Abstract
Background
The discovery that the adult heart is not a terminally differentiated organ and contains stem/progenitor cells has important implications for the development of cellular therapeutics to treat heart disease. Moreover the discovery of cardiac stem cells might be important in furthering our understanding of both normal and abnormal cardiac development and yet little is known about these cell populations in the developing human heart, which we have focused on in this study.
Methods
The presence of ABCG2 and islet-1 expressing cells in human heart was determined using immunohistochemistry and RT-PCR (and western blotting for ABCG2). Cardiac SP cells were isolated using FACS. Co-localisation immunohistochemistry was used to determine if ABCG2 positive cells expressed other known stem/progenitor cell, endothelial markers or cardiac markers.
Results
We observed that ABCG2 expressing cells show a difference in both their temporal and spatial pattern of expression from Islet-1 expressing cardiac progenitors. We identified rare cells that expressed both ABCG2 and markers of other cell lineages including CD31, CD34 and alpha actinin. We also noted the presence of cells that only expressed ABCG2. We isolated cardiac SP cells and confirmed the SP cell phenotype.
Conclusions
Our results suggest that the developing human heart contains at least two distinct cardiac stem/progenitor cell populations one of which, the ABCG2 positive cells, can be readily isolated, suggesting that this tissue could be a useful source of cardiac stem cells.
Keywords: ABCG2, islet-1, side population cells, embryonic, fetal, human heart
Introduction
The adult heart is no longer considered to be a terminally differentiated organ. The identification of both extra cardiac and cardiac stem/progenitor cell populations that give rise to new vasculature or cardiomyocytes has implications for the development of cellular therapies for the treatment of cardiac injury/disease. A greater understanding of the role of stem cells during cardiac development may also further our understanding of fetal heart abnormalities.
Cardiac stem cell populations have been isolated from both animal and human heart tissue. Selection of cardiac cell populations has been based on either the expression of distinct markers such as c-kit, Sca-1 or Islet-1 or on a specific phenotype such as cardiospheres and side population (SP) cells [1-10]. Expression of a half transporter ABCG2, a member of the ABC transporter family, is responsible for the SP cell phenotype. Tissue specific SP cells have been identified from a variety of tissues, including cardiac tissue [5, 6, 11, 13]. Cardiac side population cells (CSP) isolated from rodent hearts are capable of differentiating into cardiomyocytes both in vitro and in vivo, and studies on rodent cardiac SP cells have shown that they are capable of multi-lineage differentiation [5, 6].
The capacity to study cardiac stem cells and to develop them as therapies is limited because of difficulties in sourcing viable cells of human origin. Currently, although cardiomyocytes can be derived from human ES cells, yields are poor and there remains concern about teratoma formation. Adult bone marrow and immature skeletal muscle cells are alternate sources: the use of bone marrow derived cells that can engraft and commit to a cardiac lineage still remains questionable with bone marrow stem cell contribution to the myocardium being very low. The use of skeletal myoblasts has also yielded disappointing results and led to the development of potentially lethal cardiac arrhythmias in some patients [11]. The isolation of cardiac stem cells from adult human heart tissue may also have its limitations; the impact of myocardial disease on cardiac stem cells is unknown, as is the impact of aging. Although the adult heart contains dividing cells, with age their proliferative potential is reduced probably due in part to loss of telomeric DNA. Many of these limitations do not apply to the fetal heart [12].
In this study we have proposed that the developing human heart is a source of cardiac stem/progenitors that can contribute to cells lineages important for cardiogenesis. One such cardiac stem cell population, that has been reported to be present in the fetal human heart is c-kit expressing cells [14]. However, nothing is known about the role of ABCG2 or islet-1 in the fetal human heart. Therefore in this study we show for the first time that human fetal heart contains ABCG2 expressing cells, some of which express endothelial markers and cardiac markers; but none of which express the cardiac progenitor marker islet-1. Further ABCG2 expressing cells display spatial and temporal expression patterns distinct from islet-1 expressing cells, clearly indicating that ABCG2 expressing human cardiac cells represent a distinct cell population.
Materials and Methods
Human tissue collection and staging
Human embryonic and fetal hearts were obtained from the MRC-Wellcome Trust Human Developmental Biology Resource (http://www.hdbr.org/). Hearts were harvested from embryos/fetuses collected from women undergoing surgical termination of pregnancy. All women gave informed written consent and the study was approved by the Newcastle Research Ethics Committee (Approval 06/Q0902/28). Developmental age was based on embryo/fetal morphology using both the Carnegie staging and fetal staging system respectively [15, 16]. To determine spatial expression patterns hearts were dissected using a Stemi DV4 dissection microscope (Zeiss) into: right ventricle (RV), left ventricle (LV), right atria (RA), left atria (LA), and outflow tract (OFT). Initially the right and left sides of the hearts were identified by the apex which is in the left side of the heart, while the atria appear as ear-like appendages on the top of the ventricles. The atria were held in forceps away from the body of the heart to enable their removal, this allowed access to the OFT which has held with forceps at its base at the top of the anterior surface of the RV and severed from the heart. Finally a portion of each ventricle was cut away taking care to avoid the septum. Intact hearts where collected for immunohistochemical analysis. Thirty four morphologically normal hearts (ranging from developmental stages CS18 to F5) where used in total for this study.
