Abstract
Beyond its role in host defense, bacterial DNA methylation also plays important roles in the regulation of gene expression, virulence and antibiotic resistance. Bacterial cells in a clonal population can generate epigenetic heterogeneity to increase population-level phenotypic plasticity. Single molecule, real-time (SMRT) sequencing enables the detection of N6-methyladenine and N4-methylcytosine, two major types of DNA modifications comprising the bacterial methylome. However, existing SMRT sequencing-based methods for studying bacterial methylomes rely on a population-level consensus that lacks the single-cell resolution required to observe epigenetic heterogeneity. Here, we present SMALR (single-molecule modification analysis of long reads), a novel framework for single molecule-level detection and phasing of DNA methylation. Using seven bacterial strains, we show that SMALR yields significantly improved resolution and reveals distinct types of epigenetic heterogeneity. SMALR is a powerful new tool that enables de novo detection of epigenetic heterogeneity and empowers investigation of its functions in bacterial populations.
 Bacterial DNA methylation is involved in many processes, from host defense to antibiotic resistance, however current methods for examining methylated genomes lack single-cell resolution. Here Beaulaurier et al. present Single Molecule Modification Analysis of Long Reads, a new tool for de novo detection of epigenetic heterogeneity.
Bacterial DNA methylation is involved in many processes, from host defense to antibiotic resistance, however current methods for examining methylated genomes lack single-cell resolution. Here Beaulaurier et al. present Single Molecule Modification Analysis of Long Reads, a new tool for de novo detection of epigenetic heterogeneity.
In the bacterial kingdom, DNA methylation is catalysed by three families of DNA methyltransferases (MTases) that typically add methyl groups to DNA bases in a sequence-specific manner1,2,3. One family of MTases attaches methyl groups to adenine residues, creating N6-methyladenine (6mA), whereas the other two families introduce methyl groups to cytosine residues to create either N4-methylcytosine (4mC) or 5-methylcytosine (5mC). Many bacterial DNA MTases act in concert with and are encoded in close proximity to cognate restriction endonucleases (REs); the MTase protects DNA from digestion by the RE with which it forms a restriction-modification (R-M) system2,4,5. R-M systems are generally believed to have an ‘immune' function, protecting cells from invading foreign DNA. They are also studied as selfish elements that protect themselves against removal through the post-segregational killing of new progeny by pre-existing and stable RE molecules4,6,7. In addition, so-called ‘orphan' MTases, which occur in the genome without an associated RE, have been found to play important regulatory roles in global gene expression and other biological processes2,3,8,9,10. Furthermore, the ability of such MTases to target their recognition motifs for methylation often depends on competitive binding at the target site between several DNA-binding proteins2,3,11,12,13,14,15. These epigenetic regulators of gene expression, including both MTases and competing DNA-binding proteins, are a source of phase variation2,3,16,17 that increases the robustness of the population and provides opportunities to modulate transcription in response to changing environmental conditions18,19.
In some bacteria, the behaviour of certain MTases can vary markedly due to slipped-strand mispairing during DNA replication. Often occurring in homopolymer-rich regions, this slippage can cause frameshift mutations that result in truncated and usually inactive MTases20. Alternatively, recombination events can result in the movement of type I and III target recognition domains, which can alter the target sequence specificity of the MTase21,22,23,24,25. These mechanisms can yield heterogeneity in the methylomes of descendants from a single cell and can cause differential regulation of multiple genes, termed a phase-variable regulon (a.k.a. phasevarion)20. It has been postulated that epigenetic control of gene expression mediated by phase variation (epigenetic control of a single gene) or phasevarions (multiple genes regulated simultaneously) allows an essentially clonal population to adopt multiple distinct phenotypes26. Such heterogeneity can facilitate adaptation to diverse environmental niches, including complex host environments and the presence of antibiotics, as has been reported in several studies20,27,28,29,30.
Genomic analyses suggest that some form of DNA methylation is present in nearly all bacteria, as putative DNA MTases have been found in 94% of >3,300 sequenced bacterial genomes5. Given the large number of MTase target sites in bacterial genomes and the growing evidence suggesting complex regulatory roles of methylation by both R-M and orphan MTases2,3, the potential scope for exploring the diversity of bacterial methylation and methylation-mediated gene regulation is vast. However, the precise sequence targets and biological roles of most MTases remain largely unknown. While recent progress in bisulfite sequencing facilitates the accurate detection of 5mC methylation31, there has not been a high-throughput genome-wide sequencing methodology for efficiently detecting 6mA and 4mC methylation in bacteria.
Single molecule, real-time (SMRT) DNA sequencing technology32 enables the detection of nearly 20 different types of chemical modifications to DNA, including all three major types of DNA methylation in bacteria (6mA, 4mC and 5mC), although the reduced signal-to-noise ratio with 5mC makes detection of such events challenging. SMRT sequencing molecules consist of a double-stranded native DNA fragment that has been circularized by ligating hairpin adapters to each end. During sequencing, a DNA polymerase proceeds around the circularized DNA template multiple times, the exact number depending on fragment size and polymerase performance. The sequencing instrument monitors not only the pulse fluorescence associated with each incorporated nucleotide, but also the time between the incorporation events, termed the inter-pulse duration (IPD). Variation in IPDs (referred to as kinetic variation) is highly correlated with the presence of modifications within the DNA template33,34,35, including 6mA, 4mC, 8-oxoguanine and so on.
Many studies have used SMRT sequencing to conduct de novo identification of MTase target motifs, revealing surprising diversity in motifs among bacterial species and even among closely related strains35,36,37,38,39,40. However, the heterogeneities within bacterial populations cultured under different conditions have not been thoroughly explored. This is primarily limited by the current SMRT sequencing-based bacterial methylome analysis protocol, which relies on assessments of aggregate IPD values at each genomic position across populations of cells33,34,35,41; the individual reads (each from a single DNA molecule) are aligned to the same genomic region and then statistically modelled as an ensemble (Fig. 1a,b). This approach enhances the statistical power for methylation detection at single-nucleotide resolution, but fundamentally limits the ability to resolve epigenetic heterogeneity within the sample. Although progress beyond traditional population-level analysis has recently been reported38,42, new methods are needed to provide improved resolution on complex methylomes.
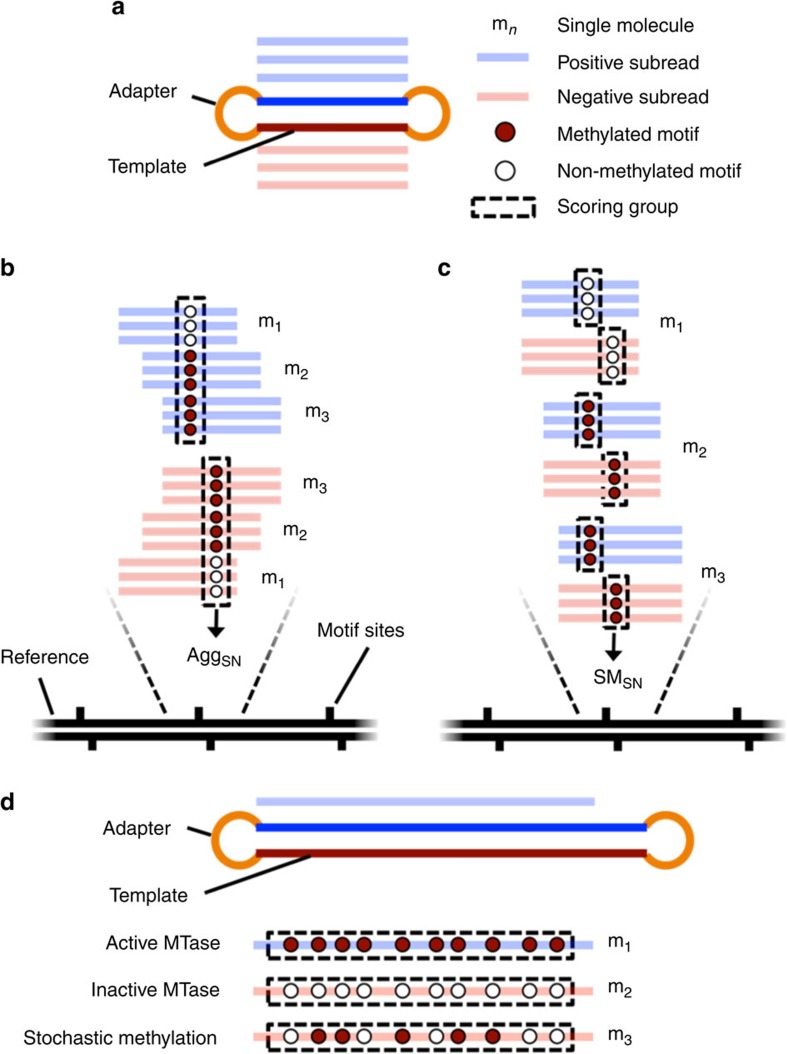
Figure 1. SMALR methods for methylation detection in SMRT reads.
