gelatin degradation in the control strain, while a large, prominent band of gelatin degradation appeared in the induced WAirR sample (Figure 3B). Importantly, both the Coomassie Blue stained protein band and gelatin degradation in the zymogram resolve at the same molecular weight from the induced WAirR, suggesting the gelatin degradation is the result of this protein. These data highly suggest that the overproduction of AirR results in the overproduction of functional cysteine endopeptidase SspB.
FIGURE 3.
Zymographic and immunoblot analysis of concentrated control and WAirR exported proteins. Equal volumes of concentrated culture supernatant were resolved by (A) 12% SDS-PAGE and proteins stained with Coomassie Blue or (B) 0.1% gelatin-12% SDS-PAGE and processed for zymography, (←, indicates point of gelatin degradation). (C) SDS-PAGE analysis of precipitated culture supernatant proteins from control and WAirR strains without and with ATc and WSspB with ATc. (D) Immunoblot detection of SspB from precipitated culture supernatants showing increased amounts of SspB with increasing induction of AirR by inducer ATc.
To test the hypothesis that the increased gelatin degradation is the result of increased production of SspB and to confirm our mass spectrometry identification data, we ethanol precipitated the exported proteins from the cell-free culture supernatants of the pYH4 control and WAirR strains without and with inducer ATc. As seen in Figure 3C, the addition of ATc had no apparent impact on the protein profile of the control strain, while the addition of ATc to WAirR resulted in a stronger detection of a protein band similar in size to the SspB zymogen at 44 kD. Additionally, many protein bands were absent in the stained SDS-PAGE from the induced WAirR supernatant, consistent with previous reports that up-regulation of SspB (and SspA) results in degradation of other exported proteins (Karlsson et al., 2001; Jones et al., 2008). As a positive control, the sspB gene was cloned into the same ATc inducible expression vector (pSspB, strain WSspB). Further confirmation of SspB up-regulation was carried out by immunoblotting using chicken egg antibody specific for SspB (Kolar et al., 2013). In the control strain, SspB production was low and appeared unaffected by the addition of ATc (Figure 3D). In an ATc dose-dependent manner, staphopain B production was up-regulated in the WAirR supernatant (Figure 3D). SspB was readily detectable in the induced positive control WSspB supernatant as well. The SspB specific antibody detected the various processed and degraded forms of the protein (Shaw et al., 2005, 2004). These data clearly indicate a regulatory link between AirSR and SspB production.
Transcription from the ssp Promoter is Regulated by AirR
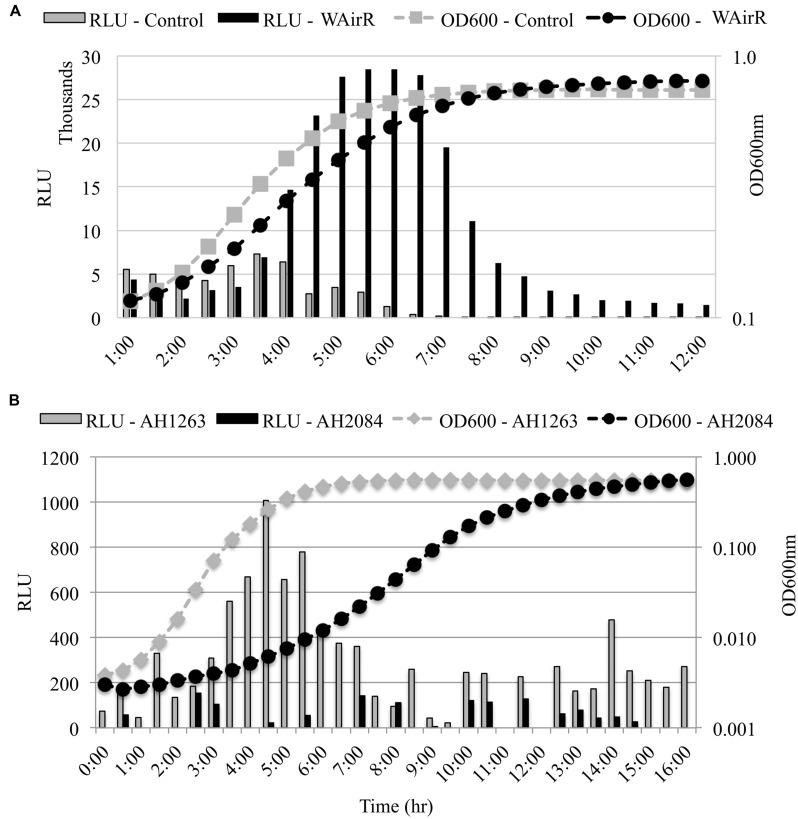
