Abstract
The arthropod-borne diseases caused by dengue virus (DENV) are a major and emerging problem of public health worldwide. Infection with DENV causes a series of clinical manifestations ranging from mild flu syndrome to severe diseases that include hemorrhage and shock. It has been demonstrated that the innate immune response plays a key role in DENV pathogenesis. However, in recent years, it was shown that DENV evades the innate immune response by blocking type I interferon (IFN-I). It has been demonstrated that DENV can inhibit both the production and the signaling of IFN-I. The viral proteins, NS2A and NS3, inhibit IFN-I production by degrading cellular signaling molecules. In addition, the viral proteins, NS2A, NS4A, NS4B, and NS5, can inhibit IFN-I signaling by blocking the phosphorylation of the STAT1 and STAT2 molecules. Finally, NS5 mediates the degradation of STAT2 using the proteasome machinery. In this study, we briefly review the most recent insights regarding the IFN-I response to DENV infection and its implication for pathogenesis.
Introduction
Dengue disease is one of the most important arthropod-borne diseases in world public health; it is caused by 4 distinct antigenically related serotypes (DENV-1, -2, -3, -4) (Halstead 2007; Sun and Kochel 2013) of dengue virus (DENV). The virus is transmitted by the bite of mosquitoes of the Aedes genus, mainly by A. aegypti, a vector that is now highly domesticated and has preferences for urban habitats (Whitehead and others 2007; WHO 2009).
The 4 serotypes of DENV are widely distributed in tropical and subtropical regions around the world. Although most infections are asymptomatic, others can also be associated with a mild flu-type disease (dengue), or evolve into a potentially severe clinical case, associated with hemorrhages, organ damage, hypovolemic shock, and in some occasions with death (severe dengue) (WHO 2009). It is estimated that among the 50 million dengue cases reported annually, 500,000 are cases of severe dengue, of which ∼25,000 are fatal. Dengue disease is endemic in close to 80 countries around the world and more that 2,500 million individuals are at risk of being infected with the virus (Simmons and others 2012; Bhatt and others 2013).
The innate immune response constitutes the first line of defense against pathogenic microorganisms, and it is particularly important in the early control of viral infections (Medzhitov 2001; Akira 2009). Some DENV components, such as single-stranded RNA (ssRNA) and double-stranded RNA (dsRNA) replication intermediates, are recognized and induce an early response in innate immune cells (Wang and others 2006; Lee and others 2012). These components are known as pathogen-associated molecular patterns (PAMPs) and are recognized by a series of innate immunity receptors known as pattern recognition receptors (PRRs) (Navarro-Sánchez and others 2005).
The interactions between PAMPs and their corresponding PRRs activate various signaling pathways that finally lead to the production of proinflammatory cytokines and type I interferon (IFN-I) (Boo and Yang 2010). However, DENV has also acquired various strategies that provide it with the capacity to inhibit IFN-I response and thereby to escape this main antiviral innate immune response of the host. In this review, an analysis is made of how DENV regulates the expression of IFN-I and evades the host antiviral response mediated by this cytokine.
DENV: Genome and Protein Production
DENV belongs to the family Flaviviridae, genus Flavivirus. These viruses are icosahedral, enveloped, and ∼30–40 nm in diameter. The genome comprises an ssRNA of positive polarity (ssRNA+) of about 11 kb. At its 5′ end, the genome possesses a 7MeGppp2′OMe structure known as type I Cap that allows ribosome recognition; the genome does not have a polyadenylated region at the 3′ end (Malet and others 2008). The genomic RNA also carries 2 noncoding regions, known as the 5′ untranslated region (UTR) and 3′UTR, that are highly structured and play an essential role in viral replication (Mukhopadhyay and others 2005). The genome contains a single open reading frame and codes for a single polyprotein of ∼3,391 amino acids that are processed by cellular and viral proteases, leading to (1) 3 structural proteins: the capsid (C), responsible of viral RNA recognition and assembly; the premembrane protein (prM), implicated in blocking immature virus fusion; and the envelope glycoprotein (E), responsible for viral interaction with the cellular receptor and for viral entry into the target cell and (2) 7 nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) involved in the regulation of protein synthesis, in viral replication, in pathogenesis, and in the control of host antiviral response (Perera and Kuhn 2008; reviewed in Urcuqui-Inchima and others 2010). Viral polyprotein translation is performed at the rough endoplasmic reticulum (ER); however, it is thought that RNA replication occurs in vesicles formed by ER membrane invagination performed, presumably by NS4A in concert with host factors involved in inducing membrane rearrangements (Miller and others 2007; Welsch and others 2009). After RNA replication, the genomic RNA associates with the C protein and buds into the ER lumen, acquiring a lipid membrane that contains heterodimers of the embedded E and prM proteins constituting immature particles. These virions are transported along the cellular secretory pathway and in the trans-Golgi network undergo a maturation step mediated by furin-dependent cleavage of prM (Yu and others 2008). Finally, the mature form of the virus is released from the cell by exocytosis (reviewed in: Urcuqui-Inchima and others 2010).
DENV and Innate Response
The innate immune response is responsible for the control of viral spread during the early stages of infection (Akira 2009; Kumar and others 2009). Therefore, DENV does not escape the host defense mechanism and the first element used for this purpose is the IFN-I cytokine. The IFN-I response represents the principal effector mechanism of innate immunity and is responsible for the control of DENV replication and also contributes to the rapid development of adaptive immune responses that completely eliminate the virus (Diamond 2003). To achieve this, IFN-I activates the expression of a series of genes known as IFN-stimulated genes (ISGs), both in infected and in nearby noninfected cells (Der and others 1998). The products of ISG expression are involved in several cellular pathways, although the most important are those generating an antiviral state in the cells affecting different steps of viral replication (Boo and Yang 2010). However, several viruses, including DENV, have evolved various strategies to evade this process, which are the main objective of this review. A better understanding of the role of the innate immune response during DENV infection and of the evasion mechanisms developed by the virus itself would be of great importance in understanding dengue pathogenesis and its severe clinical outcomes.
