Abstract.
Cancer progression has been linked to mechanics. Therefore, there has been recent interest in developing noninvasive imaging tools for cancer assessment that are sensitive to changes in tissue mechanical properties. We have developed one such method, modality independent elastography (MIE), that estimates the relative elastic properties of tissue by fitting anatomical image volumes acquired before and after the application of compression to biomechanical models. The aim of this study was to assess the accuracy and reproducibility of the method using phantoms and a murine breast cancer model. Magnetic resonance imaging data were acquired, and the MIE method was used to estimate relative volumetric stiffness. Accuracy was assessed using phantom data by comparing to gold-standard mechanical testing of elasticity ratios. Validation error was . Reproducibility analysis was performed on animal data, and within-subject coefficients of variation ranged from 2 to 13% at the bulk level and 32% at the voxel level. To our knowledge, this is the first study to assess the reproducibility of an elasticity imaging metric in a preclinical cancer model. Our results suggest that the MIE method can reproducibly generate accurate estimates of the relative mechanical stiffness and provide guidance on the degree of change needed in order to declare biological changes rather than experimental error in future therapeutic studies.
Keywords: breast cancer, computational modeling, elastography, magnetic resonance imaging, mechanical properties
1. Introduction
Elastography is an imaging technique that allows for noninvasive assessment of the mechanical stiffness in a tissue of interest.1–3 Typically, mechanical excitation (either dynamic or quasistatic) is applied and the tissue response is imaged. Model-based inverse schemes or direct solution based on an assumed constitutive equation then yields estimates of the mechanical properties of a tissue of interest. A common application of elastographic imaging is in the assessment of the stiffness of cancerous tissue.4–7
The biological basis for elastographic assessment of cancer is based on strong in vitro evidence that supports a fundamental link between mechanical stiffness and cancer. Mechanical signaling has been distinctly implicated in cancer development and progression,8–12 and the mechanobiological basis for this association continues to be uncovered. For example, the mechanical architecture of the extracellular matrix in cancer has been identified to enhance growth, differentiation and motility.13–17 The aggressive behavior of cancer cells has been shown to be strongly correlated to the mechanical nature of the extracellular matrix through mechanisms linked to invadopodia, contractility and assembly of focal adhesions.18–24
Tumor stiffness, measured ex vivo, has been positively correlated with cancer stage in breast cancer, with malignant tumors exhibiting significantly increased stiffness over benign tumors and background healthy adipose or fibroglandular tissue.25,26 Elastography has also recently been explored as a tool to monitor therapy-induced changes in tumor stiffness in response to neoadjuvant chemotherapy. For example, Falou et al. showed significant decreases beginning at four weeks after initiation of therapy in ultrasound strain imaging longitudinal strain ratio (correlated to tissue stiffness) of patients who achieved a pathological complete response (i.e., complete disappearance of tumor as determined at surgery following neoadjuvant therapy) compared to patients with residual tumor burden.27
Investigations of therapy-induced changes in tumor stiffness have continued in the preclinical setting, where Li et al. found a significant decrease in bulk stiffness as assessed by magnetic resonance (MR) elastography 24 h after administration of a vascular disrupting agent in a murine model of breast cancer.28 Interestingly, elasticity was seen to exhibit significant changes at this early imaging time point while another quantitative imaging biomarker, the apparent diffusion coefficient from diffusion-weighted MR imaging, did not. Pepin et al. have also used MR elastography to assess therapy-induced changes in tumor stiffness and have shown a significant decrease in stiffness in response to cytotoxic therapy in a murine lymphoma xenograft model.29 However, extensive study of the elasticity imaging biomarker in the preclinical setting has been limited due to association with difficulties at the much smaller length scale. Common traditional MR elasticity imaging methods employ vibratory mechanical excitation which is hampered by significant attenuation at the high frequencies required for sufficient resolution in small specimens as well as difficulties coupling the high-frequency excitation with the tissue of interest. This has been shown to result in high variability of extracted stiffness parameters as well as spatially dependent limited resolution at the tumor boundary.29
The vast majority of elastography methods previously reported in the literature are often characterized by a single enabling imaging technology, typically through ultrasound30–34 or MR imaging.4,35–38 In this work, we have utilized a different approach that is more akin to nonrigid image registration, whereby the natural image contrast available in routine anatomical imaging is used in the reconstruction. As a result, the framework is somewhat independent of any particular modality. Our approach also employs quasistatic deformation excitation, ameliorating difficulties with high-frequency vibratory excitations. The method, modality independent elastography (MIE), requires two imaging volumes to be acquired that capture the tissue of interest before and after a static/quasistatic mechanical excitation. A model-based nonrigid image registration is then performed whereby mechanical stiffness controls the registration parameters using image similarity to drive the objective function. We have previously applied this general approach to x-ray computed tomography, MR and optical imaging data from both preclinical and clinical settings.39–45 The MIE approach is a novel elastographic imaging technique that is automated, straightforward and capable of implementation across length scales and modalities.
Elasticity imaging via MIE may have applications in both indicating/predicting response to therapy and investigating/designing potential therapeutic agents. Thus, it is critical to understand the error associated with the methodology. We have previously reported an initial and preliminary reproducibility characterization of the method,46 and in this work, we present a robust assessment of the MIE elasticity biomarker for characterizing preclinical cancer mechanical stiffness. In particular, this assessment establishes the accuracy and reproducibility of the method, which is necessary for design and interpretation of future experimental studies of treatment response. For proper statistical determination of therapeutic response, we need to determine the magnitude of change required to discriminate between biological changes in mechanical stiffness and experimental error.
2. Methods
2.1. MRI Data Acquisition
Anatomical -weighted image volumes were acquired using a 7.0T MRI scanner [Agilent Technologies (formerly Varian), Palo Alto, California] with a 38-mm quadrature radiofrequency (RF) coil (Doty Scientific, Columbia, South Carolina). These data were acquired before and after compression using a fast spin echo multislice sequence and a acquisition matrix over a transverse field of view with two signal acquisitions and . Compression was applied by inflation of a 5-cc balloon catheter controlled by a syringe driver that was placed within the MR imaging coil.
2.2. Modality-Independent Elastography Reconstruction
2.2.1. Finite element model creation
MR image volumes were semiautomatically processed to segment the object of interest (phantom or murine tumor/adjacent tissue) from surrounding background using Analyze (AnalyzeDirect, Overland Park, Kansas). Smoothed boundary surfaces were generated through the use of a marching cubes extraction47 followed by radial basis function smoothing (FastRBF toolkit, FarField Technology, Christchurch, New Zealand). Tetrahedral finite element meshes were generated with a nominal edge length of using custom mesh generation software.48
2.2.2. Elasticity reconstruction
Relative volumetric elasticity is estimated from the phantom and murine image volumes by the MIE method, which analyzes anatomical image volumes under differing states of application of mechanical compression.40 A schematic of the method is shown in Fig. 1. Following the finite element model creation step (Sec. 2.2.1), a demons nonrigid image registration framework49 is used to register the postcompression image volume to the precompression image volume using multiresolution diffeomorphic demons50,51 with a sigma value of 0.8 and three sequential multiresolution steps. Boundary conditions for biomechanical simulation are then automatically extracted from the calculated nonrigid registration deformation field at the boundary surface of the finite element model mesh. As shown in Ref. 44, this automatic boundary condition extraction process eliminates the problematic and tedious process of manual designation of boundary conditions, while maintaining desirable elasticity reconstruction results. Further details on the automated boundary condition extraction process can be found in Ref. 44. Deformation between the precompression image volume and the postcompression image volume is then simulated using a computational mechanical model that assumes isotropic Hookean linear elasticity. While tissue is known to exhibit complex mechanical behavior, we approximate the system as linear as the strains observed in this work were , with an average strain . Distributions of volumetric mechanical elasticity within the domain are then iteratively reconstructed using a Polak-Ribière52 conjugate gradient53 algorithm. The adjoint method54 is used to evaluate the gradient, because it allows for a significantly more efficient calculation of the parameter sensitivity over traditional computational finite-difference based Jacobian matrix evaluation methods.55 The optimization is the minimization of the objective function, Eq. (1), which utilizes an image volume zone-based image correlation coefficient metric that determines the residual error between the model-deformed image volume and the experimentally deformed acquired image volume.
