Abstract
MicroRNAs (miRNAs) are small non-coding RNAs that have shown promise as noninvasive biomarkers in cardiac disease. This study was undertaken to investigate the miRNA expression profile in dogs with myxomatous mitral valve disease (MMVD). 277 miRNAs were quantified using RT-qPCR from six normal dogs (American College of Veterinary Internal Medicine Stage A), six dogs with MMVD mild to moderate cardiac enlargement (ACVIM Stage B1/B2) and six dogs with MMVD and congestive heart failure (ACVIM Stage C/D). Eleven miRNAs were differentially expressed (False Discovery Rate < 0.05). Dogs in Stage B1/B2 or C/D had four upregulated miRNAs, including three cfa-let-7/cfa-miR-98 family members, while seven others were downregulated, compared to Stage A. Expression of six of the 11 miRNAs also were significantly different between dogs in Stage C/D and those in Stage B1/B2. The expression changes were greater as disease severity increased. These miRNAs may be candidates for novel biomarkers and may provide insights into genetic regulatory pathways in canine MMVD.
Keywords: microRNA, dog, congestive heart failure, biomarker, myxomatous mitral valve disease, RT-qPCR
1. Introduction
MicroRNAs (miRNAs) are small (~22 nucleotide) single-stranded non-coding RNA molecules that negatively regulate gene expression by promoting degradation of mRNA transcripts or inhibition of protein translation [1]. It has been estimated that over 60% of human protein-coding genes are regulated by miRNAs [2]. According to the miRBase (www.mirbase.org) [3], 2588 and 1915 mature miRNAs have been identified in humans and mice respectively, but to date only 453 have been identified in dogs.
Myxomatous mitral valve disease (MMVD) affects approximately 9% of all dogs, increasing with age such that the overall cumulative incidence is greater than 40% [4,5]. Although the echocardiographic, pathological, and histological changes have been well documented, the molecular changes contributing to MMVD remain unclear. Although serum concentrations of natriuretic peptides increase in dogs with MMVD and CHF [6,7], additional biomarkers may enhance our knowledge about molecular changes and mechanisms in this common disease.
MiRNAs are emerging as potential biomarkers because of their specific expression in many diseases [2], their remarkable stability [8], and the fact that they are found in most tissue types and body fluids [8,9]. Gene expression studies have shown that miRNAs are differentially expressed in heart disease [10] and there is considerable evidence for an important role of miRNAs in cardiac remodeling and congestive heart failure (CHF) [11,12,13,14]. Gerling et al. documented a correlation in gene expression profiles between heart tissue and peripheral blood in a rat model of cardiac failure. Their findings supported a correlation between heart and blood transcriptomics [15]. Liew et al. showed that human blood expresses tissue-specific transcripts compared to various tissues, including cardiac [16]. Even though miR-499 is expressed almost exclusively in heart tissue, plasma miR-499 concentrations were significantly elevated in human patients with myocardial infarction compared with other groups of patients [17]. These findings suggest that circulating biomarkers can serve as appropriate surrogates for the heart tissues in the study of cardiac disease [18].
To our best knowledge, there are four reports on the expression profiling of miRNAs in canine cardiac disease and CHF to date. Only two investigated circulating miRNAs. Steudemann et al. evaluated circulating miRNA expression profiles in the serum from Doberman pinschers with and without dilated cardiomyopathy, but found no significant differences [19]. Hulanicka et al. analyzed the expression of nine preselected miRNAs in the plasma of Dachshunds with MMVD and identified two significantly downregulated miRNAs: cfa-miR-30b in Stage B and cfa-miR-133b in Stage C [20]. A third study examined time-dependent expression changes in heart tissues in an experimental canine model of CHF [21]. Despite the generally assumed roles in human ventricular remodeling, cfa-miR-1, cfa-miR-133, and cfa-miR-208 expressions were not altered in left ventricle (LV) cardiomyocytes from dogs. In a most recent study, Zhang et al. reported 16 miRNAs differentially expressed between the control dogs and dogs with atrial fibrillation and proposed a novel role of cfa-miR-206 in canine atrial fibrillation [22]. Despite this progress, more research is urgently needed to advance our understanding of the role of miRNAs in canine cardiac disease and CHF.