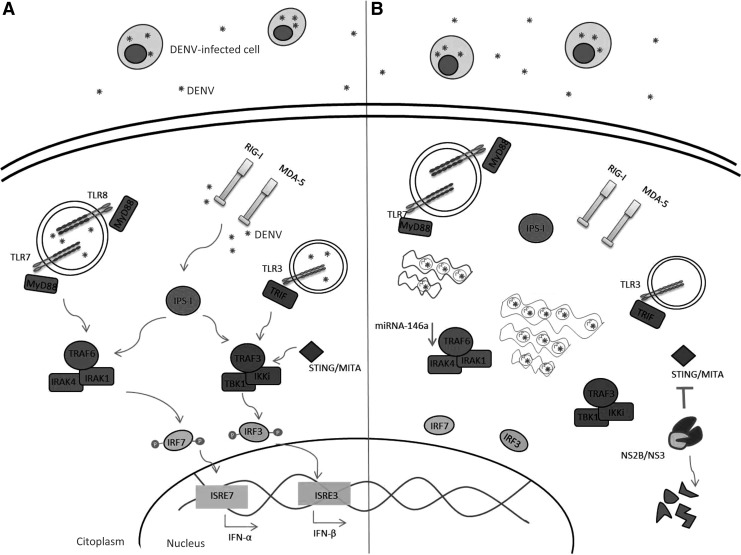
IFN-I expression requires the direct recognition of DENV-associated components that act as viral-specific PAMPs through PRR recognition. Viral antigen interaction with the different PRRs leads to activation of cells of the innate immune system and to the production of various components that help control viral infection and promote the development of the adaptive immune response (Beutler 2009). The DENV-associated antigens, ssRNA and dsRNA, that are replication intermediates are the best PAMPs described so far and are produced in the early stages of viral replication. Indeed, it has been demonstrated that DENV dsRNA can be sensed by Toll-like receptors (TLRs), which are expressed intracellularly within endosomes (TLR3, TLR7, TLR8) (Tsai and others 2009; Liang and others 2011) (Fig. 1A), but how this dsRNA reaches these TLRs is unknown, although it has been reported that TLR3 recognizes the viral nucleic acid and envelopes glycoproteins in the extracellular and endosomal compartments (de Veer and others 2001). DENV PAMPs can also be sensed by the cytoplasmic helicase receptor retinoic acid-inducible-associated protein I (RIG-I) and by the melanoma differentiation-associated gene 5 (MDA-5) (Fig. 1A), which recognize RNAs bearing a triphosphate group at the 5′ end and dsRNAs, respectively (Nasirudeen and others 2011; Qin and others 2011). However, regarding RIG-I, the results are controversial since some authors report that RIG-I inhibits both DENV and Chikungunya virus infection by an IFN-I-independent mechanism (Olagnier and others 2014), yet other authors have suggested that RIG-I activation by DENV may be involved in immunopathogenesis of DENV through the modulation of cytokine production (da Conceição and others 2013). In addition, the signaling molecules, IFN regulatory factor (IRF)-3 and IRF-7, play key roles in anti-DENV innate immunity, possibly by inducing the IFN-I response (Fig. 1A) or other innate immune mediators, such as proinflammatory cytokines (Chen and others 2013a). The activation of TLRs and of cytoplasmic helicase receptors leads to transcription initiation of IFN-I and of proinflammatory cytokines regulated by the transcription factors, IRF and nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB), respectively (Sariol and others 2011).
FIG. 1.
Dengue virus (DENV) blocks type I interferon (IFN-I) production. The innate immune system senses the pathogen microorganism through pattern recognition receptors (PRRs) that recognize patterns present in microorganisms known as pathogen-associated molecular patterns (PAMPs). (A) The PRRs involved in DENV recognition are the Toll-like receptors (TLRs), 3, 7, and 8, and also the RNA helicase-type receptors, such as retinoic acid-inducible-associated protein I (RIG-I) and melanoma differentiation-associated gene 5 (MDA-5). TLR3 senses double-stranded RNA (dsRNA) produced in DENV replication, and TLR7 and TLR8 sense single-stranded RNA (ssRNA). TLR7 and TLR8 are endosomal receptors associated with the adaptor protein MyD88. Activation of these TLRs, together with MyD88, recruits the IRAK1 and IRAK4 kinases, which associate with TNF-receptor-associated factor 6 (TRAF6), leading to its ubiquitination. TRAF6 promotes phosphorylation of the interferon regulatory factor (IRF)-7 that finally leads to transcription of IFN-α. On the other hand, phosphorylation of IRF-3 is mediated by TLR3, which is associated with the adaptor molecule TRIF and leads to the activation of TRAF3, to finally induce transcription of IFN-β. The cytoplasmic receptors, RIG-I and MDA-5, activate the mitochondria-related adaptor molecule IPS-I (also known as MAVS, CARDIF, and VISA), which in turn activates TRAF3 leading to the phosphorylation of the transcription factors, IRF-3 and IRF-7. (B) DENV inhibits IFN-I production avoiding PRR recognition through the formation of intracellular vesicles, in which the replication process is concentrated and hidden from the host innate immunity. The viral protease NS2B/NS3 degrades the signaling molecule stimulator of IFN genes (STING) involved in TRAF3 and TANK-binding kinase 1 (TBK1) activation. Finally, microRNA (miRNA)-146a induced by DENV downregulates the expression of TRAF6.
IFN-I Response in Viral Infections
Type I IFN is produced in response to viral infection and represents the most efficient antiviral mechanism (Kotenko 2011). IFN-I (α/β) is produced upon the recognition of dsRNA and its name comes from the concept that this cytokine interferes with viral replication. The most important IFN-I-producing cells are the dendritic cells and the mononuclear phagocytes, although in fact all nuclear cells are type I IFN-producing cells. When the host cells are infected with a virus, they respond by the secretion of IFN-α/β, which is responsible for the transcription of more than 100 ISGs, whose products not only generate an antiviral state in the infected cells and in nearby uninfected cells but also participate in the establishment of the adaptive immune response responsible for the complete elimination of the infection (Kato and others 2005).
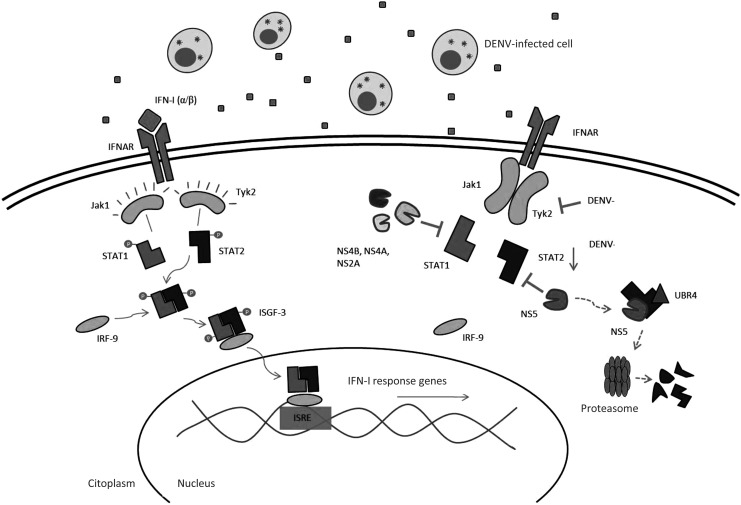
The binding of IFN-α/β with its respective receptor (IFNAR) leads to the activation of the tyrosine kinases, Jak1 and Tyk2 (janus kinase 1 and tyrosine kinase 2), associated with the receptor (Samuel 2001) (Fig. 2). In response to this event, STAT1 and STAT2 (signal transducers and activators of transcription) are phosphorylated and form a heterodimer that associates with RF-9, a DNA-binding protein, to finally form the multiproteic complex IFN-stimulated gene factor 3 (ISGF-3) (He 2006). This transcription factor complex is translocated to the nucleus and is responsible for the transcription of ISGs, whose protein products are involved in antiviral innate immunity response (Le Bon and Tough 2002) (Fig. 2). Among the most important proteins induced by IFN-I is the dsRNA-dependent kinase protein (PKR). PKR is normally present as an inactive protein, but is activated in response to interaction with dsRNA generated during viral genome replication and phosphorylates the alpha subunit of the eukaryotic initiation factor 2, promoting inhibition of translation and consequently inhibition of viral replication (Hershey 1991; Wang and others 2003; Zhang and others 2014). Although PKR regulates the innate response to virus infection, only in recent years have studies aimed at understanding the role of PKR in the replication of DENV increase (Diamond and Harris 2001; Li and others 2013).