| (1) |
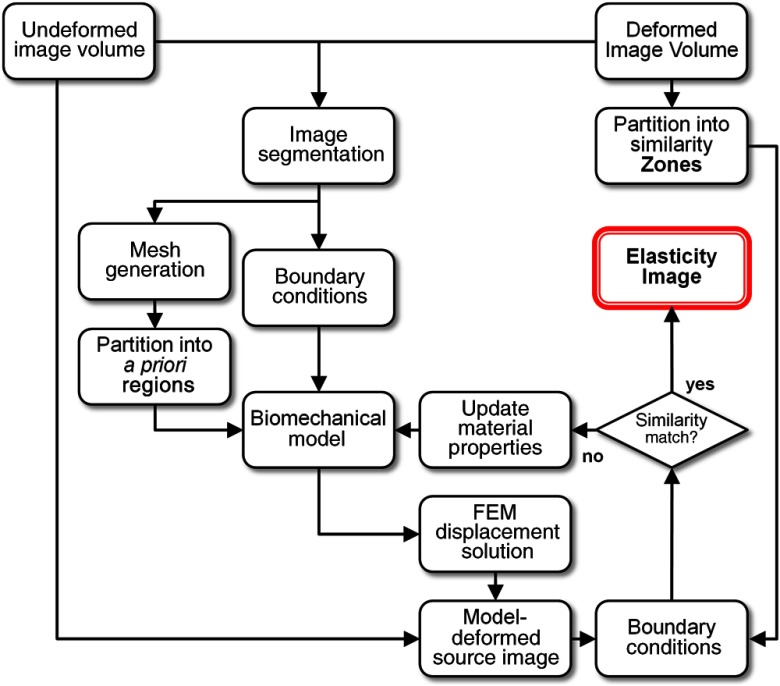
Fig. 1.
Schematic of the modality independent elastography (MIE) framework. A computer model is created from acquired magnetic resonance (MR) image volumes (pre- and postcompression), boundary conditions are extracted using nonrigid image registration, regions for reconstruction are assigned, and an estimate of the elastic properties is iteratively reconstructed by comparing the model-deformed image to the acquired deformed image inside of comparison zones using an image correlation coefficient similarity metric.
and are the similarity correlation coefficient values for comparing the experimentally deformed acquired image volume to itself and the model-deformed image volume, respectively. Following parameter reconstruction, the output metric is a volumetric spatial distribution of relative mechanical elasticity. Gaussian smoothing with an in-plane kernel width of 5 pixels and standard deviation of 1.0 is applied to smooth the resulting elasticity maps. The algorithms used in this work were custom-developed in C++ and utilize algorithms based on the Visualization Toolkit56 and Insight Segmentation and Registration Toolkit.57 Further details regarding the MIE computational methodology can be found in Refs. 42 and 44.
2.2.3. Enforcing spatial prior constraints
As acquisition of image volumes is necessary to apply the MIE methodology, it is natural to make use of the spatial information contained in the imaging data during reconstruction of the elasticity parameters. Separate tissues of interest are grouped for property estimation by a -means clustering58 algorithm that includes a Markov random field spatial constraint59 step, which classifies the -weighted anatomical MR image volume signal intensity information. The Markov random field step is used to enforce spatial continuity to the -means clustering step and produces more biologically significant tissue clusters than traditional -means clustering alone. The number of clusters designated for tissue classification is determined based on a priori information regarding the tissue composition of the sample of interest and was selected as two for both phantom (reflecting soft/hard phantom materials) and murine studies (reflecting tumor/adjacent muscle). After tissue classification based on spatial priors, geometric regions that define areas for mechanical property estimation are identified through geometrical subclustering of the identified tissue classes. The size of these geometrical subclusters, “regions,” defines the resolution of the reconstructed elasticity image. For both the murine and the phantom studies, the number of reconstruction regions was selected to correspond to a resolution with nominal spherical radius of 1 mm. This resulted in 200 regions for the tumor tissue and 150 regions for the adjacent muscle tissue based on an average tissue volume across all animals included in the study.
Similar to previous work in elasticity imaging that utilizes spatial prior constraints to improve elasticity parameter reconstruction results,60–62 we use anatomical information as a soft constraint, with a weighted penalty function applied to the calculated gradient during parameter inversion. As the penalized gradient term is used during iterative parameter estimation to calculate the spatial mechanical elasticity, this step penalizes large deviations within a tissue type via identified spatial priors. This soft prior constraint step acts to enforce regions that belong to the same tissue class to retain similar mechanical elasticity and is enforced using Eq. (2):
| (2) |
where and are the soft prior penalized gradient and calculated gradient, respectively. is defined as the magnitude of the soft prior weighting constraint and is empirically selected between 0 and 1 (where 0 provides no prior constraint and 1 provides a strict enforcement of a constant regional property, i.e., hard prior). is an spatial constraint matrix, with number of reconstruction regions, defined as
| (3) |
2.3. Phantom Validation Study
A phantom study was performed to validate the accuracy of the MIE method using independent mechanical testing as a gold standard. Three different two-material phantoms were constructed that span a range of mechanical elasticity ratios and geometrical arrangement. Phantoms were composed of polyvinyl alcohol (PVA) cryogel or agarose. Phantom A utilized 8% (weight/volume) PVA with different mechanical stiffnesses as determined by the number of freeze-thaw cycles the material underwent. Each freeze-thaw cycle consisted of 12 h of freezing at and 12 h of thawing at room temperature. The phantom was made using a layered construction with one stiff layer (two freeze-thaw cycles) that was doped with an MR contrast agent, 1% (volume/volume) gadopentetate dimeglumine, gadopentetate dimeglumine (Gd-DTPA) (Magnevist, Wayne, New Jersey), on the top of one soft layer (one freeze-thaw cycle), which was not doped with contrast agent. Phantom B utilized a different PVA cryogel formulation with a glycerol plasticizer in order to enhance the stiffness ratio between the two materials. Phantom B utilized 7% (weight/volume) PVA, 10% (volume/volume) glycerol and was made using a layered construction with two stiff layers (two freeze-thaw cycles), which were doped with 1% (volume/volume) Gd-DTPA, on the top and bottom of one soft layer (one freeze-thaw cycle) which was not doped with contrast agent. Phantom C was composed of a 2% (weight/volume) agarose gel inclusion submerged within a 1% (weight/volume) agarose gel background. All phantoms were cylindrical and constructed with a diameter of and height of .
MIE was performed on all phantoms and the relative elasticity contrast ratio between the stiff material and the soft material was calculated for each phantom based on the average value throughout the reconstructed volume. For each phantom, the reconstructed volumetric elasticity maps were normalized to the average elasticity value calculated within the soft layer. Independent evaluation of the elasticity ratio between the stiff and soft phantom materials was performed through material testing using an Enduratec Electroforce 3100 mechanical tester (Bose, Enduratec Systems Group, Minnetonka, Minnesota). Compression mechanical testing was performed on three samples of each material using a range of strain levels between 1 and 10%. Incremental unconfined step compressions to specified strain levels were applied, followed by a 60-s dwell period to dissipate viscoelastic behavior of the material. Force and displacement data were recorded at the end of the dwell, converted to Young’s modulus, and results were averaged over all samples of each material and reported with 95% confidence intervals (CI).
