Abstract
Background
Carotenoids are isoprenoid compounds synthesized by all photosynthetic organisms. Despite much research on carotenoid biosynthesis in the model plant Arabidopsis thaliana, there is a lack of information on the carotenoid pathway in Brassica rapa. To better understand its carotenoid biosynthetic pathway, we performed a systematic analysis of carotenoid biosynthetic genes at the genome level in B. rapa.
Results
We identified 67 carotenoid biosynthetic genes in B. rapa, which were orthologs of the 47 carotenoid genes in A. thaliana. A high level of synteny was observed for carotenoid biosynthetic genes between A. thaliana and B. rapa. Out of 47 carotenoid biosynthetic genes in A. thaliana, 46 were successfully mapped to the 10 B. rapa chromosomes, and most of the genes retained more than one copy in B. rapa. The gene expansion was caused by the whole-genome triplication (WGT) event experienced by Brassica species. An expression analysis of the carotenoid biosynthetic genes suggested that their expression levels differed in root, stem, leaf, flower, callus, and silique tissues. Additionally, the paralogs of each carotenoid biosynthetic gene, which were generated from the WGT in B. rapa, showed significantly different expression levels among tissues, suggesting differentiated functions for these multi-copy genes in the carotenoid pathway.
Conclusions
This first systematic study of carotenoid biosynthetic genes in B. rapa provides insights into the carotenoid metabolic mechanisms of Brassica crops. In addition, a better understanding of carotenoid biosynthetic genes in B. rapa will contribute to the development of conventional and transgenic B. rapa cultivars with enriched carotenoid levels in the future.
Electronic supplementary material
The online version of this article (doi:10.1186/s12864-015-1655-5) contains supplementary material, which is available to authorized users.
Keywords: Biosynthetic pathway, Carotenoid biosynthetic genes, Comparative genomics, Expression analysis, Brassica rapa
Background
Carotenoids represent a group of natural pigments derived from the general isoprenoid biosynthetic pathway. The reaction fundamentals and enzymes involved in the carotenoid pathway have been well studied in Arabidopsis thaliana [1]. Carotenoids are mainly synthesized from isopentenyl diphosphate (IPP) and dimethylallyl diphosphate produced by the 2-C-methyl-D-erythritol-4-phosphate (MEP) pathway. The enzymes from the upstream MEP pathway include 1-deoxy-D-xylulose-5-phosphate synthase (DXS), 1-deoxy-D-xylulose-5-phosphate reductoisomerase (DXR), 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase (CMK), 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase (MDS), 4-hydroxy-3-methylbut-2-enyl diphosphate synthase (HDS), and 4-hydroxy-3-methylbut-2-enyl diphosphate reductase (HDR).
Additionally, isopentenyl pyrophosphate isomerase (IPPI) catalyzes the isomerization of dimethylallyl diphosphate to IPP, while geranyl pyrophosphate synthase (GPS) transforms IPP to geranyl pyrophosphate (GPP) [2–12]. The first key step for carotenoid biosynthesis is the production of a 40-carbon phytoene from paired geranylgeranyl pyrophosphate (GGPP) molecules. The process is catalyzed by enzyme phytoene synthase (PSY) [13, 14]. Then, a series of desaturases and isomerases participate in the generation of lycopene (colored) from phytoene (no color), including phytoene desaturase (PDS), ζ-carotene desaturase (ZDS), 15-cis-ζ-carotene isomerase (Z-ISO), and carotenoid isomerase (CRTISO) [15–18]. Lycopene represents the first branch point of the carotenoid pathway and is catalyzed by two competing enzymes, lycopene β-cyclase and lycopene ε-cyclase, which result in the production of β-carotene and α-carotene, respectively [19]. Then, carotenoid cleavage dioxygenases (CCD) participate in the formation of apocarotenoid [20]. Cyclic carotenes are further modified by two different types of carotenoid hydroxylases in A. thaliana: non-heme di-iron enzymes (BCH type) and cytochrome P450 enzymes (CYP97 type), which include BCH1, BCH2, LUT5, LUT1, and CYP97B3 [21–23]. Zeaxanthin then enters the xanthophyll cycle through the stepwise activities of zeaxanthin epoxidase (ZEP) and violaxanthin de-epoxidase (VDE) [24, 25]. The pigments antheraxanthin and violaxanthin produced by the above processes are further converted to neoxanthin by neoxanthin synthase (NSY) [24]. Finally, the enzymes 9-cis-epoxycarotenoid dioxygenase (NCED), xanthoxin dehydrogenase (ABA2), and abscisic-aldehyde oxidase (AAO3) catalyze violaxanthin and neoxanthin to produce xanthoxin and abscisic acid, respectively [26–31].
The genus Brassica includes many vegetable crops, such as B. rapa, Brassica oleracea, Brassica napus, Brassica parachinensis, and Brassica juncea. B. rapa L. ssp. pekinensis (Chinese Cabbage) evolved in China and is an important vegetable crop in Asia. The inner leaves of several cultivars of heading B. rapa are orange and yellow, and are abundant in lutein, β-carotene, and prolycopene [32–34]. Because of the previous absence of genome information, little is known about the genes involved in the carotenoid biosynthetic pathway of B. rapa [35, 36]. The sequencing and release of the B. rapa genome [37], provides a good opportunity to systematically study the carotenoid biosynthetic genes in B. rapa. A complete understanding of the carotenogenesis genes is important for elucidating the mechanisms of carotenoid biosynthesis in B. rapa, as well as for the breeding of new B. rapa varieties with rich carotenoids, which are good for human health.
Whole-genome duplication events provide abundant amounts of genetic material for gene family expansion or the evolution of new genes in plants [38, 39]. B. rapa experienced a whole-genome triplication (WGT) event after its divergence from A. thaliana and has three subgenomes in its nucleus [40]. The level of gene loss among the three subgenomes of B. rapa is biased: fragments with the highest gene densities comprise subgenome LF, fragments with moderate gene densities comprise subgenome MF1, and those with the least genes comprise subgenome MF2 [41, 42]. The fragments of the three subgenomes in B. rapa have been well distinguished; therefore, differences in gene loss among these subgenomes can be identified unambiguously. Although the influence of WGT on some gene families has been studied [43, 44], there is presently no detailed information on the evolution of carotenoid biosynthetic genes after the WGT in B. rapa. To obtain comprehensive information on the carotenoid biosynthetic pathway in B. rapa and to explore the effect of the WGT on these genes, we performed a comparative genomic analysis between B. rapa and A. thaliana using the genome sequences and annotation information of the two species [37]. We investigated the evolution and functions of carotenoid biosynthetic genes in B. rapa by constructing phylogenic trees and analyzing their transcription patterns. The systematic analysis of carotenoid biosynthesis genes in B. rapa will improve our understanding of the genetic mechanisms of carotenoid biosynthesis and carotenoid accumulation in B. rapa crops.