Staphopain B is produced from the middle gene of a three gene operon and is bordered upstream by sspA, encoding the V8 serine endopeptidase and downstream by sspC which encodes staphostatin B, a cytoplasmic inhibitor of SspB (Rice et al., 2001). To determine if the up-regulation of SspB production occurs post-transcriptionally or if transcription from the ssp promoter is increased in the induced WAirR strain, we examined the effect of AirR overproduction and deletion of airSR on the transcription of the ssp operon using a ssp promoter-luxABCDE reporter system. The induction of AirR production with inducer ATc resulted in a fivefold maximal increase in bioluminescence intensity compared to the control (Figure 4A). Furthermore, bioluminescence driven by the ssp promoter was higher and sustained throughout the growth of WAirR, demonstrating that continued and prolonged AirR overproduction results in increased transcription from the ssp promoter (Figure 4A). To examine if the absence of AirSR impacts the ssp promoter driven bioluminescence, the ssp-lux reporter was electroporated into the wild-type AH1263 and ΔairSR, AH2084, strains. Maximum ssp-driven bioluminescence was reduced fivefold in AH2084 compared to AH1263 (Figure 4B). These data indicate AirSR is a positive transcriptional regulator of the sspABC operon.
FIGURE 4.
ssp promoter–reporter analysis. All strains harbor the Pssp-lux reporter plasmid. (A) Uninduced overnight cultures of control and WAirR were diluted 1:300, incubated at 37°C with 25 ng/ml of inducer ATc. (B) Uninduced overnight cultures AH1263 and AH2084 were diluted 1:1000 and incubated at 37°C. OD600nm and bioluminescence readings were measured every 30 min with 1 min of mixing before each reading in a BioTek Synergy II spectrophotometer. The light intensity values for each time point are given as relative light units (RLU), Lum reading/OD600nm reading for each time point. Data presented are the mean of three independent colonies.
SspABC is Not the Only Virulence Factor Involved in AirSR Mediated Survival in Human Blood
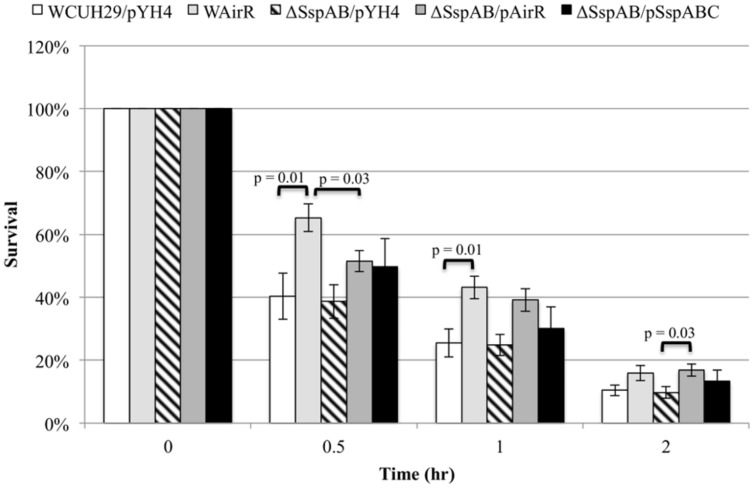
To investigate if AirR mediated enhanced survival and antiphagocytosis is due only to up-regulation of the Ssp proteases, we created a sspAB null mutant in S. aureus WCUH29 using an in-frame sspAB deletion plasmid (Mootz et al., 2013; kindly provided by Alex Horswill). We examined the survival of ATc induced wild-type WCUH29 and ΔSspAB without and with AirR overproduction in human blood. As seen previously, the overproduction of AirR increased the percentage of CFU that survived throughout the experiment (Figure 5, WCUH29/pYH4 vs. WAirR). However, deletion of sspAB did not result in decreased survival compared to wild-type WCUH29. The ΔSspAB/pAirR strain survived better compared to ΔSspAB/pYH4, but the percentage of surviving CFUs was statistically reduced when compared to induce WAirR in the first half hour of the assay only (Figure 5, WAirR vs. ΔSspAB/pAirR). After the first half hour, the deletion of sspAB had a minimal impact on the enhanced survival mediated by AirR overproduction and by two hours, WAirR and ΔSspAB/pAirR had similar percentages of surviving CFUs. Complementation of the ΔSspAB with an sspABC expression plasmid, on average, increased the percentage of bacteria that survived in human blood, but was not statistically different from WCUH29 or ΔSspAB. These data suggest, overall, SspAB contributes minimally to AirSR-mediated survival of S. aureus in human blood in the absence of AirR overproduction. Nonetheless, observing that the ATc induced ΔSspAB/pAirR strain had enhanced survival compared to ΔSspAB/pYH4 indicates additional, as yet unidentified virulence factors, are regulated by AirSR.