2.4. Murine Reproducibility Study
Four- to six-week-old female athymic nude mice (Harlan, Indianapolis, Indiana) were injected subcutaneously in the right flank with MDA-MB-231 cells in a 30% Matrigel suspension (Corning Life Sciences, Tewksbury, Massachusetts). Tumors were allowed to grow to a size of to , which occured in approximately six to eight weeks postinjection. During MRI, inhalation anesthesia was induced and maintained via a 2%/98% isoflurane/oxygen mixture. All animal procedures were approved by the Vanderbilt University Institutional Animal Care and Use Committee.
Assessment of MIE reproducibility on seven mice was performed using a test/retest approach, in which animals were removed between consecutive MIE scans and allowed to recover from anesthesia; this approach simulated the normal—and unavoidable—repositioning of an animal that occurs during a longitudinal treatment study. Each MIE scan represents two separate individual MR acquisitions (i.e., undeformed and deformed) followed by an individual offline MIE reconstruction. As the method is limited to relative measures of elasticity due to the indeterminate nature of displacement based boundary conditions, the reconstructed volumetric elasticity maps were normalized to the average tumor elasticity value to facilitate side-by-side comparisons. Therefore, elasticity maps are displayed relative to an average tumor value of one. Bulk-level reproducibility of elasticity dispersion was assessed using Bland–Altman analysis63 on extracted histogram data dispersion metrics: quartiles (25, 50, and 75%), interquartile range, peak height, and peak location. Voxel-level reproducibility was assessed through coregistration of repeated MIE scans by a demons nonrigid image registration49–51 of the -weighted MR data. The resulting deformation field is used to transform the elasticity maps from independent scans into the same image space. Following registration, prior to the assessment of voxel-level reproducibility, a kernel in-plane moving average smoothing filter was applied in order to reduce the effects of local misregistration.
2.5. Reproducibility Statistics
Reproducibility statistics were calculated as previously described,64,65 following the methods described by Bland and Altman.63 The root mean square deviation (rMSD) was calculated from the differences between repeated measurements, , as
| (4) |
where is the number of repeated measurements. The within-subject standard deviation (wSD) was calculated as
| (5) |
The within-subject coefficient of variation (wCV) was calculated by dividing wSD by the overall mean of the parameter. This value allows comparisons of precision across different analysis metrics. The 95% CI was calculated as
| (6) |
where std is the standard deviation of the differences between repeat measurements and is the appropriate -statistic for the number of degrees of freedom. 95% CIs define the range of expected variability within a group of animals; i.e., a change from a group of animals greater than this value would indicate significance at the 5% level. The coefficient of repeatability, , was calculated as
| (7) |
and is defined as the magnitude of the maximum difference that would be expected in 95% of the paired scans, i.e., the threshold for significance for an individual measurement. Therefore, an observed difference that is greater than the coefficient of repeatability between scans from an individual animal indicates a significant difference at the 5% level. Intraclass correlation coefficients (ICC) using two-way random single measures were used to calculate reliability.66
2.6. Histology
Following MRI, animals were sacrificed and tumors were excised, fixed in 10% formalin, dehydrated, embedded in paraffin, and sectioned for histology. Hematoxylin and eosin (H&E) and Masson’s trichrome blue stained sections were obtained along with collagen I immunohistochemisty and slides were scanned with a Leica SCN400 Slide Scanner (Leica Microsystems, Buffalo Grove, Illinois). For immunohistochemisty, antigen retrieval was performed using citrate buffer, pH 6, and a Decloaking Chamber (Biocare Medical, Walnut Creek, California). Anti-collagen I (Rockland Immunochemicals, Limerick, Pennsylvania, catalog # 600-401-103) was used with a dilution of 1:1600, overnight incubation at 4°C, and detected with an Envision DAB kit (Dako, Carpinteria, California).
3. Results
3.1. Phantom Validation Study
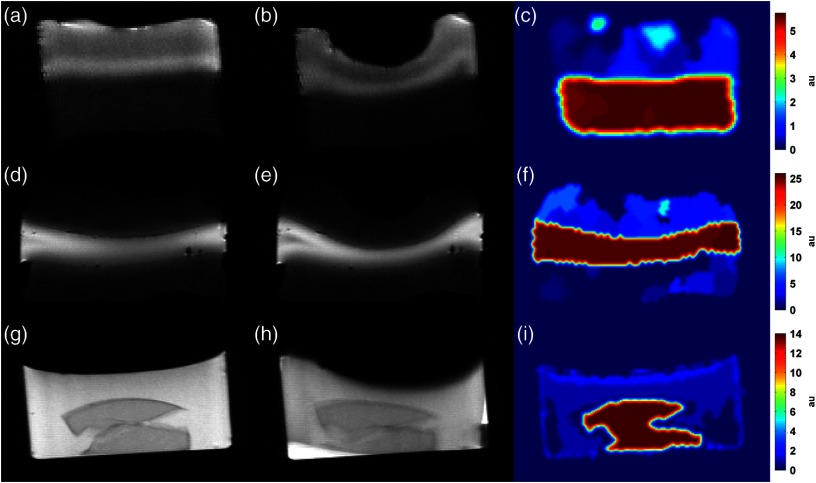
A phantom validation study was performed by comparing MIE to gold-standard material testing to assess the accuracy of the method using three multimaterial property phantoms with varying elasticity ratios between the stiff material and the soft material. MR anatomical image volumes were acquired before and after the application of deformation, and MIE was used to reconstruct estimates of mechanical elasticity throughout the volume. For phantom A, a cylindrical bilayer phantom was composed of PVA cryogel with two different freeze-thaw cycles; mechanical testing of each material determined a ground-truth elasticity ratio of 5.20 to 1. MIE yielded a ratio of 5.69 to 1, representing an error in the MIE method of 9.4%. For phantom B, a cylindrical three-layer phantom was composed of two different PVA cryogel materials with different elasticity as determined by freeze-thaw cycle; mechanical testing determined a ground-truth elasticity ratio of 27.4 to 1. MIE generated an elasticity ratio of 25.2 to 1, reflecting an error of 8.1%. Phantom C, a cylindrical two-property phantom composed of a stiff agarose gel inclusion submerged in a soft agarose gel background was found to have a ground-truth mechanical testing elasticity ratio of 14.9 to 1. The MIE method reconstructed an elasticity ratio of 13.1 to 1, representing an error of 12.1%. Central slice images of the acquired undeformed and deformed MR images and associated MIE elasticity reconstructions are shown in Fig. 2 for all phantoms. Values of the reconstructed elasticity were averaged throughout the volume within each material and the elasticity ratios are reported in Table 1 for each phantom, along with values from independent mechanical testing.
Fig. 2.
MR images and MIE reconstruction results from the phantom validation study for [(a) to (c)] phantom A, [(d) to (f)] phantom B, and [(g) to (i)] phantom C. Multiproperty gel phantoms were constructed of polyvinyl alcohol cryogel (phantoms A and B) or agarose (phantom C) with two different mechanical stiffness, as dictated by material formulation. Undeformed [(a), (d), and (g)] and deformed [(b), (e), and (h)] image volumes were used in the MIE methodology (outlined in Fig. 1) to reconstruct volumetric estimates of the relative mechanical elasticity [(c), (f), and (i)]. A balloon catheter controlled by a syringe driver was used to deform the top surface of each phantom. Note that the color bars for each phantom have different scales and represent relative elasticity (i.e., elasticity ratio), normalized to the average value estimated within the softer phantom material and expressed as arbitrary units (au).
