Abstract
Background
Cardiac rhythmic activity is initiated in functionally specialized areas of the heart. Hyperpolarization-activated and cyclic nucleotide-gated (HCN) channels are fundamental for these processes of cardiac physiology.
Results
Here we investigated transcript and protein expression patterns of HCN channels in HL-1 cardiomyocytes using a combination of quantitative PCR analysis and immunocytochemistry. Gene expression profiles of hcn1, hcn2 and hcn4 were acutely affected during HL-1 cell propagation. In addition, distinct expression patterns were uncovered for HCN1, HCN2 and HCN4 proteins.
Conclusions
Our results suggest that HCN channel isoforms might be involved in the concerted differentiation of HL-1 cells and may indirectly affect the occurrence of contractile HL-1 cell activity. We expect that these findings will promote studies on other molecular markers that contribute to cardiac physiology.
Electronic supplementary material
The online version of this article (doi:10.1186/s12860-015-0065-5) contains supplementary material, which is available to authorized users.
Keywords: Pacemaker channel, Cardiac cells, Transcript quantification, Signaling, Contractile activity
Background
Cardiac autonomous activity originates in the sinoatrial node (SAN) where primary pacemaker cells generate spontaneous rhythmic action potentials that trigger heart muscle contraction. Generation of rhythmic action potentials in the heart involves several channel proteins including T- and L-type Ca2+ channels, a K+ channel and a pacemaker channel [1]. Activation of T- and L-type Ca2+ channels results in depolarization, followed by repolarization of the membrane potential due to opening of K+ channels. Near the K+ equilibrium potential pacemaker channels open and give way to an inward Na+ current. Subsequent membrane depolarization towards the threshold voltage induces a new action potential. This pacemaker current, also termed Ih or If, is an essential component of cardiac automaticity [2, 3].
Hyperpolarization-activated and cyclic nucleotide-gated (HCN) channels are the molecular basis of If [4]. In mammals four HCN channel subunits (HCN1 – 4) with different biophysical properties and expression patterns have been identified [5, 6]. In the murine SAN the HCN4 isoform is the predominant subunit [7–9]. Although reports on expression of HCN1 – 4 in different species are not consistent, HCN1 and HCN2 have been identified consistently in murine cardiac tissue [7–10]. Transgenic studies in mice further emphasized the importance of HCN channels for cardiac function. Knockout of HCN4 as well as a point mutation in the cyclic nucleotide-binding site of HCN4 caused embryonic lethality due to cardiac malfunction [11, 12]. Conditional knockout of HCN4 in adult mice was either lethal [13] or induced cardiac arrhythmia with a reduction of If by about 75 % in the SAN [14]. Knockout of HCN2 reduced sinoatrial If by approximately 30 % [15], while a combined knockout of HCN2 and HCN4 caused a complete disruption of ventricular If [16].
However, little is known about the link between cardiac functionality and HCN channel isoform expression on the cellular level. Here we employed HL-1 cardiomyocytes, a murine atrial myocyte-based cell line that mimic properties of adult cardiomyocytes and express cardiac signature genes [17, 18]. Due to their ability to develop features similar to cardiac tissue, including spontaneous contractile activity [18], HL-1 cells are a versatile cellular model system for investigating molecular components of cardiac physiology in vitro.
In this study we focused on expression profiles of HCN channel isoforms in HL-1 cells. Differential expression of all four HCN isoform genes was examined via quantitative PCR. As an independent method, immunocytochemistry was used to analyze the presence of these proteins in HL-1 cells. Notably, HCN channel isoforms were not detected in contracting HL-1 cells. However, our results suggest a direct correlation between hcn gene expression and HL-1 cell differentiation. Thus HL-1 cells can serve as a model system for in vitro studies of cardiomyocyte development and differentiation.
Methods
HL-1 cells
HL-1 cells were obtained from Dr. W. C. Claycomb (Louisiana State University Health Science Center, New Orleans, LA, USA) and were cultured as recommended [17]. Briefly, cells were maintained in Claycomb Medium (SAFC Biosciences, Hamburg, Germany) supplemented with 10 % FBS (fetal bovine serum, SAFC Biosciences), 100 μM noradrenaline (Sigma-Aldrich, Munich, Germany), 2 mM L-glutamine (Life Technologies, Darmstadt, Germany), 300 μM ascorbic acid (Sigma-Aldrich), and 100U/ml:100 μg/ml streptomycin:penicillin (Life Technologies). Cells were cultivated on dishes coated with 0.02 % gelatin (Life Technologies) and 12.5 μg/ml fibronectin (from bovine plasma, Sigma-Aldrich) at 37 °C, 5 % CO2, and 95 % relative humidity. For imaging, cells were cultivated either on pre-coated glass coverslips or on 35 mm cell culture dishes with glass bottom (Ibidi, Martinsried, Germany).