2. Results and Discussion
2.1. Differentially Expressed miRNAs
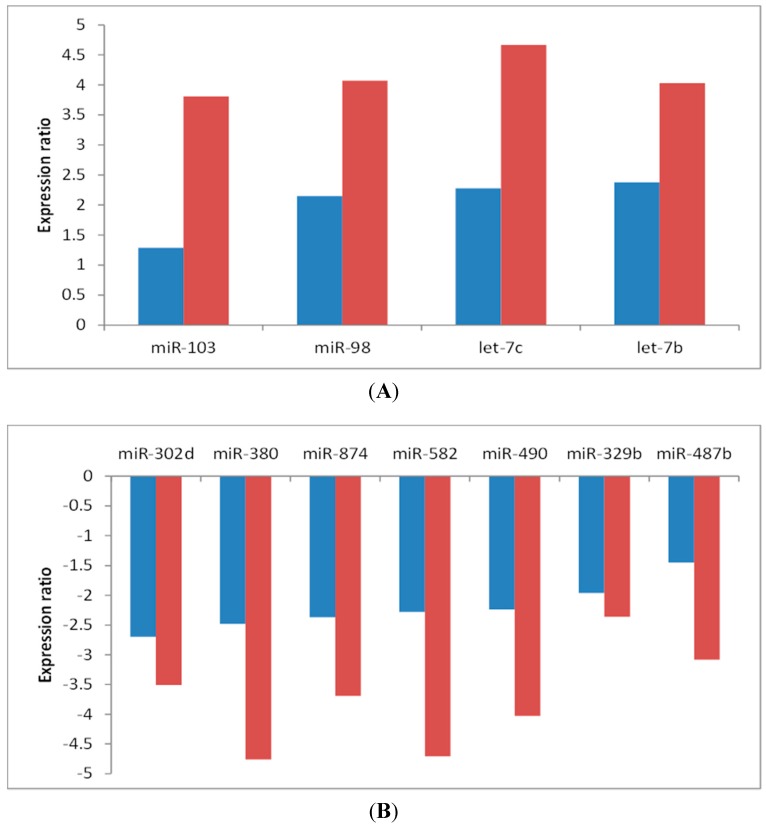
Of 277 miRNAs evaluated, Analysis of Variance (ANOVA) analysis identified 11 miRNAs with False Discovery Rate (FDR) <0.05 (Table 1). Among those, seven miRNAs (cfa-miR-302d, cfa-miR-380, cfa-miR-874, cfa-miR-582, cfa-miR-490, cfa-miR-329b, and cfa-miR-487b) displayed decreased expression, while four (cfa-miR-103, cfa-miR-98, cfa-let-7b, and cfa-let-7c) showed increased expression, in Stage B1/B2 or C/D compared with Stage A. All 11 of these differed between MMVD Stage A and Stage C/D, while nine differed between Stage A and Stage B1/B2. The expression changes were greater as disease severity increased (Figure 1). Of those 11, six (cfa-miR-582, cfa-miR-487b, cfa-miR-103, cfa-miR-98, cfa-let-7b, and cfa-let-7c) were significantly different between Stages B1/B2 and C/D (Table 1). The expression data of all 277 miRNAs are provided in Table S1.
Table 1.
Heat map of microRNAs differentially expressed between dogs in the three study groups (n = 6/group): normal dogs at risk of heart disease (Stage A), asymptomatic dogs with myxomatous mitral valve disease (MMVD) and mild to moderate cardiac enlargement (Stage B1/B2) and dogs with MMVD and congestive heart failure requiring multiple cardiac medications (Stage C/D).
