Abstract
The study of Hemipteran mitochondrial genomes (mitogenomes) began with the Chagas disease vector, Triatoma dimidiata, in 2001. At present, 90 complete Hemipteran mitogenomes have been sequenced and annotated. This review examines the history of Hemipteran mitogenomes research and summarizes the main features of them including genome organization, nucleotide composition, protein-coding genes, tRNAs and rRNAs, and non-coding regions. Special attention is given to the comparative analysis of repeat regions. Gene rearrangements are an additional data type for a few families, and most mitogenomes are arranged in the same order to the proposed ancestral insect. We also discuss and provide insights on the phylogenetic analyses of a variety of taxonomic levels. This review is expected to further expand our understanding of research in this field and serve as a valuable reference resource.
Keywords: Hemiptera, mitogenomes, rearrangement, phylogenetic relationships
1. Introduction
Entomologists first suggested that Hemiptera (true bugs) and Homoptera (planthoppers, leafhoppers, cicadas, spittlebugs, aphids, psyllids, scales, and whiteflies) are two orders according to features of the wing [1]. In 1810, Latreille suggested combining Heteroptera and Homoptera as one order called Hemiptera (s.l.) [2]. The concept of Hemiptera (s.l.) has been widely accepted since 1969 to the present [3,4,5]; therefore, in this review, Hemiptera refers to Hemiptera (s.l.). As one major order of insects, Hemiptera is the largest group of the hemimetabolous insects [6], including more than 50,000 described species [7]. They are small sap-sucking insects with body-sizes from 1 mm (0.04 in) to approximately 15 cm (6 in).
There is great variety within the order Hemiptera, more commonly known as bugs. Hemipterans have evolved an extraordinary range of body forms and lifestyles: some live on land, some live in water, some feed on plants and others are voracious carnivores or scavengers. Therefore, many species of Hemiptera are significant pests of crops and gardens. Some, as many species of aphid, cause direct damage to crop hosts and often kill the entire plants. Additionally, some delphacids cause considerable damage to grain production and have been identified as one cause of rice famine in several Asian countries [8]. Moreover, many species of Hemiptera are vectors of viruses and diseases. For example, Triatoma dimidiata is the vector of Chagas disease, a predominantly chronic disease affecting millions of people [9].
Based on the history of Hemipteran phylogeny research, we propose two controversial questions. First, how many suborders does Hemiptera include? Traditionally, Hemiptera comprised three major groups (including four suborders): Sternorrhyncha (aphids, scale bugs, whiteflies, and psyllids), Auchenorrhyncha (planthoppers, leafhoppers, spittlebugs, and cicadas), and Heteroptera (true bugs, including Coleorrhyncha) [10]. Previous morphological studies suggested that Fulgoromorpha and Cicadomorpha formed Auchenorrhyncha, and that Auchenorrhyncha is more closely related to Coleorrhyncha and Sternorrhyncha than to Heteroptera [11]. However, additional molecular and morphological evidence has challenged the monophyly of Auchenorrhyncha (summarized by [12]). The second question, what are the relationships of these suborders that have confused entomologists for many years? Cobben suggested that both Heteroptera and Fulgoromorpha form the sister clade to (Sternorrhyncha, Cicadomorpha) according to a cladistic study of morphological traits [13]. Hamilton examined the phylogenetic affiliations using features of the head and mouthparts and suggested (Fulgoromorpha, (Sternorrhyncha, Cicadomorpha)) was the sister group to the clade (Coleorrhyncha, Heteroptera) [14]. However, it has been argued that Coleorrhyncha and Heteroptera do not have an immediate common ancestor and have descended independently from separate lineages [15]. Hence, the phylogenetic relationships among the higher-level hemipteran lineages remain unclear.
Since the first insect mitogenome was published in 1985 [16], there has been a rapid accumulation of sequenced insect genomes, with representatives from all orders now available [17]. Insect mitogenomes are small, double stranded, circular DNA molecules, ranging in size from 14 to 19 kb. The mitogenome is composed of thirty-seven genes (13 protein-coding, 22 transfer RNA, and 2 ribosomal RNA genes), and contains a control region (A + T-rich region) that is thought to play a role in the initiation of transcription and replication, and is a source of length variation in the genome [18]. Particularly, mitogenome sequences can provide even more genetic information and are increasingly being utilized in insect identification, biogeographic and phylogenetic studies [19,20,21].
Here, we utilize all the mitogenomes of Hemiptera to analyze their features on the genome level and summarize the rearrangement events for the first time. In addition, all available complete mitogenomes of Hemiptera were used to reconstruct and discuss the phylogeny relationships of this order.
2. Mitogenomes of Hemiptera
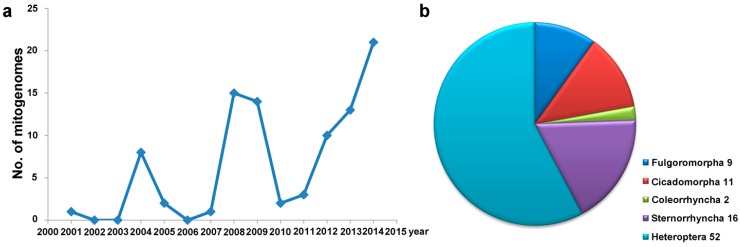
Triatoma dimidiata, the vector of Chagas disease, was the first published mitogenome of Hemipterain 2001 [9]. The sequencing history of hemipteran mitogenomes was shown (Figure 1a). There are two peaks during the past 14 years. Three years after the publication of the mitogenome of Triatoma dimidiata, Thao et al. [22] reported the complete nucleotide sequence of the mitogenomes of six species of whiteflies, one psyllid and one aphid from the suborder Sternorrhyncha. Four species of whiteflies had variations in gene order that were very different from the proposed insect ancestor (Drosophila yakuba) [16]. Subsequently, a number of studies have already proved that the rearrangements were more likely to happen in the mitogenomes of whiteflies than other insects of Hemiptera [22,23,24,25]. In 2008, Bu’s group obtained 10 complete and five nearly complete mitogenomes of Heteroptera [23] and they reported the first comparative mitogenome analysis of one suborder of Hemiptera and the phylogenetic relationships of Heteroptera [23]. With the development of PCR technology and the use of next-generation sequencing strategies [26,27,28], many complete mitogenome sequences of Hemiptera have been obtained and more will be sequenced (Figure 1a).
Figure 1.
Accumulation of mitogenome data from Hemiptera. (a) The number of species sequenced in each year is represented by the blue line; (b) The number of species sequenced in each suborder is represented by the different pieces of the pie graph.
Figure 1b and Table 1 summarize the mitogenomes of Hemiptera from the first report to the present. The total 90 complete mitogenomes can be divided into five parts by different suborders (Figure 1b) (according to the five suborder system) [12]. Heteroptera has the highest species richness of Hemiptera [29], and more than a half of the 90 complete mitogenomes are from this suborder. Coleorrhyncha, small bugs with a cryptic lifestyle, possess a mixture of cicadomorphan and bug-like characters [30], and represent a separate suborder within Hemiptera. This suborder includes a single extant family, Peloridiidae, which is currently distributed only in Patagonia and on the Australian continent [31]. Only two complete mitogenomes of Peloridiidae have been reported to date [12,32], as representatives of Coleorrhyncha. Meanwhile, the provenances and GenBank numbers of these mitogenomes were detailed set out (Table 1). Most of them as the representatives of different taxa in Hemiptera were published for the first time [12,22,23,33].
Table 1.
All available complete mitogenomes of Hemiptera.