RNA preparation and cDNA synthesis
Total RNA was isolated from HL-1 cells and mouse brain using the DNA/RNA/Protein AllPrep® Kit (Qiagen, Hilden, Germany) according to the supplier's protocol. Animal experiments were performed in accordance with institutional protocols in compliance with national and international guidelines (Directive 2010/63/EU). RNA samples were split for two independent first-strand cDNA syntheses using Oligo-dT primers (Qiagen) and Moloney Murine Leukemia Virus reverse transcriptase (M-MLV-RT, Life Technologies) according to the supplier’s protocol.
Quantification of gene expression by real-time PCR
Thermocycling was performed in a LightCycler 1.5 (Roche, Mannheim, Germany) using the QuantiTect SYBR Green PCR Kit (Qiagen) according to the supplier’s protocol. Gene-specific primers were purchased from MWG Operon (Ebersberg, Germany). Specificity and efficiency of primers (Table 1) were confirmed via BLAST analysis and PCR on cloned gene fragments. qPCR reactions were performed on 1 μl aliquots of first-strand cDNA samples in a total volume of 20 μl. The gapdh primers were designed to bind in exons separated by an intron of 134 bp to check for genomic impurities. qPCR runs were concluded by generating a melting curve to confirm homogeneity of amplified fragments. Results were analyzed using the Ct method. Gene expression levels were normalized to the housekeeping gene gapdh. Data sets generated in the course of one experimental series were normalized to values at a specified culture density (i. e. approximately 50 % confluency). Samples were grouped according to visually determined culture densities: five groups were defined, i. e. with 1–20 %, 21–40 %, 41–60 %, 61–80 %, and 81–100 % cell density. For analysis, mean ± s.e.m. values were calculated.
Table 1.
Primer pairs for qPCR on HL-1 cell and mouse brain cDNA
| Protein | Gene | Primer | Sequence (5‘→3‘) | Tm | Amplicon size |
|---|---|---|---|---|---|
| GAPDH | gapdh | Forward | GGTATCGTGGAAGGACTCATG | 62 °C | 150 bp/284 bp |
| Reverse | GCTGCCAAGGCTGTGGGC | ||||
| HCN1 | hcn1 | Forward | ACTGTGGGCGAATCCCTGG | 62 °C | 184 bp |
| Reverse | CCACCAGCAGCTGTGCAGA | ||||
| HCN2 | hcn2 | Forward | GGAGAATGCCATCATCCAGG | 62 °C | 149 bp |
| Reverse | CAGCAGGCTGTGGCCATGA | ||||
| HCN3 | hcn3 | Forward | TGGGAACCACTGGTGCACG | 62 °C | 141 bp |
| Reverse | TGAGCGTCTAGCAGATCGAG | ||||
| HCN4 | hcn4 | Forward | CACGACCTCAACTCAGGCG | 62 °C | 153 bp |
| Reverse | CAGCGGGGTCCATATAACAG |
Primer sequences and amplicon sizes are based on mouse sequences (accession numbers: NM_008084.2 (gapdh), NM_010408.3 (hcn1), NM_008226.2 (hcn2), NM_008227.1 (hcn3), and NM_001081192.1 (hcn4)). Amplicon sizes of gapdh fragments are indicated for cDNA (150 bp) and genomic DNA (284 bp).
Statistical analysis
All data are represented as mean ± s.e.m. (standard error of the mean). The two-tailed independent Student’s t test was applied for calculation of p values. One-way ANOVA (analysis of variance) was performed using GraphPad Prism v.5.04 for Windows (GraphPad Prism Software, San Diego, California, USA) for analysis of gene expression profiles. A p value of <0.05 was considered significant.
Antibodies
Primary antibodies for immunocytochemistry were anti-mHCN1 (HCN1α, guinea pig, 1:500), anti-mHCN2 (HCN2α, rabbit, 1:500), and anti-mHCN4 (PG2-1A4, rat, 1:2) (Additional file 1: Table S1). Secondary antibodies were anti-rabbit-Cy2 (polyclonal, 1:400, Dianova), anti-rat-Cy3 (polyclonal, 1:500, Dianova), and anti-guinea pig-A594 (polyclonal, 1:500, Dianova).
Immunocytochemistry
For immunostaining, cells were fixed for 10 min in 4 % (w/v) paraformaldehyde. After washing with PBS (130 mM NaCl, 70 mM Na2HPO4, 30 mM NaH2PO4, pH 7.4), unspecific binding of antibodies was blocked for 30 min in CT (5 % (v/v) chemiblocker (Chemicon, Darmstadt, Germany) and 0.5 % (v/v) Triton-X in PBS). Incubation with primary antibodies was performed for 60 min in CT. Secondary antibodies in CT were added for 60 min after washing with PBS. Samples were mounted on microscope slides with Aqua-Poly/Mount (Polysciences, Eppelheim, Germany) or preserved in PBS.