FIGURE 5.
Blood survival analysis of the wildtype and ΔSspABC without and with AirR and SspABC overproduction. Percent survival of the wild-type S. aureus WCUH29 and ΔSspAB mutant strain with pYH4 control, pAirR, and pSspABC overproduction plasmids in human blood with 250 ng/ml inducer ATc. Data is the mean and SEM of at least three experiments per strain.
Discussion
In this study, our data is the first to show that the AirSR two-component regulator is involved in pathogenesis of S. aureus. We utilized the inducible airSR antisense RNA and AirR overproduction approaches to alter the intracellular level of AirR and analyzed the impact of AirSR on bacterial survival and resistance to phagocytosis in healthy human whole blood. We revealed that the depletion of AirSR significantly inhibited the ability of the HA-MRSA isolate WCUH29 to survive in human blood during the first half hour of the assay in diverse staphylococcal genetic backgrounds, whereas the overproduction of AirR significantly enhanced survival in blood over 2 h. It was believed that airSR was essential for S. aureus growth, but this appears to a property of the WCUH29 strain. Recently, a ΔairSR mutant in the USA300 CA-MRSA lineage of S. aureus was constructed (White et al., 2014). Whereas airSR is not essential in this strain, the strain does appear to have a growth defect, as it grew much slower in the Pssp-reporter assay (Figure 3B), suggesting that AirSR, while dispensable in USA300 AH1263, is likely an important two-component system for S. aureus growth. These data indicate that the essentiality of airSR appears to be strain dependent. Similar to the WCUH29 yhcSR antisense RNA strain, the USA300 AH2084ΔairSR mutant survived significantly worse than the wild-type control (AH1263) in human blood. Furthermore, the overproduction of AirR in this mutant resulted in significantly enhanced survival of the strain in blood. These data clearly indicate, regardless of genetic background, the AirSR two-component system is important for survival in human whole blood. Additionally, analysis of a srtA mutant indicates that AirSR contributes to S. aureus survival in blood by regulating secreted factors, independent of cell wall-attached LPXTG-MSCRAMMS.
Our study indicates that one of these contributing AirR-regulated secreted factors is the cysteine endopeptidase, SspB, and possibly the serine protease, SspA, due to co-transcription. Our results show that AirSR mediates the production of staphopain B (SspB) using mass spectrometry, which was further supported by immunoblotting and gelatin zymographic assays. Moreover, using a promoter–reporter system it was demonstrated that AirR regulates the transcription of the sspABC operon, encoding V8 protease and staphopain B, both of which are known to promote survival in serum and inhibit opsonophagocytosis (Ryan et al., 2008; Smagur et al., 2009a,b; Jusko et al., 2013). This information corresponds well with the finding that the WAirR culture supernatant inhibits opsonophagocytic killing of S. aureus relative to the control extraordinarily well.
The AirSR TCS system regulates gene expression in response to the presence or absence of oxygen, and possibly reactive oxygen species (Sun et al., 2011). The regulation of sspABC by AirSR is of interest in the context of biofilm formation and stability and abscess formation, in addition to its apparent role in survival in blood. Biofilms and wound sites are known to have varying degrees of hypoxia (Sawyer et al., 1991; Beenken et al., 2004; Resch et al., 2005), thus, it is conceivable that AirSR may regulate expression of sspABC in response to the oxygen levels in the surrounding microenvironment. This regulation has implications in biofilm formation and stability, extracellular matrix destruction and wound healing, as well as neutrophil infiltration and immune response to infections (Imamura et al., 2005; Vincents et al., 2007; Smagur et al., 2009b; Ohbayashi et al., 2011; Chen et al., 2013; Jusko et al., 2013; Kolar et al., 2013; Mootz et al., 2013). Further investigation is needed to define the role of AirSR during systemic and abscess infections in relation to oxygen levels in these microenvironments and how AirSR regulation of sspABC and additional secreted virulence factors impacts the pathogenesis of S. aureus in these environments.