Results and discussion
Identification of carotenoid biosynthetic genes in B. rapa
Using the carotenoid biosynthetic enzymes as queries we searched the TAIR and KEGG pathway databases. In A. thaliana, 47 potential carotenoid biosynthetic genes were investigated, including 21 genes that participated in the MEP pathway upstream of GGPP and 26 genes encoding carotenoid biosynthetic enzymes. Based on a combination of syntenic and non-syntenic orthology analyses, 67 B. rapa carotenoid biosynthetic genes were identified, representing orthologs of 42 out of the 47 A. thaliana carotenoid biosynthetic genes (Table 1; Additional file 1: Table S1). The other five A. thaliana carotenoid biosynthetic genes (GGPS6, GGPS9, GGPS11, GGPS12 and LUT1) showed no B. rapa orthologs. Each carotenoid biosynthetic gene in B. rapa was assigned a name based on the enzymatic reaction, similar to those given in the A. thaliana carotenoid biosynthetic pathway [45].
Table 1.
Carotenoid biosynthetic genes identified in Brassica rapa
| B. rapa | |||||
|---|---|---|---|---|---|
| Enzyme | A. thaliana | Syntenic orthologs | Non-syntenic orthologs | ||
| LF | MF1 | MF2 | |||
| MEP pathway to GGPP genes | |||||
| DXS | AT4G15560 | BrDXS1 (Bra033495) | BrDXS2 (Bra012779) | - | - |
| DXR | AT5G62790 | BrDXR1 (Bra010123) | - | BrDXR2 (Bra035881) | - |
| MCT | AT2G02500 | - | BrMCT (Bra026591) | - | - |
| CMK | AT2G26930 | BrCMK (Bra012040) | - | - | - |
| MDS | AT1G63970 | - | - | BrMDS1 (Bra027672) | BrMDS2 (Bra027770) |
| HDS | AT5G60600 | BrHDS (Bra002468) | - | - | - |
| HDR | AT4G34350 | BrHDR1 (Bra011522) | - | BrHDR2 (Bra034620) | - |
| IPPI1 | AT5G16440 | - | BrIPPI1 (Bra006354) | - | - |
| IPPI2 | AT3G02780 | BrIPPI2.1 (Bra040599) | BrIPPI2.2 (Bra021411) | BrIPPI2.3 (Bra001063) | - |
| GGPS1 | AT4G36810 | BrGGPS1.1 (Bra011709) | BrGGPS1.2 (Bra017785) | BrGGPS1.3 (Bra010576) | BrGGPS1.4 (Bra028096) |
| GGPS2 (GGPS5) | AT2G23800 | BrGGPS2.1 (Bra039216) | BrGGPS2.2 (Bra032140) | - | |
| GGPS3 | AT3G14550 | BrGGPS3.1 (Bra027330) | BrGGPS3.2 (Bra021562) | BrGGPS3.3 (Bra001556) | |
| GGPS7 | AT2G18620 | - | BrGGPS4 (Bra038544) | - | |
| GGPS8 | AT3G14510 | - | BrGGPS8.1 (Bra021565) | - | |
| GGPS11 | AT3G29430 | - | - | - | |
| GGPS4 | AT2G18640 | - | BrGGPS4 (Bra038544) | - | - |
| GGPS6 | AT1G49530 | - | - | - | - |
| GGPS9 | AT3G14530 | - | - | - | - |
| GGPS10 | AT3G20160 | BrGGPS10.1 (Bra035808) | - | BrGGPS10.2 (Bra001777) | - |
| GGPS12 | AT3G32040 | - | - | - | - |
| GGR | AT4G38460 | BrGGR (Bra011898) | - | - | - |
| Carotenoid biosynthetic genes | |||||
| PSY | AT5G17230 | BrPSY1 (Bra008569) | BrPSY2 (Bra006391) | BrPSY3 (Bra023603) | - |
| PDS3 | AT4G14210 | - | BrPDS3.1 (Bra032770) | BrPDS3.2 (Bra010751) | - |
| Z-ISO | AT1G10830 | BrZ-ISO (Bra019899) | - | - | - |
| ZDS | AT3G04870 | - | BrZDS (Bra040411) | - | - |
| CRTISO | AT1G06820 | - | - | BrCRTISO (Bra031539) | - |
| CRTISO2 | AT1G57770 | - | - | BrCRTISO2 (Bra027908) | - |
| LYC | AT3G10230 | BrLYC (Bra029825) | - | - | - |
| LUT2 | AT5G57030 | BrLUT2.1 (Bra002769) | BrLUT2.2 (Bra006838) | - | BrLUT2.3 (Bra020718) |
| CHY1 | AT4G25700 | BrCHY1.1 (Bra013912) | BrCHY1.2 (Bra019145) | - | - |
| CHY2 | AT5G52570 | BrCHY2.1 (Bra003121) | - | - | - |
| LUT5 | AT1G31800 | - | BrLUT5 (Bra038437) | - | - |
| CYP97B3 | AT4G15110 | - | - | BrCYP97B3 (Bra038092) | - |
| LUT1 | AT3G53130 | - | - | - | - |
| ZEP | AT5G67030 | BrZEP1 (Bra012127) | - | BrZEP2 (Bra037130) | - |
| VDE | AT1G08550 | BrVDE (Bra018616) | - | - | - |
| NSY | AT1G67080 | - | BrNSY (Bra034026) | - | - |
| CCD7 | AT2G44990 | - | BrCCD7 (Bra040330) | - | - |
| CCD8 | AT4G32810 | BrCCD8 (Bra011384) | - | - | - |
| NCED2 | AT4G18350 | BrNCED2.1 (Bra013298) | BrNCED2.2 (Bra012603) | - | - |
| NCED3 | AT3G14440 | BrNCED3.1 (Bra027336) | BrNCED3.2 (Bra021558) | BrNCED3.3 (Bra001552) | - |
| NCED4 | AT4G19170 | BrNCED4.1 (Bra013378) | - | BrNCED4.2 (Bra020970) | - |
| NCED5 | AT1G30100 | BrNCED5 (Bra032359) | - | - | - |
| NCED6 | AT3G24220 | BrNCED6 (Bra015002) | - | - | - |
| NCED9 | AT1G78390 | BrNCED9.1 (Bra035033) | BrNCED9.2 (Bra008358) | - | - |
| ABA2 | AT1G52340 | BrABA2.1 (Bra018964) | BrABA2.2 (Bra014323) | - | - |
| AAO3 | AT2G27150 | - | BrAAO3 (Bra034325) | - | - |
Among the 67 carotenoid biosynthetic genes in B. rapa, 64 were syntenic orthologs of the 40 A. thaliana carotenoid biosynthetic genes (Fig. 1), and only three B. rapa carotenoid biosynthetic genes had no syntenic relationships. The carotenoid biosynthetic genes have expanded in the genome of B. rapa. The multiple copies of the carotenoid biosynthetic genes in B. rapa that are syntenic to genes in A. thaliana were generated from the WGT. In addition, 37 of the 42 A. thaliana carotenoid biosynthetic genes had less than three syntenic orthologs in B. rapa as a result of gene fractionation following the triplication event. GGPS is encoded by a multigene family with 12 members in A. thaliana [10], but GGPS6, GGPS9, GGPS11 and GGPS12 orthologs were not found in B. rapa. GGPS4 and GGPS7 form a tandem array in A. thaliana and correspond to one gene (Bra038544) in B. rapa. Furthermore, GGPS1, GGPS2, GGPS3, GGPS7, GGPS8, and GGPS11 share the same non-syntenic ortholog (Bra028096) in B. rapa.
