Abstract
Interleukin-1 (IL-1) cytokines, IL-1α, IL-1β, and IL-18 play a crucial role in inflammatory responses in a variety of diseases including periodontitis. In this study, the periodontopathic bacterial pathogen, Aggregatibacter actinomycetemcomitans, induced cell death and cytokine release in macrophages. Cell viability was reduced by A. actinomycetemcomitans invasion using (3-[4, 5-dimethylthiazol-2-yl]-2, 5-diphenyltetrazolium bromide assay. The production of IL-1β in A. actinomycetemcomitans-invaded macrophage cells was detected by real-time reverse transcriptase–polymerase chain reaction, western blotting, and enzyme-linked immunosorbent assay. Treatment with a caspase-1 inhibitor and silencing of the caspase-1 gene had no effect on IL-1β secretion induced by A. actinomycetemcomitans invasion. Pattern recognition receptor, NLRP3 was upregulated in A. actinomycetemcomitans-invaded macrophages. However, NLRP3 knockdown had no effect on the secretion of IL-1β in A. actinomycetemcomitans-invaded RAW 264 cells. In addition, A. actinomycetemcomitans invasion induced the generation of reactive oxygen species (ROS) and the release of cathepsin B in RAW 264 cells. Interestingly, CA074-Me, a cathepsin B inhibitor, and N-Acetyl-l-cysteine, a ROS inhibitor, prevented the production of IL-1β induced by A. actinomycetemcomitans. Taken together, these results suggest A. actinomycetemcomitans induce IL-1β production in RAW 264 cells through the production of ROS and cathepsin B, but not through the NLRP3/caspase-1 pathway.
Introduction
Aggregatibacter actinomycetemcomitans, a Gram-negative periodontopathic bacterium, is a member of the oral microbiota of humans. Recently, it was reported that periodontopathic bacteria act as reservoirs for medically important pathogens that cause systemic disorders, such as systemic infectious disease, cardiovascular diseases, respiratory diseases, diabetes mellitus, adverse pregnancy outcomes, and osteoporosis through the induction of proinflammatory cytokines (Nishihara and Koseki 2004). A. actinomycetemcomitans was also demonstrated to invade oral epithelial cells and macrophages in vitro (Sreenivasan and others 1993; Kato and others 2000; Nishihara and Koseki 2004). In a series of studies, we documented the participation of CD14 in the phagocytosis of bacterium by macrophages, resulting in the induction of cell death (Muro and others 1997; Kato and others 2000; Okinaga and others 2013).
The innate immune system represents the first line of host defense against a wide range of pathogens. When pattern recognition receptors (PRRs) that recognize danger-associated molecular patterns or pathogen-associated molecular patterns are activated, they induce immune responses that are critical for host defense. Macrophages are the major cells of the host innate immune system, and they have a critical role in several chronic inflammatory diseases. Once activated, they mediate their effects by producing and secreting proinflammatory cytokines in response to cellular stimuli (Martinon and others 2002).
The inflammasome is a large intracellular protein complex that recruits and activates caspase-1 protease, which in turn cleaves the proform of interleukin-1 (IL-1)β to its biologically active and secreted form. Inflammasome assembly is initiated by the activation and self-oligomerization of certain pyrin and HIN200 domain containing (PYHIN) or nucleotide-binding domain leucine-rich repeat containing (NLR) family PPRs in the cytoplasm (Latz 2010). The cytosolic PYHIN protein family member, absent in melanoma-2 interacts with foreign double-stranded DNA through cytosolic HIN200 and apoptosis-associated speck-like protein containing a CARD (ASC) to form a caspase-1 activating inflammasome (Burckstummer and others 2009; Fernandes-Alnemri and others 2009).
Three NLR proteins NLRP1, NLRP3, and NLRC4 have been identified as key molecules in formation of the inflammasome (Strowing and others 2012). Among them, NLRP3 is activated by pathogen-derived signals, such as viral, fungal, and bacterial infection (Muruve and others 2008; Gross and others 2009; Sahoo and others 2011). In addition, it was reported that NLRP3 oligomerization leads to pyrin domain clustering that binds to the adapter protein ASC, which in turn, recruits procaspase-1 for activation (Van de Veerdonk and others 2011). The outcome of NLRP3 inflammasome assembly is the cleavage of cytosolic pro-IL-1β, by activated caspase-1, to the mature proinflammatory cytokine IL-1β (Kanneganti and others 2006, 2007). However, some studies have demonstrated that caspase-1 is not involved in the host defense against certain types of microorganisms such as Chlamydia trachomatis, although IL-1β secretion was induced by invasion of this microorganism (Lu and others 2000; Cheng and others 2008).
In this study, we investigated the induction of cell death and IL-1β production in periodontopathic bacteria-invaded RAW 264 cells, and found that periodontopathic invasion induced the production of reactive oxygen species (ROS), and the release of cathepsin B. Moreover, IL-1β processing was downregulated by inhibition of these molecules, but not caspase-1 or NLRP3. These findings suggest that periodontopathic bacterial invasion in mouse RAW 264 cells induces IL-1β production, which is dependent upon the production of ROS and cathepsin B, but not NLRP3/caspase-1 activity.
Materials and Methods
Cells and bacterial strain
RAW 264, a murine macrophage-like cell line (RIKEN RCB0535), was cultured in α-minimum essential medium (α-MEM; Gibco Laboratories, Grand Island, NY) supplemented with 10% heat-inactivated fetal bovine serum (FBS), penicillin G (100 U/mL) (Nacalai Tesque, Kyoto, Japan), and streptomycin (100 μg/mL) (Wako Pure Chemical Industries, Osaka, Japan) at 37°C in an atmosphere of 5% CO2 and air. THP-1 and U937, human monocytic cell lines, were maintained in RPMI 1640 medium (Gibco Laboratories) containing 10% FBS, penicillin G (100 U/mL), and streptomycin (100 μg/mL) at 37°C in an atmosphere of 5% CO2 and air. THP-1- and U937 cells were activated by treatment with phorbol myristate acetate at a concentration of 10 ng/mL for 12 h. These cells were then rested for 24 h before use. Bone marrow cells (BMCs) were isolated from femurs and tibias of 6-week-old male ddY mice (Kyudo Co., Ltd., Saga, Japan) and maintained in α-MEM (Gibco Laboratories) supplemented with 10% fetal calf serum, 100 U/mL penicillin G, and 100 μg/mL streptomycin. Cells (4×104) were cultured with macrophage colony-stimulating factor (20 ng/mL; PeproTech, Rocky Hill, NJ) on 48-well plates at 37°C in 5% CO2 for 5 days. All procedures were approved by the Animal Care and Use Committee of Kyushu Dental University. Aggregatibacter actinomycetemcomitans strain Y4, ATCC 29522, and ATCC 29522 containing plasmid pNP3M (ATCC29522pNP3M; provided by Dr. Galli, Indiana University, Indianapolis, IN) (Permpanich and others 2006) were grown in Brain Heart Infusion broth (BHI; Difco Laboratories, Detroit, MI) supplemented with 1% (w/v) yeast extract at 37°C in an atmosphere of 5% CO2 and air.