| miRNA Name | ANOVA | Stage B1/B2 vs. Stage A | Stage C/D vs. Stage A | Stage C/D vs. Stage B1/B2 | ||||
|---|---|---|---|---|---|---|---|---|
| p Value | FDR a | p Value b | FC c | p Value | FC | p Value | FC | |
| cfa-miR-302d | 0.0010 | 0.0378 | 0.0050 | −2.70 | 0.0053 | −3.51 | 0.6970 | −1.30 |
| cfa-miR-380 | 0.0001 | 0.0119 | 0.0020 | −2.48 | <0.0001 | −4.76 | 0.2020 | −1.92 |
| cfa-miR-874 | 0.0016 | 0.0434 | 0.0117 | −2.37 | 0.0058 | −3.69 | 0.2547 | −1.56 |
| cfa-miR-582 | <0.0001 | 0.0004 | 0.0003 | −2.28 | <0.0001 | −4.71 | 0.0159 | −2.06 |
| cfa-miR-490 | 0.0005 | 0.0261 | 0.0101 | −2.24 | 0.0012 | −4.03 | 0.0666 | −1.80 |
| cfa-miR-329b | 0.0019 | 0.0490 | 0.0008 | −1.96 | 0.0050 | −2.36 | 0.8497 | −1.20 |
| cfa-miR-487b | 0.0012 | 0.0427 | 0.0642 | −1.45 | 0.0025 | −3.08 | 0.0023 | −2.12 |
| cfa-miR-103 | 0.0002 | 0.0140 | 0.1719 | 1.29 | 0.0011 | 3.81 | 0.0031 | 2.96 |
| cfa-miR-98 | 0.0014 | 0.0428 | 0.0090 | 2.15 | 0.0029 | 4.07 | 0.0341 | 1.90 |
| cfa-let-7c | 0.0003 | 0.0218 | 0.0309 | 2.28 | 0.0012 | 4.67 | 0.0080 | 2.05 |
| cfa-let-7b | 0.0006 | 0.0269 | 0.0123 | 2.38 | 0.0009 | 4.03 | 0.0342 | 1.69 |
a False Discovery Rate; b Student’s t-test; c Fold change: negative numbers indicate decreases in expression, positive numbers for increases in expression. Red, green, and grey colors indicate a significant increase, decrease, and non-significance in expression, respectively.
Figure 1.
Expression ratios of differentially expressed microRNAs in the serum of dogs with myxomatous mitral valve disease (MMVD). Groups of six dogs each included normal dogs at risk of heart disease (Stage A), asymptomatic dogs with MMVD and mild to moderate cardiac enlargement (Stage B1/B2) and dogs with MMVD and congestive heart failure requiring multiple cardiac medications (Stage C/D). (A) shows miRNAs upregulated in dogs with stage B1/B2 or stage C/D compared to stage A; and (B) shows miRNAs downregulated in dogs with stage B1/B2 or stage C/D compared to stage A. Blue bars indicate the changes in stage B1/B2 and red bars indicate the changes in stage C/D, compared to stage A.
2.2. Potential Role of the Cfa-let-7/cfa-miR-98 Family Members in Canine MMVD
Recent studies in humans have associated the let-7 family with the development of cardiovascular diseases, and upregulation of let-7 expression was observed in many patients with cardiovascular diseases, including cardiac hypertrophy, dilated cardiomyopathy, myocardial infarction and CHF [23,24]. Experimental evidence suggests that circulating let-7b and cellular let-7i might be biomarkers for myocardial infarction and dilated cardiomyopathy, respectively [25,26]. Remarkably, let-7c was found to be enriched in cardiac valve in dogs, monkeys and rats [27]. Angiotensin II (Ang II) plays an important role in the pathogenesis of CHF secondary to MMVD and other diseases [28]. Recent studies showed that thioredoxin (Trx1) negatively regulated AngII-induced cardiac hypertrophy by increasing the expression of let-7/miR-98 family members in human CHF and let-7/miR-98 was the downstream effector of Trx1 [29,30]. In a previous canine gene expression study (National Center for Biotechnology Information’s Gene Expression Omnibus (GEO) Accession No. GSE64544) [31], we found that Trx1 was upregulated 2.6-fold (p = 0.002) in the LV, while angiotensinogen, the precursor of Ang II, was downregulated by more than 8-fold (p ≤ 0.001) in the mitral valve (MV) of dogs with MMVD. This suggests that the Trx1-let-7/miR-98-Ang II regulatory circuitry may also exist in canine MMVD and CHF.
Target prediction analysis by TargetScan suggested that tuberous sclerosis 1 (TSC1), a gene that encodes hamartin, is a predicted gene target for cfa-let-7c (Table S2). Mutations in TSC1 cause a genetic syndrome called Tuberous sclerosis complex (TSC) [32]. Arrhythmia is relatively common in patients with TSC, and at least 50% of children with TSC developed cardiac rhabdomyomas, which in some cases can lead to CHF [32]. Mutations in TSC1 have also been associated with mitral valve prolapse [33]. Taken together, our data suggest that cfa-let-7/cfa-miR-98 family may play an important role in MMVD in dogs.
