Abstract
Infections related to Aspergillus species have emerged to become an important focus in infectious diseases, as a result of the increasing use of immunosuppressive agents and high fatality associated with invasive aspergillosis. However, laboratory diagnosis of Aspergillus infections remains difficult. In this study, by comparing the metabolomic profiles of the culture supernatants of 30 strains of six pathogenic Aspergillus species (A. fumigatus, A. flavus, A. niger, A. terreus, A. nomius and A. tamarii) and 31 strains of 10 non-Aspergillus fungi, eight compounds present in all strains of the six Aspergillus species but not in any strain of the non-Aspergillus fungi were observed. One of the eight compounds, Leu–Glu–Leu–Glu, is a novel tetrapeptide and represents the first linear tetrapeptide observed in Aspergillus species, which we propose to be named aspergitide. Two other closely related Aspergillus-specific compounds, hydroxy-(sulfooxy)benzoic acid and (sulfooxy)benzoic acid, may possess anti-inflammatory properties, as 2-(sulfooxy)benzoic acid possesses a structure similar to those of aspirin [2-(acetoxy)benzoic acid] and salicylic acid (2-hydroxybenzoic acid). Further studies to examine the potentials of these Aspergillus-specific compounds for laboratory diagnosis of aspergillosis are warranted and further experiments will reveal whether Leu–Glu–Leu–Glu, hydroxy-(sulfooxy)benzoic acid and (sulfooxy)benzoic acid are virulent factors of the pathogenic Aspergillus species.
Keywords: Aspergillus, metabolomics, metabolites, biomarkers, tetrapeptide
1. Introduction
In recent years, infections related to Aspergillus species have become an emerging focus of clinical microbiology and infectious disease as the number of patients infected with Aspergillus species rose dramatically. In immunocompetent and mildly immunocompromised hosts, Aspergillus species rarely cause serious illnesses, except for pulmonary aspergilloma in patients with pre-existing chronic lung diseases and, less commonly, chronic cavitary pulmonary aspergillosis, chronic fibrosing aspergillosis and chronic necrotizing aspergillosis [1,2,3,4,5]. On the other hand, invasive aspergillosis is one of the most important causes of morbidity and mortality in immunocompromised patients, such as those with hematological malignancies undergoing chemotherapy, hematopoietic stem cell and solid organ transplantation and HIV infection [1,6]. Among the known Aspergillus species, Aspergillus fumigatus is the most common species causing human infections in western countries, whereas Aspergillus flavus is as important as A. fumigatus in our locality and in other Asian countries, and is the second most common Aspergillus species associated with human infections in western countries [1,6]. Other Aspergillus species commonly associated with human infections include Aspergillus niger and Aspergillus terreus [1,6].
The successful management of invasive aspergillosis is hampered by difficulties in establishing timely and accurate diagnosis. The gold standard for making a diagnosis is to obtain a positive culture of A. fumigatus and to demonstrate histological evidence of mycelial invasion from tissue biopsy. Due to the very sick nature of these patients and often the presence of bleeding diathesis, tissue biopsy is often not possible or acceptable by patients. Although commercial kits for galactomannan antigen detection and our in-house developed Afmp1p and Afmp2p antigen detection assays are available for clinical use, the sensitivities of these tests are far from ideal [2,3,4,5,7]. Molecular tests such as PCR are also used for laboratory diagnosis, but such tests cannot distinguish among environmental contamination, colonization and genuine invasive infection [8,9].
Microbial metabolomics is a relatively new research field that involves the study of the unique chemical fingerprints of the metabolite profiles of microorganisms, and has been used for the characterization of a number of pathogenic microbes [10,11,12,13]. For example, urine metabolomic data can be used for the diagnosis of Streptococcus pneumoniae infections and urinary tract infections [14,15,16]. We have also reported the application of metabolomics for identification of mitorubrinol yellow pigment in the pathogenic dimorphic fungus, Penicillium marneffei, and characterization of Tsukamurella and Aspergillus strains [17,18,19], as well as detection of specific metabolites in culture supernatant of Mycobacterium tuberculosis which may have important diagnostic applications [20]. Since our previous study showed that metabolomics profiling can be used to distinguish among different Aspergillus species [17], we hypothesize that there could be Aspergillus-specific novel extracellular metabolites that may be detected in blood and body fluids for laboratory diagnosis of invasive aspergillosis. In order to look for these potential biomarkers of invasive aspergillosis, we characterized the metabolomic profiles of the culture supernatants of Aspergillus species and other clinically important fungal species, using ultrahigh performance liquid chromatography-electrospray ionization-quadrupole time-of-flight mass spectrometry (UHPLC–ESI-Q-TOF-MS). Multi- and univariate statistical analyses of the metabolomic profiles were performed to identify Aspergillus-specific metabolites.
2. Results
2.1. Visual Inspection of Total Ion Chromatograms
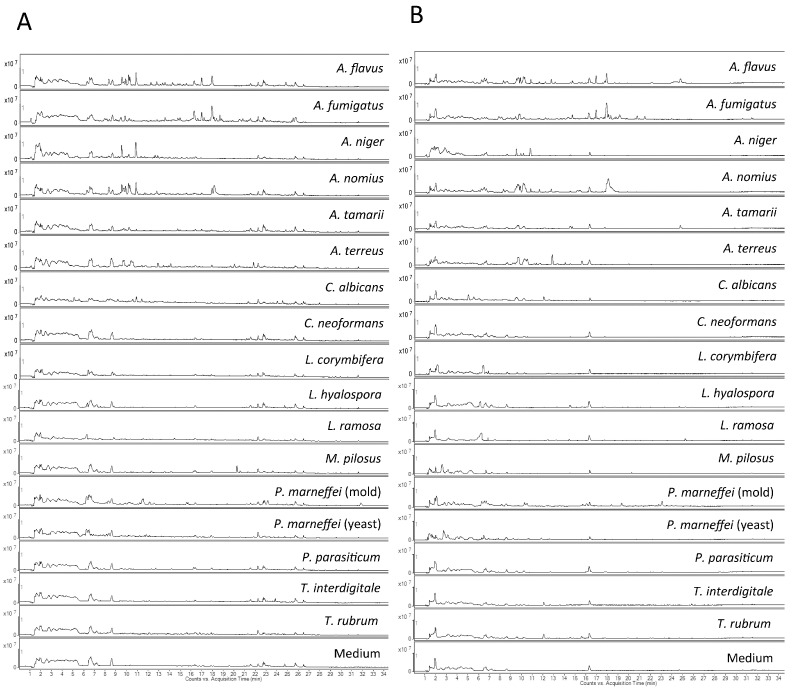
The metabolomes of culture supernatants of the 128 samples, including 30 samples of the 30 Aspergillus strains in duplicate and 34 samples from the 31 non-Aspergillus fungal strains (six samples were obtained from the mold and yeast forms of the three P. marneffei strains) in duplicate, were characterized and compared. UHPLC–ESI-Q-TOF-MS methods, operated in both positive and negative modes, for the analysis of different metabolites in fungal culture supernatant, were developed. Total ion chromatograms (TIC) from the Aspergillus strains shared considerable similarity and those of the same Aspergillus species showed high similarity, whereas significant differences were observed from the TICs of the different non-Aspergillus species. Representative examples of chromatograms from each species obtained in both positive and negative modes are shown in Figure 1A,B, respectively.
