Abstract
The ZmCBF3 gene is a member of AP2/ERF transcription factor family, which is a large family of plant-specific transcription factors that share a well-conserved DNA-binding domain. To understand the regulatory mechanism of ZmCBF3 gene expression, we isolated and characterized the ZmCBF3 promoter (PZmCBF3). Three deletion fragments of PZmCBF3 were generated, C1–C3, from the translation start codon at position −1079, −638, and −234, and fused to the GUS reporter gene. Each deletion construct was analyzed by Agrobacterium-mediated stable transformation and expression in Arabidopsis thaliana. GUS expression assays indicated that the PZmCBF3 exhibited root-specific expression activity. A 234-bp fragment upstream of the ZmCBF3 gene conferred a high level of GUS activity in Arabidopsis. Some cis-acting elements involved in the down-regulation of gene expression were detected in the promoter, encompassing positions −1079 to −234. PZmCBF3 was activated by cold stress. The MYCCONSENSUSAT elements (CANNTG) were responsible for the ability of PZmCBF3 to respond to cold stress. The results of the present study suggest that PZmCBF3 might play a role in cold tolerance in maize.
Keywords: promoter, ZmCBF3, transgenic Arabidopsis, abiotic stress
1. Introduction
Drought, high salinity, and low temperature are common stress conditions that have adverse effects on plant survival, growth, and reproduction. To continue growth and reproduction under such conditions of abiotic stresses, plants respond to unfavorable environments by using various developmental, physiological, and biochemical mechanisms. The C-repeat (CRT)/DRE (dehydration-responsive element) motif-binding transcription factors (CBFs) are identified as stress-responsive genes. Low temperatures can induce the expression of CBFs in plants, as well as enhance frost resistance [1,2]. The CBFs can specifically bind to the CRT/DRE element, which is located in the promoter region of cold-regulated genes and upregulates their expression and thus increases frost/freeze tolerance in plants [3].
The CBF gene family includes CBF1, CBF2, CBF3, and CBF4. These four genes play an important role in plants that improve resistance to cold, drought, and salt. Using the yeast one-hybrid system, AtCBF1, which specifically binds to CRT/DRE, was identified in an Arabidopsis gene library [1]. These sequences shared 88% similarity at the amino acid level. Five protein-binding DRE elements were identified in DREB1A (DRE-binding protein 1A), DREB1B, and DREB1C corresponding to CBF3, CBF1, and CBF2, respectively [4]. In 2002, the fourth member of the CBF family, CBF4, was identified [5]. In addition to Arabidopsis, CBF genes also exist in other plant species such as oilseed rape, barley, tomato, rice, rye grass, maize, and soybean [6,7,8,9,10,11,12]. Proteins belonging to the CBF family have a highly conserved DNA-binding domain called the AP2/ERF domain, which consists of ~60 amino acids. The domain is also found in CBF-like proteins of other plants [13]. The AP2/ERF domain plays an important role in recognizing and binding specifically to the CRT/DRE cis-acting elements to activate the corresponding gene expression [14].
Maize (Zea mays L.) is one of the world’s most important crops. In our previous study, the gene expression of maize leaves under drought and low-temperature stresses was analyzed by using gene chip technology (unpublished results), which showed the upregulation of ZmCBF3. Although the function of CBF proteins in plants under abiotic stresses has been previously reported, investigations on CBF promoters responding to the abiotic stress have not been performed. Studying the roles of promoters under different environmental stresses and plant hormone treatment will help us to better understand the regulation of CBF self-expression, and would be a significant guide for its actual production and transgenic breeding. In the present study, the upstream promoter sequence of the ZmCBF3 gene was isolated and its function was analyzed by using 5′-end deletion and GUS activity assays.
2. Results
2.1. Promoter Sequence Analysis
To characterize the promoter of ZmCBF3 (PZmCBF3), the 5′-upstream sequence was cloned from maize (B73) genomic DNA. The cloned DNA region was 1443 bp in length (Figure 1) and was analyzed by using the PLACE and the PlantCARE databases. The putative cis-acting elements were identified and labeled (Figure 1), which were homologous to the cis-acting elements of other plants.
Figure 1.
Nucleotide sequence and putative cis-acting elements of the ZmCBF3 promoter (PZmCBF3). The cis-acting elements are boxed, and arrows indicate their direction. The translation initiation codon is underlined.
Three kinds of promoter elements known to be regulated by hormones in some plant genes were identified in the PZmCBF3 sequence: three cis-acting elements involved in the abscisic acid responsiveness (ABREs); two cis-acting regulatory elements associated with MeJA-responsiveness (CGTCA-motif and TGACG-motif), and two cis-acting elements participating in gibberellin-responsiveness (TATC-box and GARE-motif).
Three putative regulatory motifs, which are involved in the activation of stress and defense genes in plants, were detected in the promoter region of the ZmCBF3 gene. One was the cis-acting element involved in heat stress responsiveness (HSE), the other included two MYBHv1 binding sites (CCAAT-box), and the last one comprised seven MYCCONSENSUSAT elements (CANNTG).
Four kinds of putative root-specific elements were detected within the PZmCBF3, which included four RAV1AAT (CAACA), two ROOTMOTIFPOX1 (CAACA), two RHERPATEXPA7 (KCACGW), and one OSE2ROOTNODULE (CTCTT) sites.