Table 1.
Modality independent elastography (MIE) validation results from the phantom study. MIE reconstructed mechanical elasticity ratio of phantoms A, B, and C are shown to exhibit agreement with independent mechanical testing. Mechanical testing ratios are expressed as the average value from three samples of each material with 95% confidence interval (CI) as shown.
| Phantom | MIE ratio | Mechanical testing ratio | Error |
|---|---|---|---|
| A | 5.69:1 | 5.20∶1 | 9.4% |
| B | 25.2:1 | 27.4∶1 | 8.1% |
| C | 13.1:1 | 14.9∶1 | 12.1% |
3.2. Murine Reproducibility Study
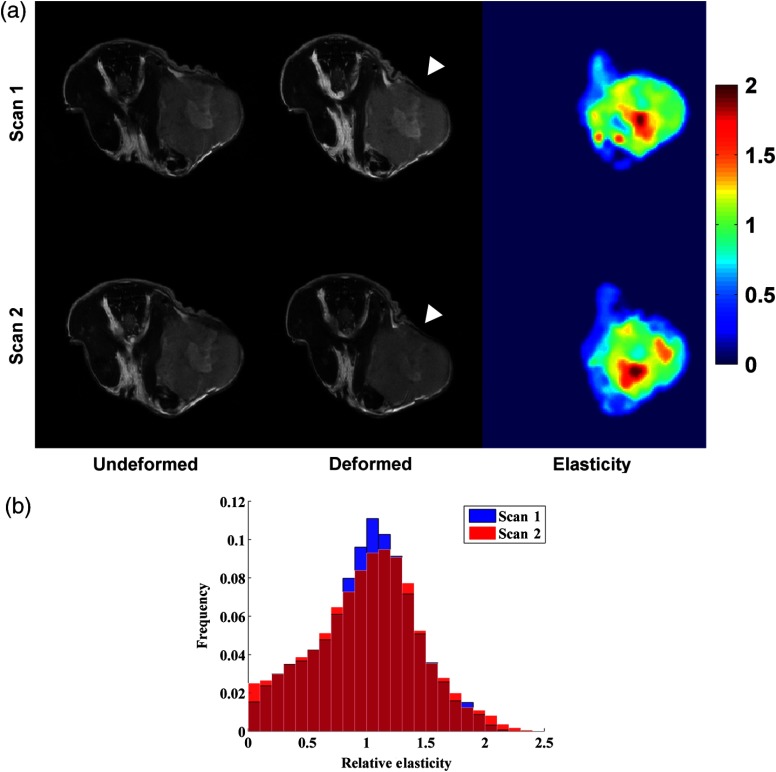
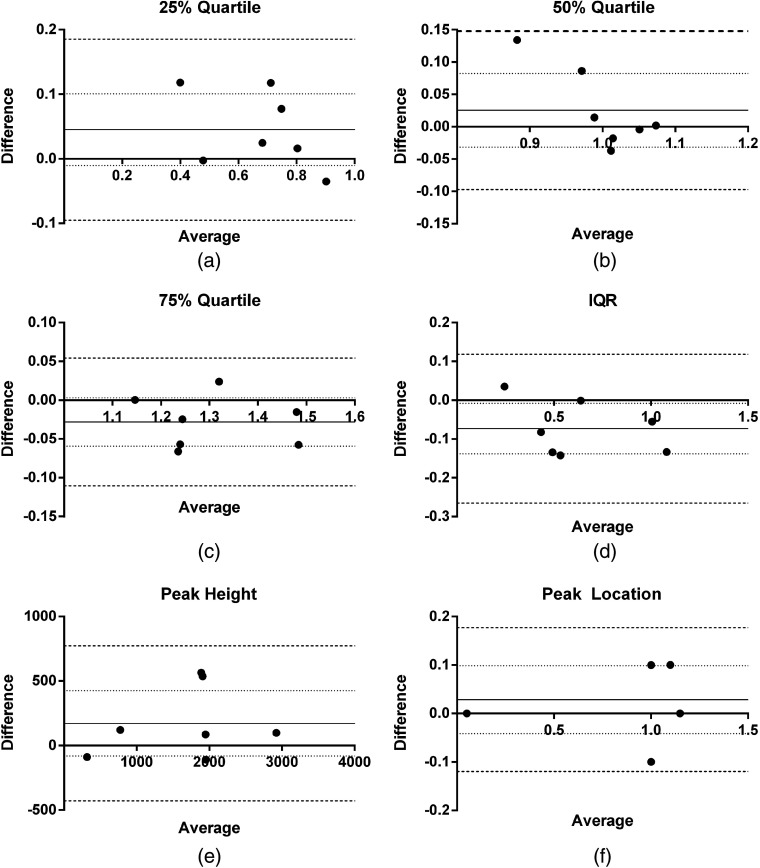
Representative anatomical MR images, acquired before and after the application of compression, associated MIE elasticity reconstructions and histograms of relative elasticity from repeated scans are shown in Fig. 3. The location of the deformation source is noted by the arrowheads in Fig. 3(a). It is important to note that while representative images are shown for the two-dimensional center slice plane, MR acquisitions and MIE reconstructions are performed for the full three-dimensional tumor volume. Each row in Fig. 3 represents a single MIE scan performed on the same animal from corresponding central slices. Qualitatively, the method appears to provide similar stiffness values and distribution within the tumor parenchyma and core in both of the test-retest scans. Table 2 reports the reproducibility statistics from the bulk-level histogram analysis and the voxel-level analysis across the full three-dimensional tumor volume. The 95% CI and wCV for all bulk-level histogram metrics was . The wCV for voxel-level analysis was 32.15%. ICCs for all metrics show substantial agreement between repeat measures with all values (note that is considered substantial agreement66). Bland–Altman plots of the difference versus the average between repeat scans for each analysis metric are shown in Fig. 4. The mean difference, 95% CI, and repeatability ranges are plotted as a solid line, dotted line, and dashed line, respectively.
Fig. 3.
(a) Reproducibility assessment from a representative murine tumor showing two separate MIE scans of the same mouse (top row and bottom row). -weighted MR image volumes in the undeformed (left column) and deformed (middle column) states are used to reconstruct volumetric estimates of the relative mechanical elasticity (right column). These data are the in vivo analogue of the data presented in Fig. 2. The MIE method produces estimates of elasticity with similar distributions emerging within the tumor region for two independent assessments of the same mouse. White arrowheads designate the position of the deformation source. Note that the color bars represent relative elasticity (i.e., elasticity ratio), normalized to the average value estimated within the tumor and expressed as arbitrary units. (b) Histogram distributions for relative elasticity from each of the MIE scans of the same mouse.
Table 2.
Repeatability statistics for region of interest histogram summary metrics (25, 50, and 75% quartiles, interquartile range, peak location, peak height) and voxel-level analyses. Mean, mean difference, 95% CI for mean difference, within-subject standard deviation, within-subject coefficient of variation, repeatability coefficient, and interclass correlation coefficient. test/retest datasets.
| Metric | Mean | Mean difference | 95% CI | wSD | wCV | Repeatability | ICC |
|---|---|---|---|---|---|---|---|
| 25% quartile | 0.67 | 0.0451 | (8.2%) | 0.0506 | 7.50% | 0.1403 | 0.96 |
| 50% quartile (median) | 0.99 | 0.0254 | (5.7%) | 0.0442 | 4.42% | 0.1225 | 0.75 |
| 75% quartile | 1.31 | (2.4%) | 0.0297 | 2.27% | 0.0824 | 0.97 | |
| Interquartile range | 0.63 | (10.3%) | 0.0693 | 10.96% | 0.1920 | 0.97 | |
| Peak location | 0.76 | 0.0286 | (9.1%) | 0.0535 | 6.99% | 0.1482 | 0.99 |
| Peak height | 1673 | 172 | (15.1%) | 216.42 | 12.94% | 599.89 | 0.97 |
| Voxel level | 1.00 | (42.0%) | 0.3215 | 32.15% | 0.8910 | 0.70 |
Note: wSD, within-subject standard deviation; wCV, within-subject coefficient of variation; ICC, intraclass correlation coefficient.