Microscopy
Fluorescent images were obtained with an inverted confocal microscope (TCS SP5II, Leica, Wetzlar, Germany) using a 63x oil immersion objective. Z-stacks (5 x 1 μm) were registered, and maximum intensity projections were generated using WCIF ImageJ 1.37c software. For identification of contracting cells, 15 sequential pictures (3frames/s) were taken using an inverted microscope (Axio Vert.A1, Zeiss, Göttingen, Germany) equipped with a digital SLR camera (Nikon D200, Nikon, Düsseldorf, Germany). Pictures were converted into video clips (Additional file 2: Video S1) (3frames/s) using ImageJ software.
Results
Cardiac contraction strongly depends on consecutive activity of different voltage-dependent ion channel classes [1]. In the heart, HCN channels not only restore the resting membrane potential from hyperpolarized values but also contribute to a novel depolarization due to their pronounced Na+ conductance. In this study we examined expression patterns of four HCN channel isoforms in HL-1 cells, both on the transcript and on the protein level.
Gene expression of HCN channel isoforms in HL-1 cells
For investigation of HCN channel isoform gene expression, specific primers were designed to amplify defined hcn fragments. The specificity of these primers was confirmed on mouse brain cDNA (Fig. 1a), where hcn1 - 4 isoforms are known to be expressed [5, 6], as well as on HL-1 cell-derived cDNA (Fig. 1b). Amplified fragments displayed expected sizes, i. e. 184 bp for hcn1, 149 bp for hcn2, 141 bp for hcn3, 153 bp for hcn4, and 150 bp for gapdh. Gene expression of hcn1 – 4 in confluent HL-1 cell culture was determined via qPCR and normalized to gapdh (Fig. 1c). While transcript levels of hcn1 and hcn2 were rather similar, hcn4 transcripts were almost twice as abundant as hcn1 or hcn2. In this quantitative analysis only marginal amounts of hcn3 transcripts were detected.
Fig. 1.
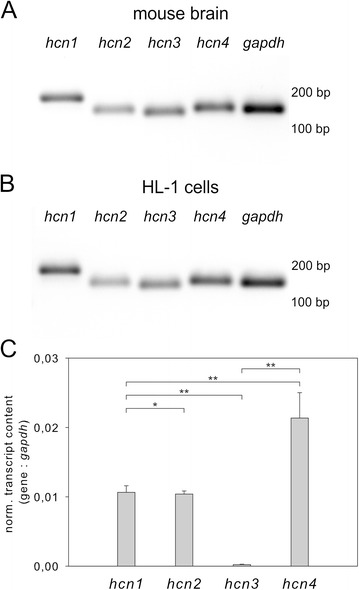
Gene expression of HCN channel isoforms in mouse brain and HL-1 cells. PCR analysis of hcn1 – 4 and gapdh gene expression on mouse brain cDNA (a) and HL-1 cell cDNA (b). Sizes of marker bands are indicated on the right. Gene expression levels were determined via qPCR on cDNA of confluent HL-1 cell cultures. Transcript expression levels for hcn 1 – 4 genes normalized to gapdh are shown in (c). Data points depict mean ± s.e.m. (n = 4) and Student's t test was performed (*p < 0,9; **p < 0,005)
HL-1 cells develop contractile activity at high culture densities, implicating cell confluency as an important factor of HL-1 cell differentiation. To examine whether the development of contractile HL-1 cell activity correlates with hcn gene expression, transcript levels of hcn1, hcn2 and hcn4 were quantified at different HL-1 culture densities. The following cultivation strategies were applied: (i) cells were plated at different densities, and mRNA was isolated after 4d in culture; (ii) cells were plated at defined densities, and mRNA was isolated after different cultivation times (1d–4d after plating). Samples were grouped according to their respective culture densities that were visually determined prior to mRNA isolation. Transcript expression was examined via qPCR and normalized to the housekeeping gene gapdh.
Striking gene-specific changes in expression profiles were observed (Fig. 2). Expression of hcn1 and hcn2 increased with increasing culture density. For both genes the highest transcript levels were measured at about 60 % cell confluency. Notably, in cultures reaching 100 % confluency hcn1 expression decreased almost to the starting level (Fig. 2a), whereas hcn2 levels remained elevated (Fig. 2b). In contrast to hcn1 and hcn2, hcn4 expression was high at low cell confluency. With increasing culture density, however, hcn4 expression decreased almost linearly (Fig. 2c). Gene expression profiles were similar when comparing data sets generated according to cultivation strategy (i) or (ii) (Additional file 1: Figure S1). Based on these transcript expression profiles one might speculate that individual hcn isoforms play a role in HL-1 differentiation as well as in establishing HL-1 cell contractile activity. However, qPCR data was obtained from the entire cell culture samples not providing information on the individual cell level. Therefore, we examined the distribution of HCN proteins via immunocytochemistry.