Other elements were also detected within the PZmCBF3 sequence, which included three cis-acting regulatory element (AREs) that were essential for anaerobic induction, one 5′-UTR Py-rich stretch (a cis-acting element conferring high transcription levels), one A-box (a cis-acting regulatory element), one CCGTCC-box (a cis-acting regulatory element related to meristem specific activation), one O2-site (cis-acting regulatory element involved in zein metabolism regulation), and several light responsive elements such as G-box, Sp1, GAG-motif, and ACE.
2.2. Basal Expression Analysis
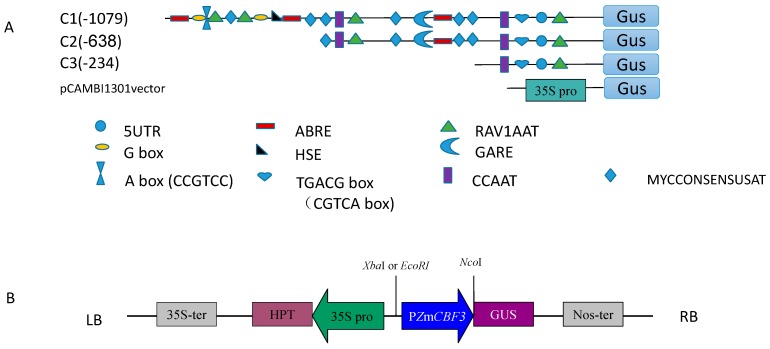
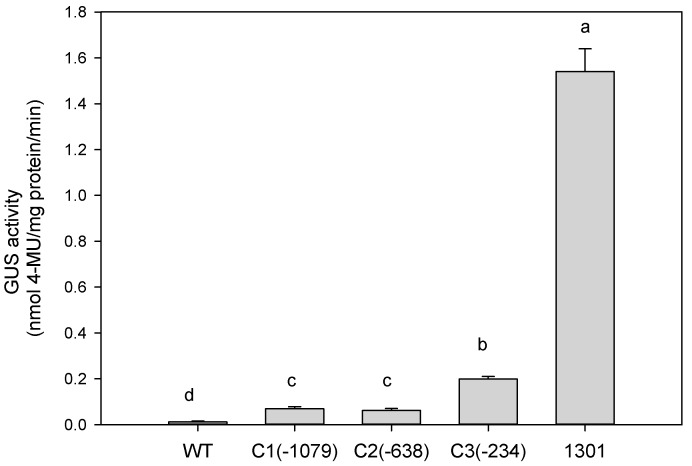
To determine the minimal necessary region of PZmCBF3, three promoter deletion fragments were fused with the GUS reporter gene in pCAMBIA1301 for agro-infiltration into Arabidopsis (Figure 2). The expression of each PZmCBF3:GUS fusion gene was examined in the leaves of 15 independent T3 transgenic Arabidopsis plants. As shown in Figure 3, deletion constructs with C3 showed much higher GUS activity than that of the C1 and C2 promoters in the leaves of transgenic Arabidopsis plants (Figure 3 and Figure 4). C1 and C2 promoter-mediated GUS activity showed no significant difference. Compared to GUS activity driven by cauliflower mosaic virus (CaMV) 35S promoter (as positive control), PZmCBF3 showed lower activity in leaves. In addition, C3-mediated GUS activity was accounting for only 13% of the CaMV 35S promoter-mediated GUS activity. These results indicated that the leaves of the T3 transgenic Arabidopsis plants had very low PZmCBF3 activity.
Figure 2.
Transformation of Arabidopsis plants using PZmCBF3:GUS constructs. (A) Schematic representation of the PZmCBF3 promoter constructs used to analyze GUS expression in Arabidopsis leaves. The serial 5′-deletion constructs of the PZmCBF3 promoter were fused to the GUS reporter gene in the vector, pCAMBIA1301; (B) Schematic representation of the PZmCBF3:GUS construct. The insertion position of the ZmCBF3 promoter in the vector is indicated with restriction enzyme sites (XbaI or EcoRI and NcoI). LB, left border; RB, right border; 35S-ter, CAMV 35S terminator; 35S pro, CAMV 35S promoter; GUS, β-glucuronidase gene; HPTII, hygromycinphosphotransferase (II) coding region; NOST, nopaline synthase terminator; PZmCBF3, ZmCBF3 promoter.
Figure 3.
Basal expression analysis of GUS activity in transgenic Arabidopsis lines. GUS activity was analyzed fluorometrically and expressed as nmoles 4-methylumbelliferone (MU)/mg protein/min. Data are expressed as the mean ± standard deviation of three independent assays of Arabidopsis leaf extracts. Six-week-old Arabidopsis T3 plants and wild type (WT) were used for this analysis. Same letters indicate no significant differences at p ≤ 0.05 by Duncan test. C1(−1,079), C2(−638), and C3(−234) represent different deletion promoters. Different letters (a–d) mean statistical differences between treatments.
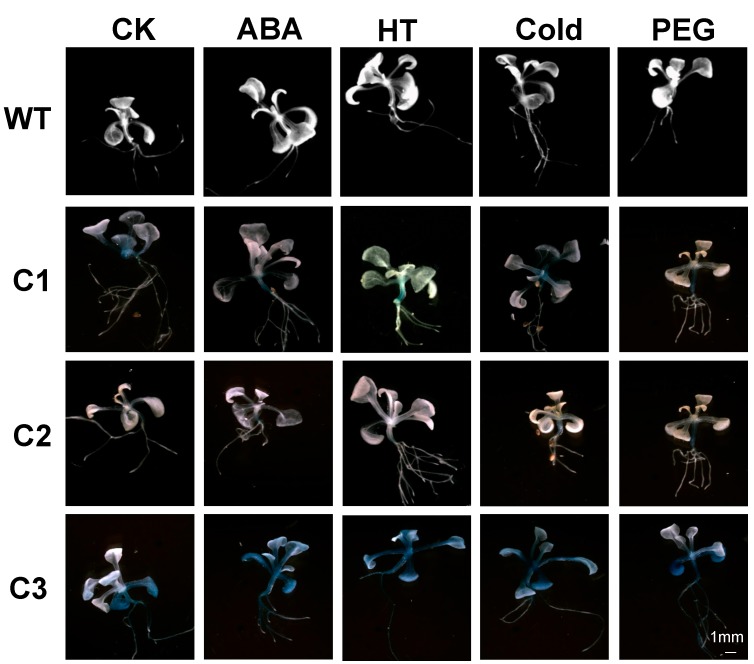
Figure 4.