To elucidate whether the enhanced bacterial survival in human blood by overexpression of AirR is attributable to its positive regulation of the sspABC operon, we determined the impact of the sspABC null mutation on AirR-mediated anti-phagocytosis. We found the deletion of the sspABC operon did not significantly alter the survival capacity of wild-type WCUH29 strain, but did significantly reduce survival for the WAirR strain in the first half of the assay. Our studies indicate, as yet unidentified secreted AirSR regulated virulence factors, contribute to the ability S. aureus to resist phagocytosis and survive in human blood.
Conclusion
The AirSR two-component system is involved in the modulation of S. aureus survival in human blood. The AirSR system positively regulates the expression of the sspABC operon at the transcriptional level and additional secreted virulence factors. Studies are ongoing to identify the additional factors that are regulated by AirSR, how oxygen impacts AirSR-mediated pathogenesis, and the contribution of these factors to anti-phagocytosis and pathogenesis of S. aureus.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Alexander R. Horswill for kindly providing critical plasmids, SspB antibody, and strains AH1263 and AH2084. This work was partially supported by a grant from the National Institutes of Health (AI057451) and by a grant from the USDA/Minnesota agriculture station.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.00682
References
- Bae T., Schneewind O. (2006). Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 55 58–63. 10.1016/j.plasmid.2005.05.005 [DOI] [PubMed] [Google Scholar]
- Beenken K. E., Dunman P. M., McAleese F., Macapagal D., Murphy E., Projan S. J., et al. (2004). Global gene expression in Staphylococcus aureus biofilms. J. Bacteriol. 186 4665–4684. 10.1128/JB.186.14.4665-4684.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begier E. M., Frenette K., Barrett N. L., Mshar P., Petit S., Boxrud D. J., et al. (2004). A high-morbidity outbreak of methicillin-resistant Staphylococcus aureus among players on a college football team, facilitated by cosmetic body shaving and turf burns. Clin. Infect. Dis. 39 1446–1453. 10.1086/425313 [DOI] [PubMed] [Google Scholar]
- Bose J. L., Fey P. D., Bayles K. W. (2013). Genetic tools to enhance the study of gene function and regulation in Staphylococcus aureus. Appl. Environ. Microbiol. 79 2218–2224. 10.1128/AEM.00136-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinster S., Lamberet G., Staels B., Trieu-Cuot P., Gruss A., Poyart C. (2009). Type II fatty acid synthesis is not a suitable antibiotic target for Gram-positive pathogens. Nature 458 83–86. 10.1038/nature07772 [DOI] [PubMed] [Google Scholar]
- Brunskill E. W., Bayles K. W. (1996). Identification and molecular characterization of a putative regulatory locus that affects autolysis in Staphylococcus aureus. J. Bacteriol. 178 611–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention [CDC]. (2003). Outbreaks of community-associated methicillin-resistant Staphylococcus aureus skin infections–Los Angeles County, California, 2002-2003. MMWR 52 88. [PubMed] [Google Scholar]
- Chen C., Krishnan V., Macon K., Manne K., Narayana S. V. L., Schneewind O. (2013). Secreted proteases control autolysin-mediated biofilm growth of Staphylococcus aureus. J. Biol. Chem. 288 29440–29452. 10.1074/jbc.M113.502039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter P. A., DiRita V. J. (2000). Bacterial virulence gene regulation: an evolutionary perspective. Annu. Rev. Microbiol. 54 519–565. 10.1146/annurev.micro.54.1.519 [DOI] [PubMed] [Google Scholar]