Reagents
Monoclonal antibodies were obtained from the following sources: anti-ASC and anti-cathepsin B (Santa Cruz Biotechnology, Santa Cruz, CA); anti-caspase-1 (Adipogen, San Diego, CA); anti-NLRP3 (Cryopyrin) (Enzo Life Sciences, Farmingdale, NY and Santa Cruz Biotechnology); anti-IL-1β (Cell Signaling Technology, Beverly, MA and Abcam, Cambridge, United Kingdom); and anti-β-actin (Sigma-Aldrich, St. Louis, MO). Dihydrorhodamine 123 was obtained from Santa Cruz Biotechnology. Cathepsin B inhibitor, CA-074Me (10 μM; Millipore Corporation, Billerica, MA), ROS inhibitor, N-acetyl-l-cysteine (NAC, 10 mM; Sigma-Aldrich), caspase-1 inhibitor, Z-YVAD-FMK (100 μM; Abcam), and cytochalasin D (1 μg/mL; Sigma-Aldrich) were used in this study. These inhibitors were added to RAW 264 cells in serum-free medium for 2 h before A. actinomycetemcomitans invasion assay.
A. actinomycetemcomitans invasion procedure
RAW 264 cells, BMCs, THP-1 cells, and U937 cells (1×106 cells/mL) were seeded into 6-well plates (Iwaki, Iwaki, Japan) at a concentration of 5×105 cells/well 1 day before the beginning of the experiment. A. actinomycetemcomitans Y4 was cultured overnight in BHI containing 1% yeast extracts. Bacterial cells were harvested by centrifugation at 1,500 g for 10 min at 4°C and suspended in α-MEM medium without antibiotics to an optical density of 0.4 at 600 nm, as measured by spectrophotometer (UV mini 1240; Shimadzu Corporation, Kyoto, Japan), which corresponded to ∼2×1010 bacteria/mL (Kato and others 2000). Bacterial suspensions were added to the wells at a multiplicity of infection (MOI) of 50 and the plates were centrifuged at 1,000 g for 10 min at 4°C before incubation at 37°C for 1 h. Cells were then washed 3 times with cell culture medium containing penicillin G (100 U/mL), streptomycin (100 μg/mL), and gentamicin (200 μg/mL) to remove extracellular bacteria. RAW264 cells, BMCs, THP-1 cells, and U937 cells were cultured in α-MEM and RPMI 1640 medium containing 5% FBS and antibiotics (Okinaga and others 2013).
Cell viability assay
RAW 264 cells and siRNA transfected RAW 264 cells were added to a 96-well plate at a concentration of 2×104 cells/well, 1 day before A. actinomycetemcomitans invasion. Caspase-1 inhibitor, Z-YVAD-FMK (100 μM) was pretreated for 1 h before A. actinomycetemcomitans invasion. The invaded RAW 264 cells were cultured in α-MEM medium containing 5% FBS for 36 h. Stock (3-[4, 5-dimethylthiazol-2-yl]-2, 5-diphenyltetrazolium bromide, 2.5 mg/Ml (MTT; Sigma Chemical Co., St. Louis, MO) solution (20 μL/well) was added to the wells and the plates were incubated for 4 h. After acid-isopropanol (100 μL of 0.04 N HCL in isopropanol) was added and mixed thoroughly, the plates were read by a Multiskan JX microplate reader (Thermo Scientific, Rockford, IL), using a test wavelength of 570 nm and a reference wavelength of 620 nm. The percentage of cell viability was calculated using the following formula: percentage of cell survival=100×(optical density at 570–620 nm with invasion/optical density at 570–620 nm without invasion).
Immunoblot analysis
A. actinomycetemcomitans-invaded RAW 264 cells, BMCs, THP-1 cells, and U937 cells were lysed in sodium dodecyl sulfate (SDS) lysis buffer (50 mM Tris-HCl, 2% SDS; pH 6.8), then the protein content of the samples was determined using protein assay reagent (Bio-Rad Laboratories, Hercules, CA). Protein samples (20 μg in each lane) were then subjected to electrophoresis on SDS-polyacrylamide gels, and electroblotted onto polyvinylidene fluoride membranes. After incubation with Blocking one (Nacalai Tesque, Kyoto, Japan) for 1 h, the membranes were reacted with primary antibodies overnight at 4°C. Immunodetection was performed using a Chemi-Lumi One super (Nacalai Tesque) and ECL Prime Western Blotting Detection Reagent (GE Healthcare UK Ltd., Amersham Place, United Kingdom). Densitometric analysis of protein bands in western blots was performed with Image Lab® (Bio-Rad, Munich, Germany).
RNA extraction and real-time reverse transcriptase–polymerase chain reaction analysis
RAW 264 cells, BMCs, THP-1 cells, and U937 cells were harvested, centrifuged at 4°C and stored at −80°C. RNA was extracted from cell pellets using a QIAshredder and RNeasy Mini Kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. RNA quality was confirmed by the presence of clearly defined ribosomal RNA bands separated by agarose gel electrophoresis. Total RNA (500 ng) was used for cDNA synthesis using Strata script reverse transcriptase (Stratagene, Cenda Creek, TX), according to the manufacturer's instructions. For real-time reverse transcriptase–polymerase chain reaction (RT-PCR), primers were designed using Primer Express 3.0 software (Applied Biosystems, Foster City, CA). The reactions were prepared using Fast SYBR® Green Master Mix (Applied Biosystems) and detection was performed with an Applied Biosystems StepOne™ Real Time PCR system (Applied Biosystems). Relative changes in gene expression were calculated using the comparative CT (ΔΔCT) method. Total cDNA abundance between samples was normalized using primers specific to the β-actin gene. The primers used for real-time RT-PCR were as follows: mouse NLRP3 (GenBank; accession no. NM_145827.3), forward 5′-CCATCG GCCGGACTAAAAT-3′ and reverse 5′-CGTCCTCGGGCTCAAACA-3′, mouse IL-1β (GenBank; accession no. NM_008361.3), forward 5′-AGTTGACGGACCCCAAAAGA-3′ and reverse 5′-GGACAGCCCAGGTCAAAGG-3′, human IL-1β (GenBank; accession no. NM_000576), forward 5′-TCAGCCAATCTTCATTGCTCAA-3′ and reverse 5′-TGGCGAGCTCAGGTACTTCTG-3′ mouse β-actin (GenBank; accession no. NM_007393.3), forward 5′-CTGAACCCTAAGGCCAACCGTG-3′ and reverse 5′-GGCATACAGGGACAGCACAGCC-3′, human 18s RNA (GenBank; accession no. M10098.1), forward 5′-CGA ACG TCT GCC CTA TCA ACT T-3′ and reverse 5′-ACC CGT GGT CAC CAT GGT A-3′.
Immunofluorescence microscopy
RAW 264 cells on glass coverslips were invaded with A. actinomycetemcomitans strain Y4 or ATCC29522pNP3M and incubated for 9 h. These cells were washed with cold phosphate buffered saline (PBS; pH 7.0), and fixed with 4% paraformaldehyde in PBS for 1 h at 4°C. After aspiration of fixative, the cells were treated with 0.2 M glycine, and rinsed with PBS. The cells were blocked with Blocking buffer (1% bovine serum albumin in PBS) for 30 min at room temperature (RT), and then incubated with primary antibody (anti-Cryopyrin antibody) and Alexa Fluor® 568 phalloidin (1/150 dilution; Invitrogen, Eugene, OR) for 20 min at RT in the dark. After rinsing in PBS, the cells were incubated with Alexa Fluor® 488 goat anti-mouse IgG (Invitrogen) for 20 min at RT in the dark and mounted with Vectashield® (Vector Laboratories, Burlingame, CA). Images were acquired using All-in-One Fluorescence microscope system (Keyence, Tokyo, Japan).