Histochemical expression of the β-glucuronidase (GUS) reporter gene in transgenic Arabidopsis and wild type (WT) in response to abiotic stresses and abscisic acid (ABA). Two-week-old Arabidopsis T3 plants were subjected to ABA, high temperature (HT), cold (4 °C), and drought (20% PEG6000) treatments for 6 h. Control (CK) plants were treated with water. C1, C2, and C3 represent different deletion promoters. Scale bar for all treatments are shown in C3-PEG. Scale bars: 1 mm.
2.3. Abiotic Stress-Induced Expression Analysis
To verify the role of stress-responsive cis-acting elements in PZmCBF3, deletion promoter constructs were fused to the GUS reporter gene (Figure 2). GUS activity of the different deletion promoters in transgenic Arabidopsis leaves after treatment with high temperature (HT), cold, ABA, and PEG6000 were then examined.
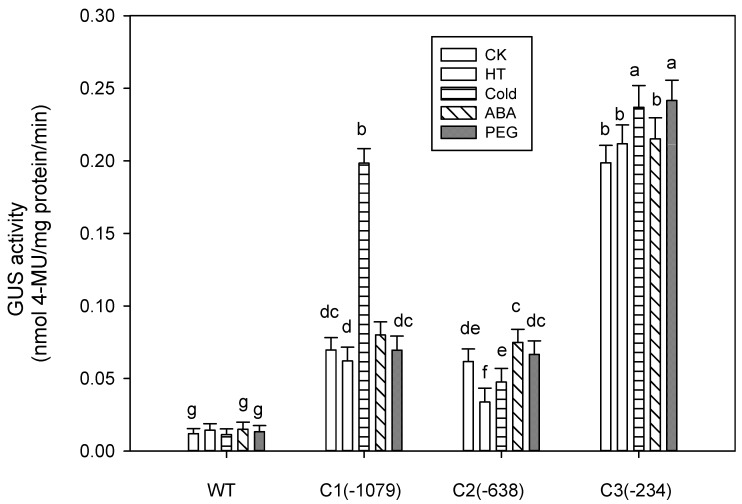
Compared to the control leaves (untreated plants), GUS activity in C2-transformed leaves decreased by 0.547-fold after HT treatment (Figure 5). However, marginal induction of GUS activity was observed in C1- and C3-transformed Arabidopsis leaves. Under cold stress, C1- and C3-mediated GUS activity increased by 2.8- and 1.19-fold, respectively, whereas GUS activity of C2-transformed plants did not significantly change (Figure 5). As shown in Figure 5, GUS activity of deletion promoters slightly increased after ABA and PEG treatment. GUS activity of the C1 and C2 promoters increased by 1.15- and 1.21-fold respectively, after ABA treatment. C3-mediated GUS activity significantly increased (by about 1.22-fold) with PEG treatment.
Figure 5.
Abiotic stress-induced GUS activity analysis of transgenic Arabidopsis lines. Six-week-old Arabidopsis T3 plants and wild type (WT) were subjected to ABA, high temperature (HT), cold (4 °C), and drought (20% PEG6000) treatments for 6 h. Control (CK) plants were treated with water. GUS activity was analyzed fluorometrically and expressed as nmoles 4-methylumbelliferone (MU)/mg protein/min. Data are expressed as the mean ± standard deviation of three independent assays involving Arabidopsis leaf extracts. The same letters indicate no significant differences at p ≤ 0.05 by Duncan’s test. Different letters (a–g) mean statistical differences between treatments.
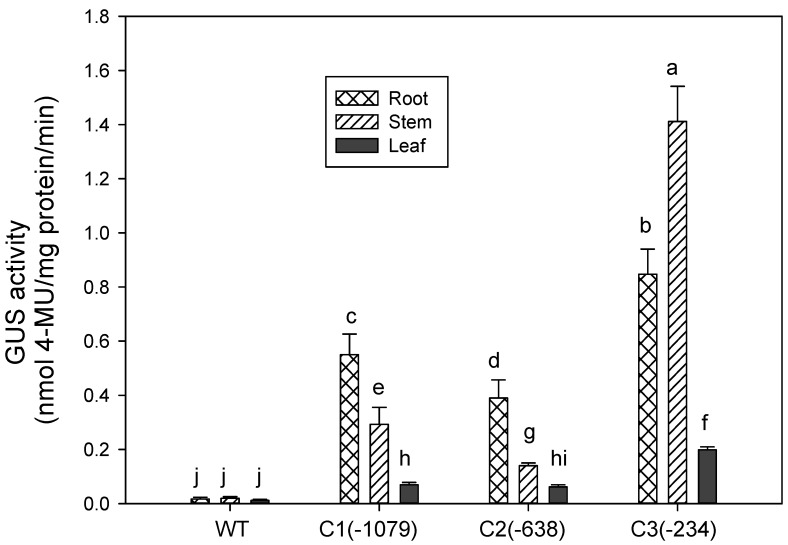
2.4. Tissue-Specific Expression Analysis
In our early experiments, histochemical staining was employed to determine GUS activity of the deletion promoters in transgenic Arabidopsis (Figure 4). In Figure 4, GUS staining was mainly detected in the stems and roots. The promoter apparently shows tissue-specific expression. To test this hypothesis, GUS activity of PZmCBF3 was determined in the transgenic Arabidopsis roots, stems, and leaves by fluorometric analyses. GUS activity of the C1 and C2 promoters was highest in the roots, followed by the stems (Figure 6). C3-mediated GUS activity was highest in the stems, which was 1.67-fold that of the root (Figure 6). These results were consistent with that of GUS staining.