FIG. 2.
DENV inhibits IFN-I signaling. IFN-α/β binding to its receptors, known as IFANR, leads to the activation of the tyrosine kinases, Janus kinase 1 (Jak1) and Tyrosine kinase 2 (Tyk2), which once activated form a heterodimer that binds to the DNA-binding protein IRF-9 to finally form the transcription factor, IFN-stimulated gene factor 3 (ISGF-3). This transcription factor translocates to the nucleus where it stimulates the expression of hundreds of IFN-I-stimulated genes (ISGs). DENV inhibits Tyk2 activation and also STAT1 phosphorylation through its proteins, NS2A, NS4A, and NS4B. DENV also downregulates the expression of STAT2 and its NS5 protein blocks STAT2 phosphorylation. Finally, NS5 degrades STAT2 through a proteosoma-dependent mechanism, binding to the cellular protein ubiquitin protein ligase E3 component recognin 4 (UBR4).
These results show that IFN blocks DENV infection by suppression of DENV RNA translation through a PKR-dependent mechanism. Interestingly, it was recently demonstrated that PKR knockdown by short interfering RNAs downregulates IFN synthesis through the RIG-I/IPS-1 pathway in human lung epithelial cells infected with DENV (Li and others 2013). Furthermore, the role of the 2′,5′-oligoadenylate synthetase (OAS) in DENV infection has also been studied. As with PKR, OAS is also activated by dsRNA and promotes the production of 5′-phosphorylated, 2′,5′-linked oligoadenylates, whose function is to activate RNase L implicated in RNA degradation (Ronni and others 1997; Castelli and others 1998). The implication of this protein in the control of DENV infection has been demonstrated in vitro and in rhesus macaques (Warke and others 2003; Sariol and others 2007). In addition, polymorphisms in the OAS1, OAS3, and OAS2 genes are associated with differential susceptibility to clinical outcomes of dengue infection (Alagarasu and others 2013; Thamizhmani and Vijayachari 2014). Taken together, these results show that both PKR and OAS, which are produced upon IFN-I stimulation, inhibit the viral replication cycle by inhibiting either DENV RNA translation or RNA degradation
The importance of IFN-I in protection against viral infections, and particularly against DENV infection, has been demonstrated in various experimental studies that show how this cytokine can limit viral replication. It has been described that in vitro treatment with IFN-α/β or IFN-γ before DENV infection protects human HepG2 cells from viral replication (Diamond and others 2000). However, if the treatment is performed after DENV infection, no difference is observed regarding viral replication (Diamond and others 2000). These studies suggest that DENV has developed an antagonist activity against the IFN-I-mediated immune response in host-infected cells.
Supporting this idea, within the last few years, it has been reported that several viruses of various families have developed specific mechanisms to inhibit IFN-I response and therefore ensure their productive replication in host-infected cells (Randall and Goodbourn 2008). To date, the main IFN-I blocking mechanisms reported are related to inhibition either of cytokine production or of the signaling pathways blocking the function of IFN-I to induce the expression of the antiviral genes, ISGs (Randall and Goodbourn 2008). However, it was recently demonstrated that the DENV-2 noncoding subgenomic flaviviral RNA interacts with RNA-binding proteins, such as G3BP1, G3BP2, and CAPRIN1, and inhibits the antiviral activity by inhibiting the ISG mRNA translation (Bidet and others 2014). It was shown that DENV encodes mechanisms that directly inhibit the antiviral activity of ISGs, as has been described for other viruses (Borrow and others 2010; Lefort and others 2010; Pierangeli and others 2011).
For this reason, it has been suggested that the innate immune evasion mechanisms, specifically IFN-I evasion, could play an essential role in viral pathogenesis and in the severity of certain viral infections. As discussed below, DENV inhibits the IFN-I-mediated innate immunity by limiting the production of IFN-α/β and also by blocking the signal induced by the cytokine in the nearby infected cells.
Inhibition of IFN-I Production by DENV
The activation of the different PRRs and RIG-I-like receptors by all the viral agonists, particularly the viral genome, leads to the production of IFN-I through the signaling of a considerable number of innate immune pathways. Nevertheless, a large number of different viruses have developed extremely efficient strategies to suppress IFN-I production and antiviral activity in infected cells (Muñoz-Jordán and Fredericksen 2010) (Fig. 1B).
The different types of viruses, and even the different types of strains, can considerably vary in their capacity to induce IFN-I production. The differences in such stimulation can be due to a variety of factors, including the amount of IFN-I inducer produced during viral infection or replication (namely ssRNA and dsRNA), the type of infected cells, and also the capacity of the virus to inhibit IFN-I production (Randall and Goodbourn 2008). In recent years, a large number of viruses have been described that can suppress IFN-I production through specific antagonists produced during replication. For instance, the Hepatitis C virus (HCV), Influenza A virus, and some paramyxoviruses can block the TLR3 and the RIG-I/MDA-5 signaling pathways (García-Sastre and others 1998; Childs and others 2007; Qashqari and others 2013). Other viruses, such as Rotavirus, Papillomavirus, and Herpes simplex virus (HSV), induce IRF-3 degradation, an essential signaling molecule for IFN-I production (Ronco and others 1998; Graff and others 2007; Melroe and others 2007).
In the specific case of DENV, 2 strategies that the virus has evolved to inhibit IFN-I production have so far been described: through active inhibition, altering signaling molecules involved in the innate immune signaling pathway responsible for IFN-I production and through passive inhibition, avoiding cellular recognition through innate immune receptors such as PRRs.
Using several experimental approaches, it has been observed that monocyte-derived dendritic cells (moDCs) infected with DENV are poor IFN-α/β producers, as opposed to what is observed with the same cells, but infected with other viruses such as the Newcastle disease virus (NDV) (Rodriguez-Madoz and others 2010a). Furthermore, these same authors demonstrated that reduced IFN-I production by moDCs was related to delayed T lymphocyte activation evaluated in coculture. Poor IFN-I production by DENV-infected DCs has also been described, even in the presence of IFN-I inducers such as TLR agonists and other types of viruses that are IFN-I inducers as are the NDV and Sendai virus (Rodriguez-Madoz and others 2010a). These results therefore suggest that indeed DENV presents an efficient antagonist mechanism limiting IFN-I production that has been suggested to depend on viral replication and on the catalytic activity of the NS2B/NS3 protease (Rodriguez-Madoz and others 2010b) (Fig. 1B). In support of this hypothesis, it was possible to identify specific elements involved in the inhibition of IFN-I production using DENV-infected moDCs as the model (Aguirre and others 2012). Using bioinformatics analysis, the authors searched for potential targets for the viral protease NS2B/NS3 in the production of the IFN-I signaling pathway, followed by in vitro verification of the interaction between NS2B/NS3 and the cellular proteins predicted; they observed that the DENV protease degrades the innate signaling molecule stimulator of IFN genes (STING), also known as mediator of IRF-3 activation (MITA), which is associated with the ER and plays an essential role in the TLR3 and RIG-I signaling pathways (Zhang and others 2012). Thus, DENV inhibits IFN-I production in moDCs through the action of its viral protease NS2B/NS3 that degrades STING. Similar results were described almost simultaneously by Yu and others (2012), who mapped the cleavage site of MITA (STING), by the DENV NS2B3, to LRR↓96Gleading to suppression of MITA (Fig. 1B). More interesting was the fact that the viral protease was unable to degrade the murine homolog of MITA, which indeed can negatively regulate DENV replication. MITA is a protein located in the mitochondrial external membrane associated with MAVS (Mitochondrial antiviral signaling), also known as VISA or IPS-1 (Zhong and others 2008; Zhang and others 2012). MITA acts as an adaptor molecule involved in the recruitment of TANK-binding kinase 1 (TBK1), IRF-3, and MAVS; moreover, silencing of MITA inhibits the antiviral response. DENV can also inhibit IFN-I production by blocking the kinase domain of the cellular signaling molecule IκB kinase ɛ (IKKɛ), thereby significantly affecting its function that therefore circumvents the proper induction and signaling of RIG-I, inhibiting the phosphorylation and nuclear translocation of IRF-3 (Angleró-Rodríguez and others 2014).