- Crosa J. H. (1997). Signal transduction and transcriptional and posttranscriptional control of iron-regulated genes in bacteria. Microbiol. Mol. Biol. Rev. 61 319–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLeo F. R., Diep B. A., Otto M. (2009). Host defense and pathogenesis in Staphylococcus aureus infections. Infect. Dis. Clin. North Am. 23 17–34. 10.1016/j.idc.2008.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fey P. D., Endres J. L., Yajjala V. K., Widhelm T. J., Boissy R. J., Bose J. L., et al. (2013). A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. MBio 4 e00537-12. 10.1128/mBio.00537-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster T. J., Geoghegan J. A., Ganesh V. K., Höök M. (2014). Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus. Nat. Rev. Microbiol. 12 49–62. 10.1038/nrmicro3161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier B., Klier A., Rapoport G. (2001). The two-component system ArlS–ArlR is a regulator of virulence gene expression in Staphylococcus aureus. Mol. Microbiol. 41 247–261. 10.1046/j.1365-2958.2001.02515.x [DOI] [PubMed] [Google Scholar]
- Fournier B., Philpott D. J. (2005). Recognition of Staphylococcus aureus by the innate immune system. Clin. Microbiol. Rev. 18 521–540. 10.1128/CMR.18.3.521-540.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandotra S., Schnappinger D., Monteleone M., Hillen W., Ehrt S. (2007). In vivo gene silencing identifies the Mycobacterium tuberculosis proteasome as essential for the bacteria to persist in mice. Nat. Med. 13 1515–1520. 10.1038/nm1683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudo A. T., Calzolari A., Cataldi A. A., Bogni C., Nagel R. (1999). The sae locus of Staphylococcus aureus encodes a two-component regulatory system. FEMS Microbiol. Lett. 177 15–22. 10.1111/j.1574-6968.1999.tb13707.x [DOI] [PubMed] [Google Scholar]
- Gordon R. J., Lowy F. D. (2008). Pathogenesis of methicillin-resistant Staphylococcus aureus infection. Clin. Infect. Dis. 46 S350–S359. 10.1086/533591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J. W., Ji Y. (2013). “Sensing and adapting to anaerobic conditions by Staphylococcus aureus,” in Advances in Applied Microbiology, eds Sariaslani S., Gadd G. M. (San Diego, CA: Academic Press; ), 1–25. [DOI] [PubMed] [Google Scholar]
- Hammerstrom T. G., Roh J. H., Nikonowicz E. P., Koehler T. M. (2011). Bacillus anthracis virulence regulator AtxA: oligomeric state, function and CO2-signalling. Mol. Microbiol. 82 634–647. 10.1111/j.1365-2958.2011.07843.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell A., Dubrac S., Andersen K. K., Noone D., Fert J., Msadek T., et al. (2003). Genes controlled by the essential YycG/YycF two-component system of Bacillus subtilis revealed through a novel hybrid regulator approach. Mol. Microbiol. 49 1639–1655. 10.1046/j.1365-2958.2003.03661.x [DOI] [PubMed] [Google Scholar]
- Huang J., O’Toole P. W., Shen W., Amrine-Madsen H., Jiang X., Lobo N., et al. (2004). Novel chromosomally encoded multidrug efflux transporter MdeA in Staphylococcus aureus. Antimicrob. Agents Chemother. 48 909–917. 10.1128/AAC.48.3.909-917.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura T., Tanase S., Szmyd G., Kozik A., Travis J., Potempa J. (2005). Induction of vascular leakage through release of bradykinin and a novel kinin by cysteine proteinases from Staphylococcus aureus. J. Exp. Med. 201 1669–1676. 10.1084/jem.20042041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y., Marra A., Rosenberg M., Woodnutt G. (1999). Regulated antisense RNA eliminates alpha-toxin virulence in Staphylococcus aureus Infection. J. Bacteriol. 181 6585–6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones R. C., Deck J., Edmondson R. D., Hart M. E. (2008). Relative quantitative comparisons of the extracellular protein profiles of Staphylococcus aureus UAMS-1 and its sarA, agr, and sarA agr regulatory mutants using one-dimensional polyacrylamide gel electrophoresis and nanocapillary liquid chromatography coupled with tandem mass spectrometry. J. Bacteriol. 190 5265–5278. 10.1128/JB.00383-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jusko M., Potempa J., Kantyka T., Bielecka E., Miller H. K., Kalinska M., et al. (2013). Staphylococcal proteases aid in evasion of the human complement system. J. Innate Immun. 6 31–46. 10.1159/000351458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapral F. A., Shayegani M. G. (1959). Intracellular survival of staphylococci. J. Exp. Med. 110 123–138. 10.1084/jem.110.1.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson A., Saravia-Otten P., Tegmark K., Morfeldt E., Arvidson S. (2001). Decreased amounts of cell wall-associated protein A and fibronectin-binding proteins in Staphylococcus aureus sarA mutants due to up-regulation of extracellular proteases. Infect. Immun. 