Schematic illustrating the general approaches of both the existing and two proposed SMALR methods for detecting DNA methylation in SMRT sequencing reads. (a) A single SMRT sequencing molecule (short DNA insert+adapters) and the subreads that are produced during sequencing. (b) The existing methylation detection method is based on a molecule-aggregated, single-nucleotide (AggSN) score. For a given strand and genomic position, the IPD values from all the subreads aligning to that strand and position are aggregated together across all molecules to infer the presence of a consensus methylated base. (c) The proposed single molecule, single nucleotide (SMSN) method for detecting DNA methylation relies instead on separate consideration of subreads from different molecules. The SMSN scores are calculated for each molecule, strand and genomic position. (d) A single SMRT sequencing molecule (long DNA insert+adapters) with a single long subread and the proposed single molecule, pooled (SMP) approach for assessing MTase activity. This approach pools together IPD values from multiple motif sites along the length of a single long subread.
Here, we present SMALR, a novel framework for single molecule-level detection and phasing of bacterial DNA methylation using SMRT sequencing data. This approach relies on two complementary methods that use molecule-specific IPD information to infer methylation states at single-molecule resolution (Fig. 1c,d). We demonstrate the effectiveness of SMALR through comprehensive and quantitative characterization of seven bacterial methylomes and by identifying distinct types of heterogeneity in these methylomes. The enhanced resolution provided by SMALR allows de novo detection of bacterial epigenetic heterogeneity, and should broaden our understanding of the diverse roles of methylation in modulating bacterial physiology. SMALR is freely available through GitHub.
Results
Single molecule-level methylation detection and phasing
We first present the rationale and description of the two complementary methods within SMALR for (i) single molecule, single nucleotide, strand-specific detection of methylation events and (ii) single molecule-level epigenetic phasing analysis. Our strategy revolves around the interrogation of circular consensus sequence data generated from short-insert SMRT sequencing libraries (∼250 bp) and continuous long-read data generated from long-insert SMRT sequencing libraries32,33. Using short insert templates, we achieved average read lengths between 4,735 and 9,023 bp (Supplementary Table 1; Methods), although recent releases of the sequencing chemistry kits promise significantly longer-read lengths (12,000 bp on average). The multiple passes of the polymerase over the same template sequence (Fig. 1a,c) allow the calculation of a single molecule, single-nucleotide (SMSN) score, which is defined as the difference between the mean site/strand-specific log (natural) IPD value from a native DNA molecule and corresponding mean site/strand-specific log IPD value from whole-genome amplified (WGA) DNA molecules (Methods). This score provides the statistical power to detect shifts in kinetic variation at a specific nucleotide position on a given DNA molecule.
Complementing the short-insert libraries are the long-insert libraries (our data generated in early 2014 achieved mean insert sizes of 3,357–7,304 bp and mean read lengths of 4,957–7,965 bp, although a mean read length of 12,000 bp can now be achieved; Supplementary Table 1; Methods). Most of the reads generated from this protocol represent long stretches of contiguous DNA (as opposed to the circular consensus of shorter libraries). The long library sequencing protocol has been applied to de novo bacterial genome assembly because of its ability to resolve genomic regions with complex repeated elements43,44,45. Here, we instead leverage the continuous long reads for methylation phasing at single-molecule resolution. The methylated motifs profiled in this study are represented in their genomes between 210 and 42,186 times (Supplementary Table 2), a number largely dependent on both the motif complexity and evolutionary selection. As such, a motif may be represented many times in a single long read, providing multiple data points revealing the kinetic variation for the motif. By pooling these kinetic variation statistics for a single read, it becomes possible to not only infer whether the cell that provided that DNA fragment contained MTase activity targeting that motif, but also to quantitatively estimate the processivity of the MTase (Fig. 1d). We therefore created the single molecule, pooled (SMP) score, which is defined as the difference between the mean log IPD value from all motif sites in a single native subread and the mean log IPD value from those same motif sites in WGA DNA molecules (Methods). Although this estimation procedure is more powerful for shorter methylation motifs than for the longer motifs that occur less frequently, the relatively longer-read lengths (reads up to 60,000 bp are now possible with recent updates to the SMRT chemistry kit) provide an opportunity to estimate the SMP scores accurately for longer motifs.
Sensitivity and specificity of SMSN detections
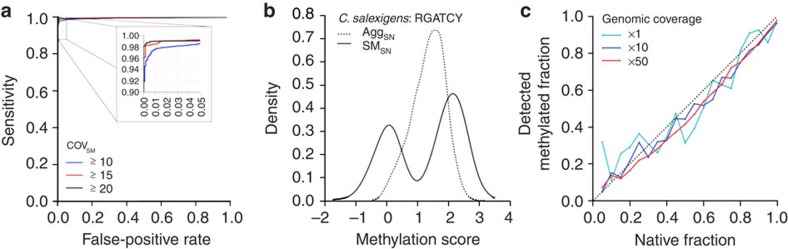
An accurate mean log IPD for each nucleotide in a single molecule permits confident estimation of the IPD ratio. The accuracy of the mean log IPD increases as the number of subreads for each molecule increases33,34, as each subread provides an independent estimate of the IPD. For a given read length, the number of subreads (that is, single-molecule coverage, covSM) is negatively correlated with the insert size of the sequencing library. To quantify the sensitivity and specificity for SMSN detection, we analysed methylated 5′-CTGCAG sites in a native E. coli O104:H4 C227-11 strain35 (referred to as C227), and a matching WGA sample that is free of methylation (Fig. 2a; Methods). As expected, the sensitivity of the method increased with increasing covSM. For the SMSN approach, covSM≥15 enables detection of 6 mA with a sensitivity of 98.5% and a specificity of 99.5%. Although smaller library sizes can provide higher values of covSM, a minimum library size of 150–200 bp is recommended to avoid loss of genomic DNA during library construction and to facilitate removal of adapter-dimer constructs during purification.
Figure 2. Performance of SMSN level detection of DNA methylation.
Multiple metrics showing the performance of the proposed single molecule, single nucleotide (SMSN) detection method. (a) Performance of the approach for detecting 6mA modifications in the 5′-CTGCAG motif of E. coli C227 using three thresholds for minimum single-molecule coverage (covSM). (b) Distribution of the aggregate, single nucleotide (AggSN) and SMSN methylation scores for the partially non-methylated 5′-RGATCY motif in C. salexigens. The bimodal distribution of the SMSN scores enables the accurate and objective estimation of this fraction. (c) Accuracy of SMSN-enabled estimations of the methylated fraction (using covSM≥10) for the 5′-CTGCAG motif of E. coli C227 at various levels of genomic-sequencing coverage.
Estimating the sensitivity and specificity of detecting 6mA at single-molecule resolution for other 6 mA motifs, we found the results to be comparable to 5′-CTGCAG in strain C227 (Supplementary Fig. 1a,b). We found that 4mC also can be accurately detected at single-molecule resolution, albeit with slightly lower sensitivity and specificity (Supplementary Fig. 1c). We did not attempt similar analysis of 5mC motifs due to the significantly lower signal-to-noise ratio24,38, even after conversion of 5mC to 5hmC with Tet enzymes46. The remainder of this report will focus solely on the characterization of 6 mA due to both its prevalence in bacteria and the substantial body of evidence supporting its important regulatory potential12,22,23; however, all of the analyses detailed also are applicable to 4mC.
SMSN scores for estimating global methylation heterogeneity
The methylome of Chromohalobacter salexigens characterized in a recent study was found to contain a substantial number of non-methylated 5′-RGATCY sites37. In particular, 23.5% of the motif sites were predicted to be non-methylated based on the standard population-level analysis, which we replicated using molecule-aggregated, single-nucleotide (AggSN) scores (Fig. 2b; Methods). As shown, the distribution of AggSN scores does not show clear separation between methylated and non-methylated 5′-RGATCY sites, indicating that quantifying the methylated fraction of 5′-RGATCY sites in the genome with AggSN scores relies on a subjective and ad hoc threshold. In contrast, there is clear bimodality in the distribution of SMSN scores, where the components centred near SMSN≈0 and SMSN≈2 represent the non-methylated and methylated fractions, respectively (Fig. 2b). A distribution of SMSN centred at zero correspond to motif sites that are not methylated because the IPDs do not differ between native and WGA DNAs; in contrast, a distribution of SMSN centred near two corresponds to motif sites that are methylated (specific to 6 mA; consistently observed across multiple bacterial species and motifs as shown in Supplementary Figs 3-8). This bimodality allows the percentage of methylated motif sites to be estimated at 60.4% using a standard expectation maximization (EM) algorithm47 (Methods) without need for a subjective input threshold.
SMSN-based quantification at low sequencing coverage
As the SMSN scores depend on covSM rather than the genomic-sequencing coverage, we conducted an in silico experiment to test the accuracy of SMSN-based estimates of global methylated fraction at varying levels of genomic coverage. By mixing native and WGA-sequencing molecules at varying proportions and by gradually reducing the total number of molecules, we found that the SMSN-based estimates of the global methylated fraction are stable even when the genomic sequencing coverage is as low as × 1 (Fig. 2c; Methods). This finding could have implications for the characterization of in vivo isolates, for which low sequencing coverage due to limited DNA input is often a challenge.