| Suborder | Family | Species | GenBank No. | Reference |
|---|---|---|---|---|
| Cicadomorpha | Aphrophoridae | Philaenus spumarius | NC_005944 | [34] |
| Cercopidae | Abidama producta | NC_015799 | [35] | |
| Cercopidae | Aeneolamia contigua | NC_025495 | [35] | |
| Cercopidae | Callitetix braconoides | NC_025497 | [35] | |
| Cercopidae | Callitetix versicolor | EU725832 | [35] | |
| Cercopidae | Callitettix biformis | NC_025496 | [35] | |
| Cercopidae | Paphnutius ruficeps | NC_021100 | [36] | |
| Cicadellidae | Empoasca vitis | NC_024838 | [37] | |
| Cicadellidae | Homalodisca coagulata | AY875213 | - | |
| Cicadellidae | Homalodisca vitripennis | NC_006899 | * | |
| Membracidae | Leptobelus gazella | NC_023219 | * | |
| Coleorrhyncha | Peloridiidae | Hackeriella veitchi | GQ884145 | [12] |
| Peloridiidae | Hemiodoecus leai | NC_025329 | [32] | |
| Fulgoromorpha | Delphacidae | Laodelphax striatella | JX880068 | [27] |
| Delphacidae | Laodelphax striatellus | NC_013706 | [38] | |
| Delphacidae | Nilaparvata lugens | NC_021748 | [27] | |
| Delphacidae | Nilaparvata muiri | NC_024627 | - | |
| Flatidae | Geisha distinctissima | NC_012617 | [39] | |
| Fulgoridae | Laternaria candelaria | NC_019576 | [40] | |
| Fulgoridae | Lycorma delicatula | NC_012835 | [19] | |
| Issidae | Sivaloka damnosa | NC_014286 | [41] | |
| Ricaniidae | Ricania marginalis | JN242415 | [40] | |
| Heteroptera | Alydidae | Riptortus pedestris | NC_012462 | [23] |
| Anthocoridae | Orius niger | NC_012429 | [23] | |
| Anthocoridae | Orius sauteri | NC_024583 | [42] | |
| Aradidae | Aradacanthia heissi | HQ441233 | [43] | |
| Aradidae | Brachyrhynchus hsiaoi | NC_022670 | [44] | |
| Aradidae | Neuroctenus parus | NC_012459 | [23] | |
| Berytidae | Yemmalysus parallelus | NC_012464 | [23] | |
| Colobathristidae | Phaenacantha marcida | NC_012460 | [23] | |
| Coreidae | Hydaropsis longirostris | NC_012456 | [23] | |
| Cydnidae | Macroscytus gibbulus | EU427338 | [23] | |
| Enicocephalidae | Stenopirates sp. | NC_016017 | [45] | |
| Gelastocoridae | Nerthra indica | NC_012838 | [19] | |
| Geocoridae | Geocoris pallidipennis | NC_012424 | [23] | |
| Gerridae | Aquarius paludum | NC_012841 | [19] | |
| Hydrometridae | Hydrometra greeni | NC_012842 | [19] | |
| Largidae | Physopelta gutta | NC_012432 | [23] | |
| Lygaeidae | Kleidocerys resedae | KJ584365 | [46] | |
| Heteroptera | Malcidae | Chauliops fallax | NC_020772 | [47] |
| Malcidae | Malcus inconspicuus | NC_012458 | [23] | |
| Miridae | Adelphocoris fasciaticollis | NC_023796 | [48] | |
| Miridae | Apolygus lucorum | NC_023083 | [49] | |
| Miridae | Lygus lineolaris | EU401991 | - | |
| Miridae | Nesidiocoris tenuis | NC_022677 | [50] | |
| Nabidae | Alloeorhynchus bakeri | HM235722 | [51] | |
| Nabidae | Gorpis annulatus | NC_019595 | [24] | |
| Nabidae | Gorpis humeralis | NC_019593 | [24] | |
| Nabidae | Nabis apicalis | NC_019594 | [24] | |
| Naucoridae | Ilyocoris cimicoides | NC_012845 | [19] | |
| Nepidae | Laccotrephes robustus | NC_012817 | [19] | |
| Notonectidae | Enithares tibialis | NC_012819 | [19] | |
| Ochteridae | Ochterus marginatus | NC_012820 | [19] | |
| Pentatomidae | Dolycoris baccarum | NC_020373 | [52] | |
| Pentatomidae | Halyomorpha halys | NC_013272 | [53] | |
| Pentatomidae | Nezara viridula | NC_011755 | [23] | |
| Plataspidae | Coptosoma bifaria | NC_012449 | [23] | |
| Plataspidae | megacopta cribraria | NC_015342 | * | |
| Pleidae | Paraplea frontalis | NC_012822 | [19] | |
| Pyrrhocoridae | Dysdercus cingulatus | NC_012421 | [23] | |
| Reduviidae | Agriosphodrus dohrni | NC_015842 | [54] | |
| Reduviidae | Brontostoma colossus | NC_024745 | [28] | |
| Reduviidae | Oncocephalus breviscutum | NC_022816 | [55] | |
| Reduviidae | Peirates arcuatus | NC_024264 | [56] | |
| Reduviidae | Sirthenea flavipes | NC_020143 | [57] | |
| Reduviidae | Triatoma dimidiata | NC_002609 | [9] | |
| Reduviidae | Valentia hoffmanni | NC_012823 | [19] | |
| Rhopalidae | Aeschyntelus notatus | NC_012446 | [23] | |
| Rhopalidae | Stictopleurus subviridis | NC_012888 | - | |
| Saldidae | Saldula arsenjevi | NC_012463 | [23] | |
| Tessaratomidae | Eusthenes cupreus | NC_022449 | [58] | |
| Tingidae | Corythucha ciliata | NC_022922 | [59] | |
| Tingidae | Pseudacysta perseae | NC_025299 | * | |
| Urostylididae | Urochela quadrinotata | NC_020144 | [60] | |
| Sternorrhyncha | Aleyrodidae | Aleurochiton aceris | NC_006160 | [22] |
| Aleyrodidae | Aleurodicus dugesii | NC_005939 | [22] | |
| Aleyrodidae | Bemisia afer | NC_024056 | [25] | |
| Aleyrodidae | Bemisia tabaci | NC_006279 | [22] | |
| Aleyrodidae | Neomaskellia andropogonis | NC_006159 | [22] | |
| Aleyrodidae | Tetraleurodes acaciae | NC_006292 | [22] | |
| Aleyrodidae | Trialeurodes vaporariorum | NC_006280 | [22] | |
| Aphididae | Acyrthosiphon pisum | NC_011594 | * | |
| Aphididae | Aphis gossypii | NC_024581 | [61] | |
| Aphididae | Cavariella salicicola | NC_022682 | [62] | |
| Aphididae | Cervaphis quercus | NC_024926 | [33] | |
| Aphididae | Diuraphis noxia | NC_022727 | [63] | |
| Aphididae | Schizaphis graminum | NC_006158 | [22] | |
| Aphididae | Sitobion avenae | NC_024683 | [64] | |
| Psyllidae | Pachypsylla venusta | NC_006157 | [22] | |
| Psyllidae | Paratrioza sinica | NC_024577 | [65] |
Legend: “-” refer to direct submission; “*” refers to submitted the data and not a published paper.
3. Features of Hemipteran Mitogenomes
3.1. Genome Organization
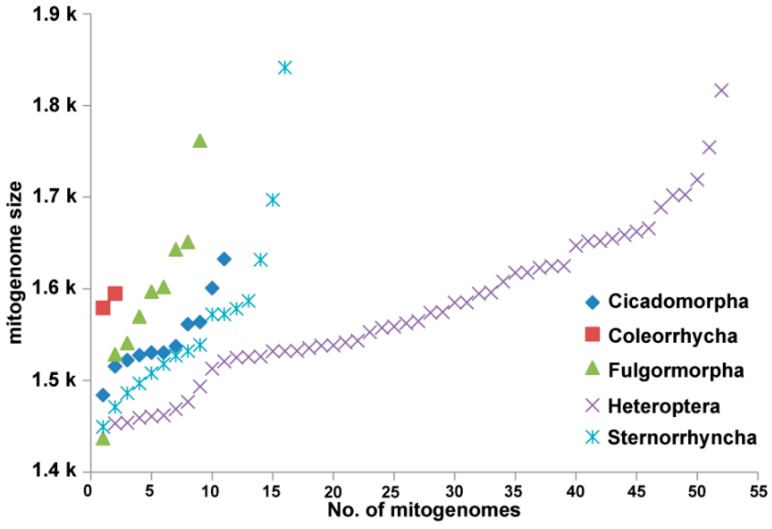
The mitogenome sizes of Hemiptera range from 14,371 bp (Nilaparvata muiri) to 18,414 bp (Trialeurodes vaporariorum) and have an average value of 15,733 bp (Figure 2). The size changes of five suborders are also shown (Figure 2). The size variation is mainly attributed to the non-coding regions, especially the control regions and repeat regions in some groups (such as the control regions of the true water bugs [23] and the repeat regions of aphids [62]).
Figure 2.
The size variation of mitogenomes from Hemiptera.
Most of the mitogenomes (76/90) resemble that of the known ancestral species (D. yakuba [16]) in structural organization and composition with 13 protein coding genes (PCGs), 22 transfer RNAs (tRNAs), and 2 ribosomal RNAs (rRNAs). The remaining mitogenomes differ only in the number of tRNAs, most likely due to gene deletion events. For example, Neomaskellia andropogonis (Sternorrhyncha) contains only 18 tRNAs [22].