Reverse transcription-polymerase chain reaction
Total RNA was isolated from the RA, LA, RV, LV, and OFT using the RNAqueous ®-Micro Kit (Ambion, Austin, USA). cDNA was synthesised using either: SuperScript TM First-Strand Synthesis System for RT-PCR (Invitrogen, Paisley, UK) or using a cDNA synthesis kit (Bioline, London, UK) according to the manufacturer’s protocol. Reverse transcription–polymerase chain reaction (RT-PCR) was prepared as follows; 12.5 μl of PCR master mix (Promega, Hampshire, UK), 0.5 μl of forward primer, 0.5 μl of reverse primer, 2.5 μl of cDNA, and 9 μl of nuclease free water. PCR primer pairs used for this study included the following: ABCG2 forward: 5′-GCTGCAAGGAAAGATCCAAG-3′; reverse: 5′-TCCAGACACACCACGGATAA-3′; Isl1 forward: 5′-AAACAAAACGCAAAACCCAG-3′; reverse; 5′-GGCAAGGCAATGACCAATA-3′; GapdH forward: 5′-CTGCCGTCTAGAAAAACC-3′; reverse: 5′-CCAAATTCGTTGTCATACC-3′. Beta-actin forward: 5’-GGACTTCGAGCAAGAGATGG-3’; reverse: 5’-AGCACTGTGTTGGCGTACAG-3’. For all reactions a no-RT control was included and placental cDNA was used as a positive control (data not shown). Nine hearts where used in total for RNA isolation, date shown represents RNA isolated from distinct anatomical regions from single hearts.
Western Blotting for Abcg2 expression
Heart tissue from each anatomical area was disrupted and lysed in sucrose homogenising buffer, consisting of 25mM Tris Base, 0.25M sucrose, and 1mM EDTA pH 7.6, supplemented with protease inhibitor cocktail (Sigma-Aldrich, St. Louis, USA) 1 μl/ 20mg of tissue. Protein levels were determined using the Lowery assay (Bio-Rad, Hercules, CA, USA). The lysates were analyzed by SDS-PAGE. Equal amounts of protein (20μg) were electrophoresed in a 7.5% SDS-polyacrylamide gel and then transferred onto polyvinylidene fluoride (PVDF) membranes. Homogenised placenta protein was used as a positive control. Membranes were blocked in 10% non-fat milk for 1 hour at room temperature (RT) and incubated in the presence of anti-mouse ABCG2 monoclonal antibody (BXP-21, ab3380; Abcam plc, Cambridge, UK) at dilution 1:100 in 1% blocking solution at 4°C overnight. The membranes were washed and incubated in horseradish peroxidase (HRP) goat anti mouse conjugated secondary antibody (Dako, Glostrup, Denmark) applied at a dilution of 1:3000 for 1 hour at RT. After washing membranes with PBS, proteins were visualized using a chemiluminescent detection method (ECL plus western blotting detection reagents; Amersham Life Sciences, GE healthcare, USA). For western analysis anatomical regions where isolated from single hearts, with the exception of CS18 (6 weeks pc,) when to generate 20μg protein, distinct anatomical regions from two CS18 hearts had to be combined.
ABCG2 gene silencing by antisense morpholino oligonucleotides
Morpholino oligonucleotides used in this study were synthesised by Gene Tools, LLC, USA. Morpholino oligonucleotides were designed to block translation of the mRNA transcript of human ABCG2 and the sequence was ACTTCGACATTACTGGAAGACATCT. An antisense morpholino oligonucleotides composed of a scrambled irrelevant sequence was also included as a control. Delivery of the morpholino oligonucleotides into MCF-7 cells was undertaken using the EndoPorter system following the manufacturer’s protocol to optimise the transfection efficiency (Gene Tools, LLC, USA). Gene silencing was confirmed by Western blotting, as described.
Immunohistochemical Staining
Embryonic/fetal hearts were immersion fixed in 4% paraformaldehyde (pH 7.2) overnight at 4°C, rinsed in PBS and paraffin embedded. 3 micron thick coronal sections were deparaffinised in xylene and rehydrated. To prevent non-specific binding to endogenous peroxidases all sections were incubated in 1% hydrogen peroxide/methanol for 15 minutes and then blocked with the respective 10% normal serum for 10 minutes. Sections were then incubated in primary antisera in a humid chamber for 1 hour at RT. Primary antisera used in this study included mouse anti-ABCG2 (clone 5D3, dilution1:50, Stem Cell Technologies, Grenoble, France), mouse anti CD34 (clone 581, dilution 1:50, BD Pharmingen, BD Biosciences, San Jose, CA), mouse anti Isl1 (clone 40.2D6, dilution 1:50, Developmental Studies Hybridoma Bank, University of Iowa, USA), mouse anti human CD31 (clone HC1/6, dilution 1: 20, Novocastra Laboratories Ltd, Newcastle Upon Tyne, UK) and mouse anti-alpha actinin, sarcomeric (Clone EA53, dilution 1:200, Sigma). Sections were then washed and incubated with the appropriate secondary antisera (for 30 minutes at RT) which was either biotinylated anti-mouse secondary antibody, using a Vectastain Elite ABC Kit (Vector Laboratories Burlingame, CA, USA) according to manufacturer’s protocol, followed by final visualisation with either 3,3′-diaminobenzidine (DAB) peroxidase subs ortrate Vector NovaRED (Vector Laboratories) or Vector SG blue/grey (Vector Laboratories) sections were then counterstained with haematoxylin or not counterstained, or incubated with polyclonal rabbit anti mouse antibody (dilution 1:100, Dako, Glostrup, Denmark) for 30 minutes followed by incubation in tertiary PAP mouse monoclonal antibody (dilution 1:50, Dako) for 30 minutes and final visualisation with chromogenic alkaline phosphatase substrate Vector blue used according to manufacturer’s instructions (Vector Laboratories). For intracellular antigens, sections were either incubated in the presence of pronase (0.01 g/mL, Calbiochem, Merck Chemicals Ltd, San Diego, USA) for 5 minutes or subjected to heat antigen retrieval using sodium citrate buffer (pH 6). Negative controls for all antibodies included omission of the primary antibodies, examples are shown in Figures 1E and 2E. Sections were then mounted in aqueous mounting media (Vector Laboratories). Images were recorded using a Zeiss AxioImager Z2 Apotome and Axiovision 4.8.1 software. Final images were generated using Adobe PhotoShop C5 software package (Adobe, San Jose, CA, USA).