Global methylation heterogeneity in six bacteria
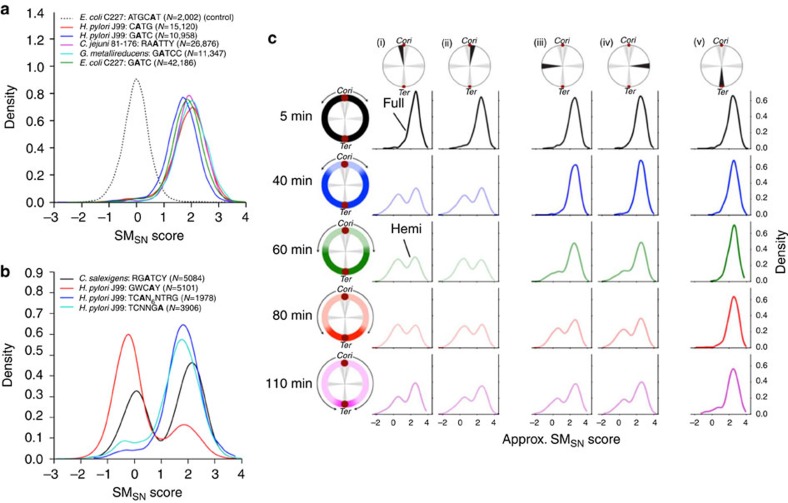
We applied the SMSN analysis to six bacterial methylomes that were recently sequenced35,37,38 or were specifically sequenced for this study (Supplementary Table 1). We first detected methylation motifs based on existing methods24,35,37 and divided them into two groups based on the global distribution of SMSN scores. In the first group (Fig. 3a), most (>95%) motif sites were methylated, with only a small proportion non-methylated, likely due to competitive binding between the MTases and other DNA-binding proteins such as transcription factors2,3,11,13,14. In the second group (Fig. 3b), a substantial (>5%) percentage of motif sites were non-methylated, suggesting the existence of alternative mechanisms that drive methylome heterogeneity. After analysing the SMSN scores for all bacterium-motif pairs (Supplementary Figs 3-8), we observed that while most motifs belong to the first group and do not show extensive non-methylation, the second group includes the 5′-RGATCY motif of C. salexigens and three motifs from Helicobacter pylori J99. Most intriguingly, the H. pylori motif 5′-GWCAY shows a very high (75.3%) percentage of non-methylated sites, which we subsequently investigated using the SMP analysis (see below).
Figure 3. SMSN score distributions reveal epigenetic heterogeneity.
(a) Single molecule, single nucleotide (SMSN) score distributions for multiple bacterium-motif pairs (and the genome-wide motif count, N, of each motif) that exhibit near complete methylation, along with a non-methylated motif for comparison. (b) SMSN score distributions for multiple bacterium-motif pairs that display significant non-methylated fractions. The H. pylori J99 motifs show minor variation in the SMSN associated with each peak due to subtle differences in the chemistry version used for SMRT sequencing of the native and WGA samples. (c) SMSN interrogation of 5′-GANTC methylation at five genomic positions (columns) in a synchronized C. crescentus culture during a single round of DNA replication. Five time points (minutes post-synchronization; rows) provide snapshots of the bidirectional progression of the replication forks from the origin of replication (Cori) to the terminus (Ter). Grey wedges in the chromosome schematics show the 200-kb genomic regions where the SMSN scores are queried for each time point. Two regions are on either side of the Cori: (i) Cori - 0.1 Mbp and (ii) Cori+0.1 Mbp. Another two are halfway between Cori and Ter: (iii) Cori - 1 Mbp and (iv) Cori+1 Mbp. The final region covers the terminus: (v) Ter. Light (hemimethylated) to dark (fully methylated) colour shading in the schematic illustrates the approximate position of the replication fork at each time point. The bimodal distributions of approximate SMSN scores (Methods) reveal the progressive hemi-methylation of 5′-GANTC sites following the passage of the replication forks. Hemimethylated sites cannot transition back to full methylation until the MTase gene, ccrM, is transcribed, which does not occur until late in the replication process.
SMSN score distributions show regional methylation dynamics
The Caulobacter crescentus genome encodes a methyltransferase (CcrM) targeting 5′-GANTC sites48. The corresponding gene, ccrM, is only expressed at late stages of the cell cycle. Consequently, fully methylated 5′-GANTC sites transition to hemi-methylated as the replication fork proceeds from the origin of replication (Cori) to the terminus (Ter)38. Recently, Kozdon et al. 38 used SMRT sequencing to study such transitions at five time points spanning a single, synchronized round of the cell cycle. This allowed us to conduct an approximate SMSN analysis (approximate due to lack of WGA sequencing in the original data; Methods) to test the use of SMSN for detecting regional methylation heterogeneity, focusing specifically on five distinct 200-kilobase regions in each of five time points (Fig. 3c).
As the replication fork proceeds from the Cori to the Ter in the five time points, the SMSN score distributions reveal an increasing fraction of the genome that has been converted from fully methylated to hemi-methylated 5′-GANTC sites. At the first time point (5 min post-synchronization), the fully methylated state of the cells is shown by the single mode in approximate SMSN scores (Methods) in all five genomic regions. As the cells begin to differentiate (40 min), there are bimodal SMSN scores in regions (i) and (ii) that are closest to the Cori, reflecting the passage of the replication fork through those regions. The next two time points reveal the transition of the single-mode approximate SMSN distributions in regions (iii) and (iv) to clear bimodal distributions, reflecting the passage of the replication fork through those regions. The unequal bimodal distribution after 80 min in regions (iii) and (iv) indicates that the molecules from that region have not all been converted from fully to hemi-methylated, likely due to stochastic variance in the position of the replication fork 80 min post-synchronization. The final time point reveals a genome that has almost completely converted to a hemi-methylated state, with the exception of the region immediately surrounding the Ter (v), which shows only a small amount of hemi-methylation. Analysis at this time of regions (i)-(iv) indicates that these sites have already begun to transition from equal bimodality (universal hemi-methylation) towards their starting single-mode distributions (universal full methylation). This likely is also due to imperfect synchronization, allowing some replication forks to progress past the ccrM gene at the LPD time point, thus activating transcription and beginning the process of re-methylating all the 5′-GANTC sites that had been rendered hemi-methylated by passage of the replication fork.
SMP identifies distinct types of epigenetic heterogeneity
We considered three potential explanations for the heterogeneous methylation observed in select bacterium-motif pairs (Fig. 3b). First, we hypothesized that methylation motifs, especially degenerate motifs, may be inaccurately annotated; the observed heterogeneity would simply be due to a mixture of truly methylated motifs and motifs falsely identified by the motif enrichment algorithm. For example, the heterogeneous methylation of the 5′-RGATCY motif in C. salexigens (Fig. 2b) may result from only three of the four explicit specifications (A/G)GATC(C/T) being truly methylated. However, we can reject this hypothesis as the SMSN score distributions of the four explicit specifications showed similar heterogeneity (Supplementary Fig. 9).
A second possibility is that the MTase is stochastically methylating only a fraction of its recognition motif sites in each cell, in which case the SMSN-based estimates of methylated fraction reflect a universally active MTase, albeit one without the ability to methylate all of its target motifs. In that case, we would expect the methylated motif sites to be interspersed with non-methylated sites throughout a single copy of the genome.
A third potential cause of heterogeneity in SMSN scores is phase variation of the MTase responsible for targeting the motif. Phase-variable MTases have been well-documented in a variety of bacteria, in which stochastic mutations in homopolymers or other simple sequence repeats in the MTase can induce changes in the transcribed product that either activate/deactivate the MTase or modify its target specificity20,22,24. Either of these two switching modes can induce cell-wide methylation patterns that might differ between neighbouring cells in a single population. In this scenario, the methylated fraction estimated by SMSN scores reflects the fraction of cells with an active MTase targeting the motif.
We can differentiate between these latter two hypotheses (intra-cellular stochastic methylation versus stochastic phase-variable MTase with different phases in different cells) by using the SMP method to phase epigenetic information across the full length of each read (that is, methylation co-occurrence on a single molecule). If some molecules within a sample are methylated at all sites while others are completely non-methylated, this supports the existence of a stochastic phase-variable MTase. In contrast, a mixture of methylated and non-methylated motif sites on a single read suggests intracellular stochastic methylation as the source of heterogeneity in the SMSN scores.
Direct detection of phasevarion in single H. pylori colonies
Phase-variable genes regulated by length variation of homopolymeric tracts have been well-documented in H. pylori20,22,49,50,51,52. Several putative MTases contain inactivating frameshift mutations that prevent the transcription of an active protein. By experimentally correcting frameshifts in several MTases and SMRT sequencing the mutated populations, Krebes et al. were able to detect methylation (using aggregation-based methods) by the re-activated MTases and identify their target motifs24. However, because most H. pylori isolates are likely not clonal and significant homopolymer length variation has been observed in MTases between closely related strains of H. pylori49, it is possible that active copies of the phase-variable MTases are already present in the wild-type isolates, but at levels too low to detect using the aggregation-based method. This problem provided an opportunity to use the SMP method for direct detection in wild-type isolates of minor subpopulations with active and inactive MTases.