Fig. 1.
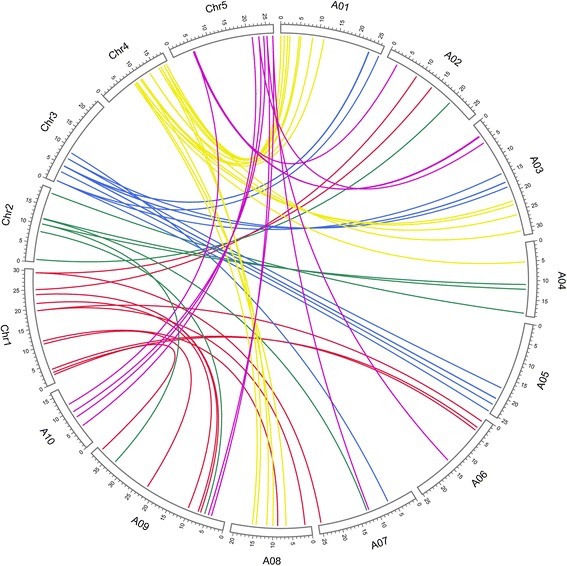
Ideogram of syntenic carotenoid biosynthetic genes in Brassica rapa and Arabidopsis thaliana. Ch1 to Ch5 are A. thaliana chromosomes, and A01 to A10 are B. rapa chromosomes. All ranges are to scale
Chromosomal and subgenomic distributions
A diagrammatic representation of the chromosomal distribution of carotenoid biosynthetic genes on the 10 B. rapa chromosomes is depicted in Fig. 2. In total, 66 of 67 B. rapa carotenoid biosynthetic genes were mapped to the 10 chromosomes, with 12, 5, 11, 4, 5, 4, 5, 7, 9, and 4 B. rapa carotenoid biosynthetic genes being located on chromosomes A01–A10 in the B. rapa genome V1.5, respectively (Fig. 2). The remaining gene, BrZDS (Bra040411), was assigned to Scaffold000203, which has not yet been assembled to any chromosome according to the B. rapa genome V1.5 (Additional file 1: Table S1). Interestingly, there are no tandem duplicated carotenoid genes in B. rapa.
Fig. 2.
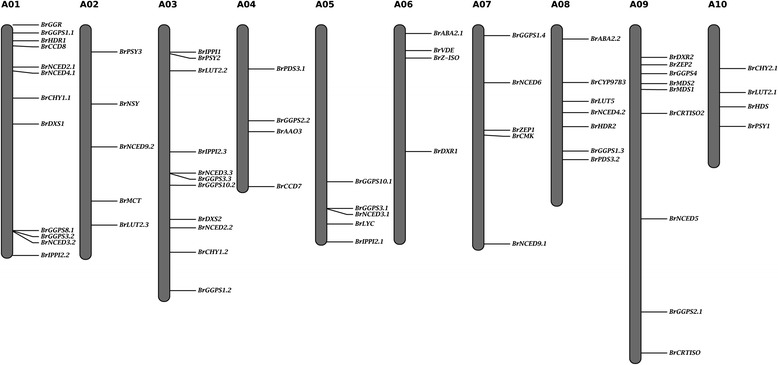
Genomic distribution of 66 carotenoid biosynthetic genes on the 10 chromosomes of Brassica rapa
The genome structure of B. rapa was shaped by the WGT event followed by extensive diploidization [46]. In the genome, fragments with the highest gene densities are in subgenome LF, fragments with moderate gene densities are in subgenome MF1, and the fragments with the least genes are in subgenome MF2 [37, 42]. With this subgenomic information, we then assigned all B. rapa carotenoid biosynthesis genes to the three subgenomes. There were 28, 23, and 16 genes located in LF, MF1, and MF2, respectively. There were more genes located in LF, while fewer genes were distributed in MF1 and even fewer genes in MF2. Of the 64 syntenic orthologs, 27 were in LF, 22 were in MF1, and 15 were in MF2. The proportion of total subgenomic B. rapa genes was used as the background to calculate the P-value using Fisher’s t-test. The P-value was 0.715 (>0.05), indicating that the proportion of carotenoid biosynthetic genes in each subgenome of B. rapa was not significantly different from the background. These results show that the distribution of carotenoid biosynthetic genes is consistent with the gene fractionation status at the whole-genome level [37, 42]. Based on the determination of these carotenoid biosynthetic genes, the carotenoid biosynthetic pathway in B. rapa was thus established.
Evolution of the PSY genes in B. rapa
The WGT event in the B. rapa genome provides a model for the study of the evolutionary fate of multi-copy genes and the effects of polyploidy in economically important crops. To investigate the evolutionary relationship of carotenoid biosynthetic genes in B. rapa, separate neighbor-joining trees were generated for the enzyme PSY by aligning the protein sequence with the corresponding orthologs in Arabidopsis and other plant species (Fig. 3). The phylogenetic analysis of PSY in B. rapa, Arabidopsis, and other monocot and dicot plant species revealed that PSYs cluster into two separate monocot- and dicot-specific clades, where most of the members show a monophyletic pattern of origin. In the Brassicaceae family, the species were separated into two specific clades. As shown in Fig. 3, PSY1, PSY2, and PSY3 sequences from B. rapa each clustered into groups with their respective B. oleracea orthologs on a separate branch. Interestingly, B. rapa, B. oleracea, and Schrenkiella parvula are clustered on one branch, which indicates that the Brassica are closer to S. parvula than to Arabidopsis and that the Brassiceae triplication event occurred near the time of the divergence between Brassiceae and Schrenkiella [46]. However, three B. napus PSYs were clustered on one specific branch with the Arabidopsis and Thellungiella PSYs.
Fig. 3.