Figure 1.
ABCG2 is expressed in the developing human heart. (A) Representative RT-PCR analysis for ABCG2 expression from CS18 (6 weeks pc) through to F5 (13 weeks pc) comparing the expression of ABCG2 mRNA in distinct anatomical cardiac regions. Note at CS18 the expression appears to be similar in all cardiac regions with the exception of the OFT where it is slightly elevated. At CS23 (8 weeks pc) expression is down regulated in the LV compared with the other anatomical regions and in the fetal stages the expression of ABCG2 is reduced in LV, LA compared with the RV, RA and OFT. GapdH was used as a loading control. (B) FACS profile of CSP cells from an F1 heart (9 weeks pc). (C) Inhibition of the SP cell phenotype after addition of FTC. Abbreviations: pc: post conception; SP, side population: RV, right ventricle; LV, left ventricle; RA, right atria; LA, left atria; OFT, outflow tract; FTC, fumitremorgin C; RT-PCR, reverse transcriptase polymerase chain reaction. IHC analysis for ABCG2, ABCG2 expressing cells are located throughout the human embryonic and fetal heart. Shown are representative images taken of cardiac tissue stained with an anti-ABCG2 antisera. (D and F) ABCG2 expressing cells in the embryonic heart at CS23 (8 weeks pc) (D) and in the ventricles at F1 (9 weeks pc) shown here in the LV (F). Note the absence of staining in the no primary negative control (E). ABCG2 was visualised with 3’-diaminobenzidine (brown), all sections were counterstained with haematoxylin (blue). Magnifications: size bar = 10 μm for all images; Abbreviations: pc: post conception; SP, side population: RV, right ventricle; LV, left ventricle; RA, right atria; LA, left atria; OFT, outflow tract; FTC, fumitremorgin C; RT-PCR, reverse transcriptase polymerase chain reaction, IHC, immunohistochemical.
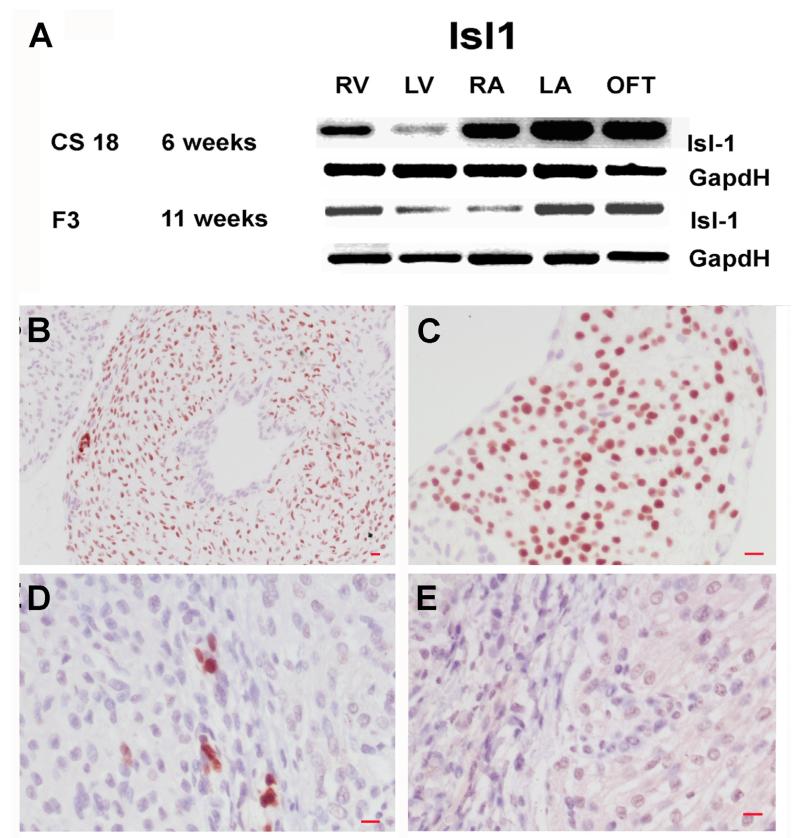
Figure 2.