3.2. Nucleotide Composition
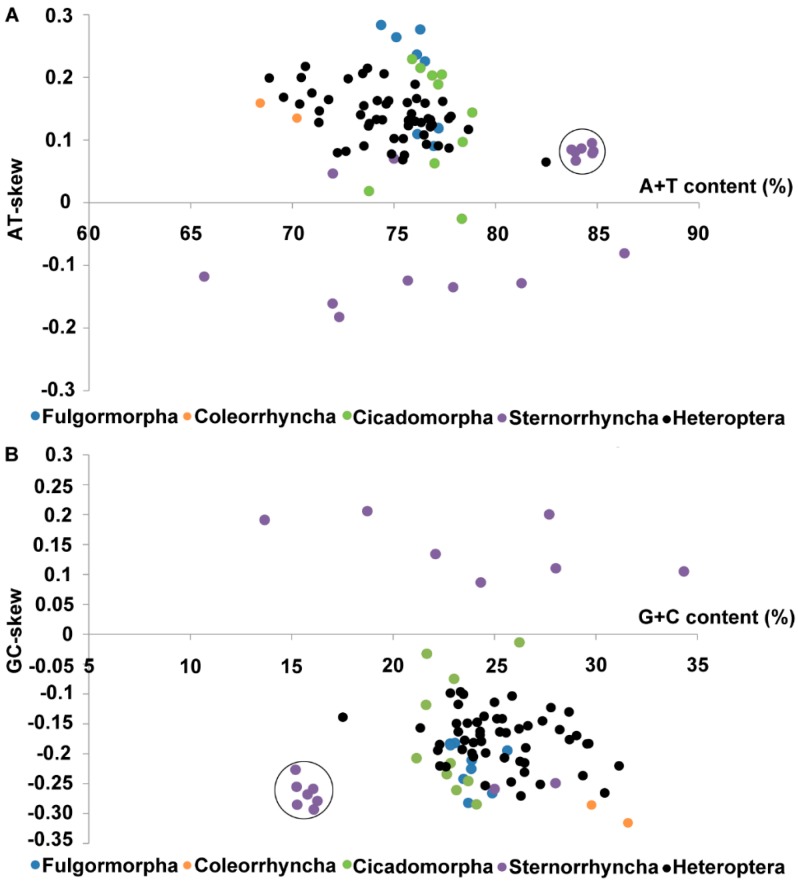
The A%, T%, C% and G% values and the AT and GC skews were calculated for all available complete mitogenomes of Hemiptera species (Figure 3). Interestingly, the lowest and the highest A + T contents of the hemipteran mitogenomes were found in the suborder Sternorrhyncha (65.67% in Bemisia afer and 86.33% in Aleurodicus dugesii). Species from the suborders Fulgoromorpha, Coleorrhyncha and Heteroptera were all A and C skewed. This was also the case for the species of Cicadomorpha, except for Empoasca vitis. For the suborder Sternorrhyncha, nine species were A and C skewed, including all aphid species. This discovery of all aphid species forming a cluster is similar to the results of previous studies (cycle in Figure 3 [62,63]). In contrast, the seven other Sternorrhynchan species (whiteflies), which had highly rearranged gene orders [22,25], were G and T skewed.
Figure 3.
Nucleotide composition across 90 complete hemipteran mitogenomes. (A) A + T content and AT skew; (B) G+C content and GC skew. Dots in the cycle represent values for all the seven aphid mitogenomes.
3.3. Protein-Coding Genes
All PCGs in the majority of hemipteran mitogenomes were initiated with familiar triplet initiation codons (as shown in the invertebrate mitochondrial genetic code table), including the commonly used ATN and some special couplet codons. For instance, in the suborder Coleorrhyncha, cox1 starts with CGA in Xenophyes cascus and with TCG in Hackeriella veitchi [12]. Furthermore, the tetranucleotide initiation codons were also found in hemipteran mitogenomes; such as in Cydnidae where nad2 was supposed to be initiated with an atypical initiation codon, ATCA [23]. In fact, atypical initiation codons are not rare in other insects; for example, the tetranucleotide TTAG is the initiation codon for cox1 of Bombyx mori (Lepidoptera: Bombycidae) [66]. Most PCGs stopped with TAA/TAG termination codons or truncated termination codons (TA or T) that are presumed to be completed via posttranscriptional polyadenylation [67].
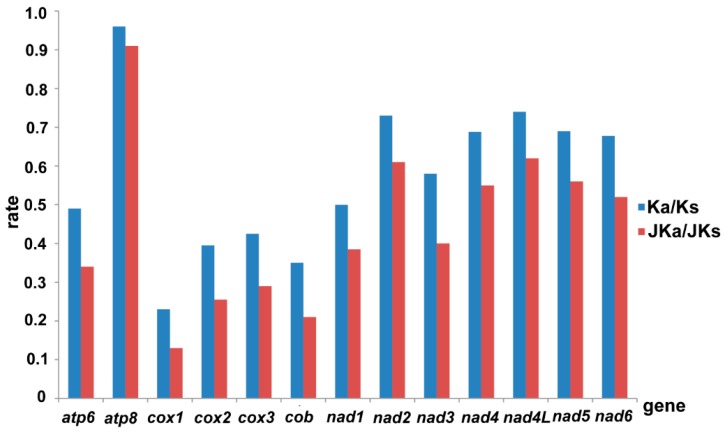
In view of the evolutionary forces acting on the mitochondrial PCGs of hemipteran species, the average rate of non-synonymous substitutions (Ka), the average rate of synonymous substitutions (Ks), the average ratio of Ka/Ks, and the Jukes-Cantor adjusted Ka/Ks (JKa/JKs) were calculated for each PCG, respectively [68]. The results showed that atp8 had the highest evolutionary rate, followed by nad2, while cox1 appeared to be the lowest (Figure 4). Notably, the ratio of Ka/Ks for each PCG was below 1, indicating that these genes are evolving under purifying selection. The uniformly low values of the Ka/Ks and JKa/JKs ratios for cox1–3 and cob indicate strong evolutionary constraints in cytochrome c oxidase [69] and also suggest a strong purifying selection in the species of Hemiptera. Therefore, a DNA barcoding approach based on cox1 sequence diversity has been utilized for identification of closely related species [70]. Similarly, cob and cox2 with relatively slow rates may also be candidate barcoding markers [24,62]. By contrast, due to the highest divergence, atp8 and nad2 can be used as an effective molecular marker to analyze intraspecific relationships and reveal relationships between populations within the same hemipteran species. This result is highly consistent with previous findings in most metazoans [71].
Figure 4.
Evolutionary rates of protein-coding genes in hemipteran mitogenomes. The blue bar indicates the gene’s Ka/Ks, and the red bar indicates the Jukes-Cantor adjusting data.
3.4. tRNAs and rRNAs
All 22 tRNA coding genes usually were found in the mitogenomes of Hemiptera and the tRNAs were between 60 and 75 bp in length. The anticodon nucleotides for the corresponding tRNA genes are identical to those of other available arthropod mitogenomes [66,72]. All tRNA genes had the typical clover-leaf structure with one exception: trnS(AGN), in which the dihydrouridine arm formed a simple loop (as in some other metazoan species, including most insects [66,72,73].
The arrangements of both rrnL and rrnS in the hemipteran mitogenomes are commonly conserved, and are generally located between trnL(CUN) and trnV, and between trnV and the control region. The lengths of rrnL and rrnS are determined to be 1192–1260 and 711–766 bp, respectively. These lengths are similar to those of other orders of Insecta [16,66,72,73].
3.5. Non-Coding Regions
There are some non-coding (NC) regions interspersed throughout the hemipteran mitogenomes, thus the mitogenomes of Hemiptera displayed a moderate size variation. Four distinct large NC regions were identified in the following gene pairs of hemipteran mitogenomes: trnI-trnQ, trnS-nad1, trnE-trnF and rrnS-trnI. The region located between rrnS and trnI, coincided with the A + T-rich region, also called the control region, including the origin of replication and promoters for transcription initiation [16,74,75]. Tandem repeats were detected in the remaining three regions, and named repeat regions.
3.5.1. Control Region
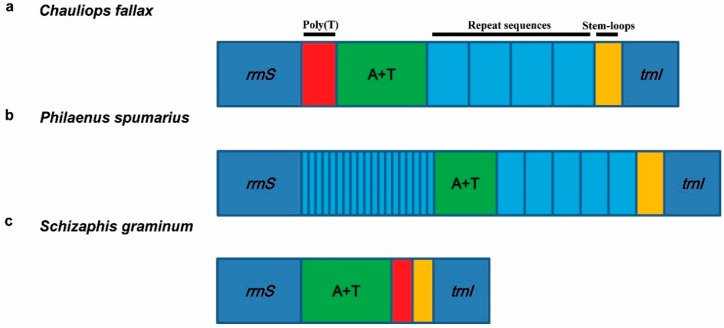
Most control regions of hemipteran mitogenomes were longer than 1 kb, with high rates of nucleotide substitution and indels, and a variable number of tandem repeats. Generally, one control region of the hemipteran mitogenome includes four parts without order: tandem repeat sequences, sequences of poly(T) stretch, a subregion with high A + T content, and stem-loop structures (for example, Chauliops fallax Figure 5a). This feature of the control region was summarized by Cook for arthropods [76]. There are some interesting exceptions in the hemipteran mitogenomes. For example, in some species of Cicadomorpha (Philaenus spumarius) [34], Fulgoromorpha (Geisha distinctissima, Sivaloka damnosa, Laodelphax striatella and Laodelphax striatellus) [27,38,39,41] and Heteroptera (Alloeorhynchus bakeri) [51], two fragments of tandem repeat sequences insert into the control region separately (for example, Philaenus spumarius Figure 5b). A few of the control regions of hemipteran species did not contain all four parts (for example, Schizaphis graminum Figure 5c) [22]. The conserved sequences, stem-loop structures and tandem repeat sequences found in the present study can provide useful information for research of the phylogeny of specific groups [34,35,45,47,62]. For example, in the systematic research of Aphidinae, the phylogenetic tree based on PCGs is similar to the clusters of the stem-loop structures [62]. Another interesting question is how functionality is retained under such great variations in both length and sequence. Considering the high nucleotide substitution rate, both the secondary structures and the conserved segments might be key clues in determining the function of the control region.
Figure 5.