69 4742–4748. 10.1128/IAI.69.8.4742-4748.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. H., Yu J., Nahm M. H. (2003). Efficiency of a pneumococcal opsonophagocytic killing assay improved by multiplexing and by coloring colonies. Clin. Vaccine Immunol. 10 616–621. 10.1128/CDLI.10.4.616-621.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klevens R. M., Morrison M. A., Nadle J., Petit S., Gershman K., Ray S., et al. (2007). Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA 298 1763–1771. 10.1001/jama.298.15.1763 [DOI] [PubMed] [Google Scholar]
- Kolar S. L., Antonio Ibarra J., Rivera F. E., Mootz J. M., Davenport J. E., Stevens S. M., et al. (2013). Extracellular proteases are key mediators of Staphylococcus aureus virulence via the global modulation of virulence-determinant stability. Microbiol. Open 2 18–34. 10.1002/mbo3.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreiswirth B. N., Löfdahl S., Betley M. J., O’Reilly M., Schlievert P. M., Bergdoll M. S., et al. (1983). The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305 709–712. 10.1038/305709a0 [DOI] [PubMed] [Google Scholar]
- Kuroda M., Kuroda H., Oshima T., Takeuchi F., Mori H., Hiramatsu K. (2003). Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol. Microbiol. 49 807–821. 10.1046/j.1365-2958.2003.03599.x [DOI] [PubMed] [Google Scholar]
- Liang X., Yu C., Sun J., Liu H., Landwehr C., Holmes D., et al. (2006). Inactivation of a two-component signal transduction system, SaeRS, eliminates adherence and attenuates virulence of Staphylococcus aureus. Infect. Immun. 74 4655–4665. 10.1128/IAI.00322-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X., Zheng L., Landwehr C., Lunsford D., Holmes D., Ji Y. (2005). Global regulation of gene expression by ArlRS, a two-component signal transduction regulatory system of Staphylococcus aureus. J. Bacteriol. 187 5486–5492. 10.1128/JB.187.15.5486-5492.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowy F. D. (1998). Staphylococcus aureus infections. N. Engl. J. Med. 339 520–532. 10.1056/NEJM199808203390806 [DOI] [PubMed] [Google Scholar]
- Maki D. G., Stolz S. M., Wheeler S., Mermel L. A. (1997). Prevention of central venous catheter-related bloodstream infection by use of an antiseptic-impregnated cathetera randomized, controlled trial. Ann. Intern. Med. 127 257–266. 10.7326/0003-4819-127-4-199708150-00001 [DOI] [PubMed] [Google Scholar]
- Mandell G. L. (1974). Bactericidal activity of aerobic and anaerobic polymorphonuclear neutrophils. Infect. Immun. 9 337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meehl M., Herbert S., Götz F., Cheung A. (2007). Interaction of the grars two-component system with the vrafg abc transporter to support vancomycin-intermediate resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 51 2679–2689. 10.1128/AAC.00209-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk I. R., Shah I. M., Xu M., Tan M.-W., Foster T. J. (2012). Transforming the untransformable: application of direct transformation to manipulate genetically Staphylococcus aureus and Staphylococcus epidermidis. MBio 3 e00277-11. 10.1128/mBio.00277-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery C. P., Boyle-Vavra S., Daum R. S. (2010). Importance of the global regulators Agr and SaeRS in the pathogenesis of CA-MRSA USA300 infection. PLoS ONE 5:e15177 10.1371/journal.pone.0015177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootz J. M., Malone C. L., Shaw L. N., Horswill A. R. (2013). Staphopains modulate Staphylococcus aureus biofilm integrity. Infect. Immun. 81 3227–3238. 10.1128/IAI.00377-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick R. P., Ross H. F., Projan S. J., Kornblum J., Kreiswirth B., Moghazeh S. (1993). Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 12 3967–3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohbayashi T., Irie A., Murakami Y., Nowak M., Potempa J., Nishimura Y., et al. (2011). Degradation of fibrinogen and collagen by staphopains, cysteine proteases released from Staphylococcus aureus. Microbiology 157 786–792. 10.1099/mic.0.044503-0 [DOI] [PubMed] [Google Scholar]