Enzyme-linked immunosorbent assay analysis
Supernatant from RAW 264 cells were collected 0–24 h following invasion. Secreted cytokine levels were assessed using mouse enzyme-linked immunosorbent assay (ELISA) kit for IL-1β (Uscn Life Science, Inc., Wuhan, China) according to the manufacturer's instructions.
Detection of ROS
The intracellular production of ROS was assayed using by flow cytometry (Beckman Coulter, Inc., Pasadena, CA). The substrate, dihydrorhodamine (DHR) 123, diffuses into cells and is oxidized by ROS to fluorescent Rhodamine 123. The supernatant in A. actinomycetemcomitans-invaded RAW 264 cells was replaced with DHR (100 μM) and incubated for 1 h. The reaction was stopped on ice, and the fluorescence intensity of the cells was analyzed immediately by flow cytometry and MultiCycle for Windows (Phoenix Flow Systems, San Diego, CA).
Silencing of NLRP3 and caspase-1 expression by specific siRNA
siRNA targeting was used to knock down NLRP3 and caspase-1 expression in RAW 264 cells. siRNAs against mouse NLRP3 and caspase-1, and siRNA control were purchased from Nacalai Tesque. A NEPA21 Super Electroporator (Nepa Gene Co., Ltd., Chiba, Japan) was used to deliver siRNA into cells according to the manufacturer's instructions. In brief, 1×106 cells were suspended in 100 μL of α-MEM and transfected with siRNA at a final concentration of 300 nM. Specific gene knockdowns were assessed by real-time RT-PCR.
Statistical analysis
All data are expressed as the mean±standard deviation of 3 experiments, with similar results obtained in each experiment. Statistical differences were determined using the unpaired Student's t-test. P<0.05 was considered statistically significant.
Results
A. actinomycetemcomitans-invaded RAW 264 cells
Confocal fluorescence microscopic analysis revealed that green fluorescence protein-expressing A. actinomycetemcomitans strain ATCC29522pNP3M was localized in the cytoplasm of RAW 264 cells 1 h after co-incubation at MOI 50 (Fig. 1A). Internalization of A. actinomycetemcomitans into RAW 264 cells was confirmed by flow cytometry analysis as shown in Fig. 1B. To distinguish between adherent and intracellular bacteria, trypan blue was added to all samples to quench extracellular fluorescence. Invasion of A. actinomycetemcomitans Y4, ATCC 29522, and ATCC 29522 pNP3M (data not shown) at MOI 50 reduced cell viability after 36 h of incubation in RAW 264 cells using the MTT assay (Fig. 1C).
FIG. 1.
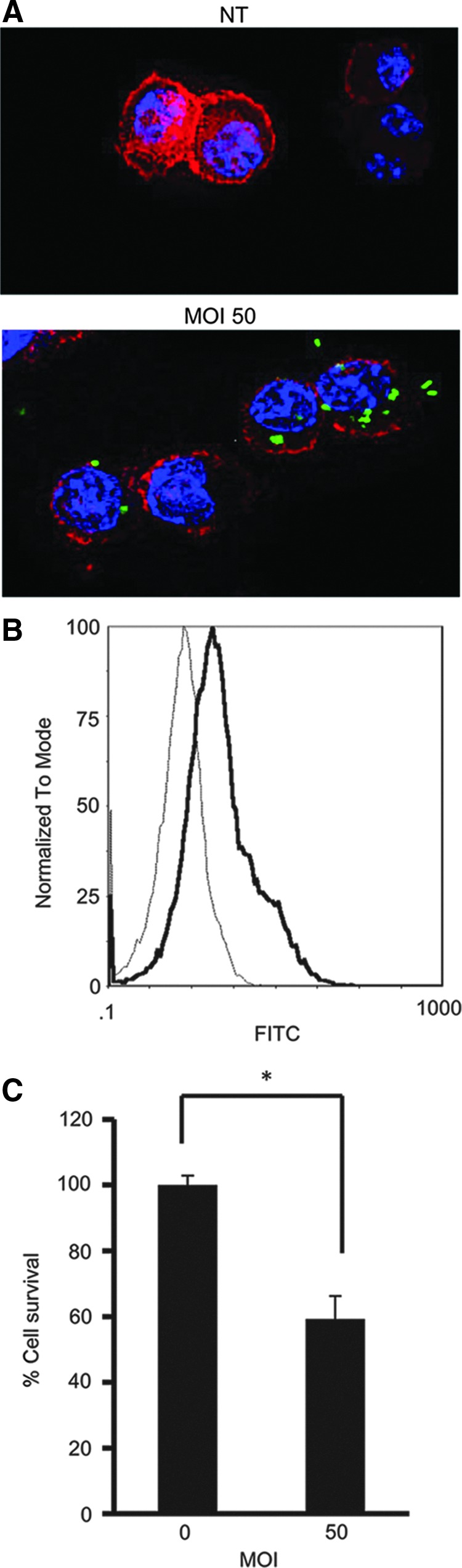
Aggregatibacter actinomycetemcomitans invasion of RAW 246.7 cells. RAW 264 cells, a mouse macrophage cell line, were treated with A. actinomycetemcomitans at multiplicity of infection (MOI) 50. (A) After treatment with green fluorescence protein (GFP)-labeled A. actinomycetemcomitans ATCC29522pNP3M for 1 h, invasion of RAW 264 cells by A. actinomycetemcomitans was confirmed by immunofluorescence staining. A. actinomycetemcomitans-invaded cells were stained with phalloidin and mounted with 4′,6-diamidino-2-phenylindole (DAPI)-containing buffer. Blue, DAPI staining of nucleus; green, GFP-labeled A. actinomycetemcomitans; red, phalloidin staining of actin filament. Images (magnification ×80) are representative of 3 independent experiments. (B) The invasion of RAW 264 cells by A. actinomycetemcomitans was confirmed by flow cytometric analysis. Data are representative of 3 independent experiments, with similar results obtained for each. Thin histograms, control cells; thick histograms, A. actinomycetemcomitans-invaded cells (MOI 50). (C) Cell viability was assessed in A. actinomycetemcomitans-invaded RAW 264 cells (MOI 50) by (3-[4, 5-dimethylthiazol-2-yl]-2, 5-diphenyltetrazolium bromide (MTT) assay. Data are representatives of 3 independent experiments performed in triplicate (*P<0.05, Student's t-test).