Control regions of mitogenomes from some representative species of Hemiptera. (a) the control region of Chauliops fallax includes four parts; (b) the control region of Philaenus spumarius includes two fragments of tandem repeat sequences; (c) the control region of Schizaphis graminum includes three parts without repeat sequences.
3.5.2. Repeat Region
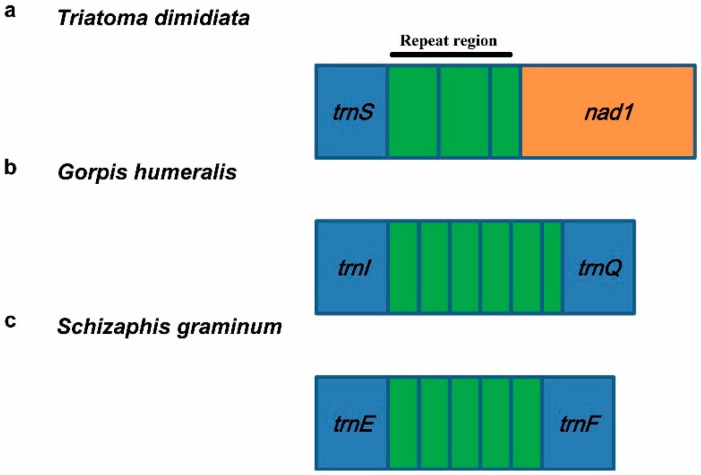
In general, the NC regions of an insect mitogenome consist of a control region and short intergenic spacers. However, some special species of Hemiptera include one repeat region (Figure 6). These repeat regions mainly are located into different positions (trnE-trnF, trnI-trnQ and trnS-nad1) in three families (Aphididae, Nabidae and Reduviidae), and differ in repeat unit sequence and copy number, suggesting that they are highly species-specific (Table 2). These repeat regions are not similar to any known sequences in GenBank. We speculate that this region, full of tandem repeats, has a function similar to the intergenic spacer in Apis mellifera that is thought to be another origin of replication [77].
Figure 6.
Repeat regions of mitogenomes from some representative species of Hemiptera. (a) the repeat region of Triatoma dimidiata (Hemiptera: Heteroptera: Reduviidae); (b) the repeat region of Gorpis humeralis (Hemiptera: Heteroptera: Nabidae); (c) the repeat region of Schizaphis graminum (Hemiptera: Sternorrhyncha: Aphididae).
Table 2.
Repeat regions of hemipteran mitogenomes.
| Species | Classification | Location | Repeat Number | Repeat Unit Size | Reference |
|---|---|---|---|---|---|
| Agriosphodrus dohrni | Heteroptera: Reduviidae | trnS-nad1 | two and a partial | 58 bp | [54] |
| Triatoma dimidiata | Heteroptera: Reduviidae | trnS-nad1 | two and a partial | 135 bp | [9] |
| Gorpis annulatus | Heteroptera: Nabidae | trnS-nad1 | three and a partial | 179 bp | [24] |
| Gorpis humeralis | Heteroptera: Nabidae | trnS-nad1 | two and a partial | 188 bp | [24] |
| Gorpis humeralis | Heteroptera: Nabidae | trnI-trnQ | five and a partial | 244 bp | [24] |
| Himacerus nodipes | Heteroptera: Nabidae | trnI-trnQ | four | 135 bp | [24] |
| Acyrthosiphon pisum | Sternorrhyncha: Aphididae | trnE-trnF | seven and a partial | 203–206 bp | * |
| Aphis gossypii | Sternorrhyncha: Aphididae | trnE-trnF | four and a partial | 196 bp | [61] |
| Cavariella salicicola | Sternorrhyncha: Aphididae | trnE-trnF | three | 199 bp | [62] |
| Diuraphis noxia | Sternorrhyncha: Aphididae | trnE-trnF | three and a partial | 194–195 bp | [63] |
| Schizaphis graminum | Sternorrhyncha: Aphididae | trnE-trnF | four and a partial | 151–153 bp | [22] |
| Sitobion avenae | Sternorrhyncha: Aphididae | trnE-trnF | one and a partial | 202 bp | [64] |
Legend: “*” refers to submitted the data and not a published paper.
4. Hemipteran Mitogenome Arrangements and Evolution
Within Insecta, the order of the mitochondrial genes is highly conserved and has led to the proposal of an ancestral gene order [16]. The majority of hemipteran families that have been sequenced possess this ancestral insect arrangement. Gene orders of Coleorrhyncha and Cicadomorpha are mostly conserved; however, a few families of Fulgoromorpha, Sternorrhyncha and Heteroptera show extreme rearrangement (Table 3). The three rearrangement types of gene movements, transposition, inversion, and inverse transposition [78], were all found in the hemipteran mitogenomes (Table 3). Two species in the superfamily Pyrrhocoroidea share the same gene order with the inversion of trnT and trnP [23]. Whiteflies (Aleyrodidae) are the group that is most likely to have rearrangements: cox3-trnG-nad3 is inverse transposed into three different locations in the mitogenome [22]. In Stenopirates sp., the most striking features were the inversion of two tRNA genes (trnT and trnP) and the transpositions of five gene clusters (trnT-trnP-nad6, cob-trnS, nad1-trnL, rrnL-trnV-rrnS and control region) between nad4L and trnI [45]. The four hot spots of rearrangements are summarized: 1. upstream of nad2; 2. between nad2 and cox1; 3. between nad4L and nad1; and 4. downstream of rrnL (Table 3).
Table 3.
Mitogenome rearrangements found in Hemiptera.
| Classification | Species | Level | Rearrangement | Reference |
|---|---|---|---|---|
| Fulgoromorpha: Delphacidae | Laodelphax striatella | family | Inversion of trnC and trnW, inverse transposition: trnT-trnP-nad6 → nad6-trnP-trnT | [27] |
| Fulgoromorpha: Delphacidae | Laodelphax striatellus | family | Inversion of trnC and trnW, transposition of trnH, and inverse transposition: trnT-trnP-nad6 → nad6-trnP-trnT | [38] |
| Fulgoromorpha: Delphacidae | Nilaparvata lugens | family | Inversion of trnC and trnW, inverse transposition: trnT-trnP-nad6 → nad6-trnP-trnT, and insertion two trnC | [27] |
| Heteroptera: Aradidae | Aradacanthia heissi | species | Inversion of trnI and trnQ, inversion of trnC and trnW | [43] |
| Heteroptera: Aradidae | Brachyrhynchus hsiaoi | genus | Inversion of trnI and trnQ | [44] |
| Heteroptera: Aradidae | Neuroctenus parus | genus | Inversion of trnI and trnQ | [23] |
| Heteroptera: Enicocephalidae | Stenopirates sp. | species | Inversion of trnT and trnP, inverse transposition: trnT-trnP-nad6-cytB-trnS-nad1-trnL-rrnL-trnV-rrnS-control region → cytB-trnS-control region-rrnL-trnV-rrnS-nad1-trnL-trnP-trnT-nad6 | [45] |
| Heteroptera: Largidae | Physopelta gutta | superfamily | Inversion of trnT and trnP | [23] |
| Heteroptera: Pyrrhocoridae | Dysdercus cingulatus | superfamily | Inversion of trnT and trnP | [23] |
| Sternorrhyncha: Aleyrodidae | Aleurochiton aceris | genus | Inversion of trnC and trnY, inverse transposition: cox3-trnG-nad3 → insertion the location cob-nad1 | [22] |
| Sternorrhyncha: Aleyrodidae | Aleurodicus dugesii | genus | Inversion of trnC and trnY | [22] |
| Sternorrhyncha: Aleyrodidae | Bemisia afer | genus | Inversion of trnC and trnY, transposition of trnQ, and inverse transposition: cox3-trnG-nad3 → insertion the location control region-rrnS | [25] |
| Sternorrhyncha: Aleyrodidae | Bemisia tabaci | genus | Inversion of trnC and trnY, transposition of trnQ, and inverse transposition: cox3-trnG-nad3 → insertion the location control region-rrnS | [22] |
| Sternorrhyncha: Aleyrodidae | Neomaskellia andropogonis | genus | Transposition of trnH and trnK, and inverse transposition: cox3-trnG-nad3 → insertion the location rrnL-rrnS | [22] |
| Sternorrhyncha: Aleyrodidae | Tetraleurodes acaciae | genus | Inversion of trnC and trnY, transposition of trnQ and trnA, and inverse transposition: cox3-trnG-nad3 → insertion the location control region-rrnS | [22] |
| Sternorrhyncha: Aleyrodidae | Trialeurodes vaporariorum | genus | Inversion of trnI and trnQ, inversion of trnC and trnY, and transposition of trnG | [22] |
Rearrangements of the mitogenomes are relatively rare events at the evolutionary scale [17]. Therefore, they can be powerful tool to delimit deep divergences among some insect lineages. The first discovery in this aspect of Hemiptera was found in whitefly in 2004. Thao et al. determined the complete mitogenomes of six whitefly species and their results indicated a clustering of whitefly species that corresponded to the gene arrangement types [22]. Then, in 2009, a comparison of gene orders and contents revealed that Hemiptera had three conserved gene blocks shared by all 20 species [53]. Gene orders and contents of both Heteroptera and Auchenorrhyncha (Cicadomorpha and Fulgoromorpha) were mostly conserved, whereas those of Sternorrhyncha showed extreme rearrangement [53]. However, compared with Laodelphax striatella [27], Stenopirates sp. [45] and Aradacanthia heissi [43], rearrangements in species of Fulgoromorpha and other true bugs seem to occur independently of family or species (Table 3). These results suggest that mitogenome orders might lack the resolution to deduce phylogenetic relationships among infraorders within Fulgoromorpha and Heteroptera.