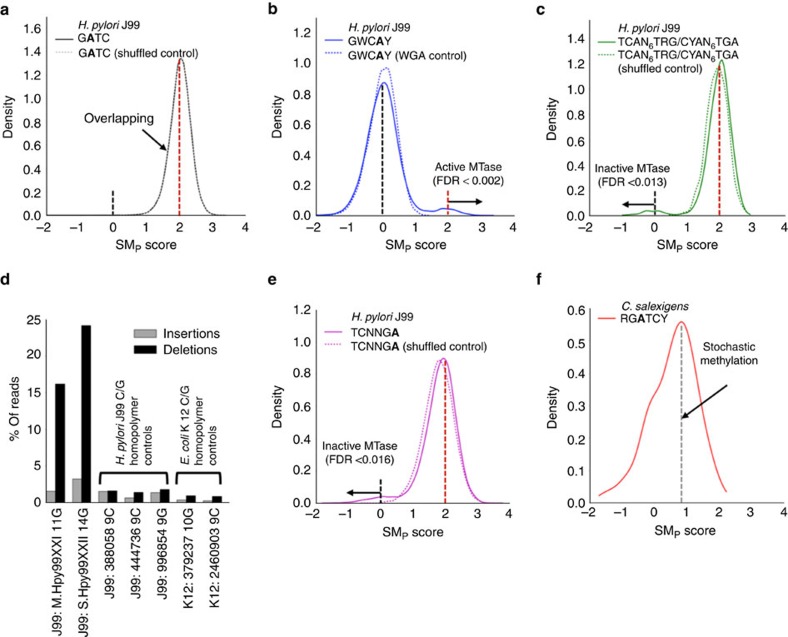
We first tested the SMP method on an H. pylori J99 isolate that was sequenced using libraries with long (∼20 kb) DNA inserts. We targeted the 4-mer 5′-GATC motif, where >95% of sites are expected to be methylated based on its SMSN distribution (Fig. 3a), and calculated a SMP score for each long read containing at least 10 GATC sites. The distribution of SMP scores for 5′-GATC was compared with a control distribution of SMP scores calculated after randomly shuffling IPD values between molecules (Fig. 4a). No bimodality is present in the 5′-GATC SMP distribution and it is nearly identical to the IPD-shuffled SMP distribution, suggesting that the MTase responsible for targeting the 5′-GATC motif in H. pylori J99 (M.Hpy99VI) is constitutively active. Through false-discovery rate (FDR) estimation (Methods), we found that only 0.07% of the molecules with at least 10 5′-GATC sites had evidence of non-methylation (maximum FDR=1%). This level of non-methylation is consistent with several other motifs for which we expect to observe near-universal methylation activity (5′-CATG, 5′-GANTC and 5′-GAGG), suggesting that the small number of non-methylated molecules may have originated from transiently hemi-methylated regions directly behind the DNA replication fork. Furthermore, phase variation of M.Hpy99VI was considered unlikely as no significant sequence variation was observed in its coding sequence (Supplementary Fig. 10).
Figure 4. SMP score distributions reveal distinct types of epigenetic heterogeneity.
(a) Single molecule, pooled (SMP) distribution for H. pylori J99 motif 5′-GATC and its corresponding IPD-shuffled control. The identical unimodal distributions suggest a fully active MTase (as expected). (b) SMP distributions for H. pylori J99 motif 5′-GWCAY and its corresponding WGA control. The major peak around SMP≈0 and minor peak around SMP≈2 suggests that the mostly inactive MTase targeting 5′-GWCAY, M.Hpy99XXI, is methylating 5′-GWCAY in a small fraction of cells. Methylated molecules with SMP scores>2 have an FDR<0.2%. (c) SMP distributions of 5′-TCAN6TRG/5′-CYAN6TGA in H. pylori J99 and its corresponding IPD-shuffled control. The major peak around SMP≈2 and minor peak around SMP≈0 indicates that the normally active MTase, Hpy99XXII, is inactive in a small fraction of cells. Non-methylated molecules with SMP scores<0 have an FDR<1.3%. (d) High-accuracy sequencing with Illumina MiSeq and read-level analysis of insertion/deletion calls shows significant variation in the lengths of two specific homopolymers in the coding sequences of M.Hpy99XXI and S.Hpy99XXII. The high percentage of deletions in these two genes stands apart from the deletion rates found in five other C/G homopolymers from H. pylori J99 and E. coli K12, suggesting that this is not simply due to lower sequencing accuracy in homopolymer regions. (e) SMP distributions of 5′-TCNNGA in H. pylori J99 and its corresponding IPD-shuffled control. The SMP scores suggest a MTases behaviour similar to that of Hpy99XXII. Non-methylated molecules with SMP scores<0 have an FDR<1.6%. (f) SMP distribution for the C. salexigens motif 5′-RGATCY. The major peak near SMP≈0.9 indicates that the IPDs sampled for each molecule reflect a mixture of both non-methylated (IPD≈0) and methylated (IPD≈2) motif sites, suggesting stochastic methylation as the primary source of epigenetic heterogeneity for this motif.
Next, we applied the same analysis to a motif, 5′-GWCAY, targeted by a known phase-variable MTase in H. pylori J99. The modification (M) subunit of the type III R-M system Hpy99XXI contains a 10-12G homopolymer locus, the exact length of which determines whether transcription downstream of the locus is in-frame (active MTase) or out-of-frame (inactive MTase). Methylation of the 5′-GWCAY motif is normally undetectable in H. pylori J99 using aggregation-based methods, suggesting that the majority of M.Hpy99XXI proteins are inactive24. However, the SMP distribution shows (Fig. 4b) a subpopulation of molecules with SMP≈2, indicative of a subpopulation of cells containing the active form of M.Hpy99XXI. Illumina MiSeq sequencing of the same DNA supported this mechanism, revealing a substantial length variation in the M.Hpy99XXI homopolymer (Supplementary Fig. 11; Methods).
Hpy99XXII is another R-M system in H. pylori J99 known to contain phase-variable components. In contrast to the Hpy99XXI system, the phase variation in this system is due to homopolymer length variation within the specificity (S) subunit that targets the 5′-TCAN6TRG/5′-CYAN6TGA motif. The SMP scores for this motif show that, although the majority of molecules have scores suggesting active methylation, there is a subpopulation of molecules with SMP≈0 (Fig. 4c), indicative of cells within the culture lacking a functional Hpy99XXII system. MiSeq sequencing confirmed the presence of significant S.Hpy99XXII homopolymer length variation (Supplementary Fig. 12).
On most platforms, there is reduced sequencing accuracy of homopolymeric regions, especially poly-C/G tracts. To ensure that the observed insertions and deletions were not due to sequencing errors, we searched for length variation in other C/G homopolymers in the H. pylori J99 and E. coli K12 genomes that are not linked to phase variation (Fig. 4d). The number of read-level deletions (Methods) observed in the M.Hpy99XXI and S.Hpy99XXII genes is markedly higher (>15%) than those observed (<3%) in the control C/G homopolymers, indicating that true length variation, rather than sequencing error, is driving the considerable read-level variation seen in these two homopolymers.
The H. pylori J99 strain used in this study has been passaged many times over the 15 years since its original isolation53. While phase variation refers to rapid state switching in response to the environment, it is also possible that, over the course of many passages, stable subpopulations have diverged within the isolate, each containing distinct and fixed homopolymer lengths. To investigate this possibility, we isolated five single colonies (HPXZ1383-1387; Methods) from the original isolate for Illumina sequencing and subjected the reads to analysis of read-level variation (Methods). Even in these clonal isolates, significant length variation was observed in the homopolymers within the M.Hpy99XXI and S.Hpy99XXII genes (Supplementary Fig. 13). This indicates that homopolymer length variation at these loci develops rapidly, generating heterogeneity in MTase activity in the descendants from a single cell. It is also worth noting that one of the five colonies revealed a large percentage of deletions in M.Hpy99XXI (that is, mostly 10G homopolymer rather than 11G), suggesting that this single colony is likely derived from a parent cell with a 10G homopolymer.
Interestingly, although a similar distribution of SMP scores was observed for both the 5′-TCAN6TRG/5′-CYAN6TGA (Fig. 4c) and 5′-TCNNGA (Fig. 4e) motifs, no sequence variation was observed in or near the coding region of the genes involved in the Hpy99XVIII R-M system that methylates 5′-TCNNGA (Supplementary Fig. 14). This finding suggests that regulatory elements outside the coding region (genetic or epigenetic) may be responsible for the observed fraction of 5′-TCNNGA non-methylated molecules. Analysis of SMP scores for other methylated motifs in H. pylori J99 did not reveal significant bimodality, suggesting full MTase activity (Supplementary Fig. 15).