A. actinomycetemcomitans invasion induces the production and secretion of IL-1β
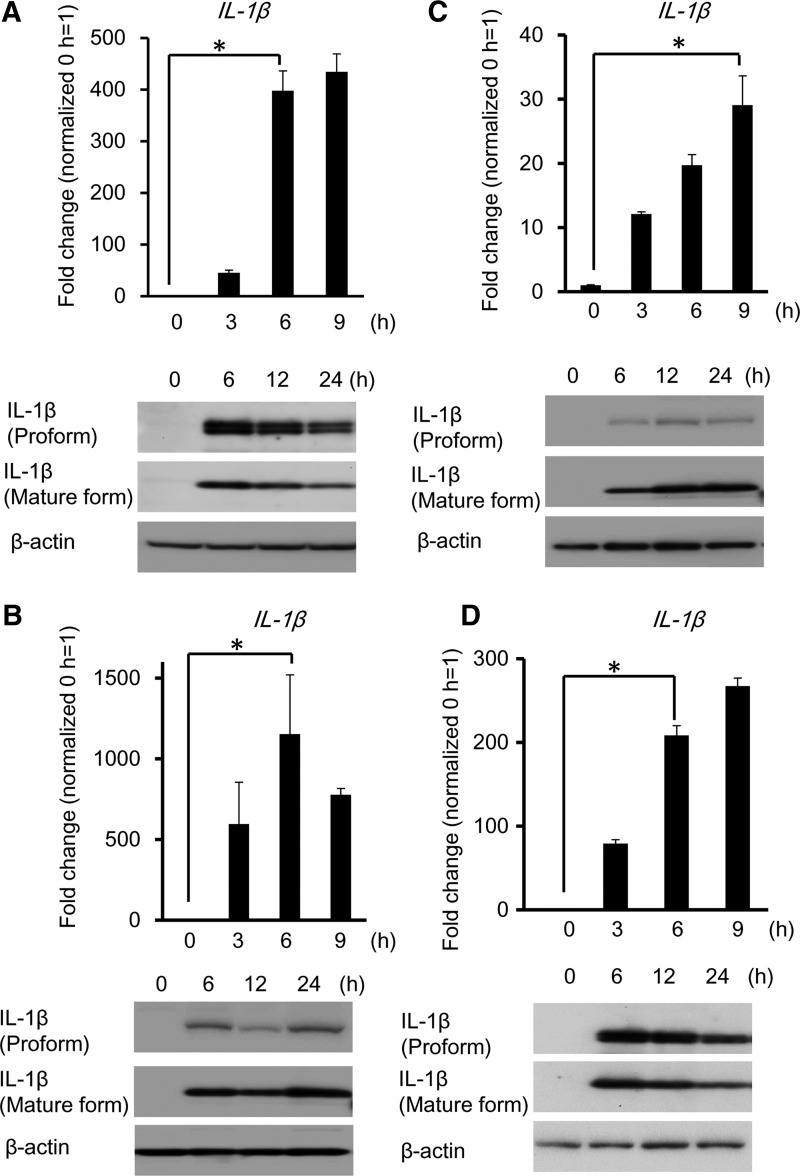
The gene expression of IL-1β was significantly increased in invaded RAW 264 cells at MOI 50 for 6 h. Pro-IL-1β and mature IL-1β expression were detected by western blotting analysis in A. actinomycetemcomitans-invaded RAW 264 cells at 6 h postinvasion (Fig. 2A). In addition, we confirmed the expression of IL-1β in A. actinomycetemcomitans-invaded primary BMCs (Fig. 2B), THP-1 cells (Fig. 2C), and U937 cells (Fig. 2D). The gene expression of IL-1β and mature IL-1β were also induced in A. actinomycetemcomitans-invaded mouse BMCs and human monocytic cells.
FIG. 2.
A. actinomycetemcomitans invasion induces the production and secretion of interleukin-1 (IL-1)β. (A) (Upper panel) IL-1β gene expression was determined by real-time reverse transcriptase–polymerase chain reaction (RT-PCR) at the indicated time points in A. actinomycetemcomitans-invaded RAW 264 cells (MOI 50). Values are presented relative to noninvaded control cells, which were arbitrarily assigned a value of 1. Data are shown as the mean of 3 independent experiments, with 3 independent samples examined in each. (*P<0.01, Student's t-test). (Lower panel) Detection of IL-1β protein by immunoblotting. Detection of IL-1β gene and protein expression in A. actinomycetemcomitans-invaded (B) bone marrow cells, (C) THP-1 cells or (D) U937 cells as for Fig. 2A. Data are representatives of 3 independent experiments performed in triplicate (*P<0.01 Student's t-test).
IL-1β production induced by A. actinomycetemcomitans invasion was independent of caspase-1
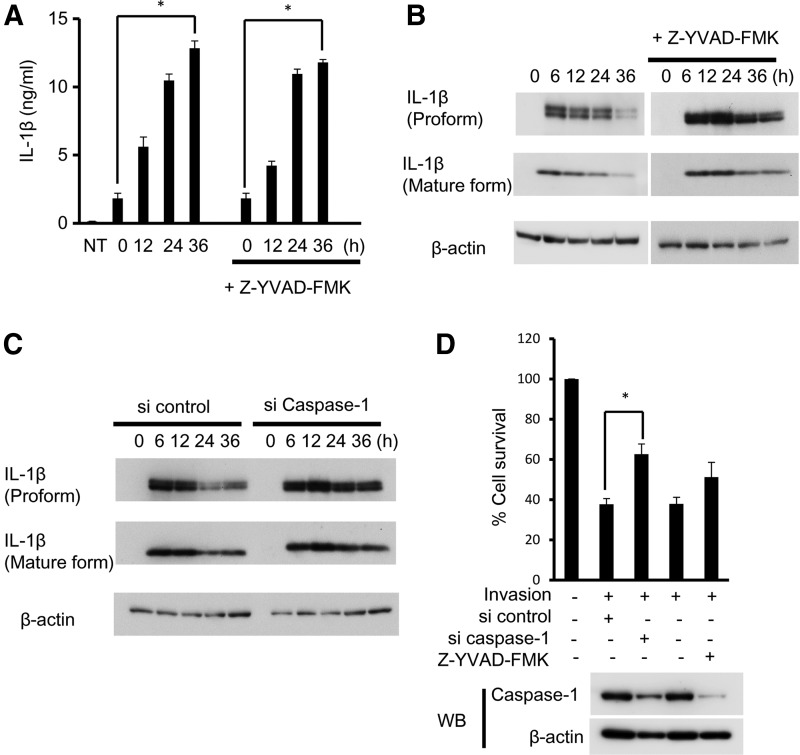
A. actinomycetemcomitans invasion caused an increase in the secretion of IL-1β at 12 h after invasion as measured by ELISA. Pan-caspase inhibitor, Z-YVAD-FMK (100 μM), had no effect on the secretion of IL-1β up to 36 h (Fig. 3A). The protein expression of proform IL-1β was decreased after 24 h in RAW 264 cells, but was not decreased in Z-YVAD-FMK-treated cells and caspase-1 knockdown cells (Fig. 3B, C). The reduction of cell viability induced by A. actinomycetemcomitans invasion was inhibited in caspase-1 knockdown cells, but not in Z-YVAD-FMK-treated RAW 264 cells (Fig. 3D).
FIG. 3.
IL-1β production induced by A. actinomycetemcomitans invasion is independent on caspase-1. (A) A. actinomycetemcomitans-invaded RAW 264 cells were cultured for the indicated times with or without pan-caspase inhibitor, Z-YVAD-FMK (100 μM) for 1 h before invasion. The secretion of IL-1β was determined by enzyme-linked immunosorbent assay (ELISA). Data are representative of 3 independent experiments performed in triplicate (*P<0.05, Student's t-test). (B) Detection of IL-1β protein by immunoblotting treated with Z-YVAD-FMK (100 μM). (C) RAW 264 cells were treated with siRNA against caspase-1. Detection of IL-1β protein by immunoblotting in Caspase-1 knockdown cells. (D) Cell viability was assessed in A. actinomycetemcomitans-invaded RAW 264 cells, caspase-1 inhibitor-treated RAW 264 cells, and caspase-1 knockdown RAW 264 cells by MTT assay. Data are representative of 3 independent experiments performed in triplicate (*P<0.05, Student's t-test). Inhibitor and knockdown of caspase-1 were confirmed by western blotting (lower panel).