2.3. Cfa-miR-302d as a Potential Negative Regulator of TGF-β Signaling
TGF-β signaling pathway has been implicated to play a central role in the pathology of canine MMVD [28,34,35]. In a recent study using human kidney mesangial cells, miR-302 expression was shown to inhibit TGF-β receptor II (TβRII) transcription [36]. As a result, miR-302d decreased TGF-β-induced epithelial mesenchymal transition and attenuated TβRII-mediated production of fibronectin and thrombospondin [36]. The current study showed decreased expression of cfa-miR-302d in the serum of dogs with MMVD. This, along with previous observations in dogs with MMVD of increased gene and protein expression of fibronectin and TβRI/TβRII, respectively in the MV [34,35] and increased thrombospondin 1 in LV and MV and thrombospondin 4 in the LV [31], suggest a potential role of cfa-miR-302d/TGF-β regulatory network in the pathology of MMVD in dogs.
2.4. Other MiRNAs
Members of the miR-103/miR-107 family have been implicated in the pathogenesis of cardiovascular disease and were recently shown to regulate insulin sensitivity in obese mice [37], the exact role of miR-103 in CHF remains unclear. TβRII and TβRIII are among predicted targets for cfa-miR-103 (Table S2 ), suggesting a potential role for cfa-miR-103 in canine MMVD.
MiR-874 was shown to cause myocardial cell death, an important reason for CHF and myocardial infarction, by targeting the caspase-8 in both in vitro and in vivo mouse models [38]. Circulating miR-380 and miR-582 were reported as a potential noninvasive biomarker for survival prediction after acute myocardial infarction [39] and for detection of deep vein thrombosis [40], respectively. A recent study has demonstrated that miR-329 is a negative regulator of angiogenesis by directly targeting CD146 [41]. Gene silencing of the four 14q32 miRNA cluster, including miR-329 and miR-487b, resulted in increased perfusion after ischemia in mice [42]. Additional research is necessary to unravel potential roles of these miRNAs in canine MMVD.
Neither cfa-miR-133 nor cfa-miR-30 reached statistical significance in our ANOVA analysis. Although this differs from the findings of Hulanicka et al. [20], the group did not report ANOVA analysis, instead employed a series of t-tests to address a multisample hypothesis [43]. Chen et al. also failed to observe changes in cfa-miR-133 expression in LV cardiomyocyte of an induced model of canine CHF [21].
Besides let-7c, both miR-125b and miR-204 were enriched in cardiac valves of rat, dog and monkey [27]. Inhibitory interaction between miR-204 and TβRII was identified in silico and demonstrated experimentally in human tissue culture cells [27]. Although neither showed significance in our overall ANOVA analysis (FDR = 0.12 and FDR = 0.43, respectively), simple t-tests suggested they were different between stages B1/B2 and C/D (p = 0.03 and p = 0.01, respectively). This suggests that the lack of statistical significance may reflect a type 2 error (false negative) due to small sample size.
2.5. Limitations
This pilot study was limited by a small sample size and heterogeneity of the dogs from which serum samples were collected. Future investigations with larger cohorts of animals are required before any clinical applications can be considered. It was also not possible to determine the effects of medications on expression due to the small number of animals and variable medications they were receiving. This warrants additional research in future studies. In addition, in situ hybridization can provide additional confirmation on some of the important miRNAs. Nonetheless, the findings reported here suggest there is an opportunity for using some of these circulating miRNAs as biomarkers for diagnosis, prognosis or monitoring response to treatment in MMVD in dogs. Our pilot study will provide a platform and knowledge for future studies.
3. Experimental Section
3.1. Animals and Sample Collection
For the current study, 18 dogs of various breeds were classified either as being healthy or with MMVD by echocardiography performed or evaluated by a board-certified veterinary cardiologist, pathological examination of the heart, or both (Table S3). Dogs were further classified into one of three groups of six dogs each using the American College of Veterinary Internal Medicine guidelines for diagnosis of MMVD [4]: normal dogs at risk of heart disease (Stage A: 2 neutered males [MN], 4 spayed females [FS]; age 8.5 ± 2.7 years; body weight 10.2 ± 13.6 kg), asymptomatic dogs with MMVD and mild to moderate cardiac enlargement (Stage B1/B2: 2 MN, 4 FS; age 10.9 ± 2.7 years; body weight 13.5 ± 8.8 kg) and dogs with MMVD and CHF requiring multiple cardiac medications (Stage C/D: 2 MN, 4 FS; age 10.3 ± 3.4 years; body weight 10.4 ± 6.3 kg). Venous blood was collected and serum was separated and frozen at −80 °C until analysis. The study protocol was reviewed and approved by Nestlé Purina’s Institutional Animal Care and Use Committee.