Islet-1 is expressed during development of the human heart. (A) RT-PCR results for islet1 mRNA expression at CS18 (6 weeks pc) and F3 (11 weeks pc). Immunohistochemical analysis for islet-1 expressing cells, large numbers of isl+ cells are present in the OFT (B) and RA (C) at CS23 (8 weeks pc). Small numbers of isl+ cells are also present in ventricles at F6 (14 weeks pc) (D). Note the absence of staining in the no primary control, F6 (14 weeks pc) ventricle (E). Islet1 was visualised with 3’-diaminobenzidine (brown) and sections were counterstained with haematoxylin (blue) Magnifications size bar = 10μm for all images. Abbreviations: RT-PCR: reverse transcription–polymerase chain reaction, pc: post conception, Isl+: islet-1 positive, RA: right atria, OFT: outflow tract.
Isolation of SP cells from heart tissue
Hearts were harvested, minced and digested in pronase 10 mg/ml (Calbiochem) at 38°C for 20 minutes, the enzyme digestion was then terminated and the tissue triturated to obtain a single cell suspension. Ten hearts where used in total for optimisation of the SP cell FACS protocol (see supplemental table 1). Cells were resuspended at 1.0 × 106 cells/ml in Hank’s buffer containing 2% fetal calf serum. The cells were incubated with Hoechst 33342 dye ranging at concentrations of 12.5, 5.0, 2.5 and 1.25 μg/ml for 60 minutes at 37°C to determine the optimal concentration that was non-toxic and enabled an identifiable SP cell profile to be visualised. In order to confirm the SP cell phenotype the ABCG2 specific inhibitor, Fumitremorgin C (FTC) 5 μg /mL (Alexis Biochemicals, Lausen, Switzerland) was added prior to the addition of Hoechst 33342 dye. Prior to flow cytometric analysis (FACS) propidium iodide at a concentration of 2μg/ml was added to the cell suspension. Cells were analysed using a Becton Dickinson FACS Diva (BD Biosciences, San Jose, CA) which contains a 350 nm ultraviolet laser used to excite Hoechst 33342 dye. The fluorescent emission was detected through 450-nm Bp (Hoechst blue) and a 675 nm LP (Hoechst red) filters to identify SP cells, while propidium iodide was excited at 488 nm (fluorescence emission being detected through 610-nm Bp filter) to allow for exclusion of dead cells.
Results
ABCG2-expression in the embryonic/fetal human heart
ABCG2 expression during cardiac development was confirmed both at the level of the transcriptome, using semi-quantitative RT-PCR, and at the protein level using immunohistochemistry and western blot analysis. The presence of an ABCG2 expressing SP cell population was also confirmed using dual wavelength FACs combined with Hoechst 33342 dye efflux.
To determine if there was a spatial pattern of ABCG2 expression during cardiac development we isolated anatomical distinct regions of the heart during cardiogenesis at CS18 through to 23 and fetal stages F3 and F5 (weeks 6 through 13 post conception). Using semi-quantitative RT-PCR ABCG2 expression was detected in both ventricles and atria and in the outflow tract at CS18 through CS22. At CS23 expression became down regulated in the LV relative to the other cardiac regions at this stage and thereafter in the fetal stages expression was down regulated in the LA and LV relative to the RV, RA and OFT (Fig. 1A).
Using dual wavelength FACS in combination with Hoechst 33342 dye efflux cardiac cells (1×106cell/ml) where incubated in the presence of 12.5, 5.0, 2.5 and 1.25 μg/ml Hoechst 33342 (data not shown). The optimal concentration that was non-toxic and enabled an identifiable SP cell profile to be visualised was 1.25 μg/ml, using this protocol we observed an SP cell population present in the developing human fetal heart (F1, 9 weeks pc) amounting to 1.1% of cells (Fig 1B). In the presence of FTC, a specific inhibitor of ABCG2, the population of cells in the SP cell gate was reduced to 0.1% (Fig. 1C). PI was included in all cell preparations prior to FACS analysis to exclude dead cells.
Using immunohistochemical analysis we observed ABCG2+ cells in all regions of the heart at all ages. Most of the ABCG2+ cells were localised to the interstitial spaces and within the myocardium (Fig. 1D and F). Small numbers of ABCG2+ cells were also located in the atria and OFT (at the peripheral region of the OFT and also associated with epithelium of the OFT at all developmental ages (data not shown). Note the absence of staining in the no primary control (Fig 1E). ABCG2 protein in developing hearts was also confirmed using western blotting and again ABCG2 was detected in all the distinct anatomical regions examined during cardiac development, data is shown for CS18 (6 weeks pc) and F5 (13 weeks pc) (Supplemental Fig. 1A). To confirm the specificity of the antibody used for detection of ABCG2 protein, translation of ABCG2 protein was blocked by antisense morpholino oligonucleotides in the breast carcinoma MCF7 cell line, post-silencing note the absence of ABCG2 protein expression (Supplemental Fig. 1B).
Islet-1 expression during human embryonic/fetal heart development
Spatial analysis revealed that at CS18 (6 weeks pc) islet-1 mRNA was expressed in the RV, LV, RA, LA and OFT with higher expression in the RA, LA, OFT, (Fig 2A). At later stages (F3, 11 weeks pc) islet-1 mRNA expression was reduced in the RV, LV, and RA compare with the LA and OFT (Fig. 2A). Immunohistochemical analysis for islet-1 revealed the presence of large numbers of isl1+ cells in the OFT and RA at CS22 (8 weeks pc) (Fig 2B and C) with a smaller number of isl1+ cells being detected in LA, numbers of isl-1+ cells in these regions decreasing with increasing age (data not shown). While at later ages a small numbers of isl1+ cells were detected in the ventricles (Fig 2D). Note the absence of staining in the no primary control (Fig 2E).