- Ollinger J., Wiedmann M., Boor K. J. (2008). SigmaB- and PrfA-dependent transcription of genes previously classified as putative constituents of the Listeria monocytogenes PrfA regulon. Foodborne Pathog. Dis. 5 281–293. 10.1089/fpd.2008.0079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Z., Deka R. K., Norgard M. V. (2011). BosR (BB0647) controls the RpoN-RpoS regulatory pathway and virulence expression in Borrelia burgdorferi by a novel DNA-binding mechanism. PLoS Pathog. 7:e1001272 10.1371/journal.ppat.1001272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resch A., Rosenstein R., Nerz C., Götz F. (2005). Differential gene expression profiling of Staphylococcus aureus cultivated under biofilm and planktonic conditions. Appl. Environ. Microbiol. 71 2663–2676. 10.1128/AEM.71.5.2663-2676.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice K., Peralta R., Bast D., de Azavedo J., McGavin M. J. (2001). Description of Staphylococcus Serine protease (ssp) operon in Staphylococcus aureus and nonpolar inactivation of sspA-encoded serine protease. Infect. Immun. 69 159–169. 10.1128/IAI.69.1.159-169.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby K. M., DeLeo F. R. (2012). Neutrophils in innate host defense against Staphylococcus aureus infections. Semin. Immunopathol. 34 237–259. 10.1007/s00281-011-0295-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan M. H., Petrone D., Nemeth J. F., Barnathan E., Björck L., Jordan R. E. (2008). Proteolysis of purified IgGs by human and bacterial enzymes in vitro and the detection of specific proteolytic fragments of endogenous IgG in rheumatoid synovial fluid. Mol. Immunol. 45 1837–1846. 10.1016/j.molimm.2007.10.043 [DOI] [PubMed] [Google Scholar]
- Saravolatz L. D., Markowitz N., Arking L., Pohlod D., Fisher E. (1982). Methicillin-resistant Staphylococcus aureus. Epidemiologic observations during a community-acquired outbreak. Ann. Intern. Med. 96 11–16. 10.7326/0003-4819-96-1-11 [DOI] [PubMed] [Google Scholar]
- Sawyer R. G., Spengler M. D., Adams R. B., Pruett T. L. (1991). The peritoneal environment during infection. The effect of monomicrobial and polymicrobial bacteria on pO2 and pH. Ann. Surg. 213 253 10.1097/00000658-199103000-00013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw L., Golonka E., Potempa J., Foster S. J. (2004). The role and regulation of the extracellular proteases of Staphylococcus aureus. Microbiology 150 217–228. 10.1099/mic.0.26634-0 [DOI] [PubMed] [Google Scholar]
- Shaw L. N., Golonka E., Szmyd G., Foster S. J., Travis J., Potempa J. (2005). Cytoplasmic control of premature activation of a secreted protease zymogen: deletion of staphostatin B (SspC) in Staphylococcus aureus 8325-4 yields a profound pleiotropic phenotype. J. Bacteriol. 187 1751–1762. 10.1128/JB.187.5.1751-1762.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A., Wilm M., Vorm O., Mann M. (1996). Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68 850–858. 10.1021/ac950914h [DOI] [PubMed] [Google Scholar]
- Smagur J., Guzik K., Bzowska M., Kuzak M., Zarebski M., Kantyka T., et al. (2009a). Staphylococcal cysteine protease staphopain B (SspB) induces rapid engulfment of human neutrophils and monocytes by macrophages. Biol. Chem. 390 361–371. 10.1515/BC.2009.042 [DOI] [PubMed] [Google Scholar]
- Smagur J., Guzik K., Magiera L., Bzowska M., Gruca M., Thøgersen I. B., et al. (2009b). A new pathway of staphylococcal pathogenesis: apoptosis-like death induced by staphopain B in human neutrophils and monocytes. J. Innate Immun. 1 98–108. 10.1159/000181014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaan A. N., Surewaard B. G. J., Nijland R., van Strijp J. A. G. (2013). Neutrophils versus Staphylococcus aureus: a biological tug of war. Annu. Rev. Microbiol. 67 629–650. 10.1146/annurev-micro-092412-155746 [DOI] [PubMed] [Google Scholar]