Fig. 4.
Bland-Altman plots (difference versus average between each repeat scan) for histogram dispersion metrics: (a) 25% quartile, (b) 50% quartile, (c) 75% quartile, (d) interquartile range, (e) peak height, and (f) peak location. Mean difference is indicated by the solid line, 95% confidence interval (CI) of mean difference is indicated by the dashed line, and Bland-Altman limits of agreement bounds (95% CI of the limits of agreement) is indicated by the dotted lines. test/retest datasets.
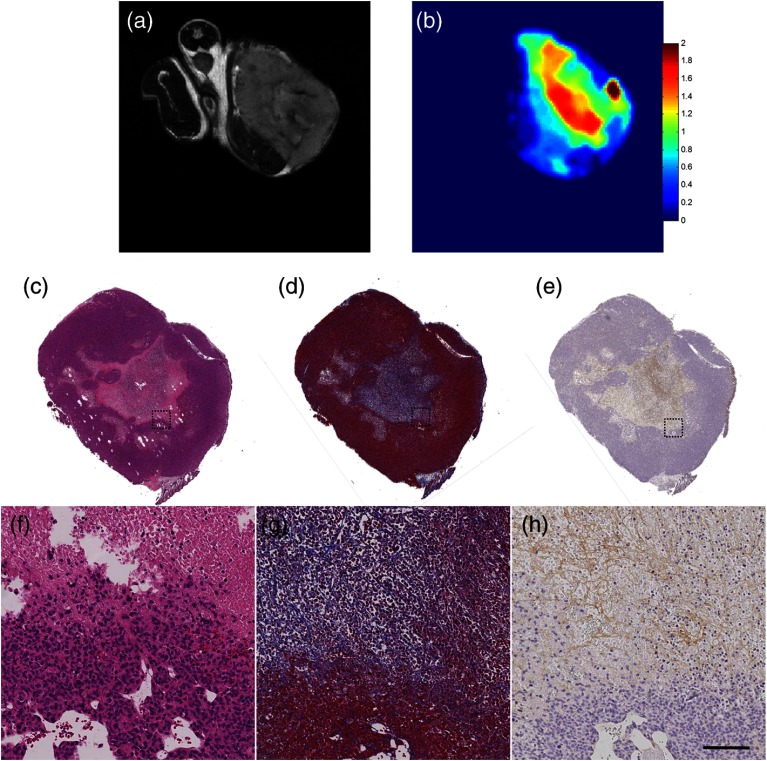
Notably, as shown in Fig. 5, there is significant focal regional heterogeneity in the MIE elasticity map within the tumor [Fig. 5(b)], with an elevated stiffness observed in the core of the tumor. The significant heterogeneity of the tumor extracellular matrix was confirmed by histological staining for H&E, Masson’s trichrome, and collagen I immunohistochemistry. H&E staining shows a relative lack of cell nuclei in the core of the tumor relative to the periphery, indicating central necrosis. Masson’s trichrome staining [Figs. 5(d) and 5(g)] shows blue staining throughout the tumor stroma, including within the tumor core, reflecting the presence of collagen in this area. Collagen I immunohistochemistry [Figs. 5(e) and 5(h)] confirms this finding as an elevation of type I collagen.
Fig. 5.
Comparison of MRI, MIE, and histology. (a) -weighted MRI, (b) MIE elasticity reconstruction, (c) H&E staining, (d) Masson’s trichrome blue staining, and (e) collagen I immunohistochemistry with magnifications as indicated by the dotted outline; (f), (g), and (h) indicate the presence of central necrosis and collagen throughout the tumor stroma. Elevated focal stiffness is seen in the elasticity map for the core of the tumor. Masson’s trichrome blue and collagen I histological staining indicate presence of collagen in the tumor core. Scale bar, in (h), equals 100 microns.
4. Discussion
While we have previously studied the use of the general MIE method,39–44 we have recently fundamentally advanced the automation, translation, and application toward the preclinical setting.45 MIE is now positioned as a noninvasive imaging methodology that can provide quantitative characterization of tissue mechanical status. In this work, we evaluate the accuracy and reproducibility of the MIE method in a preclinical model of breast cancer. This study is an initial realization toward a more systematic study of MIE in characterizing the mechanical properties of cancer. The method accurately estimates mechanical elasticity ratios, with errors over a range of elasticity ratios in phantoms, and reproducibly, with test/retest reproducibility wCV of 2 to 13% at the bulk level and 32% at the voxel level. Our results provide guidance for a future treatment response study where we would be able to declare statistical significance for chances in MIE elasticity metrics that lie beyond the indicated 95% CI for each metric using a similar sample size. For example, if we expect chemotherapeutic treatment to reduce the tumor stiffness, we would anticipate a leftward shift in the histogram, reducing the right-hand tail of the histogram and the location of the peak.
While other metrics assessed from quantitative MRI in preclinical cancer models have recently progressed to providing validated and repeatable measurements, to our knowledge, this study is the first to assess the reproducibility of an elasticity imaging biomarker in a preclinical murine cancer model, so direct comparisons to other results are unavailable. However, our results are in agreement with previously reported reproducibility analyses for other elastography methods reported in the clinical setting for hepatic and brain elasticity measurements. The reproducibility of MRE has been reported with wCVs from 6.07 to 10.78% and ICCs from 0.85 to 0.94 for measurements of human hepatic elasticity in healthy and fibrotic patients67 and coefficient of variation of 1% for healthy whole brain, following edge artifact correction.68 In ultrasound elastography, the reproducibility of transient elastography has been reported with ICCs of 0.98 for clinical assessment of hepatic elasticity.69 Ultrasound shear wave elastography assessment of hepatic elasticity has been reported with wCVs from 12 to 17% and ICCs from 0.65 to 0.95.70 Our results are also in agreement with reproducibility analysis for other types of quantitative MRI data previously reported.65,71–73 In a similar mouse model of breast cancer, Barnes et al. reported wCVs from 8.3 to 15.6% and 95% CIs from 7.5 to 14.3% for metrics extracted from standard pharmacokinetic analysis of dynamic contrast enhanced MRI (DCE-MRI) data over the tumor region of interest for repeat scans.71 Similarly, Whisenant et al. reported a wCV of 12.0% and 95% CI of 11.8% for a metric extracted from repeated diffusion weighted MRI (DW-MRI) scans.65 As our animal model is similar to these studies, the MIE elasticity biomarker is shown to exhibit a precision similar to DCE-MRI and DW-MRI analyses.
In Fig. 5, we show an elasticity map alongside histological staining for collagen content from similar sections. As shown in the Masson’s trichrome and collagen I staining, there is supporting collagen present throughout the tumor stroma, with clear structure compactness and observable collagen density in the necrotic region which was shown to be stiffer by MIE. There is currently a paucity of elastography studies with independent histological confirmation, especially in murine breast cancer model systems. Consistent with the results of our study, Chamming’s et al. found elevated stiffness in the central region of murine xenograft tumors, with a significant correlation between ultrasound shear wave elasticity and fibrosis determined by histological findings of the presence of collagen in Masson’s trichrome staining and the absence of cell nuclei (, ).74 This study also confirms a finding previously reported by our group, showing that the total collagen content in murine mammary tumors is related to bulk mechanical stiffness.75 Though preliminary, this result is suggestive that elastography may be able to noninvasively assess tumor extracellular matrix properties. It will be important in future studies to further assess the molecular underpinnings of elastography in preclinical cancer models by examining the elastic properties of tumors alongside in-depth histological examination.