Gene expression in M.Hpy99XXI and S.Hpy99XXII mutants
To assess the effect of a genome-wide absence of methylated 5′-GWCAY and 5′-TCAN6TRG/5′-CYAN6TGA motifs, we compared the transcriptomes of the five clonal wild-type J99 isolates and the two mutant strains lacking M.Hpy99XXI and M.Hpy99XXII (HPXZ1401 and HPXZ1398, respectively; Methods). In the strain lacking 5′-GWCAY methylation (HPXZ1401; Supplementary Table 5), 38 genes were significantly differentially expressed, while the strain lacking 5′-TCAN6TRG/5′-CYAN6TGA methylation (HPXZ1398; Supplementary Table 6) contained 41 significantly differentially expressed genes (P<0.001; an adapted Fisher's exact test (ref 70); FDR<0.05). The most striking changes in HPXZ1401 were the upregulation of flgE, which encodes the flagellar hook protein FlgE, and two other flagellum-related genes, flgB and flaG (Supplementary Table 5). Flagella are essential for Helicobacter motility and are subject to phase variation via reversible length variation in a short homopolymeric sequence repeat in the fliP gene54. The upregulation of three flagella-related genes in HPXZ1401 may suggest that other novel mechanisms are involved in H. pylori motility switching. Two other significantly upregulated genes, groEL and groES, which reside in the same operon based on in silico prediction55 and transcription start site mapping in another strain56, showed 2.0- and 5.4-fold enrichment, respectively, of 5′-GWCAY motif sites (Supplementary Fig. 16) compared with the average genome-wide frequency. Such chaperone-related genes play important roles in bacterial stress responses to the host environment57. The expression changes in HPXZ1398 also include the flagellar hook-encoding gene, flgE, and several genes related to DNA processing and metabolism (Supplementary Table 6). These results demonstrate that genome-wide alterations of methylation patterns can significantly impact gene expression, although the specific mechanisms by which methylation regulates the expression of these genes are unknown.
Intracellular stochastic methylation in C. salexigens
In contrast to 5′-GWCAY, 5′-TCAN6TRG/5′-CYAN6TGA and 5′-TCNNGA in H. pylori J99, the SMP scores of the 5′-RGATCY motif in C. salexigens do not support the existence of phase-variable MTase activity (Fig. 4f). Instead of a peak near SMP≈2, there is a peak near SMP≈0.9, indicating that the IPD values used to calculate SMP scores for each molecule reflect a mixture of both non-methylated and methylated motif sites. This observation combined with a lack of detected sequence variation in the coding sequence of the 5′-RGATCY-targeting M.CsaI gene (Supplementary Fig. 17) and that M.CsaI is an orphan MTase without a corresponding RE37 indicates that intracellular stochastic methylation by M.CsaI is the likely mechanism driving the observed heterogeneity in SMSN scores (Fig. 3b). Therefore, M.CsaI in C. salexigens may have stochastic methylation activity (∼60%) in each cell, constituting an alternate form of epigenetic heterogeneity in a bacterial population.
Discussion
Here, we present SMALR, the first systematic framework for single molecule-level characterization of epigenetic heterogeneity in bacterial methylomes using SMRT sequencing. We demonstrate the enhanced resolving power of our approach in analyses of seven methylomes that show distinct types of epigenetic heterogeneity in bacterial populations. The short library-based SMSN method enables accurate estimation of the fractions of methylated and non-methylated motif sites without the use of subjective thresholds, even at low levels of genomic sequence coverage. Furthermore, the robust separation between methylated and non-methylated sites based on SMSN scores at low levels of genomic sequence coverage suggests the possibility of reference-free, de novo discovery of methylation motifs from SMRT sequencing data at a much lower coverage than is currently required. The long library-based SMP method uses long reads to assess the co-occurrence patterns of methylated motif sites at single-molecule resolution. By surveying MTase activity at the single molecule-level, the SMP scores can reveal small fractions of cells in a bacterial population that contain active or inactive MTases, and quantitatively estimate the processivity of an MTase of interest.
The existing, consensus-based methods for methylation detection cannot provide the resolution necessary to survey complex MTase activity. The application of SMALR and its integration with other single molecule- or single cell-level data, such as RNA and protein expression will enable a more detailed understanding of the functions of DNA methylations in bacterial physiology.
DNA methylation in promoter regions of bacteria has been extensively linked to the regulation of nearby genes2,3,12,58, but there is growing appreciation for the potential roles of methylation in the gene body31,59,60. More generally, the importance of epigenetic regulation in bacteria is emphasized in a recent study that proposed an epigenetics-driven adaptive evolution model21. In this model, heterogeneous methylomes may drive unique patterns of gene expression and cellular phenotypes, thereby serving as units of natural selection. The methods presented here make it possible to characterize the epigenetic modes that induce transcriptional diversity, analogous to the many known genetic modes17,26,49,54,61.
While SMRT sequencing enables de novo detection of a wide variety of DNA modifications, it also poses challenges unique to third-generation sequencing technology. Specifically, detection of DNA modifications from SMRT reads fundamentally depends on the number of repeated observations for each single molecule. For a given read length, there is a resulting tradeoff between library size and accuracy in single molecule, single nucleotide-level methylation detection. The methods proposed here provide examples of effectively leveraging the unique features of SMRT sequencing using a combination of short- and long-library designs. This general methodology can be modified to accommodate forthcoming third-generation real-time sequencing techniques62.
While the current study uses SMSN scores from a single site to detect DNA methylation, previous work has demonstrated that some DNA modifications (DNA methylation or damage) can create kinetic signatures that extend into surrounding bases34. Although beyond the scope of the current work, the single molecule-level approaches presented here can be expanded to include more sophisticated algorithms required for detection of these modification types.
Finally, while the current study focused on cultures of single bacterial strains, the single-molecule resolution methods proposed here can also be applied to mixed populations of bacteria. Such samples could include diverse clinical isolates, including samples that contain a high abundance pathogen or even diverse microbiome samples. The methods are also applicable outside the bacterial kingdom, such as in the analysis of human mitochondrial DNA or DNA viruses, both of which present significant genetic and epigenetic heterogeneity.
Methods
Code availability
SMALR is implemented in a stand-alone software package (written in Python) and available at https://github.com/fanglab.
Bacterial strains and culture conditions
The E. coli C227 sample was isolated from a 64-year-old woman from Hamburg, Germany, who was hospitalized in Copenhagen, Denmark after presenting with bloody diarrhoea. Rasko et al.44 isolated DNA using a Qiagen DNEasy Blood and Tissue Kit as per manufacturer's instructions. The isolate was grown overnight in standard LB broth and the extraction was performed according to the kit instructions using 1 ml overnight culture per reaction and treating with proteinase K for 2 h. The DNA was eluted in 100 μl AE buffer per column.
H. pylori J99 was originally isolated in Nashville Tennessee from an American patient with a duodenal ulcer63 and sequenced in 1999 (ref. 53). Cells (50 μl) of frozen H. pylori J99 stock from the original isolating lab were first spotted on trypticase soy agar (TSA) plates with 5% sheep blood (TSA, BBL Microbiology Systems, Cockeysville, MD) at 37 °C with 5% CO2 for 48-h incubation, then were spread on new TSA plates in the same conditions64. After 24-h incubation, H. pylori cells were collected in 1.0 ml phosphate-buffered saline (PBS, pH 7.4) and centrifuged at 800 g for 5 min. Bacterial genomic DNA was prepared using the Wizard Genomic DNA purification kit according to manufacturer's instructions (Promega, Madison WI). DNA concentration was measured by Nanadrop 1,000 spectrophotometer (Thermo Scientific, Rockford, IL).
DNA for Chromohalobacter salexigens strain 1H11 was ordered from DSM ( http://www.dsmz.de/catalogues/details/culture/DSM-3043.html).
Kozdon et al.38 grew C. crescentus cells at 28 °C in M2-glucose minimal media (M2G) and performed small-scale synchrony. To have sufficient volume to remove large aliquots at each time point, swarmer cells were pooled from 16 small-scale synchronies. Cells were allowed to proceed synchronously through the cell cycle at 28 °C in M2G. Genomic DNA was isolated using the Gentra Puregene Yeast/Bacteria Kit (Qiagen). Cells were removed from synchronous cultures growing in M2G. Genomic DNA was isolated from 4 ml of cells at each time point. The protocol provided with the kit was followed with a few modifications: 4 μl RNaseA was used instead of 1.5 μl, and after protein precipitation the samples were left on ice for 15 min and pelleted for 5 min at 4 °C. The DNA pellet was allowed to dry for 10 min and then resuspended in 25 μl 10 mM Tris, pH 8.0. Samples were incubated at 65 °C for 1 h and at room temperature overnight38.
Murray et al.37 received the DNA of G. metallireducens from an established culture collection65. Murray et al. received C. jejuni 81–176 and C. jejuni NCTC 11168 DNAs from Stuart Thompson, Medical College of Georgia.
SMRT sequencing
Long insert DNA library preparation and sequencing was performed according to the manufacturer's instructions. Upon completion of library construction, samples were assessed for quantity and insert size using an Agilent DNA 12,000 gel chip. Additional size selection was conducted using Sage Science Blue Pippin 0.75% agarose cassettes to enrich for library in the range of 7,000–50,000 bp. This selection is necessary to narrow the library distribution and maximize the SMRTbell sub-readlength. 11–23% of the input libraries was eluted from the agarose cassette and was available for sequencing. For all cases, this yield was sufficient to proceed to primer annealing and DNA sequencing on the Pacific Biosciences RSII machine. Primer was then annealed to the size-selected SMRTbells with the full-length libraries (80 °C for 2 min 30 followed by decreasing the temperature by 0.1°per second to 25 °C). The polymerase–template complex was then bound to the P5 (or P4) enzyme using a ratio of 10:1 polymerase to SMRTbell at 0.5 nM for 4 h at 30 °C and then held at 4 °C until ready for magbead loading, before sequencing. The magnetic bead-loading step was conducted at 4 °C for 60 min per manufacturer's guidelines. The magbead-loaded, polymerase-bound, SMRTbell libraries were placed onto the RSII machine at a sequencing concentration of 75 pM and configured for a 180 min continuous sequencing run.