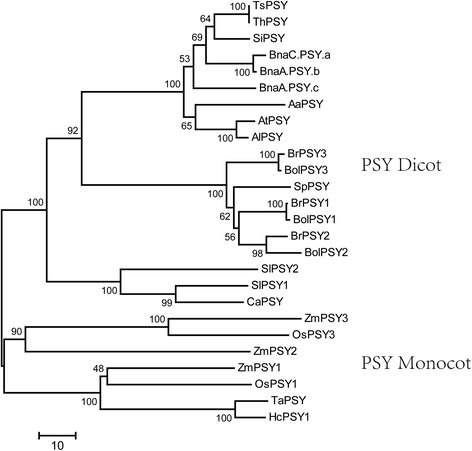
Phylogenetic relationship of the PSY gene between Brassica rapa and other species. The unrooted tree was generated using the Muscle program by the neighbor-joining method. Bootstrap values from 1000 replicates are indicated at each node. Ts: Thellungiella salsuginea; Th: Thellungiella halophila; Si: Sisymbrium irio At: Arabidopsis thaliana; Al: Arabidopsis lyrata; Aa: Aethionema arabicum Br: Brassica rapa; Bol: Brassica oleracea; Sp: Schrenkiella parvula; Bna: Brassica napus; Sl: Solanum lycopersicum; Ca: Capsicum annuum; Os: Oryza sativa; Zm: Zea mays; Ta: Triticum aestivum; and Hc: Hordeum chilense
PSY is a rate-limiting enzyme in Solanum lycopersicum fruit, B. napus seeds, Gentiana scabra Bunge flowers, and other plants [47–50], and is a key committed enzyme in the carotenoid biosynthetic pathway [51]. In A. thaliana, a single PSY gene regulates phytoene synthesis in all tissues [14], while three genes encoding PSY enzymes were retained in B. rapa and B. oleracea after the WGT event. The PSY gene family expansion preceded the speciation of B. rapa and B. oleracea, dating back to the WGT event [52]. Furthermore, six PSYs were retained in B. napus; however, we selected only three B. napus PSY genes that had complete sequences online. The evolution of carotenoid biosynthetic genes, such as PSYs, was consistent with the evolution of B. rapa.
Expression profiles of carotenoid biosynthetic genes in six organs of B. rapa
To determine the expression patterns of carotenoid biosynthetic genes in different organs and elucidate their roles in the carotenoid biosynthetic pathway, we performed next-generation transcriptome sequencing (RNA-Seq) [53], as well as a hierarchical-clustering analysis of carotenoid biosynthetic gene expression patterns using the R software [54]. As shown in Fig. 4, the expression levels of the carotenoid biosynthetic genes in the six organs were diverse and could be divided into two groups. Flowers and leaves were in group I, and the flowers had the highest overall expression level and the maximum number of expressed genes among the six tissues. Siliques, roots, stems, and calli were in group II, and calli had the lowest overall expression level, although it was only slightly lower than in the roots.
Fig. 4.
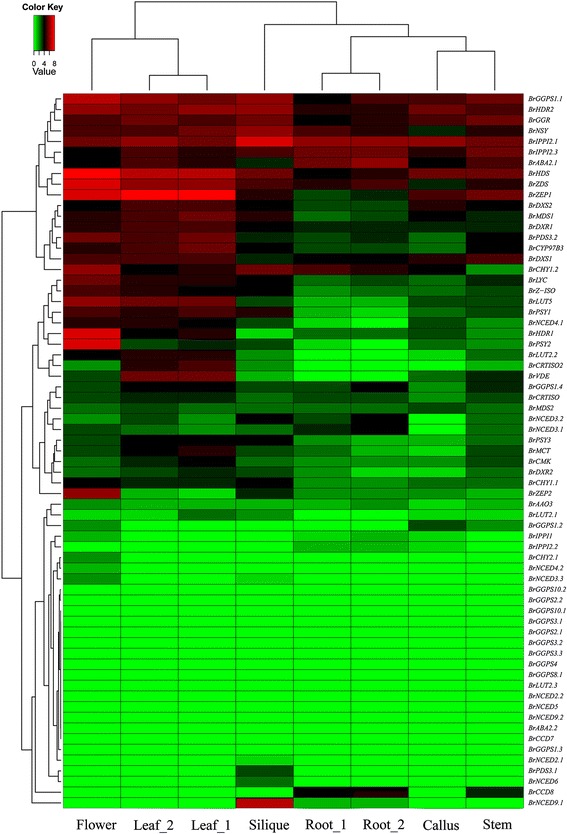
Expression profiles of Brassica rapa carotenoid biosynthetic genes in six organs. Transcriptome sequencing was employed to investigate expression patterns of B. rapa carotenoid biosynthetic genes. The color scale shown at the top represents FPKM-normalized log2-transformed counts. Green indicates low expression levels, while red indicates high levels
The B. rapa carotenoid biosynthetic genes could be divided into two clusters based on their expression patterns (Fig. 4; Additional file 2: Table S2). Cluster 1 was composed of genes that were expressed in all six organs, while Cluster 2 contained genes that had low expression levels and some organ-specific expression patterns. Cluster 1 included two expression groups. The first group showed a high expression level in all six organs. The second group exhibited a higher expression level in flowers, leaves, and siliques, indicating their roles in the carotenoid biosynthetic pathway in these tissues, than in roots, calli, and stems. The genes belonging to Cluster 2 had lower expression levels and could be subdivided into three groups. The first group was composed of 10 genes that had low expression levels in siliques, roots, calli, and stems. The genes in the second group exhibited low but stable expression levels in all six organs, except BrZEP2, which was highly expressed in flowers. This result was consistent with a previous study and indicates that BrZEP2 plays an important role in the synthesis and accumulation of carotenoids in B. rapa flowers [55]. Most genes belonging to the third group exhibited low or undetectable expression levels in all six organs. Interestingly, the gene BrCCD8 was expressed in stems and roots but could not be detected in flowers, leaves, siliques, or calli, and BrNCED9.1 was highly expressed in siliques.
According to the expression analysis results, most B. rapa carotenoid biosynthetic genes appeared to have similar roles to their orthologs in other species. For example, it has been reported that the PSY, ZDS, PDS, and ZEP genes play crucial roles in carotenoid biosynthesis in A. thaliana [1]. In B. rapa, these genes exhibited either predominant or specific expression patterns in leaves, such as BrZEP1, which was highly expressed in flowers and leaves. A. thaliana contains a family of 12 genes that are similar to GGPS, but only five GGPS genes have been shown to be expressed in different tissues during plant development [10]. Although there are 13 duplicated BrGGPS genes in B. rapa, only BrGGPS1.1 was highly expressed in the six organs we examined. Moreover, most of the BrGGPS genes did not have detectable expression levels, which may be due to functional divergence after the triplication event or because they are specifically expressed at other developmental stages.
Expression variance among duplicated carotenoid biosynthetic genes in B. rapa
Expression differentiation, which is considered the first step in the functional divergence between duplicated genes, has long been a subject of great interest to geneticists and evolutionary biologists [56–58], because it increases the probability of the retention of duplicated genes in a genome [57]. The duplicated genes that we have described here are paralogous carotenoid biosynthetic genes that originated due to the WGT event in B. rapa. The carotenoid biosynthetic genes that have more than one copy were selected to analyze expression variance in B. rapa. Analysis of RNA-Seq data generated from B. rapa root, leaf, flower, silique, callus and stem tissues suggests that WGT paralogous gene pairs show significant expression differentiation (Fig. 5; Additional file 2: Table S2). In each duplicated carotenoid biosynthetic gene pair, the expression levels change significantly. For example, in some carotenoid biosynthetic genes, such as BrDXR, BrMDS, BrHDR, BrIPPI2, BrGGPS1, BrPSY, BrPDS3, BrLUT2, BrCHY1, BrZEP, BrNCED3, BrNCED4, BrNCED9, and BrABA2, one copy is much more highly expressed than the other. In other genes, such as BrDXS, BrGGPS2, BrGGPS3, BrGGPS10, and BrNCED2, the differential expression levels between the copies were not significant. Additionally, the expression levels of each copy of BrGGPS2, BrGGPS3, and BrGGPS10 were lower. In general, the expression levels of genes in the LF subgenome were significantly higher than the corresponding syntenic genes in the MF1 and MF2 subgenomes (Additional file 3: Figure S1).