Figure 1.
Total ion chromatograms of Aspergillus species and other control fungal species used in this study. (A) Positive ionization mode; (B) Negative ionization mode.
2.2. Omics-Based Bioinformatics Analysis
There were 32,849 molecular features (MFs) detected in positive electrospray ionization (ESI) mode and 39,303 MFs detected in negative ESI mode. All MFs were subjected to the Mass Profiler Professional (MPP) software for statistical analysis. After filtering by the MPP software, total of 2404 MFs in positive mode and 1737 MFs in negative mode were obtained. Stepwise filtering procedure was used to identify significant MFs in Aspergillus strains and to reduce the dimensionality of data prior to principle component analysis (PCA) and partial least squares discriminant analysis (PLS-DA). To compare the metabolomes between Aspergillus and non-Aspergillus fungal strains, both uni- and multi-variate analyses were performed. Three-hundred-and-six features in positive mode and 44 features in negative mode were further subjected for volcano plot analysis. Using Student’s t-test with p-value <0.01 and fold-change (FC) >16 to compare the Aspergillus and non-Aspergillus groups, the number of MFs was reduced to 287 features in positive mode and 42 features in negative mode after stepwise filtering for PCA and PLS-DA analyses.
2.3. PCA and PLS-DA Modeling
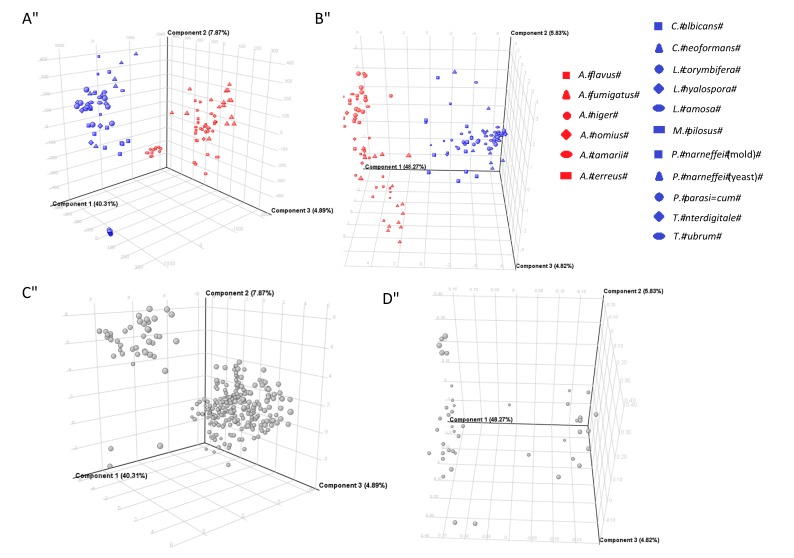
To compare the metabolomes between Aspergillus and non-Aspergillus strains, multivariate analysis was performed. In the positive ionization mode, PCA score plot showed that 48.18% of the total variance in the data was represented by the first two principal components (PCs), in which PC1 explained 40.31% of the variance and PC2 explained 7.87% in the three-dimensional PCA score plots (Figure 2A). In the negative ionization mode, PCA score plot showed that 54.10% of the total variance in the data was represented by the first two PCs, in which PC1 explained 48.27% of the variance and PC2 explained 5.83% in three-dimensional PCA score plots, which gave a better clustering than the positive ionization mode (Figure 2B). All PCA score plots revealed that the Aspergillus strains were closely related to each other and can be distinguished from the non-Aspergillus strains, based on the first two principal components, where the Aspergillus strains were clearly separated from the non-Aspergillus strains along PC1. PCA loading plots for the respective MFs, which account for the separation between the Aspergillus and non-Aspergillus strains, were also generated in either positive ionization mode (Figure 2C) or negative ionization mode (Figure 2D). MFs that showed statistically significance in the Aspergillus strains were extracted for further analysis.
Figure 2.
PCA score plots and PCA loading plots of 128 fungal culture supernatant samples. PCA score plots for (A) positive ionization mode data and (B) negative ionization mode data; PCA loading plots of molecular features for (C) positive ionization mode data and (D) negative ionization mode data.
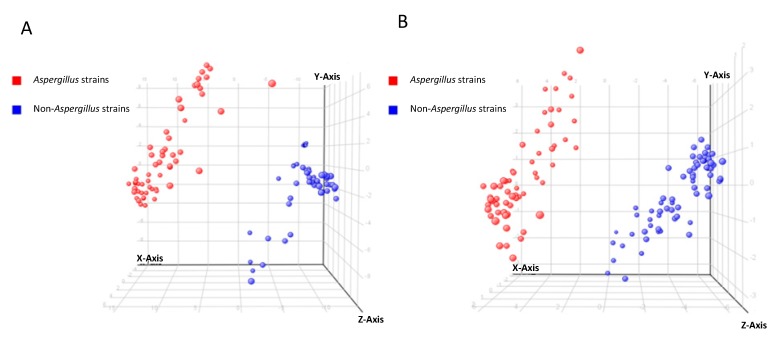
In view of the significant separation achieved using PCA, supervised PLS-DA using t-score plots was employed to maximize the separation between the Aspergillus and non-Aspergillus strains, to find potential marker variables, to validate statistical model and to predict sample class membership. In both positive and negative ionization modes, sample classifications using the PLS-DA model achieved 100% accuracy in both recognition and prediction abilities, indicating that the Aspergillus and non-Aspergillus strains were correctly classified during model training and cross-validation. Excellent separations of the Aspergillus and non-Aspergillus strains obtained by PLS-DA were shown in Figure 3.
Figure 3.
PLS-DA t-score plots of 128 fungal culture supernatant samples. (A) Positive ionization mode; (B) negative ionization mode.
2.4. Identification of Potential Biomarkers in Aspergillus Species
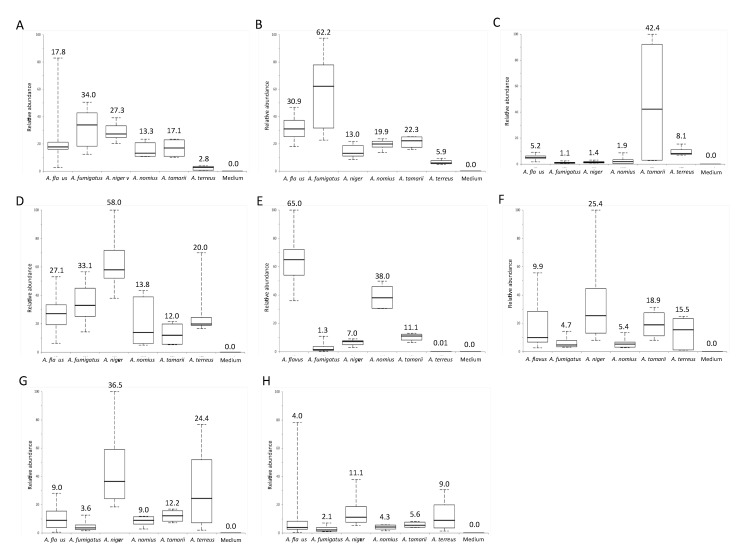
Potential biomarkers in Aspergillus species were identified according to the elution order, MS/MS fragmentation pattern, molecular formula and database search. A total of eight potential biomarkers were identified (Table 1 and Table A1). Box and whisker plots of the integral peak areas for all the eight metabolites were constructed (Figure 4).