For all short (250 bp) insert library preparations, similar methodology was used, except shearing was done using a Covaris microtube ultrasonication and all AMPure XP purification steps were done using a 1.8 × volume ratio. Libraries were completed without the size selection step used for the long-insert libraries. Similar procedures were followed for sequencing, except that diffusion-based loading was used instead of magbead loading.
Filtering subreads and preprocessing SMRT reads
An initial filtering step removes all subreads with ambiguous alignments (MapQV<240), low accuracy (<80%) or short-aligned length (<100 bases). Next, because sequencing errors in the subreads are likely to introduce noise into the IPD distribution that is being used to infer methylation status, an additional filtering step removes the subread IPD values from the positions +1:−1 on either side of any errors with respect to the reference sequence. This removes a substantial number of IPD values from consideration, but those that remain are minimally impacted by sequencing errors. Subread IPD normalization corrects for any potential slowing of polymerase kinetics over the course of an entire read (which consists of many subreads) and is done by log-transforming all subread IPD values and subtracting their mean from each individual log-transformed IPD value. Finally, the first ten and last fifteen bases from each subread are removed from analysis due to the potential for bias in the IPD values near the transition between template and adapter sequences.
SMSN detection of methylation states
Considering each native molecule separately, the IPD values (post-filtering) for a given motif are grouped by their strand and mapped genomic position. Following natural log conversion of the set of IPD values (transforming exponentially distributed IPD to follow an approximately normal distribution) for the molecule/strand/position, the mean value is calculated. Contrary to the treatment of native IPD values, the WGA IPD values are aggregated across all molecules covering each strand and mapped genomic position. This aggregation is done because all molecules are expected to be free of any DNA methylations due to the amplification process33,35. The SMSN score is calculated by subtracting the WGA strand/position-matched mean log IPD value from this native molecule/strand/position-specific mean log IPD value. This score approximately follows a normal distribution34.
Whole-genome amplification
The Qiagen REPLI-g amplification kit was used to perform whole-genome amplification to exclude epigenetically modified bases. The method produced micrograms of DNA from 50 ng of input genomic DNA, following the manufacturer's guidelines and 10 h of amplification time at 30 °C followed by deactivation at 65 °C for 3 min.
SMP detection of MTase activity
First, considering each molecule separately, the native IPD values for a given motif that survive filtering are grouped by their strand only, irrespective of their mapped genomic positions. The natural log is taken for all values in this group and the mean calculated. Second, the strand/position-matched WGA IPD values are aggregated across all molecules, as they were with the SMSN method. However, here the WGA IPD values are additionally aggregated across each motif site covered by the native molecule in question. Then, these WGA strand- and motif site-matched IPD values are natural log-converted and their mean value is subtracted from the native molecule/strand-specific mean, resulting in the SMP score.
Sensitivity and specificity of SMSN methylation detection
To quantify the sensitivity and specificity for SMSN detection, we applied the approach to methylated 5′-CTGCAG sites in a native E. coli O104:H4 C227-11 strain35 (C227) and a matching WGA sample in which the methylation sites were erased. In the analysis, we assumed 100% of the 5′-CTGCAG sites were methylated in the native DNA to leverage the large number of motif sites in the native bacterial genome. This is expected to provide a more robust estimation of sensitivity and specificity compared with the use of short DNA oligos that have limited diversity of expanded local sequence contexts flanking any given methylation motif site34,66. Given that 5′-CTGCAG sites are part of an active type-II R-M system, it is reasonable to assume the vast majority of sites are methylated to prevent restriction of the host DNA. However, as shown in recent studies35,36,38, a small number of motif sites (even those that are part of R-M systems) are sometimes found to be stably non-methylated, likely due to competitive binding between the MTase and other DNA-binding proteins or to the transient non-methylated window that follows behind the replication fork. Because the true methylated fraction of 5′-CTGCAG sites is slightly lower than 100%, the sensitivity estimated in the above analysis represents the lower bound of the actual sensitivity of the SMSN approach.
AggSN detection of methylation states
The IPD values from all native molecules were aggregated according to their strand and mapped genomic position. Similarly, the IPD values from all the whole-genome amplified (WGA) molecules were aggregated according to their strand and mapped genomic position. The AggSN score per strand/position represents the difference between the mean native log IPD value and the mean WGA log IPD value.
Modified fraction estimates with Gaussian mixture modelling
The mixture module of the Python package PyMix was used to run the EM algorithm67 for Gaussian mixture estimation.
To evaluate the ability of EM with a Gaussian mixture model to estimate the modified fraction, we applied the algorithm to distributions of SMSN scores that were generated from in silico mixing of WGA and native SMRT sequencing molecules at known proportions. 100,000 total molecules sequenced from E. coli C227 were used for each specific mixture fraction, with the native fraction of molecules ranging from 5,000 (5%) to 100,000 (100%). The SMSN-based methylated fraction for the motif 5′-CTGCAG was analysed with the EM algorithm for each in silico mixture. To assess the stability of EM-mediated estimation of methylated fraction at lower levels of genomic coverage (that is, the number of total sequenced bases in relation to the genome size), we downsampled the scores in the SMSN distribution for the in silico mixtures of WGA and native molecules. The EM algorithm was then applied to these downsampled SMSN score distributions.
The EM algorithm slightly, yet consistently, underestimates the native fraction in this mixture. To determine whether any non-normality in the methylated and non-methylated SMSN score distributions might be causing the EM algorithm to underestimate the size of the modified fraction, we created Gaussian mixtures consisting of two entirely simulated normal distributions. To represent the non-methylated and methylated SMSN scores, the distributions were simulated with α=2 and α=0, respectively (σ=0.5 for each). As an alternative, we also created mixtures using SMSN values exclusively from WGA molecules where +2 was added to a subset of the SMSN scores in order to get between 5 and 100% ‘methylated' SMSN values, the distributions of which will retain any non-normality found in the WGA SMSN distribution. The EM estimations of methylated fraction for these two types of simulated mixtures were both very similar and very accurate (Supplementary Fig. 2), indicating that non-normality of SMSN score distributions is not the reason for the observed slight underestimation of native fraction. Instead, this evidence suggests that stably non-methylated 5′-CTGCAG sites in E. coli C227 are the reason for this phenomenon.
Approximate SMSN scores for C. crescentus
WGA sequencing was unavailable for analysis in this study, so the approximate SMSN score shown in Fig. 3c is simply the mean of native molecule-, strand-, and position-specific log IPD values (rather than its difference from the mean of strand- and position-matched WGA log IPD values). This approximate SMSN score is susceptible to IPD biases introduced by local sequence contexts, but provides a sufficient ability to resolve the methylated and non-methylated components in the bimodal distributions.
FDR estimation for identifying methylated molecules
To assess the significance of observed methylated and non-methylated fractions based on SMP scores, we established negative control SMP distributions. When assessing the FDR for calling actively methylated molecules (for example, for the 5′-GWCAY motif in H. pylori J99), the negative control SMP distribution is generated by running the SMP method on WGA sequencing data. Alternatively, when assessing the FDR for calling non-methylated molecules (for example, for the 5′-TCAN6TRG/5′-CYAN6TGA and 5′-TCNNGA motifs in H. pylori J99), the negative control SMP distribution is created by randomly shuffling IPD values among molecules to disperse the non-modified IPD values for the motif of interest. Given that the number of non-methylated IPD values is relatively low, this creates a control distribution of SMP scores reflecting molecules that are mostly methylated at the motif of interest.
Illumina MiSeq/HiSeq sequencing and analysis
For C. salexigens and the H. pylori J99 culture, Illumina-based MiSeq whole-genome libraries were prepared with an insert size of 600 bp as assessed by Agilent Bioanalysis using standard Illumina adapters and 8 PCR cycles. 2 × 300 bp paired-end sequencing was then conducted using version 3 commercial kits to assure the longest readlength possible. For the five single colonies of H. pylori J99 and the two mutant strains, both DNA and RNA were sequenced on HiSeq 2,500 with 100 bp paired-end reads. RNA of the wild-type H. pylori J99 strain was also sequenced using this method. Total RNAs were first treated with RiboZero bacterial Gram Negative ribosomal removal kit (Epicentre; MRZGN126) to obtain rRNA depleted RNAs.
The E. coli K12 MiSeq reads used for analysis of homopolymer indel rates (Fig. 4d) were downloaded from http://www.illumina.com/systems/miseq/scientific_data.ilmn.
The DNA sequencing reads from the E. coli K12, C. salexigens, and H. pylori J99 MiSeq runs were aligned to their respective references using bwa mem68 using default parameters. The resulting alignments were processed with the samtools package69 to obtain pileups for each genomic position. Read-level mismatch and insertion/deletion calls were parsed from the pileups by counting the number of occurrences of each type of variant at each genomic position.
To call differentially expressed genes from RNAseq data, we first mapped raw RNA reads for each sample to the Genbank reference (Accession number: AE001439). Reads that are mapped to rRNA and tRNAs were excluded. A gene was included for differential expression analysis if it had more than one count per million reads (CPM=1) in at least two samples. Differentially expressed genes are then called by the software program edgeR70 at P<0.001, corresponding to a FDR of 5%.