Detection of PRRs on RAW 264 cells invaded with A. actinomycetemcomitans
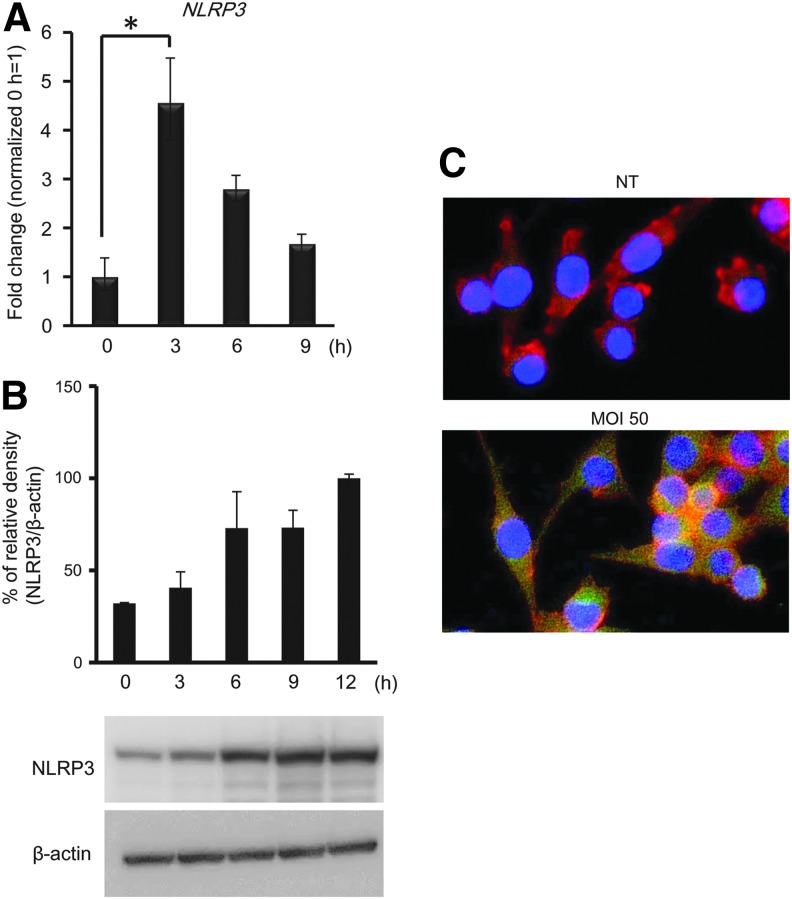
PRR mRNA expression in RAW 264 cells induced by A. actinomycetemcomitans invasion was examined by real-time RT-PCR analysis. As shown in Fig. 4A, the expression of the NLRP3 gene was significantly increased in A. actinomycetemcomitans-invaded RAW 264 cells at 3 h postinvasion. The expression of NLRP3 protein was increased in RAW 264.7 cells invaded with A. actinomycetemcomitans at 6 h. We also confirmed that NLRP3 expression was induced by A. actinomycetemcomitans invasion using immunofluorescence staining. The expression of NLRP3 protein was localized in the cytoplasm of A. actinomycetemcomitans-invaded RAW 264 cells (Fig. 4C).
FIG. 4.
A. actinomycetemcomitans invasion induces NLRP3 expression in RAW 264 cells. A. actinomycetemcomitans-invaded RAW 264.7 cells (MOI 50) were cultured for the indicated times. (A) Expression of the pattern recognition receptor gene, NLRP3, was determined by real-time RT-PCR at the indicated time points. Values are presented relative to noninvaded control cells, which were arbitrarily assigned a value of 1. Data are shown as the mean of 3 independent experiments, with 3 independent samples examined in each (*P<0.05, Student's t-test). (B) Detection of NLRP3 protein by immunoblotting. (C) A. actinomycetemcomitans-invaded RAW 264 cells were stained with FITC-anti-NLRP3, phalloidin, and mounted with DAPI-containing buffer. Blue, DAPI, nucleus; green, NLRP3; red, actin filament. Images (magnification×40) are representative of 3 independent experiments.
Role of NLRP3 in IL-1β production induced by A. actinomycetemcomitans invasion
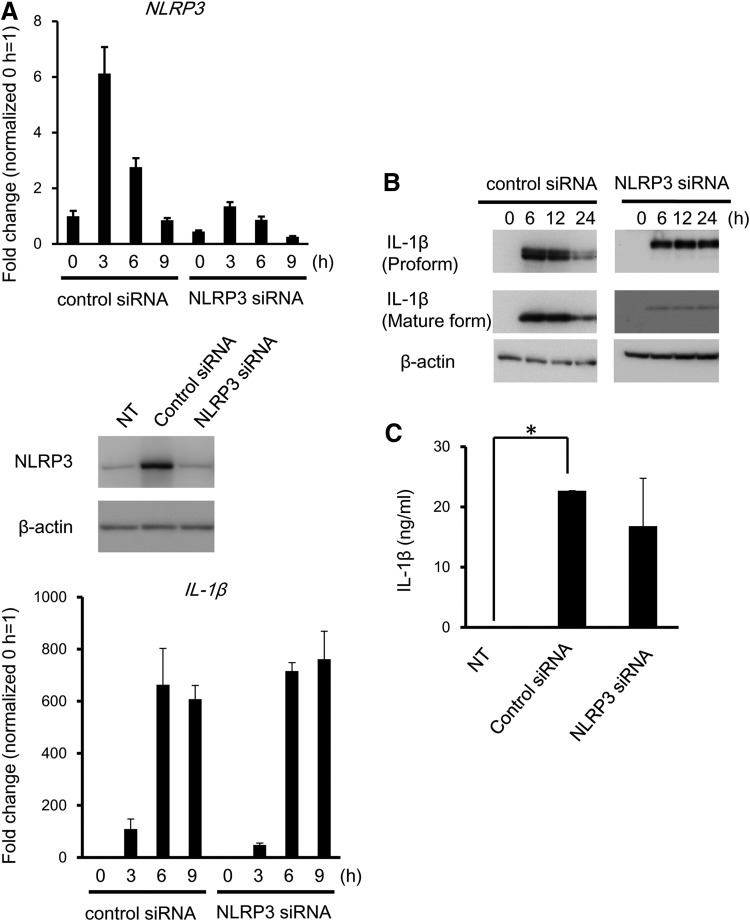
To define the role of NLRP3 in IL-1β expression and secretion of IL-1β in A. actinomycetemcomitans-invaded RAW 264 cells, NLRP3 was genetically inhibited by siRNA (Fig. 5A). No change in expression of the IL-1β gene was detected in RAW 264.7 cells, even when cells were treated with NLRP3 siRNA (data not shown). Depletion of NLRP3 downregulated the expression of the mature IL-1β (Fig. 5B). Secreted IL-1β levels were reduced; however, this was not statistically significant, due to the high standard deviation between experiments (Fig. 5C).
FIG. 5.