Table 1.
Specific metabolites in culture supernatant of Aspergillus species.
| Compound | m/z | Retention Time (min) | Ionization Mode | Ion | MS/MS Fragment Masses | p Value a | Molecular Formula | Putative Identity |
|---|---|---|---|---|---|---|---|---|
| 1 | 166.0724 | 1.86 | Positive | [M + H]+ | 107.0232, 124.0493, 149.0446, 166.0712 | <0.001 | C6H7N5O | 7-Methylguanine |
| 2 | 166.0724 | 3.60 | Positive | [M + H]+ | 110.0327, 135.0292, 149.0441, 166.0702 | <0.001 | C6H7N5O | 1-Methylguanine |
| 3 | 232.9775 | 6.20 | Negative | [M − H]− | 96.9593, 109.0288, 153.0192, 188.9861, 232.9775 | <0.001 | C7H6O7S | Hydroxy(sulfooxy)benzoic acid |
| 4 | 216.9822 | 6.80 | Negative | [M − H]− | 79.9570, 93.0346, 96.9598, 137.0245, 172.9911, 216.9816 | <0.001 | C7H6O6S | (sulfooxy)benzoic acid |
| 5 | 503.2749 | 9.98 | Positive | [M + H]+ | 96.0458, 114.0557, 131.0818, 225.1232, 243.1329, 261.1452, 503.2730 | <0.001 | C22H38N4O9 | Leu–Glu–Leu–Glu |
| 6 | 292.2645 | 19.55 | Positive | [M + H]+ | 74.0963, 203.1426, 219.1743, 292.2633 | <0.001 | C19H33NO | No match |
| 7 | 352.2466 | 28.5 | Positive | [M + H]+ | 81.0189, 94.0269, 165.1127, 194.1155, 210.1103 | <0.001 | C20H33NO4 | No match |
| 8 | 352.2466 | 28.8 | Positive | [M + H]+ | 81.0191, 94.0268, 165.1126, 194.1156, 210.1105 | <0.001 | C20H33NO4 | No match |
a p value from ANOVA analysis.
Figure 4.
Comparison of abundance of metabolites specific to the six Aspergillus species. Box-and-whisker plots of (A) 7-methylguanine; (B) 1-methylguanine; (C) hydroxyl(sulfooxy)benzoic acid; (D) (sulfooxy)benzoic acid; (E) Leu–Glu–Leu–Glu; (F) metabolite with protonated molecular ion of m/z 292.2645 eluted at 19.55 min; (G) metabolite with protonated molecular ion of m/z 352.2466 eluted at 28.5 min; (H) metabolite with protonated molecular ion of m/z 352.2466 eluted at 28.8 min. Boxes show first to third quartiles; whiskers show 5% and 95% percentiles; middle lines in boxes and numbers on the top of boxes indicate the median of the relative abundance of metabolite.
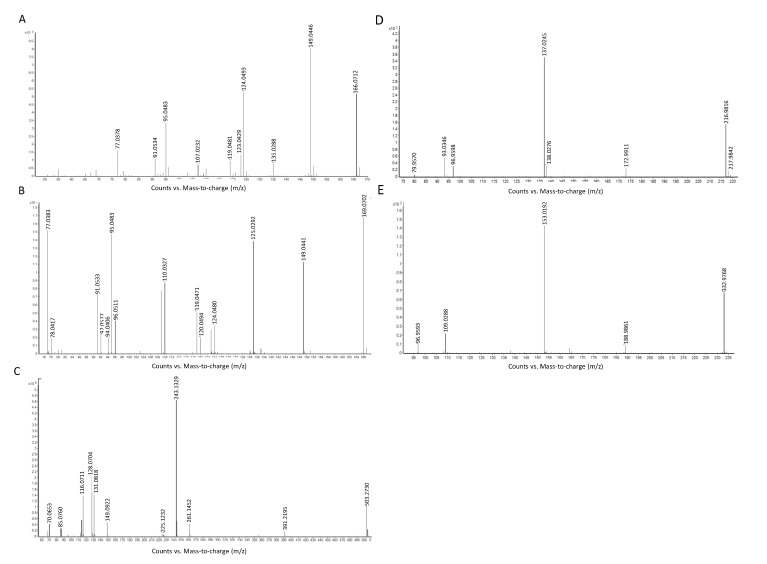
Two metabolites (compounds 1 and 2 in Table 1), at retention time 1.86 and 3.60 min, with the same theoretical [M + H]+ of m/z 166.0724 and molecular formula of C6H7N5O, were identified as 7-methylguanine and 1-methylguanine respectively by database search in METLIN. MS/MS spectra of 7-methylguanine (Figure 5A) and 1-methylguanine (Figure 5B) showed the major fragmentation peak at m/z 149, which was a result of the loss of an amino group at the C-2 position of the guanine group. Another peak at m/z 124 in the MS/MS spectra of 7-methylguanine was formed by losing the N-1, N-9 and C-8 position of 7-methylguanine, whereas the peak at m/z 135 in the MS/MS spectra of 1-methylguanine represent the loss of methyl group at N-1 position and amino group at C-2 position from 1-methylguanine. Among all the six Aspergillus species, 7-methylguanine and 1-methylguanine were both expressed in highest abundance in A. fumigatus and lowest abundance in A. terreus.
Figure 5.
MS/MS spectra of (A) 7-methylguanine; (B) 1-methylguanine; (C) Leu–Glu–Leu–Glu; (D) (sulfooxy)benzoic acid and (E) hydroxyl(sulfooxy)benzoic acid.
The metabolite (compound 5 in Table 1) with [M + H]+ of m/z 503.2749, at a retention time of 9.98 min, and molecular formula of C22H38N4O9 matched 24 tetra-peptides by database search in METLIN. The peptide sequence was revealed to be Leu–Glu–Leu–Glu by using Peaks©7 (Bioinformatics Solution Inc., Waterloo, ON, Canada). Different peaks in the MS/MS spectrum of the tetra-peptide represented different fragmented peptide ions (m/z = 96, b1-H2O ion; m/z = 114, b1 ion, m/z = 131, y1-NH3 ion; m/z = 225, b2-H2O ion; m/z = 243, y2-H2O and/or b2 ions and m/z = 261, y2 ion) (Figure 5C). Among all the Aspergillus species, Leu–Glu–Leu–Glu was expressed in highest abundance in A. flavus and lowest abundance in A. terreus.