Fig. 5.
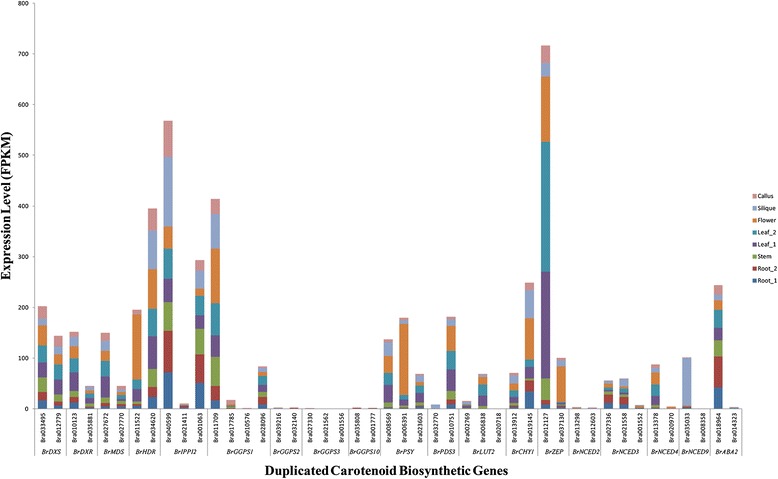
Different transcript levels of duplicated carotenoid biosynthetic genes in Brassica rapa
The expression differentiation between duplicate genes has been reported in many studies [59–62], and the functional divergence of PSY gene expression facilitates the accumulation of high levels of carotenoids in chromoplast-rich tissues in B. napus [52]. In this paper, we studied the expression variance of genes that were created during a WGT event using RNA-Seq data. A gene expression analysis revealed an extensive variance between paralogs of each carotenoid biosynthetic gene. These duplicated gene expression variations may be signs of subfunctionalization among different tissues and contribute to an increased complexity in the regulatory networks of the carotenoid pathway after polyploidization.
Conclusions
We have identified 67 carotenoid biosynthetic genes in the genome of B. rapa through a comparative analysis between A. thaliana and B. rapa, and established the carotenoid biosynthetic pathway in B. rapa. Multiple copies of carotenoid biosynthetic genes were generated and retained after the WGT event and most of them maintained their syntenic relationships with their orthologs in A. thaliana. Fisher’s t-test indicated that the proportion of carotenoid biosynthetic genes in each subgenome of B. rapa was not significantly different from the whole genome backgrounds. The composition of carotenoid biosynthetic genes could explain the metabolic profiles of carotenoid accumulation and help to elaborate the genetic mechanism of carotenoid biosynthesis in B. rapa. The expression analysis of carotenoid biosynthetic genes showed that the paralogs of these genes were differentially expressed among roots, stems, leaves, flowers, calli, and siliques, suggesting that the functional differentiation of the duplicated carotenoid biosynthetic genes occurred after polyploidization.
Our study of the complete set of carotenoid biosynthetic genes in B. rapa will provide insights into carotenoid metabolic mechanisms in Brassica crops. In addition, a better understanding of carotenoid biosynthetic genes in B. rapa will facilitate the development of conventional and transgenic B. rapa cultivars with enriched carotenoid levels in the future.
Methods
Database for carotenoid biosynthetic gene identification in B. rapa
Gene sequences of A. thaliana involved in the carotenoid biosynthetic pathway were acquired from the KEGG pathway database (http://www.genome.jp/kegg/pathway.html) and TAIR database (www.arabidopsis.org). The B. rapa genome (version 1.5) and a set of annotated gene sequences from BRAD (http://Brassicadb.org) were used to identify the carotenoid biosynthetic genes in B. rapa [41].
Identification and analysis of orthologs between B. rapa and A. thaliana
The carotenoid biosynthetic gene and protein sequences of A. thaliana were aligned with the genome and protein sequences of B. rapa using BLASTN and BLASTP, respectively, with a cut off E-value ≤ 1E−10 and coverage ≥ 0.75. Syntenic orthologs between A. thaliana and B. rapa from BRAD (http://brassicadb.org/brad/) were identified based on sequence similarity (cutoff: E ≤ 10−20) and the collinearity of flanking genes [63].
Phylogenetic analysis
The phylogenetic tree was constructed based on the full-length sequences of PSY proteins. A neighbor-joining tree was built using MEGA version 5.05 [64], adopting the Poisson correction distance. Support for the tree obtained was assessed using the bootstrap method with 1000 replicates.
Accession numbers
Sequence data for the article can be found in the TAIR, NCBI, and Brassica (BRAD; http://brassicadb.org/brad/) databases. The accession numbers for the sequences are as follows: SlPSY1: NP_001234812.1; SlPSY2: NP_001234671.1; ZmPSY1: ACY70935.1; ZmPSY2: NP_001108117.1; ZmPSY3: NP_001108125.1; HcPSY: AEH05575.1; TaPSY: ABS83386.1; CaPSY: ADH04284.1; OsPSY1: AAS18307.1; OsPSY3: ABC75828.1; BnaC.PSY.a: JF920037; BnaA.PSY.b: JF920038; BnaA.PSY.c: JF920039, and all others can be found in Brassica databases.
Expression analysis of B. rapa carotenoid biosynthetic genes
The expression patterns of carotenoid biosynthetic genes in B. rapa were measured using RNA-Seq data [53]. Six tissues, root, stem, leaf, flower, callus, and silique, of B. rapa accession Chiifu-401-42 were prepared for mRNA extraction. Plants were grown under greenhouse conditions at 22 °C. Callus tissue was obtained from tissue culture. Root, stem, and leaf tissues were collected from 7-week-old plants. Two samples of root and leaf tissues were generated from different batches of plants. Flower tissue was obtained from blooming plants on the same day and excluded the floral shoot. Silique tissue was collected from plants 15 days after pollination [53]. FPKM values are available in Additional file 2: Table S2. Gene expression FPKM values were log2 transformed, and R software was employed to normalize the expression data and calculate hierarchical clustering [54].
Availability of supporting data
The B. rapa genome sequence (version 1.5) and gene sequences were acquired from BRAD (http://brassicadb.org/brad/).
Acknowledgments
We thank Dr. Shengyi Liu (Oil Crops Research Institute of the Chinese Academy of Agricultural Sciences) for kindly providing the RNA-Seq data of B. rapa. This work was supported by the National Basic Research and Development Program (Chinese 973 Program, 2012CB113900) (to RS) and the National High Technology Research and Development Program of China (Chinese 863 Program, 2012AA100100) (to RS). Research was carried out in the Key Laboratory of Biology and Genetic Improvement of Horticultural Crops, Ministry of Agriculture, P.R. China.