We also recognize that there are several limitations with our current approach. Due to the indeterminate nature of displacement based boundary conditions, the method is limited to relative measures of elasticity rather than absolute values. To facilitate direct comparisons, we normalized our results to the average value within the tumor. As we would not expect the average elasticity to change during the time course of the present study, this normalization does not affect our results. However, when extending the method to assess longitudinal changes in elasticity, possibly in response to administration of chemotherapeutics, relative elasticity measures could potentially confound results. While changes in the data dispersion, as assessed by histogram metrics, could yield valuable results, quantitative values of reduction/increase in elasticity over time would not be possible with the current approach. We have previously investigated, with success, the use of an offset material with a known modulus of elasticity to serve as an internal control to scale elasticity values,76 and we anticipate the use of this approach when moving forward toward longitudinal studies. Another limitation is that comparisons between scans are inexact. In our experimental protocol, we simulated the repositioning that would occur during a treatment study by removing the animal and allowing for recovery from anesthesia between repeat scans. While voxel-level analyses were performed following a nonrigid image registration, inaccurate voxel-level registration remains a possibility even following correction through the use of a smoothing kernel. An additional limitation is due to the discretization of the number of tissue clusters and reconstruction regions for subsequent elasticity reconstruction. Our reproducibility statistics are valid only for the selected parameters in this study. While we expect that an increase or decrease in the number of regions used for elasticity reconstruction would affect reconstruction resolution with minimal impact on reproducibility, future studies need to be performed in order to characterize this effect.
In summary, we have assessed the accuracy and reproducibility of MIE within the context of a preclinical animal model of breast cancer. This assessment provides guidance on the magnitude of changes required to declare significant statistical differences in tissue elasticity assessed by MIE during a course of therapy in a preclinical model of breast cancer. The level of accuracy and precision reported for our elasticity imaging biomarker provides a significant degree of enthusiasm for future studies directed at investigating the potential changes in mechanical stiffness of tumors in response to cancer therapeutic agents.
Acknowledgments
This work was supported by the National Cancer Institute through R01CA138599, R25CA092043, U01CA142565, and U01CA174706. This work was also supported by the Vanderbilt Initiative in Surgery and Engineering Pilot Award Program and the Vanderbilt University School of Engineering Summer Research Program. We thank Dr. Melinda Sanders for assistance with examining histological data. We also acknowledge the services of the Vanderbilt University Translational Pathology Shared Resource and the Vanderbilt University Digital Histology Shared Resource. We thank the Kleberg Foundation for the generous support of our biomedical imaging program.
Biographies
Jared A. Weis is a research assistant professor of biomedical engineering at Vanderbilt University. He received his BS degree in biomedical engineering from Washington University, St. Louis, USA, in 2005 and his MS and PhD degrees in biomedical engineering from Vanderbilt University in 2009 and 2011, respectively. His research interests focus on integrating computational modeling and medical imaging to study the multiscale influences of mechanics in cancer progression and response to therapy.
Katelyn M. Flint received her BS degree in biomedical engineering from Vanderbilt University in 2015. Currently, she is a graduate student in biomedical engineering at Duke University.
Violeta Sanchez is a histotechnologist and specialized tissue studies technologist for the Vanderbilt Breast Specialized Program of Research Excellence in the Vanderbilt-Ingram Cancer Center at Vanderbilt University.
Thomas E. Yankeelov is an ingram professor of cancer research and a professor of radiology and radiological sciences, physics, biomedical engineering, and cancer biology. He serves as a director of cancer imaging research for the Vanderbilt-Ingram Cancer Center, where he is also the coleader of the Host–Tumor Interactions Research Program. His research focuses on the development of imaging methods for predicting treatment response in cancer and extends from mathematical modeling to implementation in human studies.
Michael I. Miga received his BS and MS degrees in mechanical engineering from University of Rhode Island in 1992 and 1994, respectively, and his PhD in biomedical engineering from Dartmouth College in 1998. He joined Vanderbilt in 2001 and is a professor of BME. He directs the Biomedical Modeling Laboratory and is cofounder of the Vanderbilt Initiative in Surgery and Engineering Center. His research is in computational modeling and inverse problems for medical applications.
References
- 1.Glaser K. J., Manduca A., Ehman R. L., “Review of MR elastography applications and recent developments,” J. Magn. Reson. Imaging 36(4), 757–774 (2012). 10.1002/jmri.23597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mariappan Y. K., Glaser K. J., Ehman R. L., “Magnetic resonance elastography: a review,” Clin. Anal. 23(5), 497–511 (2010). 10.1002/ca.21006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parker K. J., Doyley M. M., Rubens D. J., “Imaging the elastic properties of tissue: the 20 year perspective,” Phys. Med. Biol. 56(1), R1–R29 (2011). 10.1088/0031-9155/56/1/R01 [DOI] [PubMed] [Google Scholar]
- 4.McKnight A. L., et al. , “MR elastography of breast cancer: preliminary results,” Am. J. Roentgenol. 178(6), 1411–1417 (2002). 10.2214/ajr.178.6.1781411 [DOI] [PubMed] [Google Scholar]
- 5.Venkatesh S. K., et al. , “MR elastography of liver tumors: preliminary results,” AJR Am. J. Roentgenol. 190(6), 1534–1540 (2008). 10.2214/AJR.07.3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garteiser P., et al. , “MR elastography of liver tumours: value of viscoelastic properties for tumour characterisation,” Eur. Radiol. 22(10), 2169–2177 (2012). 10.1007/s00330-012-2474-6 [DOI] [PubMed] [Google Scholar]
- 7.Good D. W., et al. , “Elasticity as a biomarker for prostate cancer: a systematic review,” BJU Int. 113(4), 523–534 (2014). 10.1111/bju.12236 [DOI] [PubMed] [Google Scholar]
- 8.Paszek M. J., Weaver V. M., “The tension mounts: mechanics meets morphogenesis and malignancy,” J. Mammary Gland Biol. Neoplasia 9(4), 325–342 (2004). 10.1007/s10911-004-1404-x [DOI] [PubMed] [Google Scholar]
- 9.Paszek M. J., et al. , “Tensional homeostasis and the malignant phenotype,” Cancer Cell 8(3), 241–254 (2005). 10.1016/j.ccr.2005.08.010 [DOI] [PubMed] [Google Scholar]
- 10.Paszek M. J., et al. , “Mechano-signaling in mammary morphogenesis and tumorigenesis,” Mol. Biol. Cell 15, 241A (2004).. [Google Scholar]