Role of NLRP3 in IL-1β secretion induced by A. actinomycetemcomitans invasion. RAW 264 cells were treated with siRNA against NLRP3. (A) Knockdown of NLRP3 was confirmed by real-time RT-PCR (upper panel) and western blotting (lower panel) in A. actinomycetemcomitans-invaded RAW 264 cells. (B) The expression of IL-1β protein in A. actinomycetemcomitans-invaded NLRP3 knockdown cells was detected by immunoblotting. (C) The secretion of IL-1β was determined by ELISA. Data are representative of 3 independent experiments performed in triplicate (*P<0.05, Student's t-test).
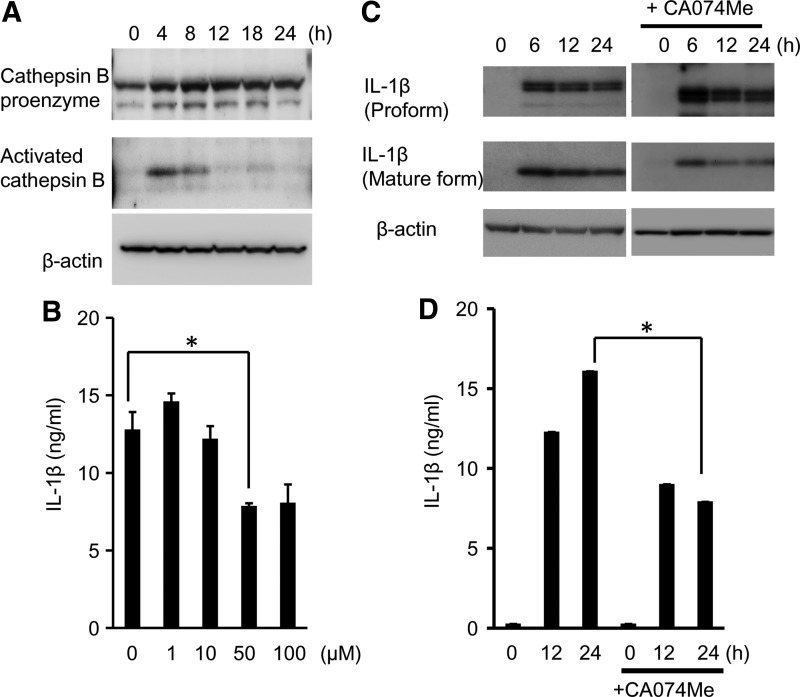
Effect of cathepsin B on IL-β production induced by A. actinomycetemcomitans invasion
NLRP3 is activated by lysosomal damage and the release of cathepsin B. In this study, we confirmed the activation of cathepsin B in A. actinomycetemcomitans-invaded RAW 264 cells. Cathepsin B pro-enzyme was detected in RAW 264 cells, and activated cathepsin B was detected at 4 h after A. actinomycetemcomitans invasion by western blotting (Fig. 6A). To explore whether cathepsin B is involved in IL-1β production induced by A. actinomycetemcomitans invasion, CA074-Me, a cell-permeable inhibitor of thiol protease that blocks cathepsin B activity, was used to pre-treat RAW 264 cells for 2 h before invasion. The relevant concentration of CA074-Me in this study was established by examination of the secretion of IL-1β. As shown in Fig. 6B, the secretion of IL-1β induced by A. actinomycetemcomitans invasion was suppressed in CA074-Me-treated (50 μM) RAW 264 cells. Treatment with CA074-Me partially downregulated the expression of mature IL-1β (Fig. 6C) and the secretion of IL-1β (Fig. 6D) in A. actinomycetemcomitans-invaded RAW 264 cells.
FIG. 6.
Involvement of cathepsin B in IL-1β secretion in A. actinomycetemcomitans-invaded RAW264 cells. (A) Detection of cathepsin B protein in A. actinomycetemcomitans-invaded RAW264 cells by immunoblotting. (B) RAW264 cells were pretreated with a cathepsin B inhibitor, CA074-Me for 2 h at the indicated concentrations before invasion. The secretion of IL-1β was determined by ELISA. Data are representative of 3 independent experiments performed in triplicate (*P<0.05, Student's t-test). (C) Detection of IL-1β protein by immunoblotting in A. actinomycetemcomitans-invaded RAW264 cells with CA074-Me pretreatment. (D) RAW264 cells were pretreated with CA074-Me (50 μM) for 2 h at indicated concentrations before invasion. The secretion of IL-1β was determined by ELISA. Data are representative of 3 independent experiments performed in triplicate (*P<0.05, Student's t-test).
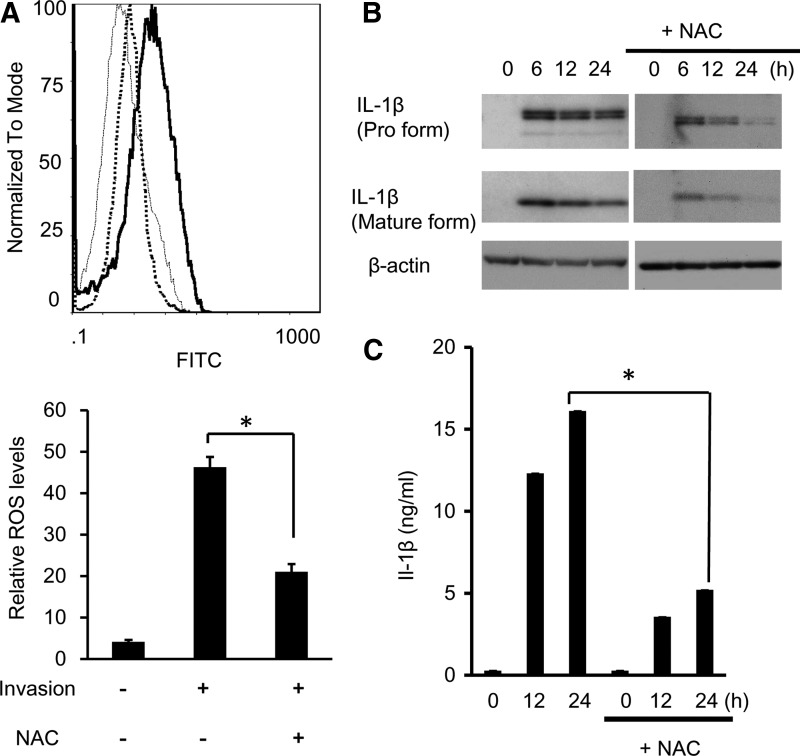
Effect of ROS on IL-1β production induced by A. actinomycetemcomitans invasion
Since several studies reported ROS has an important role in immune responses, we measured ROS production in A. actinomycetemcomitans-invaded RAW 264 cells by flow cytometry. ROS production was detected at 6 h in A. actinomycetemcomitans-invaded RAW 264 cells, and NAC treatment inhibited 43% of ROS production compared with nontreated cells (Fig. 7A). The amount of secreted IL-1β in culture supernatants and mature IL-1β in cell pellets was measured. NAC treatment significantly downregulated the expression of mature IL-1β (Fig. 7B) and the secretion of IL-1β (Fig. 7C) in A. actinomycetemcomitans-invaded RAW 264 cells.
FIG. 7.