Additional files
Gene inventory of the carotenoid pathway and the Brassica rapa orthologs [65].
FPKM values for carotenoid biosynthetic gene expression levels in Brassica rapa.
Average expression levels of carotenoid biosynthetic genes in three subgenomes of Brassica rapa.
Footnotes
Peirong Li and Shujiang Zhang contributed equally to this work.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
PL researched the carotenoid biosynthetic genes, performed the detailed analysis of carotenoid biosynthetic genes by comparative genomics, and drafted the manuscript. SZ critically reviewed the manuscript. SFZ, FL, HZ, JW, and XW participated in the data analysis. FC participated in manuscript modification. RS conceived and supervised the work. All the authors have read and approved the final manuscript.
Contributor Information
Peirong Li, Email: lprdream@126.com.
Shujiang Zhang, Email: zhangshujiang@caas.cn.
Shifan Zhang, Email: zhangshifan@caas.cn.
Fei Li, Email: lifei@mail.caas.net.cn.
Hui Zhang, Email: zhanghui05@caas.cn.
Jian Wu, Email: wujian@caas.cn.
Xiaowu Wang, Email: wangxiaowu@caas.cn.
Rifei Sun, Phone: +86-10-82109511, Email: sunrifei@caas.cn.
References
- 1.Ruiz-Sola MÁ, Rodríguez-Concepción M. Carotenoid biosynthesis in Arabidopsis: a colorful pathway. The Arabidopsis book/American Society of Plant Biologists 2012;10:28. [DOI] [PMC free article] [PubMed]
- 2.Bauer J, Hiltbrunner A, Kessler F. Molecular biology of chloroplast biogenesis: gene expression, protein import and intraorganellar sorting. Cell Mol Life Sci. 2001;58(3):420–33. doi: 10.1007/PL00000867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campbell M, Hahn FM, Poulter CD, Leustek T. Analysis of the isopentenyl diphosphate isomerase gene family from Arabidopsis thaliana. Plant Mol Biol. 1998;36(2):323–8. doi: 10.1023/A:1005935516274. [DOI] [PubMed] [Google Scholar]
- 4.Carretero-Paulet L, Ahumada I, Cunillera N, Rodriguez-Concepcion M, Ferrer A, Boronat A, Campos N. Expression and Molecular Analysis of the ArabidopsisDXR Gene Encoding 1-Deoxy-d-Xylulose 5-Phosphate Reductoisomerase, the First Committed Enzyme of the 2-C-Methyl-d-Erythritol 4-Phosphate Pathway. Plant Physiol. 2002;129(4):1581–91. doi: 10.1104/pp.003798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Estévez JM, Cantero A, Romero C, Kawaide H, Jiménez LF, Kuzuyama T, Seto H, Kamiya Y, León P. Analysis of the expression of CLA1, a gene that encodes the 1-deoxyxylulose 5-phosphate synthase of the 2-C-methyl-D-erythritol-4-phosphate pathway in Arabidopsis. Plant Physiol. 2000;124(1):95–104. doi: 10.1104/pp.124.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gil MJ, Coego A, Mauch‐Mani B, Jordá L, Vera P. The Arabidopsis csb3 mutant reveals a regulatory link between salicylic acid‐mediated disease resistance and the methyl‐erythritol 4‐phosphate pathway. Plant J. 2005;44(1):155–66. doi: 10.1111/j.1365-313X.2005.02517.x. [DOI] [PubMed] [Google Scholar]
- 7.Hsieh M-H, Chang C-Y, Hsu S-J, Chen J-J. Chloroplast localization of methylerythritol 4-phosphate pathway enzymes and regulation of mitochondrial genes in ispD and ispE albino mutants in Arabidopsis. Plant Mol Biol. 2008;66(6):663–73. doi: 10.1007/s11103-008-9297-5. [DOI] [PubMed] [Google Scholar]
- 8.Hsieh M-H, Goodman HM. The Arabidopsis IspH homolog is involved in the plastid nonmevalonate pathway of isoprenoid biosynthesis. Plant Physiol. 2005;138(2):641–53. doi: 10.1104/pp.104.058735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lange BM, Ghassemian M. Genome organization in Arabidopsis thaliana: a survey for genes involved in isoprenoid and chlorophyll metabolism. Plant Mol Biol. 2003;51(6):925–48. doi: 10.1023/A:1023005504702. [DOI] [PubMed] [Google Scholar]
- 10.Okada K, Saito T, Nakagawa T, Kawamukai M, Kamiya Y. Five geranylgeranyl diphosphate synthases expressed in different organs are localized into three subcellular compartments in Arabidopsis. Plant Physiol. 2000;122(4):1045–56. doi: 10.1104/pp.122.4.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rohdich F, Wungsintaweekul J, Eisenreich W, Richter G, Schuhr CA, Hecht S, Zenk MH, Bacher A. Biosynthesis of terpenoids: 4-diphosphocytidyl-2C-methyl-D-erythritol synthase of Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2000;97(12):6451–6. doi: 10.1073/pnas.97.12.6451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu F, Suzuki K, Okada K, Tanaka K, Nakagawa T, Kawamukai M, Matsuda H. Cloning and functional expression of a novel geranylgeranyl pyrophosphate synthase gene from Arabidopsis thaliana in Escherichia coli. Plant Cell Physiol. 1997;38(3):357–61. doi: 10.1093/oxfordjournals.pcp.a029174. [DOI] [PubMed] [Google Scholar]
- 13.Scolnik PA, Bartley GE. Nucleotide sequence of an Arabidopsis cDNA for phytoene synthase. Plant Physiol. 1994;104(4):1471. doi: 10.1104/pp.104.4.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Welsch R, Medina J, Giuliano G, Beyer P, Von Lintig J. Structural and functional characterization of the phytoene synthase promoter from Arabidopsis thaliana. Planta. 2003;216(3):523–34. doi: 10.1007/s00425-002-0885-3. [DOI] [PubMed] [Google Scholar]
- 15.Chen Y, Li F, Wurtzel ET. Isolation and characterization of the Z-ISO gene encoding a missing component of carotenoid biosynthesis in plants. Plant Physiol. 2010;153(1):66–79. doi: 10.1104/pp.110.153916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park H, Kreunen SS, Cuttriss AJ, DellaPenna D, Pogson BJ. Identification of the carotenoid isomerase provides insight into carotenoid biosynthesis, prolamellar body formation, and photomorphogenesis. Plant Cell. 2002;14(2):321–32. doi: 10.1105/tpc.010302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scolnik P, Bartley G. Nucleotide sequence of Zeta-carotene Desaturase (accession no. U38550) from Arabidopsis. Plant Physiol. 1995;109(4):1499. [Google Scholar]