Fig. 2.
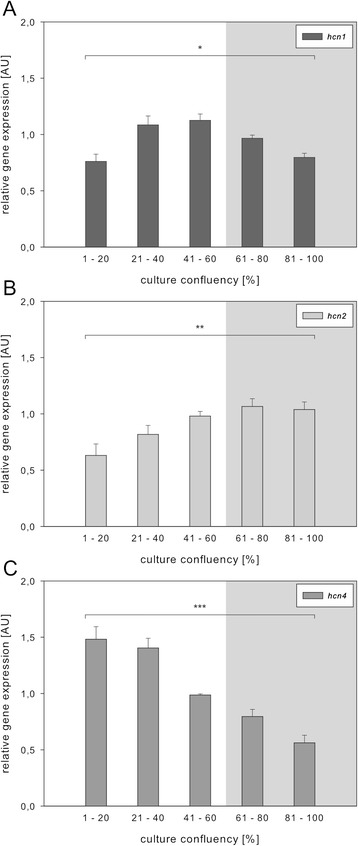
Expression of hcn genes in HL-1 cell samples obtained at different culture densities. Gene expression levels were determined via qPCR on cDNA from HL-1 cells propagated at different culture densities according to cultivation strategy (i). Quantification of genes (y-axis) is shown in relation to culture confluency (x-axis). Values for hcn1 (a), hcn2 (b), and hcn4 (c) were normalized to gapdh (see 2.3.). Samples were grouped according to visually determined culture densities. Grey inlays denote conditions where HL-1 cells displayed strong contractile activity. Data points depict mean ± s.e.m. and one-way ANOVA was performed for each group (for culture confluency: *F(4,27) = 8.298, p < 0.0002; **F(4,28) = 5.708, p < 0.002; ***F(4,25) = 31.22, p < 0.0001)
Analysis of HCN channel isoform expression in HL-1 cells
In order to gain insight into expression patterns of HCN proteins in individual HL-1 cells we performed a series of immunocytochemical stainings with HCN isoform-specific primary antibodies. Expression of HCN1, HCN2 and HCN4 was reliably detected (Fig. 3a–3c), whereas detection of HCN3 proteins failed. HCN1, HCN2 and HCN4 were primarily located at the plasma membrane (Fig. 3a–3c, arrowheads). Notably, only a fraction of HL-1 cells in culture expressed HCN channel proteins: We observed approximately 3 % HCN1-positive, 1 % HCN2-positive, and 10 % HCN4-positive cells in confluent HL-1 cell cultures (Additional file 1: Figure S2). HCN1 expression was detected in single cells as well as in small cell clusters. Expression of HCN2 was found mostly in single cells. In contrast, HCN4-expressing cells tended to form large cell clusters. Co-immunostaining revealed co-expression of HCN1 and HCN4 in approximately 0.5 % of cells (Fig. 3d3, asterisks), however, the majority of HCN1 and HCN4-positive cells expressed only one HCN isoform (Fig. 3d3, arrowheads). Co-immunolabeling of HCN2 and HCN4 showed that these proteins were expressed in different cells or cell clusters (Fig. 3e3). Similar expression patterns have been reported in vivo for these HCN isoforms [10, 19]. Our findings on cell specific and restricted expression of HCN channel proteins suggest that HL-1 cells in culture occur in various states, rather than being a homogeneous population of cells.
Fig. 3.