Figure 6.
Tissue-specific expression analysis of GUS activity among different transgenic Arabidopsis lines. GUS activity was examined in six-week-old Arabidopsis T3 plants and wild type (WT). GUS activity was analyzed fluorometrically and expressed as nmoles 4-methylumbelliferone (MU)/mg protein/min. Data are expressed as the mean ± standard deviation of three independent assays involving Arabidopsis leaf extracts. The same letters indicate no significant differences at p ≤ 0.05 by Duncan’s test. Different letters (a–j) mean statistical differences between treatments.
3. Discussion
3.1. Basal Expression Analysis of the Deletion Promoters
To determine the function of cis-acting elements, deletion fragment analysis of the PZmCBF3 was performed. The deletion promoter constructs were then transformed into Arabidopsis by using Agrobacterium-mediated transformation. Several studies have earlier shown that promoter activity gradually decreases with shorter promoter lengths [15,16]. However, in the present study, the smallest promoter fragment C3(−234) showed the highest activity among the deletion promoters (Figure 3). This finding suggests that the sequence between −1079 and −234 bp of the PZmCBF3 might contain cis-acting elements that are involved in the downregulation of gene expression. This phenomenon has also been observed in other promoters [17]. Dalal et al. reported that the strength of the tissue-specific promoter ShCYC-B drastically decreased in deletion mutants, and further deletion of the promoter sequence at 5′ end resulted in the highest activity [18].
3.2. Abiotic Stress-Induced Expression Analysis
The deletion promoters of ZmCBF3 exhibited inducible expression in response to different stress stimuli (HT, cold, ABA, and PEG; Figure 5). Heat shock elements (HSEs) are the binding sites for the trans-active heat shock factor (HSF), and three or more units are required for efficient binding [19,20]. In the present study, sequence analysis of PZmCBF3 showed that there was one HSE within the segment encompassing −691 to −701 bp. GUS activity in C1-transformed Arabidopsis leaves was almost not induced by high temperature (Figure 5). The results suggest that HSE did not affect the C1 promoter in response to high temperature. However, negative regulatory elements might be located in the promoter region, covering −638 to −1079 bp, which induced a decrease in GUS activity of C2-mediated after HT treatment.
After cold treatment, GUS activity of C1-mediated plants increased by 2.8-fold (Figure 5), which may be attributed to MYCCONSENSUSAT elements (CANNTG). Sequence analysis identified seven MYCCONSENSUSAT elements within PZmCBF3. The MYC recognition sequence was the binding site of ICE1, which encodes a MYC-like bHLH transcriptional activator that specifically binds to multiple MYC DNA regulatory elements in the CBF3 promoter, stimulating CBF3 transcription [21]. However, GUS activity of C2-transformed plants did not significantly change, which suggests that part of the seven elements played an important role in regulating gene expression during cold stress. The MYCCONSENSUSAT elements present between −638 to −234 bp might not play an important role for the promoter.
ABRE (PyACGTG/TC) is a well-studied cis-element involved in ABA-induced gene expression [22]. Promoters sequence analysis has revealed that ABA-responsive gene expression requires multiple ABA-responsive elements (ABREs or CE3) [23,24]. The ABRE-binding protein/factor (AREB/ABF) family is composed of nine members in Arabidopsis [25,26]. Sequence analysis of PZmCBF3 identified three ABREs within the promoter. We hypothesized that ABREs play an important role in increasing GUS activity under ABA treatment. ABA is one of the most important signaling molecules; furthermore, it is also a cross-talking signal molecule. In plants, osmotic stress-responsive transcriptional regulation depends mainly on two pathways: ABA-dependent and ABA-independent pathways [27]. In the present study, GUS activity of PZmCBF3 showed the same changes under ABA and PEG stress, suggesting that the expression of ZmCBF3 might be an ABA-dependent pathway under PEG stress.
3.3. Expression of PZmCBF3 Is Tissue-Specific
Most AP2/ERF transcription factors exhibit root-specific expression [28]. Research investigations on root-specific promoters and root-specific cis-acting elements are limited. For example, tissue-specific expression of TobRB7 in tobacco involves the root meristem and central cylinder regions and encodes a cytosol protein [29]. Other root-specific promoters have also been identified such as AtEXPA7, AtEXPA18 [30], EXPANSIN A [31,32], and PAL [33]. In addition, some root-specific cis-acting elements have been detected in root-specific promoters. The Atlg73160 promoter sequence [29] consists of root-specific regulatory elements such as ASF1MOTIFCAMV, ARFAT, OSE1, OSE2, SURE core, and RAV1AAT. RHERPATEXPA7 and ROOTMOTIFPOX1 have also been identified in other promoters [34,35].