Two metabolites (compounds 3 and 4 in Table 1), with undefined positional isomers, were identified by exact molecular weight, predicted molecular formula, MS/MS fragmentation pattern and literature search. Compound 4 in Table 1, with deprotonated molecular ion at m/z 216.9822 and at a retention time of 6.80 min, was identified as C7H6O6S. The MS/MS fragmentation pattern (Figure 5D) supported that the chemical structure of compound 4 is (sulfooxy)benzoic acid. The peak at m/z 173 referred to the loss of carboxylic acid from the parent ion. The peak at m/z 137 represented the ion of hydroxybenzoic acid, which was formed by losing a sulfoxy group at the o-position of the phenol group from the parent ion. A further loss of a carboxylic acid group yielded a phenol ion with m/z 93. Based on the predicted molecular formula and the MS/MS fragmentation of compound 4, we concluded that compound 4 is either 4-(sulfooxy)benzoic acid or its positional isomers (o-positional isomer, 2-(sulfooxy)benzoic acid; p-positional isomer, 3-(sulfooxy)benzoic acid). Another metabolite, compound 3, with deprotonated molecular ion at m/z 232.9775 and at a retention time of 6.20 min, was identified as C7H6O7S. The MS/MS fragmentation pattern (Figure 5E) supported that the chemical structure of compound 3 is hydroxyl-(sufooxy)benzoic acid. The peak at m/z 189 referred to the loss of carboxylic acid from the parent ion. The peak at m/z 153 represented the ion of dihydroxybenzoic acid, which was formed by losing a sulfoxy group at the o-position of the hydroxyphenol group from the parent ion. A further loss of a carboxylic acid group yielded a benzene-diol ion with m/z 109.
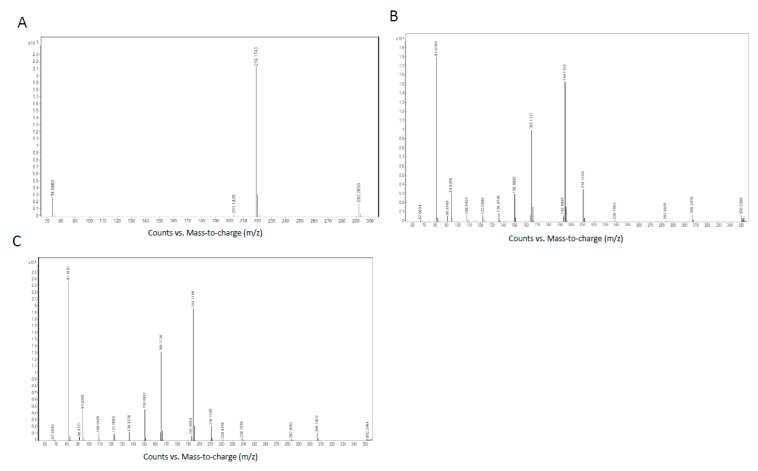
The metabolite (compound 6 in Table 1), with [M + H]+ of m/z 292.2645, at a retention time of 19.55 min, and with molecular formula C19H33NO, matched 17β-amino-5α-androstan-11β-ol by database search in METLIN. However, the MS/MS fragmentation pattern was not compatible with the chemical structure of the matched compound (Figure A1). Similar discrepancies between the matched compound by database search and MS/MS fragmentation patterns were also observed in another two metabolites, compounds 7 and 8 in Table 1. These two metabolites, at retention time of 28.5 and 28.8 min, with the same [M + H]+ of m/z 352.2466 and molecular formula of C20H33NO4, were both matched to N-3-oxo-hexadec-11(Z)-enoyl-l-Homoserine lactone by database search in METLIN. However, the MS/MS fragmentation patterns of these two metabolites were not compatible with the chemical structures of the matched compounds (Figure A1). Since compound 7 and compound 8 had the same molecular formula, as well as their similar retention times and MS/MS spectra, these two compounds were proposed to be diastereomers. Compounds 6–8 listed in Table 1 are the potentially novel metabolites.
3. Discussion
In this study, by comparing the metabolomic profiles of the six Aspergillus species and 10 non-Aspergillus fungal species, we identified at least eight Aspergillus species-specific compounds in Aspergillus culture supernatants. The six Aspergillus species used were the four most commonly used Aspergillus species encountered in clinical specimens, including A. fumigatus, A. flavus, A. niger and A. terreus, as well as A. nomius and A. tamarii, two Aspergillus species commonly misidentified as A. flavus [17]. The control non-Aspergillus fungal species include commonly encountered yeasts (Candida albicans and Cryptococcus neoformans), molds (Monascus pilosus, Phaeoacremonium parasiticum, Trichophyton rubrum, Trichophyton interdigitale and Lichtheimia species) and thermal dimorphic fungus in our locality (P. marneffei). Multivariate analyses by unsupervised PCA and supervised PLS-DA showed that the Aspergillus species and non-Aspergillus species were clustered separately into two groups, indicating that there were potential metabolite(s) that could be used to differentiate them (Figure 2 and Figure 3). Subsequent extraction, statistical analysis and manual verification of the MFs revealed eight compounds that could be detected in the culture supernatants of all the six Aspergillus species but none of the other control fungal species. Since both the Aspergillus and non-Aspergillus species were cultured in the same culture medium, these compounds were genuine metabolites of the Aspergillus species. These Aspergillus specific fungal compounds and/or their metabolites may represent specific molecules that can be potentially detected in serum and/or urine of patients with invasive aspergillosis. Further studies using serum and urine samples of patients with culture and histology documented invasive aspergillosis should be performed to investigate which of these compounds and/or their metabolites could be detected in patients with aspergillosis for use in clinical microbiology laboratories.
One of the eight metabolites, compound 5 (Leu–Glu–Leu–Glu), observed in the six pathogenic Aspergillus species but not in other fungi, is a novel tetrapeptide specific to the Aspergillus species. Tetrapeptides are ribosomally or non-ribosomally synthesized secondary metabolites with biological functions. Ribosomally synthesized tetrapeptides are synthesized by ribosomal peptide synthetic (RiPS) pathway, whereas non-ribosomally synthesized tetrapeptides are synthesized by non-ribosomal peptide synthase (NRPS) gene clusters. Structurally, tetrapeptides can be linear or cyclic. In Aspergillus species, only two tetrapeptides are known (Table 2). The first one is ustiloxin B, a ribosomally synthesized cyclic tetrapeptide with sequence Tyr–Ala–Ile–Gly, the tyrosine residue of which is modified with norvaline, found in A. flavus [21]. The other one is asperterrestide A, a cyclic tetrapeptide with sequence anthranilic acid-3-OH-N-Me-Phe–Ile–Ala, found in A. terreus [22]. The tetrapeptide Leu–Glu–Leu–Glu discovered in the present study represents the first linear tetrapeptide observed in Aspergillus species, which we propose to be named aspergitide. We searched the genomes of A. fumigatus, A. flavus, A. niger and A. terreus but found no DNA sequence that potentially encodes repetitive Leu–Glu–Leu–Glu as observed in the ustiloxin B gene cluster in the A. flavus genome [21]. Therefore, we speculate that Leu–Glu–Leu–Glu is synthesized by NRPS cluster instead of RiPS pathway. Since the six Aspergillus species are members in the Fumigati, Flavi, Nigri and Terrei sections, Leu–Glu–Leu–Glu may be a tetrapeptide conserved across many sections in this genus; or alternatively, as the six Aspergillus species in this study are all pathogenic fungi, Leu–Glu–Leu–Glu could be a tetrapeptide with virulence properties.