On the other hand, as mentioned above, DENV can passively inhibit IFN-I production, avoiding cellular recognition. A great number of viruses that replicate in the cytoplasm, in particular viruses with an RNA genome, induce in the infected cells the formation of intracellular vesicles that resemble cellular organelles, in which viral proteins and genome accumulate, thus enhancing the efficiency of viral replication and evasion of the host immune response (den Boon and others 2010). Likewise, several studies demonstrated that members of the Flaviviridae family, such as the West Nile virus, HCV, and DENV, induce the formation of intracellular vesicles within infected cells (Uchil and Satchidanandam 2003; Fredericksen and Gale 2006; Welsch and Zeuzem 2009). Using electronic microscopy techniques, it was observed that DENV infection induces the formation of vesicles and membrane complexes of ∼90 nm, in which the viral particle and viral dsRNA have been detected (Welsch and others 2009). Through electronic tomography it was established that these intracellular vesicles were derived from the cellular endomembrane complexes of the ER that contains pores believed to allow the liberation of newly synthesized RNAs (Welsch and others 2009). The formation of these types of intracellular vesicles may avoid innate immune recognition of the viral particle or of different viral components by the cellular PRRs, and hence may interfere with the production of IFN-I and proinflammatory cytokines. This mechanism could represent one of many mechanisms by which DENV induces the low production of IFN-I observed in infected cells.
Inhibition of IFN-I Signaling by DENV
Although several viruses limit IFN-I production, the most common mechanism whereby the antiviral response of this cytokine is inhibited is by blocking its signaling pathway. For instance, the Measles virus, HSV, and Japanese encephalitis virus inhibit phosphorylation of JAK, thereby blocking activation and signaling mediated by the STAT molecules (Yokota and others 2003; Chee and Roizman 2004; Lin and others 2006). Other groups of viruses code for proteins involved in directly blocking the function of the STAT signaling molecules either by blocking phosphorylation or by inducing degradation of these molecules (Elliott and others 2007; Frieman and others 2007; Chen and others 2013b).
DENV has similarly evolved the ability to alter the function of the JAK/STAT signaling pathway used by IFN-I (Fig. 2). The first report related to this problem was that of Muñoz-Jordan and others (2003); the authors transfected A549 human cells with different plasmids that independently expressed each of the 10 nonstructural proteins encoded by the DENV genome (Muñoz-Jordan and others 2003). Only the expression of the nonstructural proteins, NS2A, NS4A, and NS4B, enhanced viral replication of NDV, which is highly sensitive to the IFN-I response. Likewise, in Vero cells treated with IFN-β for 24 h, these same viral proteins induced poor activity of chloramphenicol acetyltransferase (Muñoz-Jordan and others 2003) encoded in a plasmid under the control of the IFN response element 54 (ISRE-54) and ISRE-9-27 promoters (IFN-α/β promoters present in the JAK/STAT signaling pathway), suggesting that the antagonistic role observed in the viral proteins takes place in the signaling pathway of IFN-I. Furthermore, the authors demonstrated that NS4B, and to a lesser extent NS2A and NS4A, negatively regulate the expression of IFN-β-inducible genes, specifically those involved in the signaling pathway of this cytokine. When the mechanism involved in this regulation was evaluated, it was observed that NS4B blocked the phosphorylation of STAT1 in LLCMK2 cells treated with IFN-I (Muñoz-Jordán and others 2005) (Fig. 2). The region in NS4B implicated in inhibiting STAT1 phosphorylation lies between amino acids 77 and 125, in addition to the 2K signal peptide located between NS4A and NS4B, which is responsible for the location of NS4B in the ER where its function is performed (Muñoz-Jordán and others 2005). Nevertheless, this signal peptide is not specific since the same results were obtained when different signal peptides were used.
Besides inhibiting STAT1 phosphorylation, DENV also interferes with STAT2 expression (Fig. 2). Indeed, human K562 (lymphocytes) and THP-1 (monocytes) cell lines infected with the infectious DENV-2 clone pDVWS601 express low levels of the STAT2 signaling molecule and also of other IFN-I-inducible proteins with antiviral activity, such as Myxovirus resistance protein A and PKR (Jones and others 2005). It has been suggested that the viral protein responsible for this regulation is NS5 since transfection of K562 cells with a lentiviral vector that only expresses this nonstructural protein induced IFN-I inhibition, as demonstrated by the expression of INF-I-inducible genes; the other nonstructural proteins had no effect (Mazzon and others 2009). This occurs because NS5 interacts with STAT2 through the RNA polymerase domain and inhibits phosphorylation of STAT2. Moreover, evidence exists that the expressions of all DENV nonstructural proteins simultaneously participate in inhibiting STAT2 phosphorylation (Mazzon and others 2009). On the other hand, Ashour and others (2009) demonstrated that interaction of NS5 with STAT2 was sufficient to degrade STAT2 through a mechanism that depends on its ubiquitination and proteasome complex activity, and therefore on inhibition of the IFN-I response (Fig. 2). These authors also found that the processed NS5 form resulting from viral protease cleavage (when all the nonstructural proteins were expressed simultaneously) was capable of stimulating STAT2 degradation through the proteasome activity (Ashour and others 2009). It has also been shown that the cellular protein, ubiquitin protein ligase E3 component recognin 4 (UBR4), interacts with the viral protein, NS5, mediating the mechanism whereby STAT2 is degraded by the proteasome, thus acting as a key mediator that allows NS5 binding and degradation of STAT2 (Morrison and others 2013). Moreover, the presence of UBR4 is necessary for STAT2 degradation and for viral replication in IFN-I-producing cells (Morrison and others 2013). As a result, DENV-mediated STAT2 degradation is species specific since it has been demonstrated that NS5 is able to bind and degrade human STAT2, but not mouse STAT2 (Ashour and others 2010). This becomes important because STAT2 restricts DENV replication in the first moments of mice infection in vivo (Ashour and others 2010), and even mediates innate immunity in the absence of STAT1 through type I IFN in mice (Perry and others 2011).