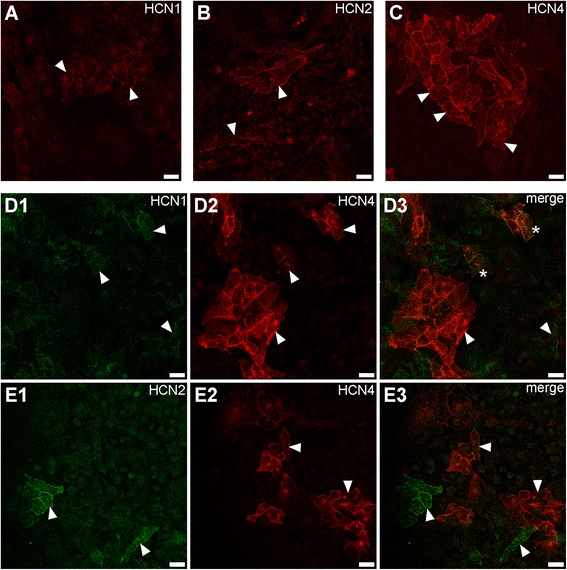
Expression of HCN channel isoforms in HL-1 cells. Immunostaining of HCN1 (a), HCN2 (b), and HCN4 (c) in confluent HL-1 cell cultures was performed with HCN isoform-specific primary antibodies (see Additional file 1: Table S1) followed by fluorescently labeled secondary antibodies. HL-1 cells were double-immunostained for HCN1 (d1) and HCN4 (d2) and for HCN2 (e1) and HCN4 (e2); merged images of double-immunostainings are shown in (d3) and (e3), respectively. Some cells specifically stained for either isoform are indicated by arrowheads; co-immunolabeled cells are marked by asterisks. Scale bars specify 20 μm
HCN isoform expression and cell contraction
To investigate whether contractile HL-1 cell activity can be attributed to the expression of HCN isoforms, spontaneously contractile cell clusters were microscopically identified on coverslips with integrated grid patterns. Subsequently, samples were fixed and stained for HCN isoforms. Images taken from live documentation and immunolabeling were superimposed and aligned based on the underlying grids (Additional file 1: Figure S3). At high culture densities HL-1 cells tend to grow in overlapping layers instead of a confluent monolayer. Superimposing live documentation images and maximal intensity projections occasionally suggested an overlap of HCN-expressing cells and spontaneously contracting areas (Additional file 1: Figure S3). However, this overlay was completely due to the layered growth of HL-1 cell cultures, since spontaneously contracting cells did not express any of the investigated proteins (Fig. 4). Nevertheless, cell clusters displaying strong contractile activity were often found in the vicinity of large HCN4-expressing cell clusters (Fig. 4c, Additional file 1: Figure S3).
Figure 4.
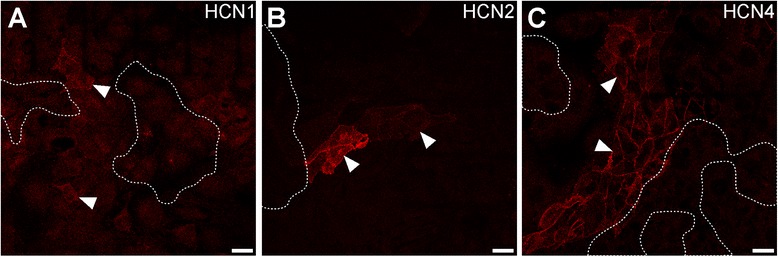
Expression of HCN isoforms in HL-1 cell cultures displaying contractile activity. Contractile activity in confluent HL-1 cell cultures was documented (dotted lines) before samples were immunostained with specific primary antibodies (see Additional file 1: Table S1) and fluorescently labeled secondary antibodies. Images of HL-1 cell contraction and immunostainings were superimposed based on underlying grids (see Additional file 1: Figure S3). Areas of contractile cell clusters are indicated by dotted lines. Immunolabeling of HCN1 (a), HCN2 (b), and HCN4 (c) is depicted. Arrowheads indicate some cells specifically stained for HCN isoforms. Scale bars specify 20 μm
During propagation, HL-1 cells acquire strikingly different morphologies. Heterogeneity is reflected both, on the phenotypic level, e. g., contractile and non-contractile cells, as well as on the molecular level as demonstrated by the highly diverse expression patterns of HCN channels.
Discussion
In recent years, knowledge of the molecular components that are instrumental in cardiac physiology has expanded significantly. Contribution of HCN channels to cardiac rhythmicity is well established and has been corroborated by several knockout studies [13, 14, 16]. However, only a few studies addressed the potential link between cardiac rhythmic activity and the expression of HCN channel isoforms on the cellular level. Here we studied an in vitro cell system of cardiac origin, the HL-1 cell line, to examine their molecular repertoire of HCN channels.
The vertebrate heart is separated into distinct, functionally specialized segments that are essential for cardiac rhythmicity. Expression of proteins in a developmentally specified manner is essential for cardiac function. Since analysis of cardiac processes in vivo is limited, a number of studies have been initiated in cellular model systems, such as cardiomyocytes or cardiac cells derived from murine embryonic stem cells (mESC) [20–22]. HL-1 cells are another versatile model for investigation of cardiac processes in vitro, due to their unique feature of retaining properties reminiscent of differentiated adult cardiomyocytes, like contractile activity, even during serial passaging [17]. Furthermore, HL-1 cells have been shown to express several cardiac marker genes, e. g., HCN channel isoforms [18, 23].
In this study, differentiation of HL-1 cells under different cultivation conditions was assessed via the emergence of spontaneous contractile activity. We noticed a marked increase of hcn1 and hcn2 gene expression with rising culture density, that ceased with the appearance of strong contractile activity in the culture. In contrast, we observed that with increasing culture confluency, the amount of hcn4 transcripts decreased to approximately 40 % of the transcript level expressed at low confluency (see Fig. 3). In native cardiac tissue, hcn4 gene expression declines with increasing age of mice [24], and the functional contribution of HCN4 to cardiac rhythmicity in vivo is successively reduced during the animal's development [12–14].