- 11.Huang S., Ingber D. E., “Cell tension, matrix mechanics, and cancer development,” Cancer Cell 8(3), 175–176 (2005). 10.1016/j.ccr.2005.08.009 [DOI] [PubMed] [Google Scholar]
- 12.Shieh A. C., “Biomechanical forces shape the tumor microenvironment,” Ann. Biomed. Eng. 39(5), 1379–1389 (2011). 10.1007/s10439-011-0252-2 [DOI] [PubMed] [Google Scholar]
- 13.Engler A. J., et al. , “Substrate compliance alters human mesenchymal stem cell morphology,” Mol. Biol. Cell 15, 298A (2004). [Google Scholar]
- 14.Engler A. J., et al. , “Cells on gels: skeletal muscle cell differentiation and adhesion on flexible substrates,” Mol. Biol. Cell 13, 63A (2002). [Google Scholar]
- 15.Engler A. J., et al. , “Substrate elasticity directs adult mesenchymal stem cell differentiation,” Biorheology 42(1–2), 33–33 (2005). [Google Scholar]
- 16.Lo C. M., et al. , “Cell movement is guided by the rigidity of the substrate,” Biophys. J. 79(1), 144–152 (2000). 10.1016/S0006-3495(00)76279-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yeung T., et al. , “Effects of substrate stiffness on cell morphology, cytoskeletal structure, and adhesion,” Cell Motil. Cytoskeleton 60(1), 24–34 (2005). 10.1002/cm.20041 [DOI] [PubMed] [Google Scholar]
- 18.Burridge K., Doughman R., “Front and back by Rho and Rac,” Nat. Cell Biol. 8(8), 781–782 (2006). 10.1038/ncb0806-781 [DOI] [PubMed] [Google Scholar]
- 19.Roovers K., Assoian R. K., “Effects of rho kinase and actin stress fibers on sustained extracellular signal-regulated kinase activity and activation of G(1) phase cyclin-dependent kinases,” Mol. Cell Biol. 23(12), 4283–4294 (2003). 10.1128/MCB.23.12.4283-4294.2003 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Helmlinger G., et al. , “Solid stress inhibits the growth of multicellular tumor spheroids,” Nat. Biotechnol. 15(8), 778–783 (1997). 10.1038/nbt0897-778 [DOI] [PubMed] [Google Scholar]
- 21.Ulrich T. A., de Juan Pardo E. M., Kumar S., “The mechanical rigidity of the extracellular matrix regulates the structure, motility, and proliferation of glioma cells,” Cancer Res. 69(10), 4167–4174 (2009). 10.1158/0008-5472.CAN-08-4859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zaman M. H., et al. , “Migration of tumor cells in 3D matrices is governed by matrix stiffness along with cell-matrix adhesion and proteolysis,” Proc. Natl. Acad. Sci. U S A 103(29), 10889–10894 (2006). 10.1073/pnas.0604460103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stein A. M., et al. , “A mathematical model of glioblastoma tumor spheroid invasion in a three-dimensional in vitro experiment,” Biophys. J. 92(1), 356–365 (2007). 10.1529/biophysj.106.093468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng G., et al. , “Micro-environmental mechanical stress controls tumor spheroid size and morphology by suppressing proliferation and inducing apoptosis in cancer cells,” PLoS One 4(2), e4632 (2009). 10.1371/journal.pone.0004632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Samani A., Plewes D., “An inverse problem solution for measuring the elastic modulus of intact ex vivo breast tissue tumours,” Phys. Med. Biol. 52(5), 1247–1260 (2007). 10.1088/0031-9155/52/5/003 [DOI] [PubMed] [Google Scholar]
- 26.Samani A., Zubovits J., Plewes D., “Elastic moduli of normal and pathological human breast tissues: an inversion-technique-based investigation of 169 samples,” Phys. Med. Biol. 52(6), 1565–1576 (2007). 10.1088/0031-9155/52/6/002 [DOI] [PubMed] [Google Scholar]
- 27.Falou O., et al. , “Evaluation of neoadjuvant chemotherapy response in women with locally advanced breast cancer using ultrasound elastography,” Transl. Oncol. 6(1), 17–24 (2013). 10.1593/tlo.12412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li J., et al. , “Tumour biomechanical response to the vascular disrupting agent ZD6126 in vivo assessed by magnetic resonance elastography,” Br. J. Cancer 110(7), 1727–1732 (2014). 10.1038/bjc.2014.76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pepin K. M., et al. , “MR elastography derived shear stiffness—a new imaging biomarker for the assessment of early tumor response to chemotherapy,” Magn. Reson. Med. 71(5), 1834–1840 (2014). 10.1002/mrm.24825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ophir J., et al. , “Elastography—a quantitative method for imaging the elasticity of biological tissues,” Ultrason. Imaging 13(2), 111–134 (1991). 10.1177/016173469101300201 [DOI] [PubMed] [Google Scholar]
- 31.de Korte C. L., et al. , “Characterization of plaque components and vulnerability with intravascular ultrasound elastography,” Phys. Med. Biol. 45(6), 1465–1475 (2000). 10.1088/0031-9155/45/6/305 [DOI] [PubMed] [Google Scholar]
- 32.Doyley M. M., Meaney P. M., Bamber J. C., “Evaluation of an iterative reconstruction method for quantitative elastography,” Phys. Med. Biol. 45(6), 1521–1540 (2000). 10.1088/0031-9155/45/6/309 [DOI] [PubMed] [Google Scholar]
- 33.Konofagou E. E., Ophir J., “Precision estimation and imaging of normal and shear components of the 3D strain tensor in elastography,” Phys. Med. Biol. 45(6), 1553–1563 (2000). 10.1088/0031-9155/45/6/311 [DOI] [PubMed] [Google Scholar]
- 34.Lorenz A., et al. , “Ultrasound elastography of the prostate: an innovative technique for tumour-detection,” Ultraschall Med. 21(1), 8–15 (2000). 10.1055/s-2000-8926 [DOI] [PubMed] [Google Scholar]
- 35.Muthupillai R., et al. , “Magnetic-resonance elastography by direct visualization of propagating acoustic strain waves,” Science 269(5232), 1854–1857 (1995). 10.1126/science.7569924 [DOI] [PubMed] [Google Scholar]
- 36.Dresner M. A., et al. , “Magnetic resonance elastography of the prostate,” Radiology 209P, 181 (1998). [Google Scholar]
- 37.Rose G. H., et al. , “‘Palpation of the brain’ using magnetic resonance elastography,” Radiology 209P, 425–425 (1998). [Google Scholar]
- 38.Manduca A., et al. , “Magnetic resonance elastography: non-invasive mapping of tissue elasticity,” Med. Image Anal. 5(4), 237–254 (2001). 10.1016/S1361-8415(00)00039-6 [DOI] [PubMed] [Google Scholar]
- 39.Miga M. I., “A new approach to elastographic imaging: modality independent elastography,” Proc. SPIE 4684, 604–611 (2002). 10.1117/12.467203 [DOI] [Google Scholar]
- 40.Miga M. I., “A new approach to elastography using mutual information and finite elements,” Phys. Med. Biol. 48(4), 467–480 (2003). 10.1088/0031-9155/48/4/304 [DOI] [PubMed] [Google Scholar]
- 41.Miga M. I., Rothney M. P., Ou J. J., “Modality independent elastography (MIE): potential applications in dermoscopy,” Med. Phys. 32(5), 1308–1320 (2005). 10.1118/1.1895795 [DOI] [PubMed] [Google Scholar]
- 42.Ou J. J., et al. , “Evaluation of 3D modality-independent elastography for breast imaging: a simulation study,” Phys. Med. Biol. 53(1), 147–163 (2008). 10.1088/0031-9155/53/1/010 [DOI] [PubMed] [Google Scholar]
- 43.Washington C. W., Miga M. I., “Modality independent elastography (MIE): a new approach to elasticity imaging,” IEEE Trans. Med. Imaging 23(9), 1117–1128 (2004). 10.1109/TMI.2004.830532 [DOI] [PubMed] [Google Scholar]