All these studies have reported that DENV inhibits INF-I by different mechanisms and with different results. On some occasions these results were contradictory. It is most likely that these differences are due to different experimental approaches, expression systems, and cellular models. Nevertheless, it is clear that the results obtained to date allow us to conclude that DENV inhibits the IFN-I response by blocking STAT1 phosphorylation and inducing STAT2 degradation or by altering the function of essential components of the IFN-I signaling pathway. Moreover, it is probable that the mechanism evolved by DENV to evade the innate immune response mediated by IFN-I is a more complex process than reported, in which the nonstructural proteins do not act as individual proteins, but rather as multiprotein complexes with each protein playing a specific role.
Conclusions and Perspectives
The innate immune response is a nonspecific and early response that allows the host to control viral infection and dispersion during the first hours of infection. In addition, innate immunity is essential to establish a proper adaptive immune response. It is well accepted that among the components of the different innate immune responses, IFN-I is the main factor responsible for protecting the host from viral infections, restricting viral replication. Nevertheless, many viruses have also evolved different strategies to evade IFN-I-mediated antiviral activity either by limiting its production or blocking and altering its signaling pathway in infected cells. In the specific case of DENV, and as discussed above, it was demonstrated that the virus can not only inhibit IFN-I production but also the signaling of this cytokine, allowing the virus to efficiently replicate in IFN-I-producing cells, such as DCs.
The direct implications of this immune evasion mechanism in DENV pathogenesis have so far been poorly explored. This is mainly because the majority of experimental approaches have been conducted with reference DENV strains or with the genetic sequences of reference strains. Moreover, the few times clinical studies were performed, inconclusive results were obtained. In 2009, a phenotypic study (Takhampunya and others 2009) was performed on 18 DENV clinical isolates from children in Bangkok, Thailand, who presented dengue fever and dengue hemorrhagic fever. The authors evaluated the ability of the strains to inhibit IFN-I response by blocking phosphorylation of STAT1 and found that the majority of the strains blocked IFN-I signaling, but they found no differences in the phenotypic characteristics between strains producing dengue fever and strains producing dengue hemorrhagic fever (Takhampunya and others 2009). Despite the fact that DENV replicates in the cytoplasm, it has been reported that NS5 is located in the nucleus of infected cells; however, this localization of NS5 is not related to DENV replication efficiency or to the inhibition ability of the IFN-I activity (Rawlinson and others 2009; Kumar and others 2013).
In recent years, microRNAs (miRNA) that are major genetic expression regulators of eukaryotic organisms have gained special attention due to the possibility that they may function as alternatives against viral infections. In particular, several miRNAs have been described with antiviral activity against viruses, such as human immunodeficiency virus (HIV), Vesicular stomatitis virus, and Influenza virus (Otsuka and others 2007; Ahluwalia and others 2008; Song and others 2010). DENV could be another such virus since it has been reported that genetically engineered viruses, meaning those that have miRNA target sequences, are highly susceptible to miRNA regulation and thus dramatically downregulate viral replication (Lee and others 2010). Moreover, cellular miRNAs, specifically miRNA-146a, enhance DENV-2 replication in human cells (Wu and others 2013). More interesting is the fact that the mechanism whereby this miRNA enhanced DENV replication was through inhibition of IFN-β production. This was accomplished by the recognition and interaction of miRNA-146a with target sequences present in the 3′UTR sequence of TNF-receptor-associated factor 6 (TRAF6), a signaling molecule involved in TLR3/7/8 and RIG-I/MDA-5 signaling, and therefore drastically downregulating expression of TRAF6 (Wu and others 2013) (Fig. 2). This evidence provides new data about how DENV manipulates several cell factors for its own benefit; in this case, cellular miRNAs encoded by the host help DENV evade the antiviral activity of IFN-I.
Likewise, some viruses indirectly block IFN-I signaling, through the upregulation of the expression of the suppressor of cytokine signaling (SOCS) cellular proteins that constitute a protein family with great capacity to regulate the JAK/STAT signaling pathway. For example, the HSV and Influenza A virus upregulate SOCS-1 and SOCS-3, thereby blocking the antiviral effect of IFN-I (Pauli and others 2008; Frey and others 2009; Jia and others 2010). Although this is a very interesting observation and although DENV is known to block the IFN-I response at different levels, SOCS regulation has not yet been studied in the case of DENV infection.
In conclusion, DENV evades the innate immune response mediated mainly by IFN-I. This immune evasion allows the virus to infect and replicate in several IFN-I-producing cells that play an important role in viral pathogenesis and host immune response such as in DCs and monocytes. A complete understanding of the innate immune evasion observed in DENV infection is necessary for the development of antiviral therapies that could be used in DENV-infected patients. Furthermore, a thorough knowledge of all the elements involved in IFN-I inhibition would allow the development of suitable animal models to study DENV infection.
Acknowledgments
The authors thank Anne-Lise Haenni for critically reviewing the manuscript and encouragement. This study was supported by COLCIENCIAS, grant number 111551928777, and Universidad de Antioquia (Estrategia de sostenibilidad 2014–2015).
Author Disclosure Statement
No competing financial interests exist.
References
- Aguirre S, Maestre AM, Pagni S, Patel JR, Savage T, Gutman D, Maringer K, Bernal-Rubio D, Shabman RS, Simon V, Rodriguez-Madoz J, Mulder LCF, Barber GN, Fernandez-Sesma A. 2012. DENV inhibits type I IFN production in infected cells by cleaving human STING. PLoS Pathog 8(10):1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahluwalia JK, Khan SZ, Soni K, Rawat P, Gupta A, Hariharan M, Scaria V, Lalwani M, Pillai B, Mitra D, Brahmachari S. 2008. Human cellular microRNA hsa-miR-29a interferes with viral nef protein expression and HIV-1 replication. Retrovirology 5:117–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akira S. 2009. Pathogen recognition by innate immunity and its signaling. Proc Jpn Acad Ser B Phys Biol Sci 85:143–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alagarasu K, Honap T, Damle IM, Mulay AP, Shah PS, Cecilia D. 2013. Polymorphisms in the oligoadenylate synthetase gene cluster and its association with clinical outcomes of dengue virus infection. Infect Genet Evol 14:390–395 [DOI] [PubMed] [Google Scholar]