3.2. Quantitative RT-PCR, Data Normalization and Statistical Analysis
Serum sample processing and real-time PCR assay were performed at a commercial laboratory using the miScript miRNA PCR Array system (Qiagen, Fredrick, MD, USA) [44]. Total RNA was extracted and purified from 200 microliters serum samples using the miRNeasy Serum/Plasma Kit from Qiagen. The cycle threshold (Ct) value was measured for each miRNA. The normalization factor for each sample was determined according to Mestdagh et al. [45]. Ct values that were undetermined or greater than 30 were reassigned with 30. Normalized Ct (ΔCt) was obtained by subtracting the normalization factor from each Ct value. The expression of each miRNA is reported as 2−∆Ct. ANOVA was performed to identify differentially expressed miRNAs among the three stages of MMVD. FDR was calculated to control multiple testing errors [46]. The miRNAs with FDR < 0.05 were deemed as significant. Significant miRNAs were subject to Student’s t-test with equal variance to compare the pairwise difference between stages. Expression fold change also was calculated.
3.3. Computational Prediction of MiRNA Targets
MiRNA targets were predicted using the software TargetScan [47]. Briefly, the software scans for the presence of conserved 7 or 8 mer that matches the miRNA’s seed region, which is the nucleotide sequence in positions 2–7 of a mature miRNA. Predicted targets of cfa-let-7b, cfa-let-7c, and cfa-miR-103 were downloaded from the TargetScan website.
4. Conclusions
The study surveyed the expression profiling of 277 circulating miRNAs in the serum of dogs at different stages of MMVD and CHF using quantitative real-time PCR array system. Eleven miRNAs were differentially expressed (FDR < 0.05). Dogs in Stage B1/B2 or C/D had four upregulated miRNAs, including three cfa-let-7/cfa-miR-98 family members, while seven others were downregulated, compared to Stage A. The expression changes were greater as disease severity increased. Our study suggests that there is an opportunity for using some of these circulating miRNAs as biomarkers for diagnosis, prognosis or monitoring response to treatment in MMVD in dogs. Further investigation of these miRNAs may also shed light on genetic regulatory pathways on canine MMVD.
Dogs and humans share some similarities in MMVD, including degenerative valvular structure, expression patterns of extracellular matrix proteins, and some common signaling pathways [48]. Therefore, results from this study, including changes in the cfa-let-7/cfa-miR-98 family members, may be relevant to the study of human MMVD.
Acknowledgments
This study was funded by Nestlé Purina Research. Lisa M. Freeman and John E. Rush report grants and personal fees from Nestlé Purina Research. Qinghong Li and Dorothy P. Laflamme are current employees of Nestlé Purina Research.
Supplementary Materials
Supplementary materials can be found at http://www.mdpi.com/1422-0067/16/06/14098/s1.
Author Contributions
Qinghong Li conceived the study, performed analysis and interpretation of the results, and wrote the manuscript. Lisa M. Freeman and John E. Rush performed examination and classification of dogs with MMDV or healthy hearts, collected the serum samples and contributed to the preparation of manuscript. Dorothy P. Laflamme contributed to the study design, and manuscript preparation.
Conflicts of Interest
Lisa M. Freeman and John E. Rush report grants and personal fees from Nestlé Purina Research. Qinghong Li and Dorothy P. Laflamme are current employees of Nestlé Purina Research.