Table 2.
Known tetrapeptides in Aspergillus species.
| Tetrapeptide | Sequence | Linear/Cyclic | Ribosomally/Non-Ribosomally Synthesized | Aspergillus Species from Which Tetrapeptide Is Found | Biological Properties |
|---|---|---|---|---|---|
| Ustiloxin B [21] | Tyr–Ala–Ile–Gly | Cyclic | Ribosomally synthesized | A. flavus | Mitotic inhibitor |
| Asperterrestide A [22] | a ABA–3-OH-N-Me-Phe–Ile–Ala | Cyclic | Unknown | A. terreus | Inhibitory effects on influenza virus H1N1 and H3N2, cytotoxicity against human carcinoma cell lines |
| Aspergitide | Leu–Glu–Leu–Glu | Linear | Unknown | A. flavus, A. niger, A. fumigatus, A. terreus, A. nomius, A. tamarii | Unknown |
a ABA—Anthranilic acid.
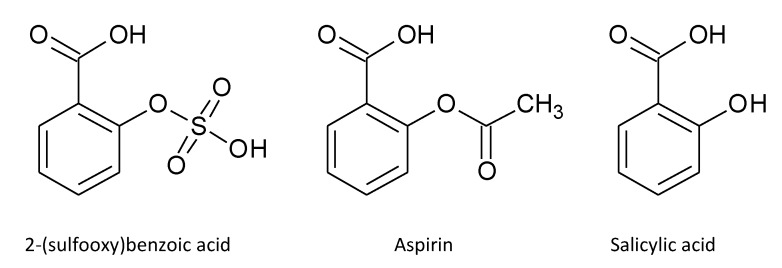
Two other metabolites, compound 3 [hydroxy-(sulfooxy)benzoic acid] and compound 4 [(sulfooxy)benzoic acid)], specific to the Aspergillus species may possess anti-inflammatory properties. 2-(sulfooxy)benzoic acid possesses a structure similar to those of aspirin [2-(acetoxy)benzoic acid] and salicylic acid (2-hydroxybenzoic acid) (Figure 6), which are potent analgesics and anti-inflammatory agents used for decades for treatment of various inflammatory conditions through inhibition of cyclooxygenases (COXs). Notably, it has been shown by molecular docking experiments, during the process of searching for analogues of gallic acid, a cyclooxygenase-2 (COX-2) inhibitor screened from medicinal plant, that 3-hydroxy-4-(sulfooxy)benzoic acid could be a potent COX-2 inhibitor [23]. However, both (sulfooxy)benzoic acid and hydroxy-(sulfooxy)benzoic acid have never been found naturally. In the present study, we showed that both of these compounds could be found in the metabolomes of the six pathogenic Aspergillus species. Since it has previously been reported that o-hydroxybenzoic acid and p-hydroxybenzoic acid were fungal secondary metabolites [24], we speculate that (sulfooxy)benzoic acid was formed by o-sulfation of hydroxybenzoic acid and hydroxy-(sulfooxy)benzoic acid was in turn formed by further oxidation of (sulfooxy)benzoic acid in the Aspergillus species. Further experiments will reveal whether these two metabolites of the pathogenic Aspergillus species are their virulent factors through suppression of the host inflammatory response.
Figure 6.
Chemical structures of 2-(sulfooxy)benzoic acid, aspirin and salicylic acid.
4. Materials and Methods
4.1. Aspergillus Strains and Culture
Thirty Aspergillus strains of A. fumigatus, A. flavus, A. niger, A. terreus, A. nomius and A. tamarii; and 34 non-Aspergillus fungal strains, belonging to C. neoformans, M. pilosus, C. albicans, P. parasiticum, P. marneffei, T. rubrum, T. interdigitale and Lichtheimia species, were included in this study (Table 3). All clinical isolates were identified by standard phenotypic tests [25]. In addition, the identities of the Aspergillus strains were confirmed by ITS, β-tubulin and calmodulin gene sequencing [17]; M. pilosus and Trichophyton species were confirmed by ITS gene sequencing; P. parasiticum was confirmed by ITS and β-tubulin gene sequencing [26]; and those of the Lichtheimia species were confirmed by ITS, β-actin, EF1α and 28S rRNA gene sequencing [27,28]. All fungal strains were grown at a concentration of 2 × 106 CFU in 10 mL RPMI 1640 medium (Gibco, Grand Island, NY, USA) supplemented with 2% glucose with shaking at 250 rpm. Strains were cultured for 48 h at 37 °C for Aspergillus species, C. neoformans, M. pilosus, C. albicans, P. parasiticum and Lichtheimia species, 108 h at 37 °C for T. rubrum and yeast form of P. marneffei and 108 h at 25 °C for mold form of P. marneffei to reach their early stationary phases to achieve comparable fungal concentrations. The culture supernatant was filtered using 0.22-μm MCE filter (Millipore, Billerica, MA, USA) and quenched immediately in liquid nitrogen for 10 min. The filtrates were lyophilized and stored at −80 °C until sample extraction and analysis. Uninoculated culture medium was used as a negative control. All strains were grown in duplicates.
Table 3.
Fungal strains used in this study.