- Angleró-Rodríguez YI, Pantoja P, Sariol CA. 2014. Dengue virus subverts the interferon induction pathway via NS2B/3 protease-IκB kinase epsilon interaction. Clin Vaccine Immunol 21(1):29–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashour J, Laurent-Rolle M, Shi PY, García-Sastre A. 2009. NS5 of dengue virus mediates STAT2 binding and degradation. J Virol 83(11):5408–5418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashour J, Morrison J, Laurent-Rolle M, Belicha-Villanueva A, Plumlee CR, Bernal-Rubio D, Williams K, Harris E, Fernandez-Sesma A, Schindler C, Garcia-Sastre A. 2010. Mouse STAT2 restricts early dengue virus replication. Cell Host Microbe 8(5):410–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beutler BA. 2009. TLRs and innate immunity. Blood 113:1399–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, Myers M, George DB, Jaenisch T, Wint GRW, Simmons C, Scott TW, Farrar JJ, Hay SI. 2013. The global distribution and burden of dengue. Nature 496:504–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidet K, Dadlani D, Garcia-Blanco MA. 2014. G3BP1, G3BP2 and CAPRIN1 are required for translation of interferon stimulated mRNAs and are targeted by a dengue virus non-coding RNA. PLoS Pathog 10(7):1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boo KH, Yang JS. 2010. Intrinsic cellular defenses against virus infection by antiviral type I interferon. Yonsei Med J 51(1):9–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrow P, Martínez-Sobrido L, de la Torre JC. 2010. Inhibition of type I interferon response during arenavirus infection. Viruses 2(11):2443–2480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelli J, Wood KA, Youle RJ. 1998. The 2-5A system in viral and apoptosis. Biomed Pharmacother 52(9):386–390 [DOI] [PubMed] [Google Scholar]
- Chee AV, Roizman B. 2004. Herpes simplex virus 1 gene products occlude the interferon signaling pathway at multiple sites. J Virol 78(8):4185–4196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HW, King K, Tu J, Sanchez M, Luster AD, Shresta S. 2013a. The roles of IRF-3 and IRF-7 in innate antiviral immunity against dengue virus. J Immunol 191:4194–4201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Wu M, Zhang X, Zhang W, Zhang Z, Chen L, He J, Zheng Y, Chen C, Wang F, Hu Y, Zhou X, Wang C, Xu Y, Lu M, Yuan Z. 2013b. Hepatitis B virus polymerase impairs interferon-α-induced STAT activation through inhibition of importin-α5 and protein kinase C-δ. Hepatology 57(2):470–482 [DOI] [PubMed] [Google Scholar]
- Childs K, Stock N, Ross C, Andrejeva J, Hilton L, Skinner M, Randall R, Goodbourn S. 2007. mda-5, but not RIG-I, is a common target for paramyxovirus V proteins. Virology 359:190–200 [DOI] [PubMed] [Google Scholar]
- da Conceição TM, Rust NM, Berbel AC, Martins NB, do Nascimiento Santos CA, da Poian AT, de Arruda LB. 2013. Essential role of RIG-I in the activation of endothelial cells by dengue virus. Virology 435(2):281–292 [DOI] [PubMed] [Google Scholar]
- de Veer MJ, Holko M, Frevel M, Walker E, Der S, Paranjape JM, Silverman RH, Williams BR. 2001. Functional classification of interferon-stimulated genes identified using microarrays. J Leukoc Biol 69(6):912–920 [PubMed] [Google Scholar]
- den Boon JA, Diaz A, Ahlquist P. 2010. Cytoplasmic viral replication complexes. Cell Host Microbe 8:77–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Der SD, Zhou A, Williams BR, Silverman RH. 1998. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc Natl Acad Sci U S A 95:15623–15628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond MS. 2003. Evasion of innate and adaptive immunity by flaviviruses. Immunol Cell Biol 81:196–206 [DOI] [PubMed] [Google Scholar]
- Diamond MS, Harris E. 2001. Interferon inhibits dengue virus infection by preventing translation of viral RNA through a PKR-independent mechanism. Virology 289(2):297–311 [DOI] [PubMed] [Google Scholar]
- Diamond MS, Roberts TG, Edgil D, Lu B, Ernst J, Harris E. 2000. Modulation of Dengue virus infection in human cells by alpha, beta, and gamma interferons. J Virol 74(11):4957–4966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott J, Lynch OT, Suessmuth Y, Qian P, Boyd CR, Burrows JF, Buick R, Stevenson NJ, Touzelet O, Gadina M, Power U, Johnston JA. 2007. Respiratory syncytial virus NS1 protein degrades STAT2 by using the Elongin-Cullin E3 ligase. J Virol 81(7):3428–3436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredericksen BL, Gale M. 2006. West Nile virus evades activation of interferon regulatory factor 3 through RIG-I-dependent and -independent pathways without antagonizing host defense signaling. J Virol 80(6):2913–2923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey KG, Ahmed CM, Dabelic R, Jager LD, Noon-Song EN, Haider SM, Johnson HM, Bigley NJ. 2009. HSV-1-induced SOCS-1 expression in keratinocytes: use of a SOCS-1 antagonist to block a novel mechanism of viral immune evasion. J Immunol 183(2):1253–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieman M, Yount B, Heise M, Kopecky-Bromberg SA, Palese P, Baric RS. 2007. Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane. J Virol 81(18):9812–9824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Sastre A, Egorov A, Matassov D, Brandt S, Levy DE, Durbin JE, Palese P, Muster T. 1998. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 252:324–330 [DOI] [PubMed] [Google Scholar]
- Graff JW, Ewen J, Ettayebi K, Hardy ME. 2007. Zinc-binding domain of rotavirus NSP1 is required for proteasome-dependent degradation of IRF3 and autoregulatory NSP1 stability. J Gen Virol 88:613–620 [DOI] [PubMed] [Google Scholar]
- Halstead SB. 2007. Dengue. Lancet 370:1644–1652 [DOI] [PubMed] [Google Scholar]
- He B. 2006. Viruses, endoplasmic reticulum stress, and interferon responses. Cell Death Differ 13:393–403 [DOI] [PubMed] [Google Scholar]
- Hershey JW. 1991. Translational control in mammalian cells. Annu Rev Biochem 60:717–755 [DOI] [PubMed] [Google Scholar]
- Jia D, Rahbar R, Chan RW, Lee SM, Chan MC, Wang BX, Baker DP, Sun B, Peiris JS, Nicholls JM, Fish EN. 2010. Influenza virus non-structural protein 1 (NS1) disrupts interferon signaling. PLoS One 5(11):1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M, Davidson A, Hibbert L, Gruenwald P, Schlaak J, Ball S, Foster GR, Jacobs M. 2005. Dengue virus inhibits alpha interferon signaling by reducing STAT2 expression. J Virol 79(9):5414–5420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Kawai T, Uematsu S, Takeuchi O, Akira S. 2005. Viral recognition and type I interferon production by Toll-like receptor and an RNA helicase. RIG-I. Int Congr Ser 1285:10–14 [Google Scholar]