References
- 1.Bartel D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Sayed D., Abdellatif M. MicroRNAs in development and disease. Physiol. Rev. 2011;91:827–887. doi: 10.1152/physrev.00006.2010. [DOI] [PubMed] [Google Scholar]
- 3.Griffiths-Jones S., Grocock R.J., van D.S., Bateman A., Enright A.J. MiRBase: MicroRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atkins C., Bonagura J., Ettinger S., Fox P., Gordon S., Haggstrom J., Hamlin R., Keene B., Luis-Fuentes V., Stepien R. Guidelines for the diagnosis and treatment of canine chronic valvular heart disease. J. Vet. Intern. Med. 2009;23:1142–1150. doi: 10.1111/j.1939-1676.2009.0392.x. [DOI] [PubMed] [Google Scholar]
- 5.Rush J.E., Cunningham S.M. Chronic valvular disease in dogs. In: Bonagura J.D., Twedt D.C., editors. Kirk’s Current Veterinary Therapy. 14th ed. Saunders; St. Louis, MO, USA: 2014. pp. 784–794. [Google Scholar]
- 6.Fox P.R., Oyama M.A., Hezzell M.J., Rush J.E., Nguyenba T.P., Defrancesco T.C., Lehmkuhl L.B., Kellihan H.B., Bulmer B., Gordon S.G., et al. Relationship of plasma N-terminal pro-brain natriuretic peptide concentrations to heart failure classification and cause of respiratory distress in dogs using a 2nd generation ELISA assay. J. Vet. Intern. Med. 2015;29:171–179. doi: 10.1111/jvim.12472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reynolds C.A., Brown D.C., Rush J.E., Fox P.R., Nguyenba T.P., Lehmkuhl L.B., Gordon S.G., Kellihan H.B., Stepien R.L., Lefbom B.K., et al. Prediction of first onset of congestive heart failure in dogs with degenerative mitral valve disease: The PREDICT cohort study. J. Vet. Cardiol. 2012;14:193–202. doi: 10.1016/j.jvc.2012.01.008. [DOI] [PubMed] [Google Scholar]
- 8.Mitchell P.S., Parkin R.K., Kroh E.M., Fritz B.R., Wyman S.K., Pogosova-Agadjanyan E.L., Peterson A., Noteboom J., O’Briant K.C., Allen A., et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen X., Ba Y., Ma L., Cai X., Yin Y., Wang K., Guo J., Zhang Y., Chen J., Guo X., et al. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 10.Bronze-da-Rocha E. MicroRNAs expression profiles in cardiovascular diseases. Biomed. Res. Int. 2014;2014:985408. doi: 10.1155/2014/985408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oliveira-Carvalho V., da Silva M.M., Guimaraes G.V., Bacal F., Bocchi E.A. MicroRNAs: New players in heart failure. Mol. Biol. Rep. 2013;40:2663–2670. doi: 10.1007/s11033-012-2352-y. [DOI] [PubMed] [Google Scholar]
- 12.Divakaran V., Mann D.L. The emerging role of microRNAs in cardiac remodeling and heart failure. Circ. Res. 2008;103:1072–1083. doi: 10.1161/CIRCRESAHA.108.183087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Condorelli G., Latronico M.V., Cavarretta E. MicroRNAs in cardiovascular diseases: Current knowledge and the road ahead. J. Am. Coll. Cardiol. 2014;63:2177–2187. doi: 10.1016/j.jacc.2014.01.050. [DOI] [PubMed] [Google Scholar]
- 14.Van R.E., Sutherland L.B., Liu N., Williams A.H., McAnally J., Gerard R.D., Richardson J.A., Olson E.N. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc. Natl. Acad. Sci. USA. 2006;103:18255–18260. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerling I.C., Ahokas R.A., Kamalov G., Zhao W., Bhattacharya S.K., Sun Y., Weber K.T. Gene expression profiles of peripheral blood mononuclear cells reveal transcriptional signatures as novel biomarkers for cardiac remodeling in rats with aldosteronism and hypertensive heart disease. JACC Heart Fail. 2013;1:469–476. doi: 10.1016/S2213-1779(13)00374-0. [DOI] [PubMed] [Google Scholar]
- 16.Liew C.C., Ma J., Tang H.C., Zheng R., Dempsey A.A. The peripheral blood transcriptome dynamically reflects system wide biology: A potential diagnostic tool. J. Lab. Clin. Med. 2006;147:126–132. doi: 10.1016/j.lab.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 17.Adachi T., Nakanishi M., Otsuka Y., Nishimura K., Hirokawa G., Goto Y., Nonogi H., Iwai N. Plasma microRNA 499 as a biomarker of acute myocardial infarction. Clin. Chem. 2010;56:1183–1185. doi: 10.1373/clinchem.2010.144121. [DOI] [PubMed] [Google Scholar]