Obtaining single colonies from H. pylori J99 stock strain
Roughly 20 μl H. pylori J99 strain frozen stock was spread on TSA plates with 5% sheep blood. After 96-h incubation at 37 °C in 5% CO2 conditions, five random single colonies were isolated and spread on new TSA plates for another 48-h incubation in the same conditions to enrich bacterial cultures for stock preparation and further tests. The five H. pylori strains obtained from the five random H. pylori J99 single colonies were named as HPXZ1383, HPXZ1384, HPXZ385, HPXZ1386 and HPXZ1387.
Construction of H. pylori J99 MTase-knockout strains
Mutations of M.Hpy99XXII (jhp1365) and M.Hpy99XXI (jhp1411) were constructed by replacing the ORFs with sacB-cat cassettes (without disrupting the ORF of the upstream or downstream genes) through homologous recombination, as described below. A 0.9 Kb fragment upstream of jhp1365 (jhp1365up) was obtained by PCR using primers JHP1365LF-SacII and JHP1365LR-SpeI (Supplementary Table 3), and a 0.6 Kb fragment downstream of jhp1365 (jhp1365down) was obtained by PCR using primers JHP1365RF-SpeI and JHP1365RR-PstI (Supplementary Table 3). Fragments jhp1365up and jhp1365down were digested with SacII/SpeI and SpeI/PstI, respectively, and then ligated together into SacII/PstI-digested pGEM-T Easy, creating pXZ577 (Supplementary Table 4). Similarly, a 0.9 Kb fragment upstream of jhp1411 (jhp1411up) was obtained by PCR using primers JHP1411LF-SacII and JHP1411LR-SpeI (Supplementary Table 3), and a 0.7 Kb fragment downstream of jhp1411 (jhp1411down) was obtained by PCR using primers JHP1411RF-SpeI and JHP1411RR-PstI (Supplementary Table 3). Fragments jhp1411up and jhp1411down were digested with SacII/SpeI and SpeI/PstI, respectively, and then ligated together into SacII/PstI-digested pGEM-T Easy, creating pXZ579 (Supplementary Table 4). The sacB-cat cassette, conferring chloramphenicol resistance (CmR), was obtained by PCR using pXZ032 (ref. 64) as template and primers SC-F-XbaI and SC-R-XbaI (Supplementary Table 3). The sacB-cat cassette was digested with XbaI, and then ligated with SpeI-digested pXZ577 and pXZ579, creating pXZ578 and pXZ580, respectively (Supplementary Table 4). All plasmid constructions were confirmed by sequencing using primers PGEMTe-seqF and PGEMTe-seqR (Supplementary Table 3).
The wild type H. pylori strain J99 was transformed to CmR with these plasmids, pXZ578 and pXZ580, to create mutant HPXZ1398 (jhp1365::sacB-cat) and HPXZ1401 (jhp1411::sacB-cat), respectively (Supplementary Table 4), as previously described64. Genomic DNA of these mutants was isolated and PCR was performed to confirm that HPXZ1398 and HPXZ1401 carry the correct mutation of jhp1365::sacB-cat and jhp1411::sacB-cat, respectively, with sacB-cat cassette-specific primer Catup and genome locus-specific primer JHP1365RR-PstI or JHP1411RR-PstI (Supplementary Table 3).
Sensitivity and specificity of SMP methylation detection
To measure the sensitivity and specificity of the SMP scores for assessing methylated molecules (and by extension MTase activity), we applied the method to the 5′-GATC motif in H. pylori J99. The responsible MTase, M.Hpy99VI, is not suspected to be phase-variable and SMSN scores revealed a very small non-methylated fraction (Fig. 3a). We therefore expect that nearly 100% of molecules in the sample will be targeted for methylation at the 5′-GATC motif. As the sensitivity and specificity curves show (Supplementary Fig. 18), the power of the SMP scores to detect methylated molecules increases with an increasing threshold on the number of motif sites present on the molecule. Although lower values of this threshold have limited ability to correctly identify methylated molecules, some methylated motifs are distributed too sparsely in the genomes to be interrogated using higher thresholds. Shorter motifs are usually present at higher densities in a genome, but there are some exceptions to this rule (Supplementary Table 2). Therefore, we suggest using the number of motif sites on each molecule (rather than motif length) as the threshold for SMP analysis.
Additional information
Accession codes: Sequence data have been deposited at the NCBI Sequence Read Archive (SRA) with BioProject under project ID PRJNA281410 and the DNA Database of Japan under accession code SRP057274. The assembled reference genomes used in this study have been deposited in GenBank under the accession codes CP011331 (E. coli C227) and CP011330 (H. pylori J99).
How to cite this article: Beaulaurier, J. et al. Single molecule-level detection and long read-based phasing of epigenetic variations in bacterial methylomes. Nat. Commun. 6:7438 doi: 10.1038/ncomms8438 (2015).
Supplementary Material
Supplementary Figures 1-18 and Supplementary Tables 1-6
Acknowledgments
We thank Jonas Korlach and Richard Roberts for providing access to raw sequencing data as well as secondary analysis of samples from Murray et al. We also thank Lucy Shapiro and Harley McAdams for providing access to raw sequencing data of samples from Kozdon et al. The work was supported in part by RO1-GM63270 from the National Institutes of Health, the Diane Belfer Program for Human Microbial Ecology, and the Daniel Ziff Fund. This work was also supported in part through the computational resources and staff expertise provided by the Department of Scientific Computing at the Icahn School of Medicine at Mount Sinai.
Footnotes
E.E.S. is on the scientific advisory board of Pacific Biosciences.
Author contributions J.B. and G.F. designed the SMSN and SMP methods; J.B. developed the SMALR package; J.B., X.-S.Z., E.E.S. and G.F. designed the experiments; X.-S.Z., R.S., C.R., G.D., N.S., D.M., M.K.W., A.C. and M.J.B. carried out sample-preparation experiments; R.S. and G.D. conducted all the sequencing; J.B., X.-S.Z., S.Z., R.S., M.K.W., A.C., M.J.B., E.E.S. and G.F. jointly analysed the data sets; J.B., E.E.S. and G.F. wrote the manuscript with input from all coauthors; G.F. conceived and supervised the project.
References
- Cheng X. Structure and function of DNA methyltransferases. Annu. Rev. Biophys. Biomol. Struct. 24, 293–318 (1995). [DOI] [PubMed] [Google Scholar]
- Casadesús J. & Low D. Epigenetic gene regulation in the bacterial world. Microbiol. Mol. Biol. Rev. 70, 830–856 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wion D. & Casadesús J. N6-methyl-adenine: an epigenetic signal for DNA-protein interactions. Nat. Rev. Microbiol. 4, 183–192 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito T., Kusano K. & Kobayashi I. Selfish behavior of restriction-modification systems. Science 267, 897–899 (1995). [DOI] [PubMed] [Google Scholar]
- Roberts R. J., Vincze T., Posfai J. & Macelis D. REBASE-a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 43, D298–D299 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handa N. & Kobayashi I. Post-segregational killing by restriction modification gene complexes: Observations of individual cell deaths. Biochimie 81, 931–938 (1999). [DOI] [PubMed] [Google Scholar]
- Pingoud A., Wilson G. G. & Wende W. Type II restriction endonucleases-a historical perspective and more. Nucleic Acids Res. 42, 7489–7527 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low D. a. & Casadesús J. Clocks and switches: bacterial gene regulation by DNA adenine methylation. Curr. Opin. Microbiol. 11, 106–112 (2008). [DOI] [PubMed] [Google Scholar]
- Seshasayee A. S. N., Singh P. & Krishna S. Context-dependent conservation of DNA methyltransferases in bacteria. Nucleic Acids Res. 40, 7066–7073 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinus M. G. & Casadesus J. Roles of DNA adenine methylation in host-pathogen interactions: mismatch repair, transcriptional regulation, and more. FEMS Microbiol. Rev. 33, 488–503 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim H. N. & van Oudenaarden A. A multistep epigenetic switch enables the stable inheritance of DNA methylation states. Nat. Genet. 39, 269–275 (2007). [DOI] [PubMed] [Google Scholar]
- Shell S. S. et al. DNA methylation impacts gene expression and ensures hypoxic survival of Mycobacterium tuberculosis. PLoS Pathog. 9, e1003419 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavazoie S. & Church G. Quantitative whole-genome analysis of DNA-protein interactions by in vivo methyl's protection in E. coli. Nat. Biotechnol. 16, 566–571 (1998). [DOI] [PubMed] [Google Scholar]
- Wang M. X. & Church G. M. A whole genome approach to invivo dna-protein interactions in Escherichia coli. Nature 360, 606–610 (1992). [DOI] [PubMed] [Google Scholar]
- Ringquist S. & Smithtt C. L. The Escherichia coli chromosome contains specific, unmethylated dam and dcm sites. Proc. Natl Acad. Sci. USA 89, 4539–4543 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallecha A., Munster V., Correnti J., Chan T. & van der Woude M. Dam- and OxyR-dependent phase variation of agn43: essential elements and evidence for a new role of DNA methylation. J. Bacteriol. 184, 3338–3347 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerdeño-Tárraga A. & Patrick S. Extensive DNA inversions in the B. fragilis genome control variable gene expression. Science 307, 1463–1466 (2005). [DOI] [PubMed] [Google Scholar]
- van der Woude M., Braaten B. & Low D. Epigenetic phase variation of the pap operon in Escherichia coli. Trends Microbiol. 4, 5–9 (1996). [DOI] [PubMed] [Google Scholar]
- van der Woude M. W. Phase variation: how to create and coordinate population diversity. Curr. Opin. Microbiol. 14, 205–211 (2011). [DOI] [PubMed] [Google Scholar]