The present study has shown that PZmCBF3 is highly expressed in the root (Figure 4 and Figure 6). Sequence analysis has identified four RAV1AAT (−954 to −949 bp, −812 to −807 bp, −403 to −397 bp, −62 to −57 bp), two ROOTMOTIFPOX1 (−734 to −729 bp, −319 to −314 bp), two RHERPATEXPA7 (−994 to −988 bp, −664 to −658 bp), and one OSE2ROOTNODULE (−973 to −968 bp) in the ZmCBF3 promoter. Two RAV1AAT (−403 to −397 bp, −62 to −57 bp) and one ROOTMOTIFPOX1 (−319 to −314 bp) were present in the C2 promoter, and only one RAV1AAT (−62 to −57 bp) was detected within the C3 promoter. Figure 6 shows that the highest C1- and C2-mediated GUS activity was observed in the roots of transgenic Arabidopsis. However, GUS activity of C3-mediated expression was lower than that of other tissues and organs, in which most of the root-specific regulatory elements of the promoter were deleted. This finding suggests that RAV1AAT and ROOTMOTIFPOX1 play an important role in the root-specific expression of PZmCBF3.
4. Experimental Section
4.1. Plant Materials, Growth Condition, and Bacterial Strains
Maize plants (cultivar B73) were raised in pots containing soil mix (soil:vermiculite, 3/1, v/v) under greenhouse conditions (16/8 h photoperiod; 26 °C), with three plants per pot. The fifth expanding leaves were sampled for genomic DNA extraction. Arabidopsis thaliana (L.) ecotype Col-0 plants were grown in a soil mix (2:1:1 mixture of vermiculite, peat moss, and perlite) and placed at 22/19 ± 2 °C (light/dark, 16/8 h photoperiod) with a light intensity of approximately 100–120 µmol·m−2·s−1 in a growth chamber. Escherichia coli strain DH5α was used to clone and propagate all recombinant plasmid vectors. Agrobacterium tumefaciens strain EHA105 was used for infiltrating A. thaliana leaves.
4.2. Genomic DNA Extraction
Total DNA was isolated from young leaves using CTAB (cetyltrimethylammonium bromide) as previously described [36], with slight modifications. Approximately 0.1 g of leaf powder was added to 0.6 mL of pre-warmed (65 °C) extraction buffer (3% (w/v) CTAB, 2% (w/v) polyvinylpyrrolidone, 1 M Tris–HCl (pH 8.0), 20 mM EDTA, 1.4 M NaCl, and 2% (v/v) β-mercaptoethanol) and incubated for 30 min at 65 °C with occasional mixing. The slurry was mixed by inversion with 0.6 mL of chloroform/isoamyl alcohol (24/1, v/v) and centrifuged for 10 min at 9600× g. The upper phase was recovered and two volumes of isopropanol was added and incubated for 2 h at −20 °C. The suspension was centrifuged for 10 min at 9600× g, and the pellet was dried and dissolved in 50 mL of deionized H2O. The integrity of genomic DNA was assessed on 1% (w/v) agarose gel by electrophoresis followed by visualization with ethidium bromide staining.
4.3. Isolation of the ZmCBF3 Gene Promoter
Using gene chip technology, we obtained the ZmCBF3 gene. The upstream sequence of the gene was obtained after aligned and analyzed at the MaizeGDB website (http://maizegdb.org/). The specific primers were designed according to the 5′ flanking sequence of ZmCBF3. The ZmCBF3 promoter (PZmCBF3) was amplified from maize genomic DNA by PCR in a mixture of 1 µL of each primer (10 µM, Pcbf-F and Pcbf-R, Table 1), 1.0 µL 5-mM dNTP, 2.5 µL 10× buffer, 0.5 µL of Taq DNA polymerase (Takara, Otsu, Japan), 0.5 µL DNA, and 18.5 µL sterile H2O. The PCR conditions were as follows: initial denaturation at 94 °C for 4 min; followed by 30 cycles of denaturation at 94 °C for 40 s, annealing at 54 °C for 40 s, and extension at 72 °C for 1 min 30 s; and a final extension at 72 °C for 10 min. PCR products were analyzed on 1% agarose/ethidium bromide gel, purified using TIANquick Midi Purification Kit (DP209, Beijing, China) [37], cloned into pMD18-T cloning vector, and then transformed into E. coli strain DH5a. The positive clones were sequenced using an automated DNA sequencer at the Beijing Genomics Institute (Beijing, China).
Table 1.
Primers used for PCR.
| Oligo Name | Sequence (5′→3′) |
|---|---|
| Pcbf-F | TGAGCCGGACGGTCTGCTAT |
| C2-638 | GGAATTCAGAGATAACGCCAGAACGAG |
| C3-234 | GGAATTCGCCTTGTTCCGTTGTTCG |
| Pcbf-R | TGCCATGGCTGCTGCTGTGACTGTGA |
| Hyg-F | GATGTTGGCGACCTCGTATT |
| Hyg-R | TCGTTATGTTTATCGGCACTTT |
4.4. Sequence Analysis
Putative cis-acting elements and their positions in the PZmCBF3 promoter were identified using PLACE [38] and PlantCARE [39].
4.5. Construction of the PZmCBF3 Deletion Constructs
To amplify the 5ʹ upstream sequence of the PZmCBF3 gene more easily, the upstream primer did not contain a restriction site, while the downstream primer contained a NcoI restriction site. The PZmCBF3 region from −1079 to +1, designated C1 (−1079), was obtained by restriction enzyme (XbaI and NcoI) digestion of the clone pMD18-T:PZmCBF3. Two deletions, P2 (−638 bp) and P3 (−234 bp) (Figure 2A), were generated by PCR amplification, as described above, using the clone pMD18-T:PZmCBF3 as template. Forward primers containing an EcoRI restriction site, and a common reverse primer containing a NcoI restriction site (Table 1). The binary vector pCAMBIA1301 was used to make all plasmid constructs (Figure 2B).