Co-expression Analysis for ABCG2 and other stem/progenitor cell markers
Using dual labelling immunohistochemical techniques we examined tissue sections ranging in ages from CS18 to F6 with antisera to both ABCG2 and Islet-1. We never observed any cells that expressed both markers (Fig. 3A & B). We also examined developing hearts for co-expression of ABCG2 and CD31 and ABCG2 and CD34. Results for immunohistochemical analysis for CD31 alone revealed that the epithelium expressed CD31 throughout all regions of the heart and at all developmental ages (data not shown). However, dual immunohistochemical analysis for ABCG2 and CD31 revealed that only a small population of cells expressed both markers (Fig 3C). Finally, a small number of cells expressed both ABCG2 and CD34 during development (Fig 3D).
Figure 3.
Representative results for co-expression analysis for ABCG2 and other stem, progenitor endothelial and cardiac markers. No cells expressed islet −1 and ABCG2 in the OFT (A) or RA (B) at CS23 (8 weeks pc). Rare cells that expressed both CD31 and ABCG2 are expressed in the embryonic heart at CS23 (8 weeks pc) in the RA, note a cell expressing ABCG2 alone and a dual labelled cell expressing both ABCG2 and CD31 (C). Small numbers of cells that express ABCG2 and CD34 are present in the developing heart, in the LV at CS23 (8 weeks pc) (D). Rare cells express both ABCG2 and alpha-actinin in the fetal heart at F1 (9weeks pc) (E). Images A,B.C, and E, ABCG2 visualised with 3’-diamionbenzidine (brown), islet-1, CD31 and alpha-actinin (sarcomeric) visualised with Vector blue (blue). Image D, ABCG2 visualised with Vector NovaRED (red), CD34 visualised with Vector SG blue/grey (grey). Magnifications: Size bar = 10μm Abbreviations: OFT: outflow tract, LV: left ventricle, pc: post conception, RA: right atria, LA: left atria, RV: right ventricle.
Immunohistochemical analysis to determine if ABCG2 expressing cells express markers of myocardial lineage cells
Heart sections were stained throughout cardiogenesis for expression of ABCG2 and alpha-actinin, rare cells were observed that expressed both (Fig 3E).
Discussion
This is the first report of the presence and localization of ABCG2 expressing cells in the human embryonic and fetal heart. Previous studies have reported the identification of SP cells from a range of human tissues, with variables in the staining protocol for these studies include Hoechst 33342 concentration, incubation time and inhibitors used [17-20]. Based on these published reports and our previous experience with SP cell isolation we have developed a protocol for the isolation of human CSP cells. We have shown the developing human heart contains a CSP cell population whose phenotype is due to expression of ABCG2. We have described the distribution of Islet-1 expressing cells and have shown that there is no co-expression of ABCG2 and Islet-1 during this period of heart development, suggesting these are different cell populations. Further their distinct spatial distribution suggests that ABCG2 expressing cells contribute to ventricular development while Islet-1 expressing cells contribute primarily to OFT and atrial development. Finally we have shown that ABCG2+ cells co-express markers of cardiomyocyte differentiation and endothelial markers, suggesting that these cells may have the ability to differentiate towards multiple cellular lineages, important for cardiogenesis.
In the mouse, ABCG2 mRNA has been detected relatively early in cardiac development in cells isolated at E7.75 from the cardiac crescent, [21], this expression being lower in adult mouse heart compared to embryonic and postnatal heart [5]. Immunohistochemical analysis for ABCG2 expression in the mouse heart revealed robust expression at E8.5 with downregulation between E11.5 – E13.5 to a subpopulation of cardiac cells [5]. Due to limited availability of very early human embryonic heart tissue we were unable to study ABCG2 mRNA expression prior to CS18 when the 4 cardiac chambers are already divided by the ventricular and atrial septi. However, we demonstrated robust ABCG2 mRNA expression at CS18 in all regions of the heart examined, with downregulation in the left side of the heart at later stages of development. These findings were confirmed at the protein level by both immunohistochemistry and western blot analysis. For the immuohistochemical analysis we used a commercially available monoclonal antibody anti-ABCG2 (clone 5D3). 5D3 has been previously used as a tool in immunohistochemistry to detect ABCG2 protein expression in human non-cardiac tissue, where it was reported to give a good signal with low background [22]; however the protocol used differed from that used in this report. The primary difference between the two protocols used was the length of time of incubation of the tissue in the presence of the primary antibody, this could be influenced by a number of factors including technical differences in fixation, the age of archival fixed materials versus newly embedded material, the types of tissue being examined, level and even cellular localisation of ABCG2. Fetsch et al [22] reported differences in staining patterns between different cell types in different tissues, for example strong cytoplasmic and membranous staining of syncytiotrophoblastic cells in placenta and granular cytoplasmic staining of the sebaceous gland in skin. For western analysis we used another commercially available anti-ABCG2 antibody (clone BXP21) which has previously been used in western analysis to detect human ABCG2 [23, 24]. BXP21has been reported to detect the presence of a 70 kDa band in both atrial and ventricular heart membrane preparations and in human placenta, a 72 kDa band in MCF7 cells and, a 72 and 140 kDa band in mature human erythrocytes, and in a study of MCF-7/AdvVP300 cells which over-express ABCG2 BXP21 also revealed a number of different molecular mass bands for ABCG2, suggesting that human ABCG2 may exist in multiple configurations [23, 24, 25]. We detected a 72 kDa band in placenta and in all regions of the heart using BXP21. In addition we observed a more abundant 280 kDa protein band in embryonic and fetal hearts that was low or absent in the placenta. To determine the specificity of BXP21, expression of ABCG2 protein was silenced in the breast carcinoma cell line MCF7post silencing no expression of ABCG2 protein could be detected in these cells, while expression of the expected 72kDa band could be detected without silencing. This suggests that the 72kDa band detected in the developing embryonic and fetal heart is indeed ABCG2, while the more robust 280 kDa band could be a different protein configuration of ABCG2. Published data supports this possibility in that multimeric ABCG2 proteins have been described [25]; this study proposed that a 285kDa protein band was likely to be a tetramer of the 72kDa ABCG2 protein. Elevated levels of ABCG2 have been reported in certain cardiomyopathies, [23, 26], and high levels of ABCG2 expression have been reported in the human placental syncytiotrophoblasts [27], this might indicate a protective role for ABCG2 against environmental toxicants. Therefore one might hypothesize that the 72kDa ABCG2 expressed in the placenta and in certain cardiomyopathies, could be acting as a transporter effluxing xenobiotics, with the need for such a system being less prevalent in the developing fetal heart.