- 18.Scolnik PA, Bartley GE. Phytoene desaturase from Arabidopsis. Plant Physiol. 1993;103(4):1475. doi: 10.1104/pp.103.4.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cunningham FX, Pogson B, Sun Z, McDonald KA, DellaPenna D, Gantt E. Functional analysis of the beta and epsilon lycopene cyclase enzymes of Arabidopsis reveals a mechanism for control of cyclic carotenoid formation. Plant Cell. 1996;8(9):1613–26. doi: 10.1105/tpc.8.9.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Auldridge ME, Block A, Vogel JT, Dabney‐Smith C, Mila I, Bouzayen M, Magallanes‐Lundback M, DellaPenna D, McCarty DR, Klee HJ. Characterization of three members of the Arabidopsis carotenoid cleavage dioxygenase family demonstrates the divergent roles of this multifunctional enzyme family. Plant J. 2006;45(6):982–93. doi: 10.1111/j.1365-313X.2006.02666.x. [DOI] [PubMed] [Google Scholar]
- 21.Kim J, DellaPenna D. Defining the primary route for lutein synthesis in plants: the role of Arabidopsis carotenoid β-ring hydroxylase CYP97A3. Proc Natl Acad Sci U S A. 2006;103(9):3474–9. doi: 10.1073/pnas.0511207103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tian L, Magallanes-Lundback M, Musetti V, DellaPenna D. Functional analysis of β-and ε-ring carotenoid hydroxylases in Arabidopsis. Plant Cell. 2003;15(6):1320–32. doi: 10.1105/tpc.011403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tian L, Musetti V, Kim J, Magallanes-Lundback M, DellaPenna D. The Arabidopsis LUT1 locus encodes a member of the cytochrome P450 family that is required for carotenoid ε-ring hydroxylation activity. Proc Natl Acad Sci U S A. 2004;101(1):402–7. doi: 10.1073/pnas.2237237100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Charrier A, Rippa S, Yu A, Nguyen P-J, Renou J-P, Perrin Y. The effect of carnitine on Arabidopsis development and recovery in salt stress conditions. Planta. 2012;235(1):123–35. doi: 10.1007/s00425-011-1499-4. [DOI] [PubMed] [Google Scholar]
- 25.Havaux M, Kloppstech K. The protective functions of carotenoid and flavonoid pigments against excess visible radiation at chilling temperature investigated in Arabidopsis npq and tt mutants. Planta. 2001;213(6):953–66. doi: 10.1007/s004250100572. [DOI] [PubMed] [Google Scholar]
- 26.Huo H, Dahal P, Kunusoth K, McCallum CM, Bradford KJ. Expression of 9-cis-EPOXYCAROTENOID DIOXYGENASE4 is essential for thermoinhibition of lettuce seed germination but not for seed development or stress tolerance. Plant Cell. 2013;25(3):884–900. doi: 10.1105/tpc.112.108902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iuchi S, Kobayashi M, Taji T, Naramoto M, Seki M, Kato T, Tabata S, Kakubari Y, Yamaguchi‐Shinozaki K, Shinozaki K. Regulation of drought tolerance by gene manipulation of 9‐cis‐epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. 2001;27(4):325–33. doi: 10.1046/j.1365-313x.2001.01096.x. [DOI] [PubMed] [Google Scholar]
- 28.Lefebvre V, North H, Frey A, Sotta B, Seo M, Okamoto M, Nambara E, Marion‐Poll A. Functional analysis of Arabidopsis NCED6 and NCED9 genes indicates that ABA synthesized in the endosperm is involved in the induction of seed dormancy. Plant J. 2006;45(3):309–19. doi: 10.1111/j.1365-313X.2005.02622.x. [DOI] [PubMed] [Google Scholar]
- 29.Nambara E, Kawaide H, Kamiya Y, Naito S. Characterization of an Arabidopsis thaliana mutant that has a defect in ABA accumulation: ABA-dependent and ABA-independent accumulation of free amino acids during dehydration. Plant Cell Physiol. 1998;39(8):853–8. doi: 10.1093/oxfordjournals.pcp.a029444. [DOI] [PubMed] [Google Scholar]
- 30.Seo M, Aoki H, Koiwai H, Kamiya Y, Nambara E, Koshiba T. Comparative studies on the Arabidopsis aldehyde oxidase (AAO) gene family revealed a major role of AAO3 in ABA biosynthesis in seeds. Plant Cell Physiol. 2004;45(11):1694–703. doi: 10.1093/pcp/pch198. [DOI] [PubMed] [Google Scholar]
- 31.Tan BC, Joseph LM, Deng WT, Liu L, Li QB, Cline K, McCarty DR. Molecular characterization of the Arabidopsis 9‐cis epoxycarotenoid dioxygenase gene family. Plant J. 2003;35(1):44–56. doi: 10.1046/j.1365-313X.2003.01786.x. [DOI] [PubMed] [Google Scholar]
- 32.Li Y, Liu Z, Wang Y, Yang N, Xin X, Yang S, Feng H. Identification of quantitative trait loci for yellow inner leaves in Chinese cabbage (Brassica rapa L. ssp. pekinensis) based on SSR and SRAP markers. Sci Hortic-Amsterdam. 2012;133:10–7. doi: 10.1016/j.scienta.2011.10.023. [DOI] [Google Scholar]
- 33.Zhang J, Li H, Zhang M, Hui M, Wang Q, Li L, Zhang L. Fine mapping and identification of candidate Br-or gene controlling orange head of Chinese cabbage (Brassica rapa L. ssp. pekinensis) Mol Breed. 2013;32(4):799–805. doi: 10.1007/s11032-013-9907-z. [DOI] [Google Scholar]
- 34.Lee S, Lee S-C, Byun DH, Lee DY, Park JY, Lee JH, Lee HO, Sung SH, Yang T-J. Association of molecular markers derived from the BrCRISTO1 gene with prolycopene-enriched orange-colored leaves in Brassica rapa. Theor Appl Genet. 2014;127(1):179–91. doi: 10.1007/s00122-013-2209-3. [DOI] [PubMed] [Google Scholar]
- 35.Kim C, Park S, Kikuchi S, Kwon S, Park S, Yoon U, Park D, Seol Y, Hahn J, Park S. Genetic analysis of gene expression for pigmentation in Chinese cabbage (Brassica rapa) BioChip J. 2010;4(2):123–8. doi: 10.1007/s13206-010-4206-9. [DOI] [Google Scholar]
- 36.Burdzinski C, Wendell DL. Mapping the anthocyaninless (anl) locus in rapid-cycling Brassica rapa (RBr) to linkage group R9. BMC Genet. 2007;8(1):64. doi: 10.1186/1471-2156-8-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang X, Wang H, Wang J, Sun R, Wu J, Liu S, Bai Y, Mun J-H, Bancroft I, Cheng F. The genome of the mesopolyploid crop species Brassica rapa. Nat Genet. 2011;43(10):1035–9. doi: 10.1038/ng.919. [DOI] [PubMed] [Google Scholar]
- 38.Hittinger CT, Carroll SB. Gene duplication and the adaptive evolution of a classic genetic switch. Nature. 2007;449(7163):677–81. doi: 10.1038/nature06151. [DOI] [PubMed] [Google Scholar]