The CaMV 35S promoter in front of the β-glucuronidase (GUS) gene was excised through digestion and replaced with one of the promoter fragments. Promoter fragments inserted into all plasmid constructs was confirmed by sequencing, and the correct plasmid constructs of PZmCBF3:GUS were transformed into Agrobacterium strain EHA105 by the freeze-thaw method [40].
4.6. Arabidopsis Plant Transformation with Different PZmCBF3:GUS Constructs
Promoter activity can be studied via instantaneous expression, which provides rapid results, but quantitative comparisons of promoter activity are error-prone because the number of infected cells is difficult to control. So in this study, we chose to study the promoter activity by transforming Arabidopsis plants to get stable expression. The pCAMBIA1301 vectors containing the promoter-GUS fusion construct was transferred into Agrobacterium strain EHA105 for transformation into Arabidopsis (Figure 2B) as described previously [41]. pCAMBIA1301 was the positive control. Arabidopsis blooms were inoculated by vacuum infiltration with Agrobacterium in Murashige and Skoog medium (MS) medium with 5% sucrose and 0.05% Silwet L-77. The transgenic Arabidopsis T0 was grown on MS agar plates supplemented with 25 mg/L hygromycin B and then the screened transgenic plants were screened again by PCR with primers from the hygromycin-coding region (Hyg-F and Hyg-R, Table 1). The transgenic Arabidopsis T1 and T2 were also screened on MS agar plates supplemented with 25 mg/L hygromycin B. At the same time, the transgenic strains, from which the ratio of resistant plants and sensitive plants was 3:1, were analyzed.
4.7. Abiotic Stress Treatments
For each treatment and control, 100 transgenic Arabidopsis T3 plants were grown on MS agar plates supplemented with 25 mg/L hygromycin B at 22 °C for 2 and 6 weeks then used for stress treatment experiments. Two-week-old transgenic Arabidopsis T3 and wild type (WT) were used for histochemical analysis and 6-week-old plants were used for the fluorescence measuring.
To characterize the induction of promoter activity by the stress-responsive and ABA, the plants were removed from MS agar plates and their roots soaked in water, solutions of abscisic acid (ABA, 100 µM) or polyethylene glycol solution (PEG6000, 20%). High temperature (HT) and cold treatment was conducted under dim light by exposing plant roots soaked in water to a temperature of 37 and 4 °C. For the dehydration experiments, the plant roots were soaked in Polyethylene glycol solution (PEG6000, 20%) to simulate drought. The transgenic plants were soaked in water as control. In each case, plants were subjected to treatment for 6 h then frozen in liquid nitrogen.
4.8. Histochemical and Fluorometric Analyses of GUS Activity
Histochemical analysis of GUS activity was performed on 2-week-old transgenic Arabidopsis T3 stress-treated and control plants. The frozen plant material was incubated with 1 mM 5-bromo-4-chloro-3-indolyl-b-d-glucuronide (X-Gluc) in 50 mM phosphate buffer (pH 7.0). After staining, chlorophyll was cleared from tissues by incubating in 70% ethanol [42]. The 6-week-old transgenic Arabidopsis T3 were used to determine GUS enzyme activity by measuring the fluorescence of 4-methylumbelliferone produced by GUS cleavage of 4-methylumbelliferyl-b-d-glucuronide (4-MUG) [42]. GUS activity is expressed as nanomoles of methylumbelliferone per minute per milligram of protein. Protein amount was determined using a Protein Assay kit (Bio-Rad, Hercules, CA, USA) using bovine serum albumin as a standard.
4.9. Data Analysis
All measurements were repeated at least three times, and the results were expressed as mean values. Error bars shown in figures represent standard deviation (SD). The means of three replicates of three independent samples were analyzed by Duncan test at the 5% probability level.
5. Conclusions
GUS expression assays indicated that the ZmCBF3 gene exhibited root-specific expression. A 234-bp fragment upstream of the ZmCBF3 gene coding sequence confers a high level of GUS expression in Arabidopsis. Some cis-acting elements involved in down-regulation of gene expression might exist in the sequence of the promoter from −1079 to −234 bp. The MYCCONSENSUSAT elements (CANNTG) were responsible for the ability of PZmCBF3 to be induced by cold stress. PZmCBF3-mediated GUS expression was an ABA-dependent pathway under PEG stress conditions, in which ABREs played an important role.
Acknowledgments
We thank Venkatesh Sivanandam and Shih-Hua Tan at University of California, Riverside for critical reading of the manuscript. This study is financed by grants from the Twelfth Five Year Plan Project of Science and Technology Support, China (2014BAD14B02, 2012BAD19B04), the Project from the Ministry of Agriculture Key Projects of GM Cultivation of New Varieties (2013ZX08004004), and the Research and Development of Industrial Technology Special at Jilin Provincial Development and Reform Commission (2013C001).