Our results show that some ABCG2+ cells also express markers associated with lymphohematopoietic stem and progenitor cells and endothelial cells. These include CD34, expressed on developmentally early lymphohematopoietic stem and progenitor cells, small vessel endothelial cells and embryonic fibroblasts and CD31 which in the heart has been reported to be expressed by all vascular endothelial cells [28, 29, 30]. We also observed rare cells that expressed both ABCG2 and alpha-actinin a marker of cardiomyocyte differentiation. This suggests that CSP are capable of multi-lineage differentiation; rodent derived CSP cells have been shown to be capable of giving rise to beatings cells that express cardiac genes in culture and can form hematopoietic colonies [6, 5]. However, it could also be that CSP cells are not a distinct cell population, our results suggest they differ from islet-1 cardiac progenitors and it has been previously reported that only a subpopulation (less that 10%) of CSP cells isolated from a Tie2-GFP mouse model express GFP suggesting that many CSP cells are distinct from endothelial cells [5]. The LIM-homeodomain transcription factor islet-1 (isl1) is thought to be a marker for cardiac progenitor cells during embryonic mouse heart development [31]. Isl1+ cells have been reported to be present in the postnatal rat, mouse and human heart located in the atria, intra-atrial septum, conus muscle and RV [3]. The number of isl+ cells markedly declining with increasing age [3]. Based on the phenotype of islet-1 null mouse, it has been proposed that isl+ cells give rise to the OFT, RV, a small population of cells in the LV and to a large percentage of atrial cells with a second islet-1 negative progenitor population giving rise to most of the LV as well as contributing to both atria [31]. Our results would lend support to Islet-1 expressing cells contributing to OFT and atrial development in the human heart with some small contribution to ventricular development. More importantly we provide evidence that ABCG2+ cells are a distinct population that may be more important in ventricular development.
It is acknowledged that the present results do not provide evidence of how ABCG2+ cells participate in cardiac development. Indeed, ABCG2 −/− mice are viable which might suggest that ABCG2 expression is not essential for normal murine embryonic development including development of the heart [32]. However, these knockout animals do not have a total loss of SP cells, when examined using FACS analysis rather there is a reduction in number of SP cells in bone marrow, muscle and heart [32, 33]. Therefore loss of ABCG2 expression, while potentially removing from CSP cells the ability to efflux xenobiotics via this specific transporter, (and therefore moving these cells outside the SP cell gate when analysed by FACS) does not lead to physically removal of the SP cells from the heart. We also observed rare ABCG2 positive cells that expressed markers of cardiomyocyte differentiation. This been not been observed in rodent studies during cardiac development [5]. However, using in vitro techniques rodent CSP cells have been reported to be able to express makers of cardiomyocyte differentiation at both the protein and transcriptional level [5-6]. Moreover, these rodent CSP when delivered iv can migrate into both the normal and injured rodent heart, where they express cardiac, endothelial or smooth muscle markers [6]. However, no studies have as yet demonstrated that CSP cells from any species can give rise to fully functional cardiomyocytes, future studies will need to focus on addressing this question.
In conclusion, we have shown that ABCG2 expressing cells are distinct from cells that express the progenitor maker islet-1. Further a small number of ABCG2+ cells express CD34, CD31 and alpha-actinin suggesting, these cells albeit at low levels, could differentiate towards multiple cellular lineages. Moreover, we have shown that we can successfully isolate CSP cell from the developing human heart, making this tissue a potential source of human cardiac derived stem cells for future therapeutic use.
Supplementary Material
Supplementary Figure 1. (A) Western blot analysis for ABCG2 protein expression in the embryonic (CS18) and fetal (F5) human heart. Note the decrease in protein expression in the ventricles in the fetal heart compared to the embryonic heart. Human placenta was used as a positive control. kDa, kilodalton; RV, right ventricle; LV, left ventricle; RA, right atria; LA, left atria; OFT, outflow tract. (B) Western analysis for ABCG2 following gene silencing in MCF7 cells. Lane 1 cells treated with control morpholino oligonucleotides. Lane 2 cells treated with translation blocking antisense morpholino oligonucleotides for ABCG2. Note the lack of expression of ABCG2 in Lane 2. GapdH is used as a loading control. MO, antisense morpholino oligonucleotide
Supplementary Table 1. Heart samples used to optimise protocol for FACS analysis for SP cell identification in the developing human heart.