- 18.Shehadeh L.A., Hare J.M. Ribonucleic acid biomarkers for heart failure is there a correlation between heart and blood transcriptomics? JACC Heart Fail. 2013;1:477–479. doi: 10.1016/j.jchf.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steudemann C., Bauersachs S., Weber K., Wess G. Detection and comparison of microRNA expression in the serum of Doberman Pinschers with dilated cardiomyopathy and healthy controls. BMC Vet. Res. 2013;9:12. doi: 10.1186/1746-6148-9-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hulanicka M., Garncarz M., Parzeniecka-Jaworska M., Jank M. Plasma miRNAs as potential biomarkers of chronic degenerative valvular disease in Dachshunds. BMC Vet. Res. 2014;10:205. doi: 10.1186/s12917-014-0205-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Y., Wakili R., Xiao J., Wu C.T., Luo X., Clauss S., Dawson K., Qi X., Naud P., Shi Y.F., et al. Detailed characterization of microRNA changes in a canine heart failure model: Relationship to arrhythmogenic structural remodeling. J. Mol. Cell. Cardiol. 2014;77C:113–124. doi: 10.1016/j.yjmcc.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y., Zheng S., Geng Y., Xue J., Wang Z., Xie X., Wang J., Zhang S., Hou Y. MicroRNA profiling of atrial fibrillation in canines: MiR-206 modulates intrinsic cardiac autonomic nerve remodeling by regulating SOD1. PLoS ONE. 2015;10:e0122674. doi: 10.1371/journal.pone.0122674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cao L., Kong L.P., Yu Z.B., Han S.P., Bai Y.F., Zhu J., Hu X., Zhu C., Zhu S., Guo X.R. MicroRNA expression profiling of the developing mouse heart. Int. J. Mol. Med. 2012;30:1095–1104. doi: 10.3892/ijmm.2012.1092. [DOI] [PubMed] [Google Scholar]
- 24.Ikeda S., Kong S.W., Lu J., Bisping E., Zhang H., Allen P.D., Golub T.R., Pieske B., Pu W.T. Altered microRNA expression in human heart disease. Physiol. Genomics. 2007;31:367–373. doi: 10.1152/physiolgenomics.00144.2007. [DOI] [PubMed] [Google Scholar]
- 25.Long G., Wang F., Li H., Yin Z., Sandip C., Lou Y., Wang Y., Chen C., Wang D.W. Circulating miR-30a, miR-126 and let-7b as biomarker for ischemic stroke in humans. BMC Neurol. 2013;13:178. doi: 10.1186/1471-2377-13-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Satoh M., Minami Y., Takahashi Y., Tabuchi T., Nakamura M. A cellular microRNA, let-7i, is a novel biomarker for clinical outcome in patients with dilated cardiomyopathy. J. Card. Fail. 2011;17:923–929. doi: 10.1016/j.cardfail.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 27.Vacchi-Suzzi C., Hahne F., Scheubel P., Marcellin M., Dubost V., Westphal M., Boeglen C., Buchmann-Moller S., Cheung M.S., Cordier A., et al. Heart structure-specific transcriptomic atlas reveals conserved microRNA–mRNA interactions. PLoS ONE. 2013;8:e52442. doi: 10.1371/journal.pone.0052442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Orton E.C., Lacerda C.M., Maclea H.B. Signaling pathways in mitral valve degeneration. J. Vet. Cardiol. 2012;14:7–17. doi: 10.1016/j.jvc.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 29.Yang Y., Ago T., Zhai P., Abdellatif M., Sadoshima J. Thioredoxin 1 negatively regulates angiotensin II-induced cardiac hypertrophy through upregulation of miR-98/let-7. Circ. Res. 2011;108:305–313. doi: 10.1161/CIRCRESAHA.110.228437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun H., Wang Y. Restriction of big hearts by a small RNA. Circ. Res. 2011;108:274–276. doi: 10.1161/CIRCRESAHA.110.239426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Q., Freeman L.M., Rush J.E., Huggings G.S., Kennedy A.D., Labuda J.A., Laflamme D.P., Hannah S.S. Veterinary medicine and multi-omics research for future nutrition targets: Metabolomics and transcriptomics of the common degenerative mitral valve disease in dogs. OMICS. 2015 doi: 10.1089/omi.2015.0057. in press. [DOI] [PubMed] [Google Scholar]