Gene expression profiles can provide important information on constitutive or differential transcriptional activity in tissues. However, they do not provide conclusive information on the corresponding protein expression patterns. This has been emphasized previously for HCN channel expression in the heart, where transcript and protein expression levels are not consistent [25]. Similar results were observed in this study (Additional file 1: Figure S2). In HL-1 cells, HCN4 was the predominant isoform, followed by HCN1, whereas HCN2 was the least expressed. Remarkably, these results are analogous to reported expression patterns of HCN isoforms in the murine cardiac conduction system [10]. Furthermore, the property of HCN4-positive cells to form cell clusters has also been found at the center of the rabbit SAN [26], as well as in mESC-derived pacemaker cell clusters, expressing either HCN1 or HCN4 [27].
HCN isoforms are expressed in a highly differential manner in the heart that correlates with cardiac physiology [8–10, 19]. Designated expression is emphasized as an essential part of functional tissue differentiation as properties of HCN channels largely depend on subunit composition [28, 29]. Co-assembly of different HCN isoforms in heterologous expression systems as well as in cardiomyocytes, results in functional HCN channels with biophysical properties that differ from homomeric channels [28, 29]. However, detailed studies addressing HCN isoform co-assembly in the heart are lacking. In this study, we observed co-expression of HCN1 and HCN4 in some HL-1 cells, similar to reports in mESC-derived pacemaker cells [27]. Although we did not perform electrophysiological measurements, it can be hypothesized that co-assembly of HCN1 and HCN4 increases the functional repertoire of pacemaking channels in HL-1 cells.
Even though HCN channels have been attributed distinct functions in cardiac rhythmicity, little is known about these aspects on the cellular level. Expression of the HCN channel-based If current has been shown in mESC-derived contracting cells, where pharmacological blocking of If caused a reduction of the contraction frequency [30]. However, reports on HCN isoform expression in mESC-derived contracting cells are not consistent [22, 27, 30]. We found that contractile HL-1 cell clusters were devoid of HCN proteins, but cell clusters expressing HCN4 were frequently located in the vicinity of contracting cells. Thus, the endogenous presence of HCN channels seems not to be required for HL-1 cells to develop contractile activity. Nevertheless, HCN channel expression in neighboring cell clusters might contribute to signaling events that culminate in cell contraction of nearby clusters. This hypothesis is supported by recent findings showing that contractility of mESC-derived cell clusters relies on the expression of specific transcription factors, e. g., Shox2 and GATA6, in adjacent, non-contractile cell clusters [20–22]. Notably, in mESC-derived model systems Shox2-positive cell clusters express HCN4 [21], and cGATA6/mink-positive cells possess If currents [22].
Conclusions
In this study, the expression patterns of HCN channels were resolved in HL-1 cells, both on the transcript and on the protein level. Notably, only a small fraction of cells in a confluent culture express these channels suggesting that HL-1 cells are present in a variety of molecular and functional states. Whether HL-1 cell contraction relies on HCN channel expression in adjacent cells and/or which additional factors contribute to the generation of contractile activity, remains to be addressed. It will also be interesting to examine whether a direct correlation between HCN channel expression and differentiation into distinct cellular phenotypes can be revealed.
Acknowledgements
We gratefully acknowledge the technical assistance of J. Schmitz in the cell culture. We thank Dr. D. Kaschuba for supplying HCN isoform-specific primers as well as Dr. F. Müller, Dr. A. Mataruga, and Dr. E. Kremmer (Helmholtz Zentrum München, Munich, Germany) for providing HCN isoform-specific antibodies. For advice on the statistical analysis we thank D. Kortzak. We thank N. Gruteser and J. Günther for discussion and comments on the manuscript. This study was not supported by third-party funding.
Abbreviations
- ESC
Embryonic stem cell
- If
Funny current
- HCN
Hyperpolarization-activated and cyclic-nucleotide gated
- SAN
Sinoatrial node
Additional files
Primary antibodies for immunocytochemistry. Figure S1. Expression of hcn genes in samples obtained at different cell culture densities. Figure S2. Relative expression of HCN isoform transcript and protein in confluent HL-1 cell cultures. Figure S3. Expression of HCN4 in cultures displaying contractile activity.
Additional file 2:
Video S1. Video of HL-1 cells displaying contractile activity in confluent culture.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
AG and AB participated in conception and design of study. AG carried out experiments, analyzed data, and performed the statistical analysis. AG and AB interpreted results and drafted the manuscript. AG prepared the figures. All authors read and approved the final manuscript.
Contributor Information
Anne Günther, Phone: +49-2461-618791, Email: a.guenther@fz-juelich.de.
Arnd Baumann, Email: a.baumann@fz-juelich.de.