- Kotenko SV. 2011. IFN-λs. Curr Opin Immunol 23(5):583–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Bühler S, Selisko B, Davidson A, Mulder K, Canard B, Miller S, Bartenschlager R. 2013. Nuclear localization of dengue virus nonstructural protein 5 does not strictly correlate with efficient viral RNA replication and inhibition of type I interferon signaling. J Virol 87(8):4545–4557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar H, Kawai T, Akira S. 2009. Toll-like receptors and innate immunity. Biochem Biophys Res Commun 388:621–625 [DOI] [PubMed] [Google Scholar]
- Le Bon A, Tough DF. 2002. Links between innate and adaptive immunity via type I interferon. Curr Opin Immunol 14:432–436 [DOI] [PubMed] [Google Scholar]
- Lee KG, Xu S, Kang ZH, Huo J, Huang M, Liu D, Takeuchi O, Akira S, Lam KP. 2012. Bruton's tyrosine kinase phosphorylates Toll-like receptor 3 to initiate antiviral response. Proc Natl Acad Sci U S A 109(15):5791–5796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TC, Lin YL, Liao JT, Su CM, Lin CC, Lin WP, Liao CL. 2010. Utilizing liver-specific microRNA-122 to modulate replication of dengue virus replicon. Biochem Biophys Res Commun 396:596–601 [DOI] [PubMed] [Google Scholar]
- Lefort S, Gravel A, Flamand L. 2010. Repression of interferon-α stimulated genes expression by Kaposi's sarcoma-associated herpesvirus K-bZIP protein. Virology 408(1):14–30 [DOI] [PubMed] [Google Scholar]
- Li Y, Xie J, Wu S, Xia J, Zhang P, Liu C, Zhang P, Huang X. 2013. Protein kinase regulated by dsRNA downregulates the interferon production in dengue virus-and dsRNA-stimulated human lung epithelial cells. PLos One 8(1):1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z, Wu S, Li Y, He L, Wu M, Jiang L, Feng L, Zhang P, Huang X. 2011. Activation of Toll-like receptor 3 impairs the dengue virus serotype 2 replication through induction of IFN-β in cultured hepatoma cells. PLoS One 6(8):1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin RJ, Chang BL, Yu HP, Liao CL, Lin YL. 2006. Blocking of interferon-induced Jak-Stat signaling by Japanese encephalitis virus NS5 through a protein tyrosine phosphatase-mediated mechanism. J Virol 80(12):5908–5918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malet H, Massé N, Selisko B, Romette JL, Alvarez K, Guillemot JC, Tolou H, Yap TL, Vasudevan S, Lescar J, Canard B. 2008. The flavivirus polymerase as a target for drug discovery. Antiviral Res 80(1):23–35 [DOI] [PubMed] [Google Scholar]
- Mazzon M, Jones M, Davidson A, Chain B, Jacobs M. 2009. Dengue virus NS5 inhibits interferon-alpha signaling by blocking signal transducer and activator of transcription 2 phosphorylation. J Infect Dis 200(8):1261–1270 [DOI] [PubMed] [Google Scholar]
- Medzhitov R. 2001. Toll-like receptors and innate immunity. Nat Rev Immunol 1(2):135–145 [DOI] [PubMed] [Google Scholar]
- Melroe GT, Silva L, Schaffer PA, Knipe DM. 2007. Recruitment of activated IRF-3 and CBP/p300 to herpes simplex virus ICP0 nuclear foci: potential role in blocking IFN-beta induction. Virology 360(2):305–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S, Kastner S, Krijnse-Locker J, Buhler S, Bartenschlager R. 2007. The non-structural protein 4A of dengue virus is an integral membrane protein inducing membrane alterations in a 2K- regulated manner. J Biol Chem 282:8873–8882 [DOI] [PubMed] [Google Scholar]
- Morrison J, Laurent-Rolle M, Maestre AM, Rajsbaum R, Pisanelli G, Simon V, Mulder LC, Fernandez-Sesma A, García-Sastre A. 2013. Dengue virus co-opts UBR4 to degrade STAT2 and antagonize type I interferon signaling. PLoS Pathog 9(3):1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S, Kuhn RJ, Rossmann MG. 2005. A structural perspective of the flavivirus life cycle. Nat Rev Microbiol 3:13–22 [DOI] [PubMed] [Google Scholar]
- Muñoz-Jordán JL, Fredericksen BL. 2010. How flaviviruses activate and suppress the interferon response. Viruses 2(2):676–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Jordán JL, Laurent-Rolle M, Ashour J, Martínez-Sobrido L, Ashok M, Lipkin WI, García-Sastre A. 2005. Inhibition of alpha/beta interferon signaling by the NS4B protein of flaviviruses. J Virol 79(13):8004–8013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Jordan JL, Sánchez-Burgos GG, Laurent-Rolle M, García-Sastre A. 2003. Inhibition of interferon signaling by dengue virus. Proc Natl Acad Sci U S A 100(24):14333–14338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasirudeen AM, Wong HH, Thien P, Xu S, Lam KP, Liu DX. 2011. RIG-I, MDA5 and TLR3 synergistically play an important role in restriction of dengue virus infection. PLoS Negl Trop Dis 5(1):1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro-Sánchez E, Desprès P, Cedillo-Barrón L. 2005. Innate immune responses to dengue virus. Arch Med Res 36:425–435 [DOI] [PubMed] [Google Scholar]
- Olagnier D, Scholte FE, Chiang C, Albulescu IC, Nichols C, He Z, Lin R, Snijder EJ, van Hemert MJ, Hiscott J. 2014. Inhibition of dengue and chikungunya virus infections by RIG-I-mediated type I interferon-independent stimulation of the innate antiviral response. J Virol 88(8):4180–4194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka M, Jing Q, Georgel P, New L, Chen J, Mols J, Kang YJ, Jiang Z, Du X, Cook R, Das SC, Pattnaik AK, Beutler B, Han J. 2007. Hypersusceptibility to vesicular stomatitis virus infection in Dicer1-deficient mice is due to impaired miR24 and miR93 expression. Immunity 27:123–134 [DOI] [PubMed] [Google Scholar]
- Pauli EK, Schmolke M, Wolff T, Viemann D, Roth J, Bode JG, Ludwig S. 2008. Influenza A virus inhibits type I IFN signaling via NF-kappaB-dependent induction of SOCS-3 expression. PLoS Pathog 4(11):1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera R, Kuhn RJ. 2008. Structural proteomics of dengue virus. Curr Opin Microbiol 11(4):369–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry S, Buck M, Lada S, Schindler C, Shresta S. 2011. STAT2 mediates innate immunity to dengue virus in the absence of STAT1 via the type I interferon receptor. PLoS Pathog 7(2):1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierangeli A, Degener AM, Ferreri ML, Riva E, Rizzo B, Turriziani O, Luciani S, Scagnolari C, Antonelli G. 2011. Interferon-induced gene expression in cervical mucosa during human papillomavirus infection. Int J Immunopathol Pharmacol 24(1):217–223 [DOI] [PubMed] [Google Scholar]
- Qashqari H, Al-Mars A, Chaudhary A, Abuzenadah A, Damanhouri G, Alqahtani M, Mahmoud M, El Sayed Zaki M, Fatima K, Qadri I. 2013. Understanding the molecular mechanism(s) of hepatitis C virus (HCV) induced interferon resistance. Infect Genet Evol 19:113–119 [DOI] [PubMed] [Google Scholar]
- Qin CF, Zhao H, Liu ZY, Jiang T, Deng YQ, Yu XD, Yu M, Qin ED. 2011. Retinoic acid inducible gene-I and melanoma differentiation-associated gene 5 are induced but not essential for dengue virus induced type I interferon response. Mol Biol Rep 38(6):3867–3873 [DOI] [PubMed] [Google Scholar]
- Randall RE, Goodbourn S. 2008. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J Gen Virol 89(Pt 1):1–47 [DOI] [PubMed] [Google Scholar]
