Abstract
Glycogen synthase kinase-3 (GSK-3) has perplexed signal transduction researchers since its detection in skeletal muscle 25 years ago. The enzyme confounds most of the rules normally associated with protein kinases in that it exhibits significant activity, even in resting, unstimulated cells. However, the protein is highly regulated and is potently inactivated in response to signals such as insulin and polypeptide growth factors. The enzyme also displays a distinct and unusual preference for substrates that have been previously phosphorylated by other protein kinases which provides obvious opportunities for cross-talk. It’s substrates are diverse and are predominantly interesting regulatory molecules. The molecular cloning of the kinase revealed it to be encoded by two related but distinct genes. Moreover, the mammalian proteins showed remarkable similarity to a fruitfly protein isolated on the basis of its role in cell fate determination. From these humble beginnings, study of the enzyme has accrued further surprises such as its inhibition by lithium, its regulation by serine and tyrosine phosphorylation and its implication in several human disorders including Alzheimers’ disease, bipolar disorder, cancer and diabetes. Most recently, small molecule inhibitors of GSK-3 have been developed and assessed for therapeutic potential in several of models of pathophysiology. The question is whether modulation of such an “involved” enzyme could lead to selective restoration of defects without multiple unwanted side-effects. This review summarizes current knowledge of GSK-3 with respect to its known functions, together with an assessment of its real-life potential as a drug target for chronic conditions such as type 2 diabetes.
Keywords: diabetes, glycogen synthase, GSK-3, insulin, Wnt signaling, Alzheimers’ disease, glucose transport, protein kinase inhibitors
INSULIN EFFECTS ON GLYCOGEN METABOLISM
Glycogen synthase (GS) is the rate-limiting enzyme in the anabolic pathway leading to glycogen deposition. Typical of regulatory enzymes, GS is the target of several post translational modifications that impact its catalytic activity. The protein is phosphorylated by at least ten different protein kinases in vitro at at least 9 serine residues[1]. These enzymes were catalogued over several years and include cyclic AMP-dependent protein kinase (PKA), phosphorylase kinase, calmodulin-dependent protein kinase II, casein kinases 1 and 2 and a novel enzyme termed glycogen synthase kinase-3 (GSK-3) [1–3]. Most of these are also phosphorylated in intact muscle although several do not change under any conditions. Phosphorylation of GS reduces catalytic efficiency (as measured +/− a cofactor, glucose-6-phosphate). Agents that promote glycogen breakdown such as epinephrine, increase phosphorylation and inactivation of GS [4]. By contrast, insulin promotes glycogen deposition by promoting dephosphorylation and activation of GS. In a classic study, Parker et al., monitored the phosphorylation status of tryptic peptides of GS following its isolation from skeletal muscles from rabbits treated with epinephine or insulin [5]. They found that insulin was selective in dephosphorylating GS; most of the decrease in phosphate was associated with sites that were specifically targeted by GSK-3. GSK-3 activity is decreased following insulin treatment in several insulin-sensitive tissues and over the years, several models have been postulated for the molecular mechanism by which insulin causes dephosphorylation of GS [6–9]. The prevailing model is via signal-dependent inactivation of GSK-3 by phosphorylation (see below). This model also predicts that other substrates of GSK-3 will also be dephosphorylated in response to insulin, along with GS. Several of these are involved in metabolic regulation and may be also important in insulin action: eIF2β [7], inhibitor-2 [10], ATP citrate lyase [11] and insulin receptor substrate 1 (IRS1)[12] (see table 1). GSK-3 has also been implicated in the transcriptional regulation of several insulin-regulated genes such as glucose-6-phosphatase and PEPCK in the liver [13] and in phosphorylation of the C/EBP transcription factor that modulates adipocyte differentiation [14].
Table 1.