| Fungal Species | Strains | Source of Strains |
|---|---|---|
| Aspergillus fumigatus | AF293, QC5096, PW1353, PW1354, PW1355, PW3460, PW3461, PW3462 | AF293, purchased from a FGSC; QC5096, QC strain (specimen 5096) from b UKNEQAS; others, clinical isolates from Hong Kong |
| Aspergillus flavus | ATCC204304, PW2952, PW2953, PW2954, PW2956, PW2957, PW2960, PW2961, PW2962 | ATCC204304, purchased from c ATCC; others, clinical isolates from Hong Kong [17] |
| Aspergillus niger | ATCC10577, PW3463, PW793, PW3464, PW3465 | ATCC10577, purchased from c ATCC; others, clinical isolates from Hong Kong |
| Aspergillus terreus | PW3466, PW3467, PW3468 | Clinical isolates from Hong Kong |
| Aspergillus nomius | PW2955, PW2959, CBS260.88 | CBS260.88, purchased from d CBS Fungal Biodiversity Centre; others, clinical isolates from Hong Kong [17] |
| Aspergillus tamarii | PW2958, CBS104.13 | PW2958, clinical isolate from Hong Kong [17]; CBS104.13 purchased from d CBS Fungal Biodiversity Centre |
| Trichophyton rubrum | PW3469 | Clinical isolate from Hong Kong |
| Trichophyton interdigitale | PW3470, PW3471 | Clinical isolates from Hong Kong |
| Cryptococcus neoformans | CBS132, PW275 | PW275, clinical isolate from Hong Kong; CBS132, purchased from d CBS Fungal Biodiversity Centre |
| Monascus pilosus | PW2536 | Clinical isolate from Hong Kong |
| Phaeoacremonium parasiticum | PW2367, PW2483 | Clinical isolates from Hong Kong [26] |
| Candida albicans | ATCC90028, PW904, PW905, PW906 | ATCC90028, purchased from c ATCC; others, clinical isolates from Hong Kong |
| Penicillium marneffei | PM1, PM35, PM41 | Clinical isolates from Hong Kong [29] |
| Lichtheimia corymbifera | PW1473, PW1449, PW1471, HKU25, PW1450, PW1472 | HKU25, QC strain (specimen 4727) from b UKNEQAS; others, clinical isolates from Hong Kong |
| Lichtheimia ramosa | PW2398, PW2399, HKU21, HKU22, HKU23, PW1493, PW1494, PW1495, PW1496 | HKU23, QC strain (specimen 8273) from b UKNEQAS; others, clinical isolates from Hong Kong [27] |
| Lichtheimia hyalospora | CBS173.67 | Purchased from d CBS Fungal Biodiversity Centre |
a FGSC—Fungal Genetics Stock Centre; b UKNEQAS—United Kingdom National External Quality Assessment Service; c ATCC—American Type Culture Collection; d CBS—Centraal Bureau voor Schimmelcultures.
4.2. Chemicals and Reagents
LC–MS grade water, methanol and acetonitrile were purchased from J.T. Baker (Center Valley, PA, USA). Formic Acid with ACS reagent grade was purchased from Sigma-Aldrich, Inc. (Saint Louis, MO, USA).
4.3. Sample Preparation
The samples were prepared according to our previous publications with minor modifications [17,19,30]. Lyophilized samples were reconstituted in 600 µL solvent mixture containing water/methanol/acetonitrile (2:4:4) (v/v/v). The sample mixtures were vortexed for 1 min followed by 10 min sonication at room temperature. After centrifugation at 14,000 rpm for 10 min at 4 °C, the supernatants were transferred to UHPLC–ESI-Q-TOF-MS analysis. To prevent batch effect, all specimens were analyzed in random manner.
4.4. UHPLC–ESI-Q-TOF-MS Analysis
UHPLC–ESI-Q-TOF-MS analysis was performed according to our previous publications with minor modifications [17,19,30]. For liquid chromatography, separations were performed using Agilent 1290 Infinity UHPLC (Agilent Technologies, Waldronn, Germany) and Waters Acquity UPLC HSS T3 (3.0 × 100 mm, 1.8 µm) column with Waters Acquity UPLC HSS T3 (2.1 × 5.0 mm, 1.8 µm) VanGuard Pre-column, The injection volume was 5 µL of each sample. The column and autosampler temperature were maintained at 45 and 15 °C respectively. Separation was performed at a flow rate of 0.4 mL/min under a gradient program. Mobile phase A was 0.1% formic acid (v/v) in water. Mobile phase B was pure methanol. The gradient program was applied as follows: t = 0 min, 1% B; t = 2.0 min, 1% B; t = 12 min, 38% B; t = 30 min, 99.5% B; t = 35 min, 99.5% B; t = 35.1 min, 1% B; t = 38 min, 1% B. Mass spectrometry was operated in both positive and negative ESI mode using Agilent 6540 Q-TOF mass spectrometer (Agilent Technologies, Santa Clara, CA, USA) with Agilent Jet Stream Electrospray ionization (ESI) source. The capillary voltage was kept at +3800 V with the nozzle voltage of +0 V in positive mode and −3500 V with the nozzle voltage of −0 V in negative mode. Other conditions were kept constant in all experiment. The gas temperature was kept at 320 °C. The drying gas (nitrogen) was set at 8 L/min. The pressure of the nebulizer gas (nitrogen) was maintained at 40 psi. The sheath gas was kept at a flow rate of 10 L/min at 380 °C. The voltages of the Fragmentor, Skimmer 1 and OctopoleRFPeak were kept at 135, 50 and 500 V, respectively. The scan range was adjusted to m/z 80–1700 at the acquisition rate of 2 spectra/s. Ultra-high purity nitrogen was used in product ion scanning (PIC) experiments. Product ion scanning was conducted using ultra-high purity nitrogen. MS/MS acquisition was operated in the same parameter as in MS acquisition. Collision Energy (CE) was set at 10, 20 or 40 eV for fragmentation of the targeted compounds to generate the best MS/MS spectra.
4.5. Data Processing and Statistical Data Analysis
The raw data from UHPLC–ESI-Q-TOF-MS and MS/MS were analyzed using Agilent Masshunter Qualitative Analysis software (version B.05.00, Agilent Technologies, Santa Clara, CA, USA). Multivariate analysis was used to examine the LC–MS data. Using molecular feature extraction (MFE) algorithm for automated baseline correction, noise calculation, peak detection and chromatogram deconvolution, molecular features (MFs) characterized by retention time (RT), chromatographic peak intensity and accurate mass were obtained. Data were converted into compound exchange file format (.cef) and analyzed using Mass Profiler Professional (MPP) software package (version B.02.02, Agilent Technologies, Santa Clara, CA, USA) for data filtering, peak alignment and statistical analysis. For the data filtering process, MFs with abundance either lower than 8000 counts per second (cps) or less than two isobaric mass peaks were removed. Alignment of RT and m/z values was performed across the sample sets with a tolerance window of ±0.2 min and ±10 mDa respectively. Data were normalized and baseline transformation was performed according to the median expression level of all data. For biomarker discovery and noise reduction, MFs having more than 50% occurrence either in Aspergillus strains or in non-Aspergillus strains were included for statistical analysis.
To identify the biomarkers in culture supernatant, univariate analysis was used. One-way analysis of variance (ANOVA) with Tukey’s post-hoc test was used to identify statistical significant biomarkers specifically present in the Aspergillus strains but not in non-Aspergillus fungal strains, where p-value <0.01 and fold-change >16 were considered as statistically significant. MFs were further filtered using Volcano Plot for Aspergillus and non-Aspergillus groups’ comparison. To reduce non-significant MFs, Student’s t-test with Benjamini-Hochberg correction was used, and MFs with p-value <0.01 were selected for further processing. MFs are sequentially filtered by fold-change (FC) analysis (FC > 16) to find MFs that have high abundance ratios between the two groups.
Stepwise reduction of the data dimensionality was followed by multivariate statistical analysis for PCA. PCA was performed by unsupervised pattern recognition technique. MFs specific to the Aspergillus strains were extracted from the PCA loading plot. PLS-DA was performed by supervised algorithm for the identification of important variables with discriminative power to explore the structure of the data and construct classification models. Box-and-whisker plots were generated using Analyse-it (Analyse-it Software, Leeds, UK). Abundance of metabolites specific to the Aspergillus strains with p-values <0.01, calculated by student’s t-test, was re-extracted from the raw data files of all the samples to re-confirm the metabolites were unique to the Aspergillus strains. The unique MFs of Aspergillus species were further studied for putative identification.