- Rawlinson SM, Pryor MJ, Wright PJ, Jans DA. 2009. CRM1-mediated nuclear export of dengue virus RNA polymerase NS5 modulates interleukin-8 induction and virus production. J Biol Chem 284(23):15589–15597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Madoz JR, Bernal-Rubio D, Kaminski D, Boyd K, Fernandez-Sesma A. 2010a. Dengue virus inhibits the production of type I interferon in primary human dendritic cells. J Virol 84(9):4845–4850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Madoz JR, Belicha-Villanueva A, Bernal-Rubio D, Ashour J, Ayllon J, Fernandez-Sesma A. 2010b. Inhibition of the type I interferon response in human dendritic cells by dengue virus infection requires a catalytically active NS2B3 complex. J Virol 84(19):9760–9774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronco LV, Karpova AY, Vidal M, Howley PM. 1998. Human papillomavirus 16 E6 oncoprotein binds to interferon regulatory factor-3 and inhibits its transcriptional activity. Genes Dev 12(13):2061–2072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronni T, Matikainen S, Sareneva T, Melen K, Pirhonen J, Keskinen P, Julkunen I. 1997. Regulation of IFN-alpha/beta, MxA, 2′,5′-oligoadenylate synthetase, and HLA gene expression in influenza A-infected human lung epithelial cells. J Immunol 158(5):2363–2374 [PubMed] [Google Scholar]
- Samuel CE. 2001. Antiviral actions of interferons. Clin Microbiol Rev 14(4):778–809, table of contents [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sariol CA, Martínez MI, Rivera F, Rodríguez IV, Pantoja P, Abel K, Arana T, Giavedoni L, Hodara V, White LJ, Anglero YI, Montaner LJ, Kraiselburd 2011. Decreased dengue replication and an increased anti-viral humoral response with the use of combined Toll-like receptor 3 and 7/8 agonists in macaques. PLoS One 6(4):1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sariol CA, Muñoz-Jordan JL, Abel K, Rosado LC, Pantoja P, Giavedoni L, Rodriguez IV, White LJ, Martinez M, Arana T, Kraiselburd EN. 2007. Transcriptional activation of interferon-stimulated genes but not of cytokine genes after primary infection of rhesus macaques with dengue virus type 1. Clin Vaccine Immunol 14(6):756–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons CP, Farrar JJ, Nguyen v, Wills B. 2012. Dengue. N Engl J Med 366:1423–1432 [DOI] [PubMed] [Google Scholar]
- Song L, Liu H, Gao S, Jiang W, Huang W. 2010. Cellular microRNAs inhibit replication of the H1N1 influenza A virus in infected cells. J Virol 84(17):8849–8860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun P, Kochel TJ. 2013. The battle between infection and host immune responses of dengue virus and its implication in dengue disease pathogenesis. ScientificWorldJournal 2013:843469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takhampunya R, Palmer DR, McClain S, Barvir DA, Lynch J, Jarman RG, Thomas S, Gibbons RV, Burgess TH, Sun P, Kamau E, Putnak R, Zhang C. 2009. Phenotypic analysis of dengue virus isolates associated with dengue fever and dengue hemorrhagic fever for cellular attachment, replication and interferon signaling ability. Virus Res 145:31–38 [DOI] [PubMed] [Google Scholar]
- Thamizhmani R, Vijayachari P. 2014. Association of dengue virus infection susceptibility with polymorphisms of 2′-5′-oligoadenylate synthetase genes: a case-control study. Braz J Infect Dis 18(5):548–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai YT, Chang SY, Lee CN, Kao CL. 2009. Human TLR3 recognizes dengue virus and modulates viral replication in vitro. Cell Microbiol 11(4):604–615 [DOI] [PubMed] [Google Scholar]
- Uchil PD, Satchidanandam V. 2003. Architecture of the flaviviral replication complex. Protease, nuclease, and detergents reveal encasement within double-layered membrane compartments. J Biol Chem 278(27):24388–24398 [DOI] [PubMed] [Google Scholar]
- Urcuqui-Inchima S, Patiño C, Torres S, Haenni AL, Díaz FJ. 2010. Recent developments in understanding dengue virus replication. Adv Virus Res 77:1–39 [DOI] [PubMed] [Google Scholar]
- Wang C, Pflugheber J, Sumpter R, Jr, Sodora DL, Hui D, Sen GC, Gale M., Jr 2003. Alpha interferon induces distinct translational control programs to suppress hepatitis C virus RNA replication. J Virol 77(7):3898–3912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JP, Liu P, Latz E, Golenbock DT, Finberg RW, Libraty DH. 2006. Flavivirus activation of plasmacytoid dendritic cells delineates key elements of TLR7 signaling beyond endosomal recognition. J immunol 177(10):7114–7121 [DOI] [PubMed] [Google Scholar]
- Warke RV, Xhaja K, Martin KJ, Fournier MF, Shaw SK, Brizuela N, de Bosch N, Lapointe D, Ennis FA, Rothman AL, Bosch I. 2003. Dengue virus induces novel changes in gene expression of human umbilical vein endothelial cells. J Virol 77(21):11822–11832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsch C, Zeuzem S. 2009. RNA-binding activity of hepatitis C virus NS4B: a novel target for small molecule inhibitors. Gastroenterology 137:2170–2172 [DOI] [PubMed] [Google Scholar]
- Welsch S, Miller S, Romero-Brey I, Merz A, Bleck CK, Walther P, Fuller SD, Antony C, Krijnse-Locker J, Bartenschlager R. 2009. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe 5(6):365–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead SS, Blaney JE, Durbin AP, Murphy BR. 2007. Prospects for a dengue virus vaccine. Nat Rev Microbiol 5(7):518–528 [DOI] [PubMed] [Google Scholar]
- WHO. 2009. Dengue. Guidelines for Diagnosis, Treatment, Prevention and Control [PubMed]
- Wu S, He L, Li Y, Wang T, Feng L, Jiang L, Zhang P, Huang X. 2013. miR-146a facilitates replication of dengue virus by dampening interferon induction by targeting TRAF6. J Infect 67(4):329–341 [DOI] [PubMed] [Google Scholar]
- Yokota S, Saito H, Kubota T, Yokosawa N, Amano K, Fujii N. 2003. Measles virus suppresses interferon-alpha signaling pathway: suppression of Jak1 phosphorylation and association of viral accessory proteins, C and V, with interferon-alpha receptor complex. Virology 306(1):135–146 [DOI] [PubMed] [Google Scholar]
- Yu CY, Chang TH, Liang JJ, Chiang RL, Lee YL, Liao CL, Lin YL. 2012. Dengue virus targets the adaptor protein MITA to subvert host innate immunity. PLoS Pathog 8(6):1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu IM, Zhang W, Holdaway HA, Li L, Kostyuchenko VA, Chipman PR, Kuhn RJ, Rossmann MG, Chen J. 2008. Structure of the immature dengue virus at low pH primes proteolytic maturation. Science 319:1834–1837 [DOI] [PubMed] [Google Scholar]
- Zhang J, Hu MM, Wang YY, Shu HB. 2012. TRIM32 protein modulates type I interferon induction and cellular antiviral response by targeting MITA/STING protein for K63-linked ubiquitination. J Biol Chem 287(34):28646–28655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Sun Y, Chen H, Dai Y, Zhan Y, Yu S, Qiu X, Tan L, Song C, Ding C. 2014. Activation of the PKR/eIF2α signaling cascade replication of Newcastle disease virus. Virol J 11:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong B, Yang Y, Li S, Wang YY, Li Y, Diao F, Lei C, He X, Zhang L, Tien P, Shu HB. 2008. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity 29(4):538–550 [DOI] [PubMed] [Google Scholar]