5. Phylogenetic Inferences by Hemipteran Mitogenomes
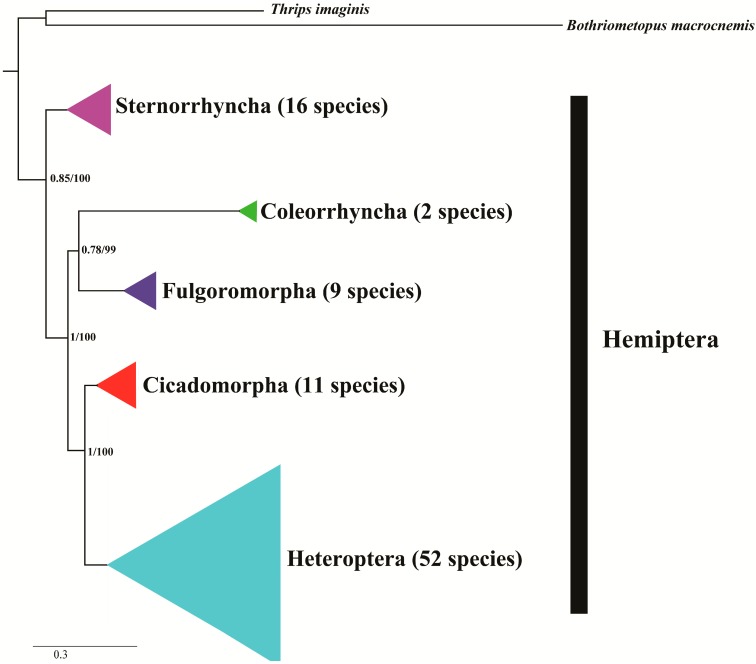
As mentioned in the introduction, the phylogenic relationship of the Hemiptera has been controversial for many years and two questions remain unanswered. Here, we reviewed the research history of hemipteran phylogenetic relationships based on mitogenomes and combine our phylogenetic analyses to discuss the most reliable results. In 2009, a study clarified the relationships of the three phylogenetically controversial suborders, Auchenorrhyncha, Sternorrhyncha, and Heteroptera [53]. Heteroptera constituted a monophyletic group, and a sister relationship was proposed for Auchenorrhyncha and Sternorrhyncha [53]. However, only one species (Cicadomorpha: Philaenus spumarius) was chosen representing Auchenorrhyncha, and no taxa of Fulgoromorpha were discussed. Therefore, in the same year, Song and Liang [38] increased the samplings of taxa and proposed the inferred genealogical proximities of hemipteran lineages of (Heteroptera + (Cicadomorpha + (Fulgoromorpha + Sternorrhyncha))). In their research, Auchenorrhyncha was clearly separated into two parts, and Fulgoromorpha and Cicadomorpha were not a monophyletic group [38]. In fact, in their reports (in 2010 and 2012), the paraphyly of Auchenorrhyncha was also supported [40,41], and their phylogenetic reconstruction supported a sister relationship between Fulgoromorpha and Sternorrhyncha [40]. The suborder Coleorrhyncha (Hemiptera) has only one extant family, Peloridiidae, comprising 36 species in 17 genera [79]. Species of this group live in the wet mosses of South America (Chile, Argentina), New Zealand, New Caledonia and eastern Australia (from North Queensland to Tasmania) [80]. Complete or nearly complete mitogenomes of Peloridiidae were not obtained until 2013 [12]. Cui’s research was the first phylogenomic study of hemipterans with complete suborder samplings. Their results supported the paraphyly of Auchenorrhyncha and proposed the close relationship between Cicadomorpha and Heteroptera [12]. Meanwhile, our result displayed the similar result (Figure 7): Sternorrhyncha located as the basal suborder and Cicadomorpha and Heteroptera clustered as sister-group. Summarizing all these viewpoints, we can make three conclusions. First, the phylogenetic relationships among suborder-level hemipteran linages remain unclear by using mitogenome inference. Most viewpoints supported that Auchenorrhyncha is not a monophyletic group; Second, whether a monophyletic group or a sister-group to Cicadomorpha, the suborder Heteroptera is the most evolved group of Hemiptera; Third, the positions of other suborders remain confused and require further investigation.
Figure 7.
ML and BI Phylogenetic tree inferred from 90 hemipteran mitogenome sequences based on 13 PCGs. The node support values are the Bayesian posterior probabilities (BPP) and the bootstrap (BS) values.
Under the suborder taxa, the phylogenetic research also was involved. We summarized all the research results of these years (Table 4). All the phylogenetic issues of every taxonomic category were considered. For example, in the relationships among the intraorders of Heteroptera, Enicocephalomorpha was the most basal sister-group of the majority of Heteroptera [45]; the position of Cimicomorpha was unclear and it is possible that it is not a monophyletic group [45,47]; and Pentatomomorpha was the most evolved group of Heteroptera [45,47,49,54,60]. Regarding interfamily relationships, Hua et al. conducted phylogenomic studies on the mitogenomes of Pentatomomorpha [23] and Nepomorpha [19], and resolved some superfamily phylogenetic problems (Table 4). In Sternorrhyncha, the mitochondrial gene rearrangements among whiteflies corresponded to the phylogenetic tree [22]. The intrasubfamily relationships of Aphids (a group with special regions [33,63]), were also discussed [62] (Table 4).
Table 4.
The phylogenetic analyses under the suborder taxa of Hemiptera.
| Classification | Level | Viewpoint | Reference |
|---|---|---|---|
| Cicadomorpha: Cercopidae and Aphrophoridae | family | The monophyly of five Callitettixini species. | [35] |
| Sternorrhyncha: Aleyrodidae, whiteflies | genus | Four types of the mitochondrial gene rearrangements among whiteflies were corresponding to the branches of phylogenetic tree. | [22] |
| Sternorrhyncha: Aphididae, aphids | subfamily | Treat pterocommatines as members of Macrosiphini. | [62] |
| Heteroptera: Reduviidae | subfamily | The monophyly of Reduviidae and the Peiratinae presents a sister position to the Triatominae + (Salyavatinae + Harpactorinae). | [57] |
| Heteroptera: Pentatomomorpha | superfamily | The monophyly of Pentatomoidea, Pyrrhocoroidea, Lygaeoidea, and Coreoidea; Aradoidea and the Trichophora are sister groups. | [23] |
| Heteroptera: Nepomorpha | superfamily | Pleoidea is not a member of the Nepomorpha and Aphelocheiroidea should be grouped back into Naucoroidea. | [19] |
| Heteroptera: Nabidae | subfamily | Three tribes from two subfamilies of Nabidae. | [24] |
| Heteroptera | intraorder | The paraphyly of Cimicomorpha, and within Reduviidae, Harpactorinae is a sister group to the Salyavatinae + Triatominae. | [54] |
| Heteroptera | intraorder | The paraphyly of Cimicomorpha, and Reduviidae was paraphyletic with respect to Anthocoridae and Miridae. | [60] |
| Heteroptera | intraorder | The sister-relationship within the individual infraorders are supported for the Pentatomomorpha, Nepomorpha, Leptopodomorpha and Gerromorpha; Stenopirates sp. (Enicocephalomorpha) is the sister group to all the remaining Heteroptera. | [45] |
| Heteroptera | intraorder | Two Gerromorpha superfamilies were monophyletic in the basal position of these five infraorders. Within Cimicomorpha, Reduviidae was paraphyletic with respect to Anthocoridae and Miridae. | [47] |
| Heteroptera | intraorder | Stenopirates sp. was the sister group to all the remaining Heteroptera; the sister relationships within Nepomorpha and Gerromorpha. | [49] |
In conclusion, the present study shows that mitogenomes may be good molecular markers for phylogenetic inference at a variety of taxonomic levels of Hemiptera (such as suborders, intraorders and families). However, some relationships have not been resolved based solely on mitogenomes. Nuclear genes evolve more slowly, and are effective for the analysis of deeper phylogenetic relationships. Moreover, some endosymbionts co-evolve with their hosts, and symbiont-derived data, in principle, could be used to reconstruct the evolutionary history of hosts [81]. So, with the development of sequencing technology, more available genetic resources are expected to provide more effective information of phylogenetic trees.
6. Experimental Section
6.1. Sampling
A total of 90 taxa were sampled in this study (Table 1). In the phylogeny analyses, the outgroups were sampled from Phthiraptera (Bothriometopus macrocnemis, GenBank accession number: NC_009983) and Thysanoptera (Thrips imaginis, GenBank accession number: NC_004371).