Author Contributions
Jinliang Liu: responsible for article writing, contributions to the manuscript and experiments. Fengting Wang: contributions to experiments and statistical interpretation of test results. Gang Yu: contributions to manuscript. Xianghui Zhang: contributions in plant cultivation. Chengguo Jia: contributions to the experiment design ideas. Jianchun Qin and Hongyu Pan: corresponding author, responsible for the experiment design ideas and funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Stockinger E.J., Gilmour S.J., Thomashow M.F. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. USA. 1997;94:1035–1040. doi: 10.1073/pnas.94.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gilmour S.J., Zarka D.G., Stockinger E.J., Salazar M.P., Houghton J.M., Thomashow M.F. Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J. 1998;16:433–442. doi: 10.1046/j.1365-313x.1998.00310.x. [DOI] [PubMed] [Google Scholar]
- 3.Thomashow M.F. Molecular basis of plant cold acclimation: Insights gained from studying the CBF cold response pathway. Plant Physiol. 2010;154:571–577. doi: 10.1104/pp.110.161794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Q., Kasuga M., Sakuma Y., Abe H., Miura S., Yamaguchi-Shinozaki K., Shinozaki K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell Online. 1998;10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haake V., Cook D., Riechmann J., Pineda O., Thomashow M.F., Zhang J.Z. Transcription factor CBF4 is a regulator of drought adaptation in Arabidopsis. Plant Physiol. 2002;130:639–648. doi: 10.1104/pp.006478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jaglo K.R., Kleff S., Amundsen K.L., Zhang X., Haake V., Zhang J.Z., Deits T., Thomashow M.F. Components of the Arabidopsis C-repeat/dehydration-responsive element binding factor cold-response pathway are conserved in Brassica napus and other plant species. Plant Physiol. 2001;127:910–917. doi: 10.1104/pp.010548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi D.-W., Rodriguez E.M., Close T.J. Barley CBF3 gene identification, expression pattern, and map location. Plant Physiol. 2002;129:1781–1787. doi: 10.1104/pp.003046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Skinner J.S., von Zitzewitz J., Szűcs P., Marquez-Cedillo L., Filichkin T., Amundsen K., Stockinger E.J., Thomashow M.F., Chen T.H., Hayes P.M. Structural, functional, and phylogenetic characterization of a large CBF gene family in barley. Plant Mol. Biol. 2005;59:533–551. doi: 10.1007/s11103-005-2498-2. [DOI] [PubMed] [Google Scholar]
- 9.Dubouzet J.G., Sakuma Y., Ito Y., Kasuga M., Dubouzet E.G., Miura S., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt-and cold-responsive gene expression. Plant J. 2003;33:751–763. doi: 10.1046/j.1365-313X.2003.01661.x. [DOI] [PubMed] [Google Scholar]
- 10.Xiong Y., Fei S.Z. Functional and phylogenetic analysis of a DREB/CBF-like gene in perennial ryegrass (Lolium perenne L.) Planta. 2006;224:878–888. doi: 10.1007/s00425-006-0273-5. [DOI] [PubMed] [Google Scholar]
- 11.Zhang G., Chen M., Li L., Xu Z., Chen X., Guo J., Ma Y. Over-expression of the soybean GmERF3 gene, an AP2/ERF type transcription factor for increased tolerances to salt, drought, and diseases in transgenic tobacco. J. Exp. Bot. 2009;60:3781–3796. doi: 10.1093/jxb/erp214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qin F., Sakuma Y., Li J., Liu Q., Li Y.-Q., Shinozaki K., Yamaguchi-Shinozaki K. Cloning and functional analysis of a novel DREB1/CBF transcription factor involved in cold-responsive gene expression in Zea mays L. Plant Cell Physiol. 2004;45:1042–1052. doi: 10.1093/pcp/pch118. [DOI] [PubMed] [Google Scholar]
- 13.Riechmann J.L., Meyerowitz E.M. The AP2/EREBP family of plant transcription factors. Biol. Chem. 1998;379:633–646. doi: 10.1515/bchm.1998.379.6.633. [DOI] [PubMed] [Google Scholar]
- 14.Sakuma Y., Liu Q., Dubouzet J.G., Abe H., Shinozaki K., Yamaguchi-Shinozaki K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration-and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 2002;290:998–1009. doi: 10.1006/bbrc.2001.6299. [DOI] [PubMed] [Google Scholar]
- 15.Tao Y., Wang F., Jia D., Li J., Zhang Y., Jia C., Wang D., Pan H. Cloning and functional analysis of the promoter of a stress-inducible gene (ZmRXO1) in maize. Plant Mol. Biol. Rep. 2014;33:1–9. doi: 10.1007/s11105-014-0741-1. [DOI] [Google Scholar]
- 16.Yang Q., Yuan D., Shi L., Capell T., Bai C., Wen N., Lu X., Sandmann G., Christou P., Zhu C. Functional characterization of the Gentiana lutea zeaxanthin epoxidase (GlZEP) promoter in transgenic tomato plants. Transgenic Res. 2012;21:1043–1056. doi: 10.1007/s11248-012-9591-5. [DOI] [PubMed] [Google Scholar]
- 17.Wang F., Liu J., Li J., Zhang S., Pan H. Functional analyses of the maize CKS2 gene promoter in response to abiotic stresses and hormones. Acta Physiol. Plant. 2014;36:1867–1878. doi: 10.1007/s11738-014-1563-3. [DOI] [Google Scholar]
- 18.Dalal M., Chinnusamy V., Bansal K.C. Isolation and functional characterization of lycopene β-cyclase (CYC-B) promoter from Solanum habrochaites. BMC Plant Biol. 2010;10:61. doi: 10.1186/1471-2229-10-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hübel A., Schöffl F. Arabidopsis heat shock factor: Isolation and characterization of the gene and the recombinant protein. Plant Mol. Biol. 1994;26:353–362. doi: 10.1007/BF00039545. [DOI] [PubMed] [Google Scholar]