Acknowledgements
This work was supported by a grant from The Newcastle upon Tyne Hospitals Trust. We also wish to acknowledge Mr Ian Harvey (Flow Cytometry Core Facility, Newcastle University) for help with Flow cytometry and Dr. Trevor Booth for help with imaging (Bio-imaging Unit, Newcastle University) The authors of this manuscript have certified that they comply with the Principles of Ethical Publishing in the International Journal of Cardiology [34]
References
- [1].Beltrami A, Barlucchi L, Torella D, et al. Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell. 2003;114:763–776. doi: 10.1016/s0092-8674(03)00687-1. [DOI] [PubMed] [Google Scholar]
- [2].Oh H, Bradfute SB, Gallardo TD, et al. Cardiac progenitor cells from adult myocardium: homing, differentiation, and fusion after infarction. PNAS. 2003;100:12313–12318. doi: 10.1073/pnas.2132126100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Laugwitz KL, Moretti A, Lam J, et al. Postnatal isl1+ cardioblasts enter fully differentiated cardiomyocyte lineages. Nature. 2005;433:647–653. doi: 10.1038/nature03215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Messina E, De Angelis L, Frati G, et al. Isolation and expansion of adult cardiac stem cells from human and murine heart. Circ Res. 2004;95:911–921. doi: 10.1161/01.RES.0000147315.71699.51. [DOI] [PubMed] [Google Scholar]
- [5].Martin CM, Meeson AP, Roberston SM, et al. Persistent expression of the ATP-binding cassette transporter, Abcg2, Identifies cardiac SP cells in the developing and adult heart. Dev Biol. 2004;265:262–275. doi: 10.1016/j.ydbio.2003.09.028. [DOI] [PubMed] [Google Scholar]
- [6].Oyama T, Nagai T, Wada H, et al. Cardiac side population cells have a potential to migrate and differentiate into cardiomyocytes in vitro and in vivo. JCB. 2007;176(3):329–341. doi: 10.1083/jcb.200603014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Bearzi C, Rota M, Hosoda T, et al. Human cardiac stem cells. PNAS. 2007;104(35):14068–14073. doi: 10.1073/pnas.0706760104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Smits AM, van Vliet P, Metz CH, et al. Human cardiomyocyte progenitor cells differentiate into functional mature cardiomyocytes: an in vitro model for studying human cardiac physiology and path physiology. Nature protocols. 2009;4(2):232–243. doi: 10.1038/nprot.2008.229. [DOI] [PubMed] [Google Scholar]
- [9].Smith RR, Barile L, Cho HC, et al. Regenerative potential of cardiosphere-derived cells expanded from percutaneous endomyocardial biopsy specimens. Circulation. 2007;115:896–908. doi: 10.1161/CIRCULATIONAHA.106.655209. [DOI] [PubMed] [Google Scholar]
- [10].Hierlihy AM, Seale P, Lobe CG, Rudnicki MA, Megeney LA. The post-natal heart contains a myocardial stem cell population. FEBS Letters. 2002;530:239–243. doi: 10.1016/s0014-5793(02)03477-4. [DOI] [PubMed] [Google Scholar]
- [11].Franco D, Moreno N, Ruiz-Lozano P. Non-resident stem cell populations in regenerative cardiac medicine. Cell Mol Life Sci. 2007;64(6):683–691. doi: 10.1007/s00018-007-6521-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Anversa P, Rota M, Urbanek K, et al. Myocardial aging-a stem cell problem. Basic Res Cardiol. 2005;100(6):482–93. doi: 10.1007/s00395-005-0554-3. [DOI] [PubMed] [Google Scholar]
- [13].Meeson AP, Hawke TJ, Graham S, et al. Cellular and Molecular Regulation of Skeletal Muscle Side Population Cells. Stem Cells. 2004;22:1305–1320. doi: 10.1634/stemcells.2004-0077. [DOI] [PubMed] [Google Scholar]
- [14].Limana F, Zacheo A, Mocini D, et al. Identification of myocardial and vascular precursor cells in human and mouse epicardium. Circ Res. 2007;101:1255–1265. doi: 10.1161/CIRCRESAHA.107.150755. [DOI] [PubMed] [Google Scholar]
- [15].O’Rahilly R. Early human development and the chief sources of information on staged human embryos. Eur J Obstet Gynecol Reprod Biol. 1979;9(4):273–80. doi: 10.1016/0028-2243(79)90068-6. [DOI] [PubMed] [Google Scholar]
- [16].Hern WM. Correlation of fetal age and measurements between 10 and 26 weeks of gestation. Obstet Gynecol. 1984;63(1):26–32. [PubMed] [Google Scholar]
- [17].Samuel S, Walsh R, Webb J, Robins A, Potten C, Mahida YR. Characterization of putative stem cells in isolated human colonic crypt epithelial cells and their interactions with myofibroblasts. Am J Physiol Cell Physiol. 2009;296:296–305. doi: 10.1152/ajpcell.00383.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hackett T, Shaheen F, Johnson A, et al. Characterisation of side population cells from human airway epithelium. Stem Cells. 2008;26:2576–2585. doi: 10.1634/stemcells.2008-0171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Terrace JD, Hay DC, Samuel K, et al. Side population cells in developing human liver are primarily haematopoietic progenitor cells. Exp Cell Res. 2009;315(13):2141–53. doi: 10.1016/j.yexcr.2009.02.004. [DOI] [PubMed] [Google Scholar]