References
- 1.Kaupp UB, Seifert R. Molecular diversity of pacemaker ion channels. Annu Rev Physiol. 2001;63:235–257. doi: 10.1146/annurev.physiol.63.1.235. [DOI] [PubMed] [Google Scholar]
- 2.Brown H, DiFrancesco D. Voltage-clamp investigations of membrane currents underlying pace-maker activity in rabbit sino-atrial node. J Physiol. 1980;308:331–351. doi: 10.1113/jphysiol.1980.sp013474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DiFrancesco D. Pacemaker mechanisms in cardiac tissue. Annu Rev Physiol. 1993;55:455–472. doi: 10.1146/annurev.ph.55.030193.002323. [DOI] [PubMed] [Google Scholar]
- 4.Biel M, Wahl-Schott C, Michalakis S, Zong X. Hyperpolarization-activated cation channels: from genes to function. Physiol Rev. 2009;89:847–885. doi: 10.1152/physrev.00029.2008. [DOI] [PubMed] [Google Scholar]
- 5.Ludwig A, Zong X, Jeglitsch M, Hofmann F, Biel M. A family of hyperpolarization-activated mammalian cation channels. Nature. 1998;393:587–591. doi: 10.1038/31255. [DOI] [PubMed] [Google Scholar]
- 6.Santoro B, Liu DT, Yao H, Bartsch D, Kandel ER, Siegelbaum SA, Tibbs GR. Identification of a gene encoding a hyperpolarization-activated pacemaker channel of brain. Cell. 1998;93:717–729. doi: 10.1016/S0092-8674(00)81434-8. [DOI] [PubMed] [Google Scholar]
- 7.Liu J, Noble PJ, Xiao G, Abdelrahman M, Dobrzynski H, Boyett MR, Lei M, Noble D. Role of pacemaking current in cardiac nodes: insights from a comparative study of sinoatrial node and atrioventricular node. Prog Biophys Mol Biol. 2008;96:294–304. doi: 10.1016/j.pbiomolbio.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 8.Marionneau C, Couette B, Liu J, Li H, Mangoni ME, Nargeot J, Lei M, Escande D, Demolombe S. Specific pattern of ionic channel gene expression associated with pacemaker activity in the mouse heart. J Physiol. 2005;562(Pt 1):223–234. doi: 10.1113/jphysiol.2004.074047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moosmang S, Stieber J, Zong X, Biel M, Hofmann F, Ludwig A. Cellular expression and functional characterization of four hyperpolarization-activated pacemaker channels in cardiac and neuronal tissues. Eur J Biochem FEBS. 2001;268:1646–1652. doi: 10.1046/j.1432-1327.2001.02036.x. [DOI] [PubMed] [Google Scholar]
- 10.Herrmann S, Layh B, Ludwig A. Novel insights into the distribution of cardiac HCN channels: an expression study in the mouse heart. J Mol Cell Cardiol. 2011;51:997–1006. doi: 10.1016/j.yjmcc.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 11.Harzheim D, Pfeiffer KH, Fabritz L, Kremmer E, Buch T, Waisman A, Kirchhof P, Kaupp UB, Seifert R. Cardiac pacemaker function of HCN4 channels in mice is confined to embryonic development and requires cyclic AMP. EMBO J. 2008;27:692–703. doi: 10.1038/emboj.2008.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stieber J, Herrmann S, Feil S, Löster J, Feil R, Biel M, Hofmann F, Ludwig A. The hyperpolarization-activated channel HCN4 is required for the generation of pacemaker action potentials in the embryonic heart. Proc Natl Acad Sci U S A. 2003;100:15235–15240. doi: 10.1073/pnas.2434235100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baruscotti M, Bucchi A, Viscomi C, Mandelli G, Consalez G, Gnecchi-Rusconi T, Montano N, Casali KR, Micheloni S, Barbuti A, DiFrancesco D. Deep bradycardia and heart block caused by inducible cardiac-specific knockout of the pacemaker channel gene Hcn4. Proc Natl Acad Sci U S A. 2011;108:1705–1710. doi: 10.1073/pnas.1010122108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herrmann S, Stieber J, Stöckl G, Hofmann F, Ludwig A. HCN4 provides a “depolarization reserve” and is not required for heart rate acceleration in mice. EMBO J. 2007;26:4423–4432. doi: 10.1038/sj.emboj.7601868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ludwig A, Budde T, Stieber J, Moosmang S, Wahl C, Holthoff K, Langebartels A, Wotjak C, Munsch T, Zong X, Feil S, Feil R, Lancel M, Chien KR, Konnerth A, Pape H-C, Biel M, Hofmann F. Absence epilepsy and sinus dysrhythmia in mice lacking the pacemaker channel HCN2. EMBO J. 2003;22:216–224. doi: 10.1093/emboj/cdg032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hofmann F, Fabritz L, Stieber J, Schmitt J, Kirchhof P, Ludwig A, Herrmann S. Ventricular HCN channels decrease the repolarization reserve in the hypertrophic heart. Cardiovasc Res. 2012;95:317–326. doi: 10.1093/cvr/cvs184. [DOI] [PubMed] [Google Scholar]