Involvement of ROS in inflammasome activation in A. actinomycetemcomitans-invaded RAW264 cells. (A) RAW264 cells were pretreated with N-acetyl-l-cysteine (NAC; 10 mM), ROS inhibitor, for 2 h before invasion. Untreated RAW264 cells (thin histogram), NAC-treated A. actinomycetemcomitans-invaded cells (dot histogram) and A. actinomycetemcomitans-invaded cells (thick histogram) were exposed to the mitochondrial probe; rhodamine 123 for 1 h. Fluorescent intensity was measured by flow cytometry. The upper panel shows the frequency histogram. The lower panel represents the mean values of rhodamine 123 fluorescence of RAW264.7 cells. Data are representative of 3 independent experiments performed in triplicate (*P<0.05, Student's t-test). (B) Effect of NAC treatment on the expression of IL-1β protein in A. actinomycetemcomitans-invaded RAW264 cells was measured by immunoblotting. (C) Effect of NAC treatment on the secretion of IL-1β in A. actinomycetemcomitans-invaded RAW264 cells. The secretion of IL-1β was determined by ELISA. Data are representative of 3 independent experiments performed in triplicate (*P<0.05, Student's t-test).
Discussion
In this study, we observed the significant invasion of RAW 264 cells by A. actinomycetemcomitans using immunofluorescence staining and flow cytometric analysis (Fig. 1). Muro and others (1997) reported the invasion of A. actinomycetemcomitans in another macrophage cell line, J774.1, and that cytochalasin D, a potent microfilament inhibitor, strongly suppressed the invasion of A. actinomycetemcomitans Y4. We also confirmed the inhibitory effects of cytochalasin D on the invasion of A. actinomycetemcomitans in RAW 264 cells by immunofluorescence staining (data not shown). These findings indicated that A. actinomycetemcomitans invasion occurred through a microfilament-dependent phagocytotic process in RAW 264 cells, and J774.1 cells, suggesting A. actinomycetemcomitans Y4 is cytotoxic only when present in the cytoplasm (Kato and others 2000; Nishihara and Koseki 2004).
Caspase-1-dependent plasma membrane pores dissipate cellular ionic gradients, producing a net increased osmotic pressure, water influx, cell swelling, and release of inflammatory intracellular contents such as IL-1β and IL-18 in Salmonella-infected macrophages, indicating that pyroptosis enhances the inflammation through the induction of proinflammatory cytokines (Fink and Cookson 2006). In this study, we found that invasion of RAW 264 cells by A. actinomycetemcomitans-induced IL-1β secretion at 6 h (Fig. 2). There was a decrease of the IL-1β proform and mature form in A. actinomycetemcomitans-invaded RAW 264 cells during 36 h culture. However, treatment with caspase-1 inhibitor and silencing of caspase-1 had no effect on the secretion of IL-1β and confirmed the partial prevention of cell viability observed using MTT assay (Fig. 3). These findings indicate that A. actinomycetemcomitans invasion induces IL-1β production independent of caspase-1 activation in RAW 264 cells.
Secretion of IL-1β is tightly controlled by a diverse class of cytosolic complexes known as the inflammasome. NLR family and PYHIN protein families form an inflammasome with ASC and caspase-1 (Latz 2010). In this study, A. actinomycetemcomitans invasion induced the activation of inflammasome-associated molecules, NLRP3 and the secretion of IL-1β in RAW 264 cells (Figs. 2–4). It was reported that the NLRP3 inflammasome activates caspase-1 via its interaction with ASC, and that a large number of NLRP3 inflammasomes are triggered by several mechanisms, such as microbial stimuli (Kanneganti and others 2006; Kankkunen and others 2010; Meixenberger and others 2010), crystalline molecules (Dostert and others 2008), or pore-forming toxins (Mariathasan and others 2006). However, knockdown of NLRP3 and caspase-1 had no effect on the downregulation of IL-1β expression and secretion (Figs. 3 and 5). This result confirms the findings that caspase-1 had no effect on intracellular pro-caspase-1 in A. actinomycetemcomitans challenged-mononuclear leukocytes (Kelk and others 2003; Belibasakis and Johansson 2012). In this study, we investigated another mechanism by which A. actinomycetemcomitans induces IL-1β production in RAW 264 cells, and we examined the role of other classes of NLR-inflammasomes in the production of IL-1β.
Here, we focused on the involvement of 3 signaling pathways, including potassium efflux, cathepsin B release, and the generation of ROS, because these pathways were found to be crucial trigger for NLRP-dependent IL-1β secretion (Halle and others 2008; Hornung and others 2008; Tschopp and Schroder 2010). Cathepsin B was required for A. actinomycetemcomitans-induced and NLRP3-dependent IL-1β secretion (Fig. 6). In addition, CA074-Me, a potent cathepsin B inhibitor, partially suppressed IL-1β secretion induced by A. actinomycetemcomitans invasion (Fig. 6), indicating the possible involvement of cathepsin B in A. actinomycetemcomitans-induced IL-1β secretion. It was clearly demonstrated that ROS is required for IL-1β secretion during bacterial infection (Abdul-Sater and others 2009). As shown in Fig. 7, NAC significantly downregulated IL-1β secretion triggered by A. actinomycetemcomitans invasion, in accordance with previous reports (Abdul-Sater and others 2009; Kankkunen and others 2010).
In conclusion, this study demonstrated that the invasion of RAW 264cells with periodontopathic bacteria, A. actinomycetemcomitans, induced IL-1β production. The conversion of the proform to the mature form and secretion of IL-1β was not downregulated in NLRP3 and caspase-1 knockdown cells, indicating IL-1β secretion is independent of NLRP3 and caspase-1 in A. actinomycetemcomitans-invaded RAW 264 cells. Although inflammasome/caspase-1-independent IL-1β maturation was previously reported in some murine models (Mayer-Barber and others 2010; Provoost and others 2011), this is the first report demonstrating the possible independent pathway of NLRP3 and caspase-1 in the production of inflammatory cytokines during parasitic periodontopathic bacterial invasion. In addition, treatment with a cathepsin B inhibitor or ROS inhibitor partially prevented the production of mature IL-1β and the secretion of IL-1β in A. actinomycetemcomitans-invaded RAW 264 cells, suggesting cathepsin B and ROS are also essential for IL-1β maturation and secretion in RAW 264 cells.
Acknowledgment
This study was supported by a Grant-in-Aid for Scientific Research (23792149), from the Ministry of Education, Culture, and Science of Japan.
Author Disclosure Statement
No competing financial interests exist.