- 20.Schöffl F., Prändl R., Reindl A. Regulation of the heat-shock response. Plant Physiol. 1998;117:1135–1141. doi: 10.1104/pp.117.4.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chinnusamy V., Ohta M., Kanrar S., Lee B.-H., Hong X., Agarwal M., Zhu J.-K. ICE1: A regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 2003;17:1043–1054. doi: 10.1101/gad.1077503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hobo T., Asada M., Kowyama Y., Hattori T. ACGT-containing abscisic acid response element (ABRE) and coupling element 3 (CE3) are functionally equivalent. Plant J. 1999;19:679–689. doi: 10.1046/j.1365-313x.1999.00565.x. [DOI] [PubMed] [Google Scholar]
- 23.Shen Q., Zhang P., Ho T. Modular nature of abscisic acid (ABA) response complexes: Composite promoter units that are necessary and sufficient for ABA induction of gene expression in barley. Plant Cell Online. 1996;8:1107–1119. doi: 10.1105/tpc.8.7.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hobo T., Kowyama Y., Hattori T. A bZIP factor, TRAB1, interacts with VP1 and mediates abscisic acid-induced transcription. Proc. Natl. Acad. Sci. USA. 1999;96:15348–15353. doi: 10.1073/pnas.96.26.15348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi H.I., Hong J.H., Ha J.O., Kang J.Y., Kim S.Y. ABFs, a family of ABA-responsive element binding factors. J. Biol. Chem. 2000;275:1723–1730. doi: 10.1074/jbc.275.3.1723. [DOI] [PubMed] [Google Scholar]
- 26.Corrêa L.G.G., Riaño-Pachón D.M., Schrago C.G., dos Santos R.V., Mueller-Roeber B., Vincentz M. The role of bZIP transcription factors in green plant evolution: Adaptive features emerging from four founder genes. PLoS ONE. 2008;3:e2944. doi: 10.1371/journal.pone.0002944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoshida T., Mogami J., Yamaguchi-Shinozaki K. ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr. Opin. Plant Biol. 2014;21:133–139. doi: 10.1016/j.pbi.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 28.Lata C., Mishra A.K., Muthamilarasan M., Bonthala V.S., Khan Y., Prasad M. Genome-wide investigation and expression profiling of AP2/ERF transcription factor superfamily in foxtail millet (Setaria italica L.) PLoS ONE. 2014;9:e113092. doi: 10.1371/journal.pone.0113092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamamoto Y.T., Taylor C.G., Acedo G.N., Cheng C.L., Conkling M.A. Characterization of cis-acting sequences regulating root-specific gene expression in tobacco. Plant Cell Online. 1991;3:371–382. doi: 10.1105/tpc.3.4.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim D.W., Lee S.H., Choi S.B., Won S.K., Heo Y.K., Cho M., Park Y.I., Cho H.T. Functional conservation of a root hair cell-specific cis-element in angiosperms with different root hair distribution patterns. Plant Cell Online. 2006;18:2958–2970. doi: 10.1105/tpc.106.045229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cho H.T., Cosgrove D.J. Regulation of root hair initiation and expansin gene expression in Arabidopsis. Plant Cell Online. 2002;14:3237–3253. doi: 10.1105/tpc.006437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin C., Choi H.-S., Cho H.-T. Root hair-specific EXPANSIN A7 is required for root hair elongation in Arabidopsis. Mol. Cells. 2011;31:393–397. doi: 10.1007/s10059-011-0046-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu Y., Wen F., Zhao X., Hawes M. Isolation of the promoter of a root cap expressed pectinmethylesterase gene from Pisum sativum L. (RCPME1) and its use in the study of gene activity. Plant Soil. 2004;265:47–59. doi: 10.1007/s11104-005-8468-2. [DOI] [Google Scholar]
- 34.Sunitha S., Mahajan N., Veluthambi K. The TrAP/REn monodirectional promoter of Mungbean yellow mosaic geminivirus (MYMV) displays root-specific expression in transgenic tobacco. Plant Cell Tissue Organ Cult. 2012;109:535–545. doi: 10.1007/s11240-012-0120-2. [DOI] [Google Scholar]
- 35.Kagaya Y., Ohmiya K., Hattori T. RAV1, a novel DNA-binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in higher plants. Nucleic Acids Res. 1999;27:470–478. doi: 10.1093/nar/27.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bousquet J., Simon L., Lalonde M. DNA amplification from vegetative and sexual tissues of trees using polymerase chain reaction. Can. J. For. Res. 1990;20:254–257. doi: 10.1139/x90-037. [DOI] [Google Scholar]
- 37.TIANquick Midi Purification Kit. [(accessed on 1 March 2015)]. Available online: http://www.tiangen.com/?productShow/t1/1/id/48.html.
- 38.PLACE. [(accessed on 1 March 2015)]. Available online: http://www.dna.affrc.go.jp/PLACE/
- 39.PlantCARE. [(accessed on 1 March 2015)]. Available online: http://bioinformatics.psb.ugent.be/webtools/plantcare/html/
- 40.Chen H., Nelson R., Sherwood J. Enhanced recovery of transformants of Agrobacterium tumefaciens after freeze-thaw transformation and drug selection. Biotechniques. 1994;16:664–668. [PubMed] [Google Scholar]
- 41.Clough S.J., Bent A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 42.Jefferson R.A. Plant reporter genes: The GUS gene fusion system. Genet. Eng. 1988;10:247–263. [Google Scholar]