- Sun F., Ji Q., Jones M. B., Deng X., Liang H., Frank B., et al. (2011). AirSR, a [2Fe-2S] cluster-containing two-component system, mediates global oxygen sensing and redox signaling in Staphylococcus aureus. J. Am. Chem. Soc. 134 305–314. 10.1021/ja2071835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Zheng L., Landwehr C., Yang J., Ji Y. (2005). Identification of a novel essential two-component signal transduction system, YhcSR, in Staphylococcus aureus. J. Bacteriol. 187 7876 10.1128/JB.187.22.7876-7880.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Arana A., Merino N., Vergara-Irigaray M., Débarbouillé M., Penadés J. R., Lasa I. (2005). Staphylococcus aureus develops an alternative, ica-independent biofilm in the absence of the arlRS two-component system. J. Bacteriol. 187 5318–5329. 10.1128/JB.187.15.5318-5329.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaras A. P., Flagler M. J., Dorsey C. W., Gaddy J. A., Actis L. A. (2008). Characterization of a two-component regulatory system from Acinetobacter baumannii that controls biofilm formation and cellular morphology. Microbiology 154 3398–3409. 10.1099/mic.0.2008/019471-0 [DOI] [PubMed] [Google Scholar]
- Torres V. J., Stauff D. L., Pishchany G., Bezbradica J. S., Gordy L. E., Iturregui J., et al. (2007). A Staphylococcus aureus regulatory system that responds to host heme and modulates virulence. Cell Host Microbe 1 109–119. 10.1016/j.chom.2007.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincents B., Önnerfjord P., Gruca M., Potempa J., Abrahamson M. (2007). Down-regulation of human extracellular cysteine protease inhibitors by the secreted staphylococcal cysteine proteases, staphopain A and B. Biol. Chem. 388 437–446. 10.1515/BC.2007.042 [DOI] [PubMed] [Google Scholar]
- Watkins R. L., Pallister K. B., Voyich J. M. (2011). The SaeR/S gene regulatory system induces a pro-inflammatory cytokine response during staphylococcus aureus infection. PLoS ONE 6:e19939 10.1371/journal.pone.0019939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins R. L., Zurek O. W., Pallister K. B., Voyich J. M. (2013). The SaeR/S two-component system induces interferon-gamma production in neutrophils during invasive Staphylococcus aureus infection. Microbes Infect. 15 749–754. 10.1016/j.micinf.2013.05.004 [DOI] [PubMed] [Google Scholar]
- White M. J., Boyd J. M., Horswill A. R., Nauseef W. M. (2014). Phosphatidylinositol-specific phospholipase C contributes to survival of Staphylococcus aureus USA300 in human blood and neutrophils. Infect. Immun. 82 1559–1571. 10.1128/IAI.01168-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisplinghoff H., Bischoff T., Tallent S. M., Seifert H., Wenzel R. P., Edmond M. B. (2004). Nosocomial bloodstream infections in us hospitals: analysis of 24179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 39 309–317. 10.1086/421946 [DOI] [PubMed] [Google Scholar]
- Xue T., You Y., Hong D., Sun H., Sun B. (2011). The Staphylococcus aureus KdpDE two-component system couples extracellular K+ sensing and agr signaling to infection programming. Infect. Immun. 79 2154–2167. 10.1128/IAI.01180-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan M., Hall J. W., Yang J., Ji Y. (2012). The essential YhcSR two-component signal transduction system directly regulates the lac and opuCABCD operons of Staphylococcus aureus. PLoS ONE 7:e50608 10.1371/journal.pone.0050608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan M., Yu C., Yang J., Ji Y. (2009). Development of shuttle vectors for evaluation of essential regulator regulated gene expression in Staphylococcus aureus. Plasmid 61 188–192. 10.1016/j.plasmid.2009.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan M., Yu C., Yang J., Ji Y. (2011). The essential two-component system YhcSR is involved in regulation of the nitrate respiratory pathway of Staphylococcus aureus. J. Bacteriol. 193 1799–1805. 10.1128/JB.01511-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zecconi A., Scali F. (2013). Staphylococcus aureus virulence factors in evasion from innate immune defenses in human and animal diseases. Immunol. Lett. 150 12–22. 10.1016/j.imlet.2013.01.004 [DOI] [PubMed] [Google Scholar]
- Zheng L., Yu C., Bayles K., Lasa I., Ji Y. (2007). Conditional mutation of an essential putative glycoprotease eliminates autolysis in Staphylococcus aureus. J. Bacteriol. 189 2734–2742. 10.1128/JB.01806-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.