6.2. Analysis of Sequence Data
The nucleotide sequences of PCGs were translated based on the invertebrate mtDNA genetic code. A + T content were calculated using MEGA version 6.0 [82]. Strand asymmetry was calculated using the formulae AT skew = [A − T]/[A + T] and GC skew = [G − C]/[G + C], for the strand encoding the majority of the protein-coding genes. The software packages DnaSP 5.0 [83] was used to calculate the synonymous substitution rate (Ks) and the nonsynonymous substitution rate (Ka) for each PCG as well as Jukes-Cantor adjusted Ka/Ks (JKa/JKs).
6.3. Phylogenetic Analysis
Each of the 13 PCGs of all 92 species were aligned individually using MEGA v6.0 [82] with default parameters. Before alignments, the stop codons were all removed from those sequences. Maximum likelihood (ML) and Bayesian inference (BI) analyses were implemented by PHYML 3.0 [84] and MrBayes version 3.1.2 [85], respectively. Model selection was based on jModeltest v0.1.1 [86]. According to the AIC, the GTR + I + G model was optimal for analysis with nucleotide alignments. MrBayes version 3.1.2 and PHYML were employed to reconstruct the phylogenetic trees. In the ML analysis, the parameters were estimated during analysis and the node support values were assessed by bootstrap re-sampling (BP) calculated using 100 replicates. In Bayesian inference, runs of ten million generations were conducted. Trees were sampled every 1000 generations with a burn-in of 25%.
7. Conclusions and Perspectives
Generally, the complete mitogenomes of Hemiptera were 14–17K bp in size and encoded all 37 genes typical for insects. These genes were arranged in the same order as the proposed ancestral insect, except in a few particular species. Notably, the mitogenomes of three families possessed a large repeat region located at three different positions. We speculate that this region, full of tandem repeats, is another origin of replication. The mitogenomes have been successfully used to reconstruct the phylogenetic relationships within a variety of taxonomic levels of Hemiptera.
Future work should focus on four goals. First, the comparative genomics of different categories need more taxon samplings and more mitogenome sequences to further describe the comprehensive characteristics of Hemiptera mitogenomes; Second, the research of various populations and phylogeographic structures of hemipteran species based on mitogenomes require more mitogenome sequences about the same species or similar species; Third, the functional and evolutionary significance of different rearrangement types should be examined to open the view of the evolutionary dynamics of Hemiptera mitogenomes; Finally, phylogenetic inference with more resource data will provide greater insight into the evolution of Hemiptera.
Acknowledgments
We want to thank Li Hu and Cui Ying for the help of data analysis and viewpoints discussion with this review. The work was supported by National Natural Sciences Foundation of China (Nos. 31430078, 31372237), the External Cooperation Program of Chinese Academy of Sciences (No. 152111KYSB20130012), the Ministry of Science and Technology of the China (MOST Grant Nos. 2011FY120200, 2014FY210200), and National Science Fund for Fostering Talents in Basic Research (No. J1210002).
Author Contributions
All authors contributed to writing this review.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Linnaeus C. Systema naturae per regna tria naturae secundum classes ordines genera species, cum characteribus differentiis synonymis locis; Proceedings of the 10th edn Reformata, Salviae Holmiae; Stockholm, Sweden. 1758. [Google Scholar]
- 2.Latreille P.A. Considérations Generales sur l’ordre naturel des Animaux Composant les Classes des Crustaces, des Arachnides, et des Insectes; avec un Tableau Methodique de Leurs Genres, disposes en Familles. Chez F. Schoell; Paris, France: 1810. [Google Scholar]
- 3.Hennig W. Die Stammesgeschichte der Insekten. Waldemar Kramer; Frankfurt am Main, Germany: 1969. [Google Scholar]
- 4.Kristensen N.P. Insects of Australia. 2nd ed. Cornell University Press; Ithaca, NY, USA: 1991. Phylogeny of Extant Hexapods; pp. 125–140. [Google Scholar]
- 5.Yoshizawa K., Saigusa T. Phylogenetic analysis of paraneopteran orders (Insecta: Neoptera) based on forewing base structure, with comments on monophyly of Auchenorrhyncha (Hemiptera) Syst. Entomol. 2001;26:1–13. doi: 10.1046/j.1365-3113.2001.00133.x. [DOI] [Google Scholar]
- 6.Schuh R.T., Slater J.A. True Bugs of the World (Hemiptera: Heteroptera) Classifiation and Natural History. Cornell University Press; Ithaca, NY, USA: 1995. [Google Scholar]
- 7.Jon M., Mick W. Hemiptera It’s A Bug’s Life. Natural History Museum; Visitado em, Germany: 2010. [Google Scholar]
- 8.Dyck V.A., Thomas B. The brown planthopper problem. IRRI. Los. Banos. Philippines. 1979;369:3–17. [Google Scholar]
- 9.Dotson E.M., Beard C.B. Sequence and organization of the mitochondrial genome of the Chagas disease vector, Triatoma dimidiata. Insect Mol. Biol. 2001;10:205–215. doi: 10.1046/j.1365-2583.2001.00258.x. [DOI] [PubMed] [Google Scholar]
- 10.Carver M., Gross G.F., Woodward T.E. Hemipter (Bugs, Leafhoppers, Cicadas, Aphids, Scale Insects, etc.) Melbourne University Press; Victoria, Australia: 1991. pp. 429–509. [Google Scholar]
- 11.Evans J.W. The phylogeny of the Homoptera. Annu. Rev. Entomol. 1963;8:77–94. doi: 10.1146/annurev.en.08.010163.000453. [DOI] [Google Scholar]
- 12.Cui Y., Xie Q., Hua J.M., Dang K., Zhou J., Liu X., Wang G., Yu X., Bu W.J. Phylogenomics of Hemiptera (Insecta: Paraneoptera) based on mitochondrial genomes. Syst. Entomol. 2013;38:233–245. doi: 10.1111/j.1365-3113.2012.00660.x. [DOI] [Google Scholar]
- 13.Cobben R.H. Evolutionary Trends in Heteroptera II Mouthpartstructures and Feeding Strategies. Mededelingen Landbouwhogeschool; Wageningen, The Netherlands: 1978. [Google Scholar]
- 14.Hamilton K.G.A. Morphology and evolution of the rhynchotan head (Insecta: Hemiptera, Homoptera) Can. Entomol. 1981;113:953–974. doi: 10.4039/Ent113953-11. [DOI] [Google Scholar]
- 15.Popov Y.A., Shcherbakov D.E. Mesozoic Peloridioidea and their ancestors (Insecta: Hemiptera, Coleorrhyncha) Geol. Palaeontol. 1991;25:215–235. [Google Scholar]
- 16.Clary D.O., Wolstenholme D.R. The mitochondrial DNA molecular of Drosophila yakuba: Nucleotide sequence, gene organization and genetic code. J. Mol. Evol. 1985;22:252–271. doi: 10.1007/BF02099755. [DOI] [PubMed] [Google Scholar]
- 17.Cameron S.L. Insect mitochondrial genomics: Implications for evolution and phylogeny. Annu. Rev. Entomol. 2014;59:95–117. doi: 10.1146/annurev-ento-011613-162007. [DOI] [PubMed] [Google Scholar]
- 18.Boore J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999;27:1767–1780. doi: 10.1093/nar/27.8.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hua J.M., Li M., Dong P.Z., Cui Y., Xie Q., Bu W.J. Phylogenetic analysis of the true water bugs (Insecta: Hemiptera: Heteroptera: Nepomorpha): Evidence from mitochondrial genomes. BMC Evol. Biol. 2009;9:134. doi: 10.1186/1471-2148-9-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma C., Yang P.C., Jiang F., Chapuis M.-P., Shall Y., Sword G.A., Kang L. Mitochondrial genomes reveal the global phylogeography and dispersal routes of the migratory locust. Mol. Ecol. 2012;21:4344–4458. doi: 10.1111/j.1365-294X.2012.05684.x. [DOI] [PubMed] [Google Scholar]
- 21.Nelson L.A., Lambkin C.L., Batterham P., Wallman J.F., Dowton M., Whiting M.F., Yeates D.K., Cameron S.L. Beyond barcoding: a mitochondrial genomics approach to molecular phylogenetics and diagnostics of blowflies (Diptera: Calliphoridae) Gene. 2012;511:131–142. doi: 10.1016/j.gene.2012.09.103. [DOI] [PubMed] [Google Scholar]