- Srikhanta Y. N., Fox K. L. & Jennings M. P. The phasevarion: phase variation of type III DNA methyltransferases controls coordinated switching in multiple genes. Nat. Rev. Microbiol. 8, 196–206 (2010). [DOI] [PubMed] [Google Scholar]
- Furuta Y. & Kobayashi I. Mobility of DNA sequence recognition domains in DNA methyltransferases suggests epigenetics-driven adaptive evolution. Mob. Genet. Elements 2, 292–296 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta Y. et al. Methylome diversification through changes in DNA methyltransferase sequence specificity. PLoS Genet. 10, e1004272 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manso A. S. et al. A random six-phase switch regulates pneumococcal virulence via global epigenetic changes. Nat. Commun. 5, 5055 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebes J. et al. The complex methylome of the human gastric pathogen Helicobacter pylori. Nucleic Acids Res. 42, 1–18 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croucher N. J. et al. Diversification of bacterial genome content through distinct mechanisms over different timescales. Nat. Commun. 5, 1–12 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J. & Blaser M. J. Bacterial populations as perfect gases: genomic integrity and diversification tensions in Helicobacter pylori. Nat. Rev. Microbiol. 4, 826–836 (2006). [DOI] [PubMed] [Google Scholar]
- Srikhanta Y. N. et al. Phasevarion mediated epigenetic gene regulation in Helicobacter pylori. PLoS ONE 6, e27569 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadesús J. & Low D. a. Programmed heterogeneity: epigenetic mechanisms in bacteria. J. Biol. Chem. 288, 13929–13935 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jen F. E.-C., Seib K. L. & Jennings M. P. Phasevarions mediate epigenetic regulation of antimicrobial susceptibility in Neisseria meningitidis. Antimicrob. Agents Chemother. 58, 4219–4221 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seib K. L. et al. Specificity of the ModA11, ModA12 and ModD1 epigenetic regulator N6-adenine DNA methyltransferases of Neisseria meningitidis. Nucleic Acids Res. 43, 4150–4162 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahramanoglou C. et al. Genomics of DNA cytosine methylation in Escherichia coli reveals its role in stationary phase transcription. Nat. Commun. 3, 886 (2012). [DOI] [PubMed] [Google Scholar]
- Eid J. et al. Real-time DNA sequencing from single polymerase molecules. Science 323, 133–138 (2009). [DOI] [PubMed] [Google Scholar]
- Flusberg B. a. et al. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat. Methods 7, 461–465 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt E. E. et al. Modeling kinetic rate variation in third generation DNA sequencing data to detect putative modifications to DNA bases. Genome Res. 23, 129–141 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang G. et al. Genome-wide mapping of methylated adenine residues in pathogenic Escherichia coli using single-molecule real-time sequencing. Nat. Biotechnol. 30, 1232–1239 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lluch-Senar M. et al. Comprehensive methylome characterization of Mycoplasma genitalium and Mycoplasma pneumoniae at single-base resolution. PLoS Genet. 9, e1003191 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray I. a. et al. The methylomes of six bacteria. Nucleic Acids Res. 40, 11450–11462 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozdon J. B. et al. Global methylation state at base-pair resolution of the Caulobacter genome throughout the cell cycle. Proc. Natl Acad. Sci. USA 110, E4658–E4667 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendall M. L. et al. Exploring the roles of DNA methylation in the metal-reducing bacterium Shewanella oneidensis MR-1. J. Bacteriol. 195, 4966–4974 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Romero M. a., Cota I. & Casadesús J. DNA methylation in bacteria: from the methyl group to the methylome. Curr. Opin. Microbiol. 25, 9–16 (2015). [DOI] [PubMed] [Google Scholar]
- Clark T. a. et al. Characterization of DNA methyltransferase specificities using single-molecule, real-time DNA sequencing. Nucleic Acids Res. 40, e29 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao B. et al. Genomic mapping of phosphorothioates reveals partial modification of short consensus sequences. Nat. Commun. 5, 3951 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin C.-S. et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 10, 563–569 (2013). [DOI] [PubMed] [Google Scholar]
- Rasko D. et al. Origins of the E. coli strain causing an outbreak of hemolytic–uremic syndrome in Germany. N. Engl. J. Med. 365, 709–717 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin C. et al. The origin of the Haitian cholera outbreak strain. N. Engl. J. Med. 364, 33–42 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark T. a. et al. Enhanced 5-methylcytosine detection in single-molecule, real-time sequencing via Tet1 oxidation. BMC Biol. 11, 4 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilmes J. A. A gentle tutorial of the EM algorithm and its application to parameter estimation for gaussian mixture and hidden Markov models. Int. Comput. Sci. Inst. 4, 126 (1998). [Google Scholar]
- Kahng L. S. & Shapiro L. The CcrM DNA methyltransferase of Agrobacterium tumefaciens is essential, and its activity is cell cycle regulated. J. Bacteriol. 183, 3065–3075 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salaun L. The diversity within an expanded and redefined repertoire of phase-variable genes in Helicobacter pylori. Microbiology 150, 817–830 (2004). [DOI] [PubMed] [Google Scholar]
- De Vries N. et al. Transcriptional phase variation of a type III restriction-modification system in Helicobacter pylori. J. Bacteriol. 184, 6615–6623 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanabria-Valentín E., Colbert M. T. C. & Blaser M. J. Role of futC slipped strand mispairing in Helicobacter pylori Lewis phase variation. Microbes Infect. 9, 1553–1560 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Kang J. & Blaser M. J. Antimutator role of the DNA glycosylase mutY gene in Helicobacter pylori. J. Bacteriol. 188, 6224–6234 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alm R. A. et al. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397, 176–180 (1999). [DOI] [PubMed] [Google Scholar]
- Josenhans C., Eaton K. a., Thevenot T. & Suerbaum S. Switching of flagellar motility in Helicobacter pylori by reversible length variation of a short homopolymeric sequence repeat in fliP, a gene encoding a basal body protein. Infect. Immun. 68, 4598–4603 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price M. N., Huang K. H., Alm E. J. & Arkin A. P. A novel method for accurate operon predictions in all sequenced prokaryotes. Nucleic Acids Res. 33, 880–892 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma C. M. et al. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature 464, 250–255 (2010). [DOI] [PubMed] [Google Scholar]
- Neckers L. & Tatu U. Molecular chaperones in pathogen virulence: emerging new targets for therapy. Cell Host Microbe 4, 519–527 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadbent S. E., Davies M. R. & Van Der Woude M. W. Phase variation controls expression of Salmonella lipopolysaccharide modification genes by a DNA methylation-dependent mechanism. Mol. Microbiol. 77, 337–353 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers K. S. et al. Genome-scale analysis of Escherichia coli FNR reveals complex features of transcription factor binding. PLoS Genet. 9, 11–13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald D. M., Bonocora R. P. & Wade J. T. Comprehensive mapping of the Escherichia coli flagellar regulatory network. PLoS Genet. 10, e1004649 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citti C., Nouvel L.-X. & Baranowski E. Phase and antigenic variation in mycoplasmas. Future Microbiol. 5, 1073–1085 (2010). [DOI] [PubMed] [Google Scholar]
- Manrao E. a. et al. Reading DNA at single-nucleotide resolution with a mutant MspA nanopore and phi29 DNA polymerase. Nat. Biotechnol. 30, 349–353 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atherton J. C., Peek R. M. J., Tham K. T., Cover T. L. & Blaser M. J. Clinical and pathological importance of heterogeneity in vacA, the vacuolating cytotoxin gene of Helicobacter pylori. Gastroenterology 112, 92–99 (1997). [DOI] [PubMed] [Google Scholar]
- Zhang X. S. & Blaser M. J. Natural transformation of an engineered Helicobacter pylori strain deficient in type II restriction endonucleases. J. Bacteriol. 194, 3407–3416 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovley D. R. & Phillips E. J. Novel mode of microbial energy metabolism: organic carbon oxidation coupled to dissimilatory reduction of iron or manganese. Appl. Environ. Microbiol. 54, 1472–1480 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z. et al. Detecting DNA modifications from SMRT sequencing data by modeling sequence context dependence of polymerase kinetic. PLoS Comput. Biol. 9, e1002935 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempster A., Laird N. & Rubin D. Maximum likelihood from incomplete data via the EM algorithm. J. R. Stat. Soc. 39, 1–38 (1977). [Google Scholar]
- Li H. & Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. et al. The sequence alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. D., McCarthy D. J. & Smyth G. K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures 1-18 and Supplementary Tables 1-6