4.6. Metabolite Identification
Accurate masses of the unique MFs in Apergillus group were selected for product ion scanning. MS/MS spectra of the potential biomarkers were processed using Agilent MassHunter Qualitative Analysis software (version B.05.00, Agilent Technologies, Santa Clara, CA, USA) to generate potential molecular formula, based on the accurate mass and isotopic pattern recognitions of their parent and fragment ions. All putative biomarkers were confirmed by matching METLIN database (http://metlin.scripps.edu/), Human Metabolome Database (HMDB) (http://www.hmdb.ca/), MassBank (http://www.massbank.jp/), E. coli Metabolome Database (ECMDB) (http://www.ecmdb.ca/), LipidMaps (http://www.lipidmaps.org/), and KEGG database (http://www.genome.jp/kegg) search using exact molecular weights or MS/MS fragmentation pattern data and literature search. Efforts were made to distinguish metabolites from the other isobaric compounds whenever possible by their elution order and virtue of difference in fragmentation pattern corresponding to their structural characteristics.
Acknowledgments
This work was partly supported by the Research Fund for the Control of Infectious Diseases (Commissioned study) of the Health, Welfare and Food Bureau of the Hong Kong SAR Government; the Strategic Research Theme Fund and University Development Fund of The University of Hong Kong; and Croucher Senior Medical Research Fellowship.
Appendix
Table A1.
Median of the relative abundance of metabolites produced in Aspergillus species.
| Metabolites | A. flavus | A. fumigatus | A. niger | A. nomius | A. tamarii | A. terreus |
|---|---|---|---|---|---|---|
| 7-Methylguanine | 17.772 | 33.947 | 27.289 | 13.295 | 17.071 | 2.750 |
| 1-Methylguanine | 30.889 | 62.172 | 13.046 | 19.916 | 22.300 | 5.938 |
| Hydroxy(sulfooxy)benzoic acid | 5.244 | 1.059 | 1.379 | 1.866 | 42.364 | 8.141 |
| (sulfooxy)benzoic acid | 27.090 | 33.055 | 58.020 | 13.822 | 11.993 | 19.945 |
| Leu–Glu–Leu–Glu | 64.978 | 1.332 | 7.029 | 38.083 | 11.068 | 0.0145 |
| Protonated molecular ion of m/z 292.3 | 9.885 | 4.691 | 25.409 | 5.392 | 18.912 | 15.520 |
| Protonated molecular ion of m/z 352.2 | 9.035 | 3.567 | 36.472 | 8.992 | 12.176 | 24.447 |
| Protonated molecular ion of m/z 352.2 | 3.965 | 2.126 | 11.142 | 4.273 | 5.556 | 9.046 |
Figure A1.
MS/MS spectra of metabolites with protonated molecular ions with (A) m/z 292.2645 eluted at 19.55 min; (B) m/z 352.2466 eluted at 28.5 min and (C) m/z 352.2466 eluted at 28.8 min.
Author Contributions
Susanna K. P. Lau and Patrick C. Y. Woo conceived and supervised the study. Kim-Chung Lee, Emily W. T. Tam, Ka-Ching Lo and Candy C. Y. Lau performed the study. Kelvin K. W. To, Jasper F. W. Chan, Kwok-Yung Yuen, Susanna K. P. Lau and Patrick C. Y. Woo supplied the fungal strains. Ching-Wan Lam, Kwok-Yung Yuen, Susanna K. P. Lau and Patrick C. Y. Woo provided reagents. Kim-Chung Lee, Emily W. T. Tam, Ka-Ching Lo, Alan K. L. Tsang, Susanna K. P. Lau and Patrick C. Y. Woo analyzed the results. Kim-Chung Lee, Emily W. T. Tam and Patrick C. Y. Woo wrote the manuscript. All authors corrected the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Latge J.P. Aspergillus fumigatus and aspergillosis. Clin. Microbiol. Rev. 1999;12:310–350. doi: 10.1128/cmr.12.2.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yuen K.Y., Chan C.M., Chan K.M., Woo P.C., Che X.Y., Leung A.S., Cao L. Characterization of AFMP1: A novel target for serodiagnosis of aspergillosis. J. Clin. Microbiol. 2001;39:3830–3837. doi: 10.1128/JCM.39.11.3830-3837.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Woo P.C., Chan C.M., Leung A.S., Lau S.K., Che X.Y., Wong S.S., Cao L., Yuen K.Y. Detection of cell wall galactomannoprotein Afmp1p in culture supernatants of aspergillus fumigatus and in sera of aspergillosis patients. J. Clin. Microbiol. 2002;40:4382–4387. doi: 10.1128/JCM.40.11.4382-4387.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woo P.C., Chong K.T., Leung A.S., Wong S.S., Lau S.K., Yuen K.Y. Aflmp1 encodes an antigenic cell wall protein in Aspergillus flavus. J. Clin. Microbiol. 2003;41:845–850. doi: 10.1128/JCM.41.2.845-850.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chong K.T., Woo P.C., Lau S.K., Huang Y., Yuen K.Y. Afmp2 encodes a novel immunogenic protein of the antigenic mannoprotein superfamily in Aspergillus fumigatus. J. Clin. Microbiol. 2004;42:2287–2291. doi: 10.1128/JCM.42.5.2287-2291.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yuen K.Y., Woo P.C., Ip M.S., Liang R.H., Chiu E.K., Siau H., Ho P.L., Chen F.F., Chan T.K. Stage-specific manifestation of mold infections in bone marrow transplant recipients: Risk factors and clinical significance of positive concentrated smears. Clin. Infect. Dis. 1997;25:37–42. doi: 10.1086/514492. [DOI] [PubMed] [Google Scholar]
- 7.Chan C.M., Woo P.C., Leung A.S., Lau S.K., Che X.Y., Cao L., Yuen K.Y. Detection of antibodies specific to an antigenic cell wall galactomannoprotein for serodiagnosis of Aspergillus fumigatus aspergillosis. J. Clin. Microbiol. 2002;40:2041–2045. doi: 10.1128/JCM.40.6.2041-2045.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barton R.C. Laboratory diagnosis of invasive aspergillosis: From diagnosis to prediction of outcome. Scientifica. 2013;2013:459405. doi: 10.1155/2013/459405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ruping M.J., Gerlach S., Fischer G., Lass-Florl C., Hellmich M., Vehreschild J.J., Cornely O.A. Environmental and clinical epidemiology of Aspergillus terreus: Data from a prospective surveillance study. J. Hosp. Infect. 2011;78:226–230. doi: 10.1016/j.jhin.2011.01.020. [DOI] [PubMed] [Google Scholar]
- 10.Olivier I., Loots D. An overview of tuberculosis treatments and diagnostics. What role could metabolomics play. J. Cell Tissue Res. 2011;11:2655–2671. [Google Scholar]
- 11.Olivier I., Loots du T. A metabolomics approach to characterise and identify various mycobacterium species. J. Microbiol. Methods. 2012;88:419–426. doi: 10.1016/j.mimet.2012.01.012. [DOI] [PubMed] [Google Scholar]