- 44.Pheiffer T. S., et al. , “Automatic generation of boundary conditions using demons nonrigid image registration for use in 3-D modality-independent elastography,” IEEE Trans. Biomed. Eng. 58(9), 2607–2616 (2011). 10.1109/TBME.2011.2159791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weis J. A., et al. , “A consistent pre-clinical/clinical elastography approach for assessing tumor mechanical properties in therapeutic systems,” Proc. SPIE 8672, 86721F (2013). 10.1117/12.2007425 [DOI] [Google Scholar]
- 46.Weis J. A., et al. , “Validation and reproducibility assessment of modality independent elastography in a pre-clinical model of breast cancer,” Proc. SPIE 9038, 90381I (2014). 10.1117/12.2042796 [DOI] [Google Scholar]
- 47.Lorensen W. E., Cline H. E., “Marching cubes: a high resolution 3D surface construction algorithm,” ACM SIGGRAPH Comput. Graph. 21, 163–169 (1987). 10.1145/37402.37422 [DOI] [Google Scholar]
- 48.Sullivan Jr J. M., Charron G., Paulsen K. D., “A three-dimensional mesh generator for arbitrary multiple material domains,” Finite Elem. Anal. Des. 25(3), 219–241 (1997). 10.1016/S0168-874X(96)00027-3 [DOI] [Google Scholar]
- 49.Thirion J. P., “Image matching as a diffusion process: an analogy with Maxwell’s demons,” Med. Image Anal. 2(3), 243–260 (1998). 10.1016/S1361-8415(98)80022-4 [DOI] [PubMed] [Google Scholar]
- 50.Vercauteren T., et al. , “Diffeomorphic demons: efficient non-parametric image registration,” Neuroimage 45(1 Suppl), S61–72 (2009). 10.1016/j.neuroimage.2008.10.040 [DOI] [PubMed] [Google Scholar]
- 51.Vercauteren T., et al. , “Non-parametric diffeomorphic image registration with the demons algorithm,” Med. Image Comput. Comput. Assist. Interv. 10(Pt 2), 319–326 (2007). 10.1007/978-3-540-75759-7_39 [DOI] [PubMed] [Google Scholar]
- 52.Polak E., Ribiere G., “Note surla convergence des méthodes de directions conjuguées,” Rev. Fr. Imform. Rech. Oper. 16, 35–43 (1969). [Google Scholar]
- 53.Fletcher R., Reeves C. M., “Function minimization by conjugate gradients,” Comput. J. 7(2), 149–154 (1964). 10.1093/comjnl/7.2.149 [DOI] [Google Scholar]
- 54.Chavent G., “Identification of functional parameters in partial differential equations,” in Goodson R. E., Polis M., Eds., Identification of Parameters in Distributed Systems, ASME Conference on Automatic Control, 17–21 June 1974, Austin, Texas, pp. 31–48, American Society of Mechanical Engineers, New York: (1974). [Google Scholar]
- 55.Oberai A. A., Gokhale N. H., Feijoo G. R., “Solution of inverse problems in elasticity imaging using the adjoint method,” Inverse Probl. 19, 297–313 (2003). 10.1088/0266-5611/19/2/304 [DOI] [Google Scholar]
- 56.Schroeder W., Martin K., Lorensen B., The Visualization Toolkit: An Object-Oriented Approach to 3-D Graphics, Prentice Hall PTR, Upper Saddle River, N.J. (1996). [Google Scholar]
- 57.Yoo T. S., et al. , “Engineering and algorithm design for an image processing API: a technical report on ITK—the Insight Toolkit,” Stud. Health Technol. Inform. 85, 586–592 (2002). [PubMed] [Google Scholar]
- 58.Gersho A., Gray R. M., Vector Quantization and Signal Compression, Springer, Norwell, Massachusetts: (1992). [Google Scholar]
- 59.Besag J., “On the statistical analysis of dirty pictures,” J. R. Stat. Soc. Series B Methodol. 48, 259–302 (1986). [Google Scholar]
- 60.McGarry M., et al. , “Including spatial information in nonlinear inversion MR elastography using soft prior regularization,” IEEE Trans. Med. Imaging 32(10), 1901–1909 (2013). 10.1109/TMI.2013.2268978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Doyley M. M., et al. , “Enhancing the performance of model-based elastography by incorporating additional a priori information in the modulus image reconstruction process,” Phys. Med. Biol. 51(1), 95–112 (2006). 10.1088/0031-9155/51/1/007 [DOI] [PubMed] [Google Scholar]
- 62.Richards M. S., Doyley M. M., “Investigating the impact of spatial priors on the performance of model-based IVUS elastography,” Phys. Med. Biol. 56(22), 7223–7246 (2011). 10.1088/0031-9155/56/22/014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bland J. M., Altman D. G., “Measuring agreement in method comparison studies,” Stat. Methods Med. Res/ 8(2), 135–160 (1999). 10.1191/096228099673819272 [DOI] [PubMed] [Google Scholar]
- 64.Galbraith S. M., et al. , “Reproducibility of dynamic contrast-enhanced MRI in human muscle and tumours: comparison of quantitative and semi-quantitative analysis,” NMR Biomed. 15(2), 132–142 (2002). 10.1002/nbm.731 [DOI] [PubMed] [Google Scholar]
- 65.Whisenant J. G., et al. , “Assessing reproducibility of diffusion-weighted magnetic resonance imaging studies in a murine model of HER2+ breast cancer,” Magn. Reson. Imaging 32(3), 245–249 (2014). 10.1016/j.mri.2013.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Landis J. R., Koch G. G., “The measurement of observer agreement for categorical data,” Biometrics 33(1), 159–174 (1977). 10.2307/2529310 [DOI] [PubMed] [Google Scholar]
- 67.Shire N. J., et al. , “Test-retest repeatability of MR elastography for noninvasive liver fibrosis assessment in hepatitis C,” J. Magn. Reson. Imaging 34(4), 947–955 (2011). 10.1002/jmri.22716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Murphy M. C., et al. , “Measuring the characteristic topography of brain stiffness with magnetic resonance elastography,” PLoS One 8(12), e81668 (2013). 10.1371/journal.pone.0081668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fraquelli M., et al. , “Reproducibility of transient elastography in the evaluation of liver fibrosis in patients with chronic liver disease,” Gut 56(7), 968–973 (2007). 10.1136/gut.2006.111302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ferraioli G., et al. , “Reproducibility of real-time shear wave elastography in the evaluation of liver elasticity,” Eur. J. Radiol. 81(11), 3102–3106 (2012). 10.1016/j.ejrad.2012.05.030 [DOI] [PubMed] [Google Scholar]
- 71.Barnes S. L., et al. , “Assessing the reproducibility of dynamic contrast enhanced magnetic resonance imaging in a murine model of breast cancer,” Magn. Reson. Med. 69(6), 1721–1734 (2013). 10.1002/mrm.24422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.O'Flynn E. A. M., et al. , “Diffusion weighted imaging of the normal breast: reproducibility of apparent diffusion coefficient measurements and variation with menstrual cycle and menopausal status,” Eur. Radiol. 22(7), 1512–1518 (2012). 10.1007/s00330-012-2399-0 [DOI] [PubMed] [Google Scholar]
- 73.Zhang X., et al. , “Reproducibility of magnetic resonance perfusion imaging,” PLoS One 9(2), e89797 (2014). 10.1371/journal.pone.0089797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chamming’s F., et al. , “Shear wave elastography of tumour growth in a human breast cancer model with pathological correlation,” Eur. Radiol. 23(8), 2079–2086 (2013). 10.1007/s00330-013-2828-8 [DOI] [PubMed] [Google Scholar]
- 75.Barnes S. L., Young P. P., Miga M. I., “A novel model-gel-tissue assay analysis for comparing tumor elastic properties to collagen content,” Biomech. Model. Mechanobiol. 8(4), 337–343 (2009). 10.1007/s10237-009-0150-9 [DOI] [PubMed] [Google Scholar]
- 76.Kim D. K., et al. , “Utilizing a reference material for assessing absolute tumor mechanical properties in modality independent elastography,” Proc. SPIE 9038, 90381F (2014). 10.1117/12.2044195 [DOI] [Google Scholar]