- 17.Claycomb WC, Lanson NA, Jr, Stallworth BS, Egeland DB, Delcarpio JB, Bahinski A, Izzo NJ., Jr HL-1 cells: a cardiac muscle cell line that contracts and retains phenotypic characteristics of the adult cardiomyocyte. Proc Natl Acad Sci U S A. 1998;95:2979–2984. doi: 10.1073/pnas.95.6.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.White SM, Constantin PE, Claycomb WC. Cardiac physiology at the cellular level: use of cultured HL-1 cardiomyocytes for studies of cardiac muscle cell structure and function. Am J Physiol Heart Circ Physiol. 2004;286:H823–829. doi: 10.1152/ajpheart.00986.2003. [DOI] [PubMed] [Google Scholar]
- 19.Liu J, Dobrzynski H, Yanni J, Boyett MR, Lei M. Organisation of the mouse sinoatrial node: structure and expression of HCN channels. Cardiovasc Res. 2007;73:729–738. doi: 10.1016/j.cardiores.2006.11.016. [DOI] [PubMed] [Google Scholar]
- 20.Hashem SI, Claycomb WC. Genetic isolation of stem cell-derived pacemaker-nodal cardiac myocytes. Mol Cell Biochem. 2013;383:161–171. doi: 10.1007/s11010-013-1764-x. [DOI] [PubMed] [Google Scholar]
- 21.Hashem SI, Lam ML, Mihardja SS, White SM, Lee RJ, Claycomb WC. Shox2 regulates the pacemaker gene program in embryoid bodies. Stem Cells Dev. 2013;22:2915–2926. doi: 10.1089/scd.2013.0123. [DOI] [PubMed] [Google Scholar]
- 22.White SM, Claycomb WC. Embryonic stem cells form an organized, functional cardiac conduction system in vitro. Am J Physiol Heart Circ Physiol. 2005;288:H670–679. doi: 10.1152/ajpheart.00841.2004. [DOI] [PubMed] [Google Scholar]
- 23.Sartiani L, Bochet P, Cerbai E, Mugelli A, Fischmeister R. Functional expression of the hyperpolarization-activated, non-selective cation current If in immortalized HL-1 cardiomyocytes. J Physiol. 2002;545:81–92. doi: 10.1113/jphysiol.2002.021535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yasui K, Liu W, Opthof T, Kada K, Lee JK, Kamiya K, Kodama I. I(f) current and spontaneous activity in mouse embryonic ventricular myocytes. Circ Res. 2001;88:536–542. doi: 10.1161/01.RES.88.5.536. [DOI] [PubMed] [Google Scholar]
- 25.Calejo AI, Reverendo M, Silva VS, Pereira PM, Santos MAS, Zorec R, Gonçalves PP. Differences in the expression pattern of HCN isoforms among mammalian tissues: sources and implications. Mol Biol Rep. 2014;41:297–307. [DOI] [PubMed]
- 26.Brioschi C, Micheloni S, Tellez JO, Pisoni G, Longhi R, Moroni P, Billeter R, Barbuti A, Dobrzynski H, Boyett MR, DiFrancesco D, Baruscotti M. Distribution of the pacemaker HCN4 channel mRNA and protein in the rabbit sinoatrial node. J Mol Cell Cardiol. 2009;47:221–227. doi: 10.1016/j.yjmcc.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 27.Barbuti A, Crespi A, Capilupo D, Mazzocchi N, Baruscotti M, DiFrancesco D. Molecular composition and functional properties of f-channels in murine embryonic stem cell-derived pacemaker cells. J Mol Cell Cardiol. 2009;46:343–351. doi: 10.1016/j.yjmcc.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 28.Whitaker GM, Angoli D, Nazzari H, Shigemoto R, Accili EA. HCN2 and HCN4 isoforms self-assemble and co-assemble with equal preference to form functional pacemaker channels. J Biol Chem. 2007;282:22900–22909. doi: 10.1074/jbc.M610978200. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Q, Huang A, Lin Y-C, Yu H-G. Associated changes in HCN2 and HCN4 transcripts and I(f) pacemaker current in myocytes. Biochim Biophys Acta. 2009;1788:1138–1147. doi: 10.1016/j.bbamem.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qu Y, Whitaker GM, Hove-Madsen L, Tibbits GF, Accili EA. Hyperpolarization-activated cyclic nucleotide-modulated “HCN” channels confer regular and faster rhythmicity to beating mouse embryonic stem cells. J Physiol. 2008;586:701–716. doi: 10.1113/jphysiol.2007.144329. [DOI] [PMC free article] [PubMed] [Google Scholar]