- [20].Nilsson J, Ali S, Harvey I, Kirby JA, Meeson AP. Stem cell therapy: A role for CXCR4 in homing bone marrow side population cells to areas of myocardial damage. Int J Cardiol. 2010 doi: 10.1016/j.ijcard.2010.05.021. doi:10.1016/j.ijcard.2010.05.021. [DOI] [PubMed] [Google Scholar]
- [21].Masino AM, Gallardo TD, Wilcox CA, Olson EN, Williams RS, Garry DJ. Transcriptional regulation of cardiac progenitors cell populations. Circ Res. 2004;95:389–397. doi: 10.1161/01.RES.0000138302.02691.be. [DOI] [PubMed] [Google Scholar]
- [22].Fetsch PA, Abati A, Litman T, et al. Localization of the ABCG2 mitoxantrone resistance-associated protein in normal tissue. Cancer Lett. 2006;235:84–92. doi: 10.1016/j.canlet.2005.04.024. [DOI] [PubMed] [Google Scholar]
- [23].Meissner K, Heydrich B, Jedlitschky G, et al. The ATP-binding cassette transporter ABCG2 (BCRP), a marker for side population cells, is expressed in human heart. J Histochem Cytochem. 2006;54(2):215–221. doi: 10.1369/jhc.5A6750.2005. [DOI] [PubMed] [Google Scholar]
- [24].Leimanis ML, Georges E. ABCG2 membrane transporter in mature human erythrocytes is exclusively homodimer. Biochem Biophys Res Commun. 2007;354:345–350. doi: 10.1016/j.bbrc.2006.12.219. [DOI] [PubMed] [Google Scholar]
- [25].Xu J, Liu Y, Yang Y, Bates S, Zhang J. Characterization of Oligomeric Human Half-ABC transporter ATP-binding cassette G2. The Journal of Biological Chemistry. 2004;279:19781–19789. doi: 10.1074/jbc.M310785200. [DOI] [PubMed] [Google Scholar]
- [26].Solbach TF, Paulus B, Weyland M, Eschenhagen T, Zolk O, Fromm MF. ATP-binding cassette transporters in human heart failure. Naunyn-Schmiedeberg’s Arch Pharmacol. 2008;377(3):231–243. doi: 10.1007/s00210-008-0279-6. [DOI] [PubMed] [Google Scholar]
- [27].Maliepaard M, Scheffer GL, Faneyte IF, et al. Subcellular Localization and Distribution of the Breast Cancer Resistance Protein Transporter in Normal Human Tissues. Cancer Res. 2001;61:3458–3464. [PubMed] [Google Scholar]
- [28].Krause DS, Fackler MJ, Civin CI, May WS. CD34: structure, biology, and clinical utility. Blood. 1996;87(1):1–13. [PubMed] [Google Scholar]
- [29].Grafe M, Grafe K, Auch-Schwelk W, Terbeek D, Hertel H, Fleck E. Cultivation and characterization of micro-and macrovascular endothelial cells from the human heart. Eur Heart Journal. 1993;14(suppl I):74–81. [PubMed] [Google Scholar]
- [30].Scholz D, Schaper J. Platelet/endothelial cell adhesion molecule-1 (PECAM-1) is localized over the entire plasma membrane of endothelial cells. Cell Tissue Res. 1997;290(3):623–31. doi: 10.1007/s004410050968. [DOI] [PubMed] [Google Scholar]
- [31].Cai CL, Liang X, Shi Y, et al. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell. 2003;5:877–889. doi: 10.1016/s1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Zhou S, Morris JJ, Barnes Y, Lan L, Schuetz JD, Sorrentino BP. Bcrp1 gene expression is required for normal numbers of side population cells in mice, and confers relative protection to mitoxantrone in hematopoietic cells in vivo. PNAS. 2002;99(19):12339–12344. doi: 10.1073/pnas.192276999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Pfister O, Oikonomopoulos A, Sereti K-I, et al. Role of the ATP-Binding Cassette Transporter Abcg2 in the Phenotype and Function of Cardiac Side Population Cells. Circ Res. 2008;103:825–835. doi: 10.1161/CIRCRESAHA.108.174615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Coats AJ. Ethical authorship and publishing. Int J Cardiol. 2009;131:149–50. doi: 10.1016/j.ijcard.2008.11.048. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. (A) Western blot analysis for ABCG2 protein expression in the embryonic (CS18) and fetal (F5) human heart. Note the decrease in protein expression in the ventricles in the fetal heart compared to the embryonic heart. Human placenta was used as a positive control. kDa, kilodalton; RV, right ventricle; LV, left ventricle; RA, right atria; LA, left atria; OFT, outflow tract. (B) Western analysis for ABCG2 following gene silencing in MCF7 cells. Lane 1 cells treated with control morpholino oligonucleotides. Lane 2 cells treated with translation blocking antisense morpholino oligonucleotides for ABCG2. Note the lack of expression of ABCG2 in Lane 2. GapdH is used as a loading control. MO, antisense morpholino oligonucleotide
Supplementary Table 1. Heart samples used to optimise protocol for FACS analysis for SP cell identification in the developing human heart.