- 22.Thao M.L., Baumann L., Baumann P. Organization of the mitochondrial genomes of whiteflies, aphids, and psyllids (Hemiptera, Sternorrhyncha) BMC Evol. Biol. 2004;4:25. doi: 10.1186/1471-2148-4-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hua J.M., Li M., Dong P.Z., Cui Y., Xie Q., Bu W.J. Comparative and phylogenomic studies on the mitochondrial genomes of Pentatomomorpha (Insecta: Hemiptera: Heteroptera) BMC Genomics. 2008;9:610. doi: 10.1186/1471-2164-9-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H., Liu H.Y., Song F., Shi A.M., Zhou X.G., Cai W.Z. Comparative mitogenomic analysis of damsel bugs representing three tribes in the family Nabidae (Insecta: Hemiptera) PLoS ONE. 2012;7:e45925. doi: 10.1371/journal.pone.0045925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang H.L., Xiao N., Yang J., Wang X.W., Colvin J., Liu S.S. The complete mitochondrial genome of Bemisia afer (Hemiptera: Aleyrodidae) Mitochondrial DNA. 2014 doi: 10.3109/19401736.2013.873921. [DOI] [PubMed] [Google Scholar]
- 26.The International Aphid Genomics Consortium Genome sequence of the pea aphid Acyrthosiphon pisum. PLoS Biol. 2010;8:e1000313. doi: 10.1371/journal.pbio.1000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang K.J., Zhu W.C., Rong X., Zhang Y.K., Ding X.L., Liu J., Chen D.S., Du Y., Hong X.Y. The complete mitochondrial genomes of two rice planthoppers, Nilaparvata. lugens and Laodelphax striatellus: Conserved genome rearrangement in Delphacidae and discovery of new characteristics of atp8 and tRNA genes. BMC Genomics. 2013;14:417. doi: 10.1186/1471-2164-14-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kocher A., Kamilari M., Lhuillier E., Coissac E., Péneau J., Chave J., Murienne J. Shotgun assembly of the assassin bug Brontostoma colossus mitochondrial genome (Heteroptera, Reduviidae) Gene. 2014;552:184–194. doi: 10.1016/j.gene.2014.09.033. [DOI] [PubMed] [Google Scholar]
- 29.Weirauch C., Schuh R.T. Systematics and evolution of Heteroptera: 25 years of progress. Annu. Rev. Entomol. 2011;56:487–510. doi: 10.1146/annurev-ento-120709-144833. [DOI] [PubMed] [Google Scholar]
- 30.Bechly G., Szwedo J. Coleorrhyncha: Moss bugs. In: Martill D.M., Bechly G., Loveridge R.F., editors. The Crato. Fossil Beds of Brazil: Window into an Ancient World. Cambridge University Press; Cambridge, UK: 2007. pp. 313–317. [Google Scholar]
- 31.Burckhardt D., Bochud E., Damgaard J., W.gibbs G., Hartung V., Larivière M.C., Wyniger D., Zuercher I. A review of the moss bug genus Xenophyes (Hemiptera: Coleorrhyncha: Peloridiidae) from New Zealand: Systematics and biogeography. Zootaxa. 2011;2923:1–26. [Google Scholar]
- 32.Gao J., Liang A.P. The complete mitochondrial genome of Hemiodoecus leai (Hemiptera: Coleorrhyncha: Peloridiidae) Mitochondrial DNA. 2014 doi: 10.3109/19401736.2014.953074. [DOI] [PubMed] [Google Scholar]
- 33.Wang Y., Huang X.L., Qiao G.X. The complete mitochondrial genome of Cervaphis quercus (Insecta: Hemiptera: Aphididae: Greenideinae) Insect Sci. 2014;21:278–290. doi: 10.1111/1744-7917.12112. [DOI] [PubMed] [Google Scholar]
- 34.Stewart J.B., Beckenbach A.T. Insect mitochondrial genomics: The complete mitochondrial genome sequence of the meadow spittlebug Philaenus spumarius (Hemiptera: Auchenorrhyncha: Cercopoidae) Genome. 2005;48:46–54. doi: 10.1139/g04-090. [DOI] [PubMed] [Google Scholar]
- 35.Liu J., Bu C.P., Wipfler B., Liang A.P. Comparative analysis of the mitochondrial genomes of callitettixini spittlebugs (Hemiptera: Cercopidae) confirms the overall high evolutionary speed of the at-rich region but reveals the presence of short conservative elements at the tribal level. PLoS ONE. 2014;9:e109140. doi: 10.1371/journal.pone.0109140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu J., Liang A.P. The complete mitochondrial genome of spittlebug Paphnutius ruficeps (Insecta: Hemiptera: Cercopidae) with a fairly short putative control region. Acta Biochim. Biophys. Sin. 2013;45:309–319. doi: 10.1093/abbs/gmt009. [DOI] [PubMed] [Google Scholar]
- 37.Zhou N.N., Wang M.X., Cui L., Chen X.X., Han B.Y. Complete mitochondrial genome of Empoasca vitis (Hemiptera: Cicadellidae) Mitochondrial DNA. 2014 doi: 10.3109/19401736.2014.928863. [DOI] [PubMed] [Google Scholar]
- 38.Song N., Liang A.P. Complete mitochondrial genome of the small brown planthopper, Laodelphax striatellus (Delphacidae: Hemiptera), with a novel gene order. Zool. Sci. 2009;26:851–860. doi: 10.2108/zsj.26.851. [DOI] [PubMed] [Google Scholar]
- 39.Song N., Liang A.P. The complete mitochondrial genome sequence of Geisha distinctissima (Hemiptera: Flatidae) and comparison with other Hemipteran insects. Acta Biochim. Biophys. Sin. 2009;41:206–216. doi: 10.1093/abbs/gmp003. [DOI] [PubMed] [Google Scholar]
- 40.Song N., Liang A.P., Bu C.P. A molecular phylogeny of Hemiptera inferred from mitochondrial genome sequences. PLoS ONE. 2012;7:e48778. doi: 10.1371/journal.pone.0048778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Song N., Liang A.P., Ma C. The complete mitochondrial genome sequence of the planthopper, Sivaloka damnosus. J. Insect. Sci. 2010;10:76. doi: 10.1673/031.010.7601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Du B.Z., Niu F.F., Wei S.J. The complete mitochondrial genome of the predatory bug Orius sauteri (Poppius) (Hemiptera: Anthocoridae) Mitochondrial DNA. 2014 doi: 10.3109/19401736.2014.915537. [DOI] [PubMed] [Google Scholar]
- 43.Shi A.M., Li H., Bai X.S., Dai X., Chang J., Guilbert E., Cai W.Z. The complete mitochondrial genome of the flat bug Aradacanthia heissi (Hemiptera: Aradidae) Zootaxa. 2012;3238:23–38. [Google Scholar]
- 44.Li H., Shi A.M., Song F., Cai W.Z. Complete mitochondrial genome of the flat bug Brachyrhynchus. hsiaoi (Hemiptera: Aradidae) Mitochondrial DNA. 2014 doi: 10.3109/19401736.2013.867437. [DOI] [PubMed] [Google Scholar]
- 45.Li H., Liu H., Shi A.M., Stys P., Zhou X.G., Cai W.Z. The complete mitochondrial genome and novel gene arrangement of the unique-headed bug Stenopirates sp. (Hemiptera: Enicocephalidae) PLoS ONE. 2012;7:e29419. doi: 10.1371/journal.pone.0029419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li T., Yi W.B., Zhang H.G., Xie Q., Bu W.J. Complete mitochondrial genome of the birch catkin bug Kleidocerys resedae resedae, as the first representative from the family Lygaeidae (Hemiptera: Heteroptera: Lygaeoidea) Mitochondrial DNA. 2014 doi: 10.3109/19401736.2014.908372. [DOI] [PubMed] [Google Scholar]
- 47.Li T., Gao C., Cui Y., Xie Q., Bu W.J. The complete mitochondrial genome of the stalk-eyed bug Chauliops fallax scott, and the monophyly of malcidae (Hemiptera: Heteroptera) PLoS ONE. 2013;8:e55381. doi: 10.1371/journal.pone.0055381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Y., Li H., Xun H.Z., Cai W.Z. Complete mitochondrial genome sequence of the plant bug Adelphocoris fasciaticollis (Hemiptera: Heteroptera: Miridae) Mitochondrial DNA. 2014 doi: 10.3109/19401736.2014.880898. [DOI] [PubMed] [Google Scholar]
- 49.Wang P., Li H., Wang Y., Zhang J.H., Dai X., Chang J., Hu B.W., Cai W.Z. The mitochondrial genome of the plant bug Apolygus lucorum (Hemiptera: Miridae): Presently known as the smallest in Heteroptera. Insect Sci. 2014;21:159–173. doi: 10.1111/1744-7917.12029. [DOI] [PubMed] [Google Scholar]
- 50.Dai X., Xun H.Z., Chang J., Zhang J.H., Hu B.W., Li H., Yuan X.Q., Cai W.Z. The complete mitochondrial genome of the plant bug Nesidiocoris tenuis (Reuter) (Hemiptera: Miridae: Bryocorinae: Dicyphini) Zootaxa. 2012;3554:30–44. [Google Scholar]
- 51.Li H., Liu H.Y., Cao L.M., Shi A.M., Yang H.L., Cai W.Z. The complete mitochondrial genome of the damsel bug Alloeorhynchus bakeri (Hemiptera: Nabidae) Int. J. Biol. Sci. 2012;8:93–107. doi: 10.7150/ijbs.8.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Q.L., Yuan M.L., Shen Y.Y. The complete mitochondrial genome of Dolycoris baccarum (Insecta: Hemiptera: Pentatomidae) Mitochondrial DNA. 2013 doi: 10.3109/19401736.2013.766182. [DOI] [PubMed] [Google Scholar]