- 12.Meissner-Roloff R.J., Koekemoer G., Warren R.M. A metabolomics investigation of a hyper-and hypo-virulent phenotype of beijing lineage M. tuberculosis. Metabolomics. 2012;8:1194–1203. doi: 10.1007/s11306-012-0424-6. [DOI] [Google Scholar]
- 13.Schoeman J.C., Loots D.T. Improved disease characterisation and diagnostics using metabolomics: A review. J. Cell Tissue Res. 2011;11:2673–2683. [Google Scholar]
- 14.Mahadevan S., Shah S.L., Marrie T.J., Slupsky C.M. Analysis of metabolomic data using support vector machines. Anal. Chem. 2008;80:7562–7570. doi: 10.1021/ac800954c. [DOI] [PubMed] [Google Scholar]
- 15.Van Q.N., Klose J.R., Lucas D.A., Prieto D.A., Luke B., Collins J., Burt S.K., Chmurny G.N., Issaq H.J., Conrads T.P., et al. The use of urine proteomic and metabonomic patterns for the diagnosis of interstitial cystitis and bacterial cystitis. Dis. Markers. 2003;19:169–183. doi: 10.1155/2004/530647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lv H., Hung C.S., Chaturvedi K.S., Hooton T.M., Henderson J.P. Development of an integrated metabolomic profiling approach for infectious diseases research. Analyst. 2011;136:4752–4763. doi: 10.1039/c1an15590c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tam E.W., Chen J.H., Lau E.C., Ngan A.H., Fung K.S., Lee K.C., Lam C.W., Yuen K.Y., Lau S.K., Woo P.C. Misidentification of Aspergillus nomius and Aspergillus tamarii as Aspergillus flavus: Characterization by internal transcribed spacer, beta-tubulin, and calmodulin gene sequencing, metabolic fingerprinting, and matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 2014;52:1153–1160. doi: 10.1128/JCM.03258-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.To K.K., Fung A.M., Teng J.L., Curreem S.O., Lee K.C., Yuen K.Y., Lam C.W., Lau S.K., Woo P.C. Characterization of a tsukamurella pseudo-outbreak by phenotypic tests, 16s rRNA sequencing, pulsed-field gel electrophoresis, and metabolic footprinting. J. Clin. Microbiol. 2013;51:334–338. doi: 10.1128/JCM.02845-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woo P.C., Lam C.W., Tam E.W., Leung C.K., Wong S.S., Lau S.K., Yuen K.Y. First discovery of two polyketide synthase genes for mitorubrinic acid and mitorubrinol yellow pigment biosynthesis and implications in virulence of Penicillium marneffei. PLoS Negl. Trop. Dis. 2012;6:e1871. doi: 10.1371/journal.pntd.0001871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lau S.K., Lam C.-W., Curreem S.O., Lee K.-C., Lau C.C., Chow W.-N., Ngan A.H., To K.K., Chan J.F., Hung I.F., et al. Identification of specific metabolites in culture supernatant of mycobacterium tuberculosis using metabolomics: Exploration of potential biomarkers. Emerg. Microbes Infect. 2015;4:e6. doi: 10.1038/emi.2015.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Umemura M., Nagano N., Koike H., Kawano J., Ishii T., Miyamura Y., Kikuchi M., Tamano K., Yu J., Shin-ya K., et al. Characterization of the biosynthetic gene cluster for the ribosomally synthesized cyclic peptide ustiloxin B in Aspergillus flavus. Fungal Genet. Biol. 2014;68:23–30. doi: 10.1016/j.fgb.2014.04.011. [DOI] [PubMed] [Google Scholar]
- 22.He F., Bao J., Zhang X.Y., Tu Z.C., Shi Y.M., Qi S.H. Asperterrestide A, a cytotoxic cyclic tetrapeptide from the marine-derived fungus Aspergillus terreus SCSGAF0162. J. Nat. Prod. 2013;76:1182–1186. doi: 10.1021/np300897v. [DOI] [PubMed] [Google Scholar]
- 23.Amaravani M., Prasad N.K., Ramakrishna V. COX-2 structural analysis and docking studies with gallic acid structural analogues. SpringerPlus. 2012;1:58. doi: 10.1186/2193-1801-1-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nielsen K.F., Smedsgaard J. Fungal metabolite screening: Database of 474 mycotoxins and fungal metabolites for dereplication by standardised liquid chromatography–UV–mass spectrometry methodology. J. Chromatogr. A. 2003;1002:111–136. doi: 10.1016/S0021-9673(03)00490-4. [DOI] [PubMed] [Google Scholar]
- 25.Brandt M.E., Warnock D.W. Taxonomy and classification of fungi. In: Versalovic J., Carroll K., Funke G., Jorgensen J., Landry M., Warnock D., editors. Manual of Clinical Microbiology. Volume 2 ASM Press; Washington, DC, USA: 2011. [Google Scholar]
- 26.To K.K., Lau S.K., Wu A.K., Lee R.A., Ngan A.H., Tsang C.C., Ling I.W., Yuen K.Y., Woo P.C. Phaeoacremonium parasiticum invasive infections and airway colonization characterized by agar block smear and its and beta-tubulin gene sequencing. Diagn. Microbiol. Infect. Dis. 2012;74:190–197. doi: 10.1016/j.diagmicrobio.2012.06.014. [DOI] [PubMed] [Google Scholar]
- 27.Woo P.C., Lau S.K., Ngan A.H., Tung E.T., Leung S.Y., To K.K., Cheng V.C., Yuen K.Y. Lichtheimia hongkongensis sp. Nov., a novel lichtheimia spp. associated with rhinocerebral, gastrointestinal, and cutaneous mucormycosis. Diagn. Microbiol. Infect. Dis. 2010;66:274–284. doi: 10.1016/j.diagmicrobio.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 28.Leung S.Y., Huang Y., Lau S.K., Woo P.C. Complete mitochondrial genome sequence of lichtheimia ramosa (syn. Lichtheimia hongkongensis) Genome Announc. 2014;2:e00644–00614. doi: 10.1128/genomeA.00644-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woo P.C., Lau C.C., Chong K.T., Tse H., Tsang D.N., Lee R.A., Tse C.W., Que T.L., Chung L.M., Ngan A.H., et al. Mp1 homologue-based multilocus sequence system for typing the pathogenic fungus Penicillium marneffei: A novel approach using lineage-specific genes. J. Clin. Microbiol. 2007;45:3647–3654. doi: 10.1128/JCM.00619-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Woo P.C., Lam C.W., Tam E.W., Lee K.C., Yung K.K., Leung C.K., Sze K.H., Lau S.K., Yuen K.Y. The biosynthetic pathway for a thousand-year-old natural food colorant and citrinin in Penicillium marneffei. Sci. Rep. 2014;4:6728. doi: 10.1038/srep06728. [DOI] [PMC free article] [PubMed] [Google Scholar]