Substrates of GSK-3 and the effect of phosphorylation on function (where known)
| Substrate | Cell Function | Effect of GSK-3 | References |
|---|---|---|---|
| Neurofilament H | Neuronal cell function | Unknown | [106] |
| Presenilin 1 (PS1) | Transmembrane protein linked to Alzheimer’s disease; also binds β-catenin | Increases degradation of C-terminal PS1 fragment | [107] |
| Tau | Microtubule-associated protein; stabilizes microtubules | Reduced microtubule binding; decreased microtubule stability | [82] |
| β-catenin | Transcriptional regulator | Targets for ubiquitination | [108] |
| Axin | Scaffold protein in Wnt Pathway | Increased β-catenin binding, increased axin stability | [60–62] |
| Adenomatous Polyposis Coli | Wnt pathway component | Increased β-catenin binding; decreased microtubule binding | [109] |
| Cubitus interuptus | Component of Hedgehog pathway | Targeted for degradation | [103, 104] |
| NF-ATc | Transcription factor; IL2 and other genes | Increased nuclear export | [95] |
| Heat Shock Factor-1 | Transcription factor; regulates genes in response to potentially lethal stressors | Inactivates transcriptional activity | [110] |
| C/EBP | Transcription factor | Interferes with function | [14] |
| c-Jun | Transcription factor; component of activator protein-1 (AP-1) that regulates many diverse genes | Decreased DNA binding and transactivation | [111] |
| c-Myc | Transcription factor; regulates genes involved in cell growth, differentiation and apoptosis | Targets for degradation | [112] |
| cAMP response element binding protein | Transcription factor; regulates cAMP-responsive genes | Increased transcription factor activity | [113] |
| Cyclin D1 | Transcription factor; cell cycle regulation | Promotesnuclear export and ubiquitination | [92] |
| eIF-2B translation factor | Critical for translation initiation | Inhibits translational initiation | [7] |
| Inhibitor-2 | Regulatory subunit of phosphatase | Activates protein phosphatase 1 | [10] |
| Glycogen synthase | Glycogen metabolism | Inhibits enzyme activity | [1] |
| Insulin receptor substrate 1 | Insulin signaling | Inhibits insulin receptor signaling | [3] |
| ATP-citrate Lyase | Fatty acid synthesis | Inactivates enzyme | [11] |
In type 2 diabetes, muscle tissues are less sensitive towards insulin and biopsy studies have shown insulin-dependent activation of GS to be impaired compared with biopsies from normal subjects [15–17]. Similar findings have been observed measuring the flux of glycogen synthesis by NMR spectroscopy [18]. Another potentially important target of GSK-3 involved in insulin signaling is IRS-1 [12]. This protein is one of a family of proteins that are the major substrates of the insulin receptor and become heavily tyrosine phosphorylated following insulin treatment. IRS proteins then act as docking sites for signaling proteins that harbour SH2 domains that recognize phosphotyrosine, including phosphatidylinositol 3′ kinase (see below), Grb-2 and the tyrosine phosphatase SHP2. IRS1 is also phosphorylated on serine and threonine residues. Several protein kinases phosphorylate the protein in vitro including GSK-3 [12]. As for other GSK-3 substrates, phosphorylation of IRS1 has an inhibitory effect on function and reduces the activity of the insulin receptor by an, as yet, unclear mechanism. In muscle and adipose tissues that form the bulk of insulin-sensitive body mass, GSK-3 thus appears to play a selective and important role in inhibiting the functions of several proteins that are activated by insulin.
GSK-3 AND DIABETES
Analysis of various diabetic models has provided evidence for a role for GSK-3 in the disease. In a mouse model of dietary induced obesity in C57Black6 mice that become insulin-insensitive, GSK-3 activity in adipocytes was reported to be double that of animals fed a control diet [19]. Higher levels of GSK-3 activity have also been observed in ob/ob mice compared to lean animals [20]. Measurements of GSK-3 activity in skeletal muscle have also been reported to be elevated compared with non-diabetic patients [21, 22]. While the specific activity of the kinase was similar between the two groups, diabetic patients exhibited a two-fold increase in GSK-3 protein and activity.
Of course, such data are correlative and changes in GSK-3 levels and activity may simply reflect a consequential effect due to physiological adaptation of tissues to the elevated blood glucose and insulin levels that typify type 2 diabetes. The critical question is whether specific modulation of GSK-3 activity can reverse the effects of insulin insensitivity. In this respect, the unusual properties of GSK-3 offer a significant advantage. As proven in practice, it is far easier to develop a small molecule that inhibits an enzyme than one that activates it. If the inability of insulin to inhibit GSK-3 is an important element of type 2 diabetes, then a synthetic GSK-3 inhibitor should hold promise by restoring one of the major effects of insulin action. The first promising data that supported this idea came from experiments with lithium. In 1996, Klein and Melton were investigating the molecular mechanism by which lithium modulated dorsal/ventral axis formation in Xenopus embryo development [23]. Lithium was known to inhibit inositol phosphatase and it was believed that accumulation of inositol phosphate in lithium-treated embryos disrupted the phosphoinositide cycling. However, a potent and selective inhibitor of inositol 1-phosphatase did not phenocopy the effects of lithium on embryonic polarity. Expression of dominant-negative GSK-3 in Xenopus embryos had previously been shown to ventralize the embryos [24]. GSK-3 was then shown to be inhibited by millimolar concentrations of lithium in vitro [23]. Treatment of cells with lithium was then shown to decrease GSK-3 activity as judged by its effect on reducing the phosphorylation of known GSK-3 substrates [25]. While not particularly potent, and certainly not specific (lithium ions alter the activity of many proteins, albeit few protein kinases), lithium offered an easy way to repress GSK-3 activity and therefore to assess its involvement in various cellular processes.