- 32.Hinton R.B., Prakash A., Romp R.L., Krueger D.A., Knilans T.K. Cardiovascular manifestations of tuberous sclerosis complex and summary of the revised diagnostic criteria and surveillance and management recommendations from the international tuberous sclerosis consensus group. J. Am. Heart Assoc. 2014;3:e001493. doi: 10.1161/JAHA.114.001493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sessa A., Righetti M., Battini G. Autosomal recessive and dominant polycystic kidney diseases. Minerva Urol. Nefrol. 2004;56:329–338. [PubMed] [Google Scholar]
- 34.Oyama M.A., Chittur S.V. Genomic expression patterns of mitral valve tissues from dogs with degenerative mitral valve disease. Am. J. Vet. Res. 2006;67:1307–1318. doi: 10.2460/ajvr.67.8.1307. [DOI] [PubMed] [Google Scholar]
- 35.Disatian S., Orton E.C. Autocrine serotonin and transforming growth factor beta 1 signaling mediates spontaneous myxomatous mitral valve disease. J. Heart Valve Dis. 2009;18:44–51. [PubMed] [Google Scholar]
- 36.Faherty N., Curran S.P., O’Donovan H., Martin F., Godson C., Brazil D.P., Crean J.K. CCN2/CTGF increases expression of miR-302 microRNAs, which target the TGFbeta type II receptor with implications for nephropathic cell phenotypes. J. Cell Sci. 2012;125:5621–5629. doi: 10.1242/jcs.105528. [DOI] [PubMed] [Google Scholar]
- 37.Trajkovski M., Hausser J., Soutschek J., Bhat B., Akin A., Zavolan M., Heim M.H., Stoffel M. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature. 2011;474:649–653. doi: 10.1038/nature10112. [DOI] [PubMed] [Google Scholar]
- 38.Wang K., Liu F., Zhou L.Y., Ding S.L., Long B., Liu C.Y., Sun T., Fan Y.Y., Sun L., Li P.F. MiR-874 regulates myocardial necrosis by targeting caspase-8. Cell Death Dis. 2013;4:e709. doi: 10.1038/cddis.2013.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matsumoto S., Sakata Y., Nakatani D., Suna S., Mizuno H., Shimizu M., Usami M., Sasaki T., Sato H., Kawahara Y., et al. A subset of circulating microRNAs are predictive for cardiac death after discharge for acute myocardial infarction. Biochem. Biophys. Res. Commun. 2012;427:280–284. doi: 10.1016/j.bbrc.2012.09.039. [DOI] [PubMed] [Google Scholar]
- 40.Qin J., Liang H., Shi D., Dai J., Xu Z., Chen D., Chen X., Jiang Q. A panel of microRNAs as a new biomarkers for the detection of deep vein thrombosis. J. Thromb. Thrombolysis. 2015;39:215–221. doi: 10.1007/s11239-014-1131-0. [DOI] [PubMed] [Google Scholar]
- 41.Wang P., Luo Y., Duan H., Xing S., Zhang J., Lu D., Feng J., Yang D., Song L., Yan X. MicroRNA 329 suppresses angiogenesis by targeting CD146. Mol. Cell Biol. 2013;33:3689–3699. doi: 10.1128/MCB.00343-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Welten S.M., Bastiaansen A.J., de Jong R.C., de Vries M.R., Peters E.A., Boonstra M.C., Sheikh S.P., Monica N.L., Kandimalla E.R., Quax P.H., et al. Inhibition of 14q32 microRNAs miR-329, miR-487b, miR-494, and miR-495 increases neovascularization and blood flow recovery after ischemia. Circ. Res. 2014;115:696–708. doi: 10.1161/CIRCRESAHA.114.304747. [DOI] [PubMed] [Google Scholar]
- 43.Zar J.H. Multisample hypotheses: The analysis of aariance. In: Ryu T., editor. Biostatistical Analysis. 4th ed. Prentice-Hall; Upper Saddle River, NJ, USA: 1999. pp. 177–207. [Google Scholar]
- 44.Mestdagh P., Hartmann N., Baeriswyl L., Andreasen D., Bernard N., Chen C., Cheo D., D’Andrade P., DeMayo M., Dennis L., et al. Evaluation of quantitative miRNA expression platforms in the microRNA quality control (miRQC) study. Nat. Methods. 2014;11:809–815. doi: 10.1038/nmeth.3014. [DOI] [PubMed] [Google Scholar]
- 45.Mestdagh P., van V.P., de W.A., Muth D., Westermann F., Speleman F., Vandesompele J. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol. 2009;10:R64. doi: 10.1186/gb-2009-10-6-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Benjamini Y., Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B. 1995;57:289–300. [Google Scholar]
- 47.Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 48.Aupperle H., Disatian S. Pathology, protein expression and signaling in myxomatous mitral valve degeneration: Comparison of dogs and humans. J. Vet. Cardiol. 2012;14:59–71. doi: 10.1016/j.jvc.2012.01.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.