- 53.Lee W., Kang J., Jung C., Hoelmer K., Lee S., Lee S. Complete mitochondrial genome of brown marmorated stink bug Halyomorpha halys (Hemiptera: Pentatomidae), and phylogenetic relationships of hemipteran suborders. Mol. Cells. 2009;28:155–165. doi: 10.1007/s10059-009-0125-9. [DOI] [PubMed] [Google Scholar]
- 54.Li H., Gao J.Y., Liu H.Y., Liu H., Liang A.P., Cai W.Z. The architecture and complete sequence of mitochondrial genome of an assassin bug Agriosphodrus dohrni (Hemiptera: Reduviidae) Int. J. Biol. Sci. 2011;7:792–804. doi: 10.7150/ijbs.7.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li H., Gao J.Y., Cai W.Z. Complete mitochondrial genome of the assassin bug Oncocephalus breviscutum (Hemiptera: Reduviidae) Mitochondrial DNA. 2013 doi: 10.3109/19401736.2013.840602. [DOI] [PubMed] [Google Scholar]
- 56.Zhao G.Y., Li H., Song F., Cai W.Z. The complete mitochondrial genome of an assassin bug Peirates Arcuatus (Hemiptera: Reduviidae) Mitochondrial DNA. 2014 doi: 10.3109/19401736.2013.867438. [DOI] [PubMed] [Google Scholar]
- 57.Gao J.Y., Li H., Truong X.L., Dai X., Chang J., Cai W.Z. Complete nucleotide sequence and organization of the mitochondrial genome of Sirthenea flavipes (Hemiptera: Reduviidae: Peiratinae) and comparison with other assassin bugs. Zootaxa. 2013;3699:1–16. doi: 10.11646/zootaxa.3669.1.1. [DOI] [PubMed] [Google Scholar]
- 58.Song W., Li H., Song F., Liu L., Wang P., Xun H., Dai X., Chang J., Cai W.Z. The complete mitochondrial genome of a tessaratomid bug, Eusthenes cupreus (Hemiptera: Heteroptera: Pentatomomorpha: Tessaratomidae) Zootaxa. 2013;3620:260–272. doi: 10.11646/zootaxa.3620.2.4. [DOI] [PubMed] [Google Scholar]
- 59.Yang W., Yu W.W., Du Y.Z. The complete mitochondrial genome of the sycamore lace bug Corythucha ciliata (Hemiptera: Tingidae) Gene. 2013;532:27–40. doi: 10.1016/j.gene.2013.08.087. [DOI] [PubMed] [Google Scholar]
- 60.Dai Y.T., Li H., Jiang P., Song F., Ye Z., Yuang X., Dai X., Chang J., Cai W.Z. Sequence and organization of the mitochondrial genome of an urostylidid bug, Urochela. quadrinotata Reuter (Hemiptera: Urostylididae) Entomotaxonomia. 2012;34:613–623. [Google Scholar]
- 61.Zhang S., Luo J., Wang C., Lv L., Li C., Jiang W., Cui J., Rajput L.B. Complete mitochondrial genome of Aphis gossypii Glover (Hemiptera: Aphididae) Mitochondrial DNA. 2014 doi: 10.3109/19401736.2014.919474. [DOI] [PubMed] [Google Scholar]
- 62.Wang Y., Huang X.L., Qiao G.X. Comparative analysis of mitochondrial genomes of five aphid species (Hemiptera: Aphididae) and phylogenetic implications. PLoS ONE. 2013;8:e77511. doi: 10.1371/journal.pone.0077511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang B., Ma C., Edwards O., Fuller S., Kang L. The mitochondrial genome of the Russian wheat aphid Diuraphis noxia: Large repetitive sequences between trnE and trnF in aphids. Gene. 2014;533:253–260. doi: 10.1016/j.gene.2013.09.064. [DOI] [PubMed] [Google Scholar]
- 64.Zhang B., Zheng J., Liang L., Fuller S., Ma C.S. The complete mitochondrial genome of Sitobion avenae (Hemiptera: Aphididae) Mitochondrial DNA. 2014 doi: 10.3109/19401736.2014.926498. [DOI] [PubMed] [Google Scholar]
- 65.Zhang Q.L., Guo Z.L., Yuan M.L. The complete mitochondrial genome of Poratrioza Sinica (Insecta: Hemiptera: Psyllidae) Mitochondrial DNA. 2014 doi: 10.3109/19401736.2014.913164. [DOI] [PubMed] [Google Scholar]
- 66.Yukuhiro K., Sezutsu H., Itoh M., Shimizu K., Banno Y. Significant levels of sequence divergence and gene rearrangements have occurred between the mitochondrial genomes of the wild mulberry silkmoth, Bombyx mandarina, and its close relative, the domesticated silkmoth, Bombyx mori. Mol. Biol. Evol. 2002;19:1385–1389. doi: 10.1093/oxfordjournals.molbev.a004200. [DOI] [PubMed] [Google Scholar]
- 67.Ojala D., Montoya J., Attardi G. tRNA punctuation model of RNA processing in human mitochondria. Nature. 1981;290:470–474. doi: 10.1038/290470a0. [DOI] [PubMed] [Google Scholar]
- 68.Nei M., Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 69.Schmidt T.R., Wu W., Goodman M., Grossman L.I. Evolution of nuclearand mitochondrial-encoded subunit interaction in cytochrome c oxidase. Mol. Biol. Evol. 2001;18:563–569. doi: 10.1093/oxfordjournals.molbev.a003836. [DOI] [PubMed] [Google Scholar]
- 70.Hebert P.D.N., Ratnasingham S., de Waard J.R. Barcoding animal life: Cytochrome c oxidase subunit 1 divergences among closely related species. Proc. R. Soc. Lond. B. 2003;270:S96–S99. doi: 10.1098/rsbl.2003.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Castellana S., Vicario S., Saccone C. Evolutionary patterns of the mitochondrial genome in Metazoa: Exploring the role of mutation and selection in mitochondrial protein coding genes. Genome Biol. Evol. 2011;3:1067–1079. doi: 10.1093/gbe/evr040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wei S.J., Shi M., Sharkey M.J., van Achterberg C., Chen X.X. Comparative mitogenomics of Braconidae (Insecta: Hymenoptera) and the phylogenetic utility of mitochondrial genomes with special reference to holometabolous insects. BMC Genomics. 2010;11:371. doi: 10.1186/1471-2164-11-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim I., Cha S.Y., Yoon M.H., Hwang J.S., Lee S.M., Sohn H.D., Jin B.R. The complete nucleotide sequence and geneorganization of the mitochondrial genome of the oriental mole cricket, Gryllotalpa orientalis (Orthoptera: Gryllotalpidae) Gene. 2005;353:155–168. doi: 10.1016/j.gene.2005.04.019. [DOI] [PubMed] [Google Scholar]
- 74.Zhang D.X., Szymura J.M., Hewitt G.M. Evolution and structural conservation of the control region of insect mitochondrial DNA. J. Mol. Evol. 1995;40:382–391. doi: 10.1007/BF00164024. [DOI] [PubMed] [Google Scholar]
- 75.Fenn J.D., Cameron S.L., Whiting M.F. The complete mitochondrial genome sequence of the Mormon cricket (Anabrus simplex: Tettigoniidae: Orthoptera) and an analysis of control region variability. Insect Mol. Biol. 2007;16:239–252. doi: 10.1111/j.1365-2583.2006.00721.x. [DOI] [PubMed] [Google Scholar]
- 76.Cook C.E., Yue Q., Akam M. Mitochondrial genomes suggest that hexapods and crustaceans are mutually paraphyletic. Proc. Biol. Sci. 2005;272:1295–1304. doi: 10.1098/rspb.2004.3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gillespie J.J., Johnston J.S., Cannone J.J., Gutell R.R. Characteristics of the nuclear (18S, 5.8S, 28S and 5S) and mitochondrial (12S and 16S) rRNA genes of Apis mellifera (Insecta: Hymenoptera): Structure, organization and retrotransposable elements. Insect Mol. Biol. 2006;15:657–686. doi: 10.1111/j.1365-2583.2006.00689.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dowton M., Castro L.R., Austin A.D. Mitochondrial gene rearrangements as phylogenetic characters in the invertebrates: The examination of genome “morphology”. Invertebr. Syst. 2002;16:345–356. doi: 10.1071/IS02003. [DOI] [Google Scholar]
- 79.Burckhardt D. Taxonomy and phylogeny of the Gondwanan moss bugs or Peloridiidae (Hemiptera, Coleorrhyncha) Dtsch. Entomol. Z. 2009;56:173–235. doi: 10.1002/mmnd.200900019. [DOI] [Google Scholar]
- 80.Kuechler S.M., Gibbs G., Burckhardt D., Dettner K., Hartung V. Diversity of bacterial endosymbionts and bacteria-Host co-evolution in Gondwanan relict moss bugs (Hemiptera: Coleorrhyncha: Peloridiidae) Environ. Microbiol. 2013;15:2031–2042. doi: 10.1111/1462-2920.12101. [DOI] [PubMed] [Google Scholar]
- 81.Koga R., Bennett G., Cryan J.R., Moran N.A. Evolutionary replacement of obligate symbionts in an ancient and diverse insect lineage. Environ. Microbiol. 2013;5:2073–2081. doi: 10.1111/1462-2920.12121. [DOI] [PubMed] [Google Scholar]
- 82.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Librado P., Rozas J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 84.Guindon S., Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 85.Ronquist F., Huelsenbeck J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 86.Posada D. jModelTest: Phylogenetic model averaging. Mol. Biol. Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]