References
- Abdul-Sater AA, Koo E, Hacker G, Ojcius DM. 2009. Inflammasome-dependent caspase-1 activation in cervical epithelial cells stimulates growth of the intracellular pathogen Chlamydia trachomatis. J Biol Chem 284:26789–26796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belibasakis G, Johansson A. 2012. Aggregatibacter actinomycetemcomitans targets NLRP3 and NLRP6 inflammasome expression in human mononuclear leukocytes. Cytokine 59:124–130 [DOI] [PubMed] [Google Scholar]
- Burckstummer T, Baumann C, Bluml S, Dixit E, Durnberger G, Jahn H, Planyavsky M, Bilban M, Colinge J, Bennett KL, Superti-Fruga G. 2009. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol 10:266–272 [DOI] [PubMed] [Google Scholar]
- Cheng W, Shivshankar P, Li Z, Chen L, Yeh I-T, Zhong G. 2008. Caspase-1 contributes to Chlamydia trachomatis-induced upper urogenital tract inflammatory pathologies without affecting the course of infection. Infect Immun 76:515–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostert C, Petrilli V, Van Bruggen R, Steele C, Nossman BT, Tschopp J. 2008. Innate immune activation through Nalp3 inflammasome sensing of asbestos and silica. Science 320:647–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes-Alnemri T, Yu JW, Wu J, Datta P, Alnemri ES. 2009. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature 458:509–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink SL, Cookson BT. 2006. Caspase-1-dependent pore formation during pyroptosis leads to osmotic lysis of infected host macrophages. Cell Microbiol 8:1812–1825 [DOI] [PubMed] [Google Scholar]
- Gross O, Poeck H, Bscheider M, Dostert C, Hannesschlager N, Endres S, Hartmann G, Tardivel A, Schweighoffer E, Tybluewicz V, Mocsai A, Tschopp J, Ruland J. 2009. Syk kinase signaling couples to the Nlrp3 inflammasome for anti-fungal host defense. Nature 459:433–436 [DOI] [PubMed] [Google Scholar]
- Halle A, Hornung V, Petzold GC, Stewart CR, Monks BG, Reinheckel T, Fitzgerald KA, Latz E, Moore KJ, Golenbock DT. 2008. The NALP3 inflammasome is involved in the innate immune response to amyloid-beta. Nat Immunol 9:857–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V, Bauernfeind F, Halle A, Samstad EO, Kono H, Rock KL, Fitzgerald KA, Latz E. 2008. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat Immunol 9:847–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kankkunen P, Teirila L, Rintahaka J, Alenius H, Wolff H, Matikainen S. 2010. (1,3)-β-glucans activate both dectin-1 and NLRP3 inflammasome in human macrophages. J Immunol 184:6335–6342 [DOI] [PubMed] [Google Scholar]
- Kanneganti TD, Body-Malapel M, Amer A, Park JH, Whitfield J, Franchi L, Taraporewala ZF, Miller D, Patton JT, Inohara N, Nunez G. 2006. Critical role for cryopyrin/Nalp3 in activation of caspase-1 in response to viral infection and double-stranded RNA. J Biol Chem 281:36560–36568 [DOI] [PubMed] [Google Scholar]
- Kanneganti TD, Lamlanfi M, Nunez G. 2007. Intracellular NOD-like receptors in host defense and disease. Immunity 27:549–559 [DOI] [PubMed] [Google Scholar]
- Kato S, Nakashima K, Inoue M, Tomioka J, Nonaka K, Nishihara T, Kowashi Y. 2000. Human epithelial cell death caused by Actinobacillus actinomycetemcomitans infection. J Med Microbiol 49:739–745 [DOI] [PubMed] [Google Scholar]
- Kelk P, Johansson A, Claesson R, Hanstrom L, Kalfas S. 2003. Caspase-1 involvement in human monocyte lysis induced by Actinobacillus actinomycetemcopmitans leukotoxin. Infect Immun 71:4448–4455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latz E. 2010. The inflammasome: Mechanisms of activation and function. Curr Opin Immunol 22:28–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Shen C, Brunham RC. 2000. Chlamydia trachomatis infection of epitherial cells induces the activation of caspase-1 and release of mature IL-18. J Immunol 165:1463–1469 [DOI] [PubMed] [Google Scholar]
- Mariathasan S, Weiss DS, Newton K, McBride J, O'Rourke K, Roose-Girma M, Lee WP, Weinrauch Y, Monack DM, Dixit VM. 2006. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature 440:228–232 [DOI] [PubMed] [Google Scholar]
- Martinon F, Burns K, Tschopp J. 2002. The inflammasome: A molecular platform triggering activation of inflammatory caspases and processing of pro IL-β. Mol Cell 10:417–426 [DOI] [PubMed] [Google Scholar]
- Mayer-Barber KD, Barber DL, Shenderov K, White SD, Wilson MS, Cheever A, Kugler D, Hieny S, Caspar P, Nunez G, Schluetet D, Flavell RA, Sutterwala G, Sher A. 2010. Caspase-1 independent IL-1β production is critical for host resistance to Mycobacterium tuberculosis and dose not require TLR signaling in vivo. J Immunol 184:3326–3330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meixenberger K, Pache F, Eitel J, Schmeck B, Hippenstiel S, Slevogt H, N'Guessan P, Witzenrath M, Netea MG, Chakraborty T, Suttorp N, Opitz B. 2010. Listeria monocytogenes-infected human peripheral blood mononuclear cells produce IL-1β, depending on listeriolysin O and NLRP3. J Immunol 184:922–930 [DOI] [PubMed] [Google Scholar]
- Muro M, Koseki T, Akifusa S, Kato S, Kowashi Y, Ohsaki Y, Yamato K, Nishijima M, Nishihara T. 1997. Role of CD14 molecules in internalization of Actinobacillus actinomycetemcomitans by macrophages and subsequent induction of apoptosis. Infect Immun 65:1147–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muruve DA, Petrilli V, Zaiss AK, White LR, Clark SA, Ross PJ, Parks RJ, Tschopp J. 2008. The inflammasome recognizes cytosolic microbial and host DNA and triggers an innate immune response. Nature 452:103–107 [DOI] [PubMed] [Google Scholar]
- Nishihara T, Koseki T. 2004. Microbial etiology of periodontitis. Periodontology 2000. 36:14–26 [DOI] [PubMed] [Google Scholar]
- Okinaga T, Ariyoshi W, Akifusa S, Nishihara T. 2013. Essential role of JAK/STAT pathway in the induction of cell cycle arrest in macrophages infected with periodontopathic bacterium Aggregatibacter actinomycetemcomitans. Med Microbiol Immunol 202:167–174 [DOI] [PubMed] [Google Scholar]
- Permpanich P, Kowolik MJ, Galli DM. 2006. Resistance of fluorescent-labelled Actinobacillus actinomycetemcomitans strains to phagocytosis and killing by human neutrophils. Cell Microbiol 8:72–84 [DOI] [PubMed] [Google Scholar]
- Provoost S, Maes T, Pauwels NS, Vanden Berqhe T, Vandenabeele P, Lambrecht BN, Joos GF, Tournoy KG. 2011. NLRP3/caspase-1-independent IL-1beta production mediates diesel exhaust particle-induced pulmonary inflammation. J Immunol 187(6):3331–3337 [DOI] [PubMed] [Google Scholar]
- Sahoo M, Ceballos-Olvera I, Barrio LD, Re F. 2011. Role of the inflammasome, IL-1β, and IL-18 in bacterial infections. ScientificWorldJournal 11:2037–2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreenivasan PK, Meyer DH, Fives-Taylor PM. 1993. Requirements for invasion of epithelial cells by Actinobacillus actinomycetemcomitans. Infect Immun 61:1239–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strowing T, Henao-Mejia J, Elinav E, Flavell R. 2012. Inflammasomes in health and disease. Nature 481:278–286 [DOI] [PubMed] [Google Scholar]
- Tschopp J, Schroder K. 2010. NLRP3 inflammasome activation: The convergence of multiple signaling pathways on ROS production? Nat Rev Immunol 10:210–215 [DOI] [PubMed] [Google Scholar]
- Van de Veerdonk FL, Netea MG, Dinarello CA, Joosten LA. 2011. Inflammasome activation and IL-1β and IL-18 processing during infection. Trends Immunol 32:110–116 [DOI] [PubMed] [Google Scholar]