- 39.Spillane C, Schmid KJ, Laoueillé-Duprat S, Pien S, Escobar-Restrepo J-M, Baroux C, Gagliardini V, Page DR, Wolfe KH, Grossniklaus U. Positive darwinian selection at the imprinted MEDEA locus in plants. Nature. 2007;448(7151):349–52. doi: 10.1038/nature05984. [DOI] [PubMed] [Google Scholar]
- 40.Freeling M, Thomas BC. Gene-balanced duplications, like tetraploidy, provide predictable drive to increase morphological complexity. Genome Res. 2006;16(7):805–14. doi: 10.1101/gr.3681406. [DOI] [PubMed] [Google Scholar]
- 41.Cheng F, Liu S, Wu J, Fang L, Sun S, Liu B, Li P, Hua W, Wang X. BRAD, the genetics and genomics database for Brassica plants. BMC Plant Biol. 2011;11(1):136. doi: 10.1186/1471-2229-11-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheng F, Wu J, Fang L, Sun S, Liu B, Lin K, Bonnema G, Wang X. Biased gene fractionation and dominant gene expression among the subgenomes of Brassica rapa. PLoS One. 2012;7(5):e36442. doi: 10.1371/journal.pone.0036442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo N, Cheng F, Wu J, Liu B, Zheng S, Liang J, Wang X. Anthocyanin biosynthetic genes in Brassica rapa. BMC Genomics. 2014;15(1):426. doi: 10.1186/1471-2164-15-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang H, Wu J, Sun S, Liu B, Cheng F, Sun R, Wang X. Glucosinolate biosynthetic genes in Brassica rapa. Gene. 2011;487(2):135–42. doi: 10.1016/j.gene.2011.07.021. [DOI] [PubMed] [Google Scholar]
- 45.DellaPenna D, Pogson BJ. Vitamin synthesis in plants: tocopherols and carotenoids. Annu Rev Plant Biol. 2006;57:711–38. doi: 10.1146/annurev.arplant.56.032604.144301. [DOI] [PubMed] [Google Scholar]
- 46.Cheng F, Mandáková T, Wu J, Xie Q, Lysak MA, Wang X. Deciphering the diploid ancestral genome of the mesohexaploid Brassica rapa. Plant Cell. 2013;25(5):1541–54. doi: 10.1105/tpc.113.110486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bramley P, Teulieres C, Blain I, Bird C, Schuch W. Biochemical characterization of transgenic tomato plants in which carotenoid synthesis has been inhibited through the expression of antisense RNA to pTOM5. Plant J. 1992;2(3):343–9. doi: 10.1111/j.1365-313X.1992.00343.x. [DOI] [Google Scholar]
- 48.Fraser PD, Truesdale MR, Bird CR, Schuch W, Bramley PM. Carotenoid biosynthesis during tomato fruit development (evidence for tissue-specific gene expression) Plant Physiol. 1994;105(1):405–13. doi: 10.1104/pp.105.1.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moehs CP, Tian L, Osteryoung KW, DellaPenna D. Analysis of carotenoid biosynthetic gene expression during marigold petal development. Plant Mol Biol. 2001;45(3):281–93. doi: 10.1023/A:1006417009203. [DOI] [PubMed] [Google Scholar]
- 50.Shewmaker CK, Sheehy JA, Daley M, Colburn S, Ke DY. Seed‐specific overexpression of phytoene synthase: increase in carotenoids and other metabolic effects. Plant J. 1999;20(4):401–12. doi: 10.1046/j.1365-313x.1999.00611.x. [DOI] [PubMed] [Google Scholar]
- 51.Zhu C, Yamamura S, Koiwa H, Nishihara M, Sandmann G. cDNA cloning and expression of carotenogenic genes during flower development in Gentiana lutea. Plant Mol Biol. 2002;48(3):277–85. doi: 10.1023/A:1013383120392. [DOI] [PubMed] [Google Scholar]
- 52.Cárdenas PD, Gajardo HA, Huebert T, Parkin IA, Iniguez-Luy FL, Federico ML. Retention of triplicated phytoene synthase (PSY) genes in Brassica napus L. and its diploid progenitors during the evolution of the Brassiceae. Theor Appl Genet. 2012;124(7):1215–28. doi: 10.1007/s00122-011-1781-7. [DOI] [PubMed] [Google Scholar]
- 53.Tong C, Wang X, Yu J, Wu J, Li W, Huang J, Dong C, Hua W, Liu S. Comprehensive analysis of RNA-seq data reveals the complexity of the transcriptome in Brassica rapa. BMC Genomics. 2013;14(1):689. doi: 10.1186/1471-2164-14-689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9(1):559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tuan PA, Park NI, Park WT, Kim YB, Kim JK, Lee J, Lee S-h, Yang T-J, Park SU. Carotenoids accumulation and expression of carotenogenesis genes during seedling and leaf development in Chinese cabbage (Brassica rapa subsp. pekinensis. Plant Omics. 2012;5(2):143–8. [Google Scholar]
- 56.Ferris SD, Whitt GS. Evolution of the differential regulation of duplicate genes after polyploidization. J Mol Evol. 1979;12(4):267–317. doi: 10.1007/BF01732026. [DOI] [PubMed] [Google Scholar]
- 57.Ohno S. Evolution by gene duplication. London: George Alien & Unwin Ltd. Berlin, Heidelberg and New York: Springer-Verlag; 1970. [Google Scholar]
- 58.Li WH, Yang J, Gu X. Expression divergence between duplicate genes. Trends Genet. 2005;21(11):602–7. doi: 10.1016/j.tig.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 59.Duarte JM, Cui L, Wall PK, Zhang Q, Zhang X, Leebens-Mack J, Ma H, Altman N. Expression pattern shifts following duplication indicative of subfunctionalization and neofunctionalization in regulatory genes of Arabidopsis. Mol Biol Evol. 2006;23(2):469–78. doi: 10.1093/molbev/msj051. [DOI] [PubMed] [Google Scholar]
- 60.Blanc G, Wolfe KH. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell. 2004;16(7):1679–91. doi: 10.1105/tpc.021410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Casneuf T, De Bodt S, Raes J, Maere S, Van de Peer Y. Nonrandom divergence of gene expression following gene and genome duplications in the flowering plant Arabidopsis thaliana. Genome Biol. 2006;7(2):R13. doi: 10.1186/gb-2006-7-2-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science. 2006;313(5793):1596–604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- 63.Cheng F, Wu J, Fang L, Wang X. Syntenic gene analysis between Brassica rapa and other Brassicaceae species. Front Plant Sci 2012, 3. [DOI] [PMC free article] [PubMed]
- 64.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–9. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schranz ME, Lysak MA, Mitchell-Olds T. The ABC’s of comparative genomics in the Brassicaceae: building blocks of crucifer genomes. Trends Plant Sci. 2006;11(11):535–42. doi: 10.1016/j.tplants.2006.09.002. [DOI] [PubMed] [Google Scholar]