Lithium had, in fact, been previously recognized for its insulin-mimetic effects on liver and fat cells [26–29]. However, as mentioned above, the pleiotropic actions of lithium precluded this as definitive evidence that the insulin-like effects could be ascribed to GSK-3 inhibition. Over the past two years, new small molecules that selectively inhibit GSK-3 have been reported by groups at Glaxo Smith Kline (no relation) and Chiron [30-33]. The Glaxo compounds are maleimides, SB-216763 and SB-415286, that inhibit both forms of GSK-3 with K(i)s of 9 nM and 31 nM respectively but do not significantly interfere with many other protein kinases that were tested [30]. Treatment of isolated human liver cells with the compounds stimulated glycogen synthesis. The Chiron compounds are derivatives of aminopyrimidine, CT98014 and CT98023. These compounds increased glycogen synthase activity in human skeletal muscle cells without affecting glucose transport [32]. In animal studies, however, such as the Zucker diabetic fatty (ZDF) rat model, treatment of isolated soleus and epitrochlearis muscles increased GS activity and oral administration of the drugs resulted in increased insulin-stimulated glucose clearance from the blood [33]. Lithium treatment yielded similar effects. By contrast, a second study using the ZDF model found that although the inhibitors activated GS in both muscle and liver, significant changes in glycogen deposition were found in the liver and this synthesis accounted for most of the glucose clearance [31]. The Chiron compounds enhanced insulin-induced glucose transport in insulin-resistant ZDF rats but had no effect on glucose levels in lean ZDF animals [34]. While these studies differ in some respects, they all demonstrate that in vivo inhibition of GSK-3 leads to activation of glycogen synthase which, depending on the specifics of the study, leads to enhanced glycogen synthesis and better glucose clearance.
Given that the compounds used in these studies are the first to become available, there is likely much room for refinement in pharmacodynamic, potency and selectivity. Even so, the inhibitors have demonstrated proof-of-principle to the idea that inhibition of GSK-3 may be a therapeutic strategy for increasing insulin sensitivity and improving glucose clearance in type 2 diabetes.
OTHER FUNCTIONS OF GSK-3
The nature of type 2 diabetes is such that therapeutic interventions must be chronic and used over a period of many years. Improvement in insulin sensitivity will likely require life-long administration of drugs, following or concomitant with meals. In the case of GSK-3 inhibitors, this raises the issue of the consequences of regular down-regulation of this enzyme. Given the role of GSK-3 in many processes other than insulin signaling, it is important to understand those roles in assessing the possible risks associated with GSK-3 targeted therapy [35–37].
As mentioned above, GSK-3 is a highly conserved enzyme and homologs have been identified in every eukaryote investigated to date [Table 2, 37]. Several species harbor multiple isoforms or related genes but all share high similarity in their protein kinase domain including the presence of a phosphorylated tyrosine in the so-called “T loop” that acts as a gateway for substrate access to the active site. Direct functional conservation has been explicitly demonstrated for several homologs by testing for functional rescue of an inactive mutant by a mammalian cDNA [38, 39]. Micro-injection of a kinase-dead form of GSK-3 into Xenopus embryos mimics the effect of injection of the cognate Xenopus mutant [24]. This high degree of functional conservation has been exploited in analysis of the physiological functions of the kinase in genetically tractable organisms such as fruit flies, yeast and slime mold. For example, study of GSK-3 in Dictyostelium discoideum revealed the first evidence of regulation by a G-protein coupled receptor (GPCR) and cyclic AMP during differentiation of initially identical amoeba into functionally distinct cells of an aggregated slime mold fruiting body [40, 41]. Subsequently, mammalian GSK-3 was shown to be negatively regulated by GPCRs and cyclic AMP [42, 43]. Dictyostelium also has the distinction of the only organism in which the tyrosine kinase responsible for phosphorylation of the T loop has been identified (ZAK1; [44, 45]).
Table 2. GSK-3 genes.
In addition to the species in this table, GSK-3 family members have also identified in other invertebrates (nematode, fruit fly, ascidians, ticks, sea urchin and malaria parasite), plants (Arabidopsis, Hydra, garden petunia, rice, clover, sunflower, alfalfa, chickpea and tobacco) and fungi (baker’s yeast, fission yeast and slime mold). Adapted from [37].
| Species | Name | Identify of catalytic domain to human GSK-3α |
|---|---|---|
|
| ||
| Homo sapiens | GSK-3α | 100 |
| Homo sapiens | GSK-3β | 91 |
| Homo sapiens | GSK-3β2 | 90 |
| Rattus norvegicus | GSK-3α | 99 |
| Rattus norvegicus | GSK-3β | 90 |
| Mus musculus | GSK-3β | 91 |
| Danio rerio | ZGSK-3α | 92 |
| Danio rerio | ZGSK-3β | 90 |
| Xenopus laevis | XGSK-3β | 90 |
| Ciona intestinalis | GSK-3 | 90 |
| Paracentrotus lividus | SUGSK-3 | 84 |
| Drosophila melanogaster | zw3sgg | 82 |
| Hydra vulgaris | GSK-3 | 78 |
| Caenorhabditis elegans | gsk-3 | 80 |
| Petunia hybrida | shaggy | 80 |
| Nicotiana tabacum | shaggy | 71 |
| Arabidopsis thaliana | ASK/ATK1 | 70 |
| Oryza sativa | OSK | 71 |
| Medicago sativa | MSK1-3 | 70–71 |
| Dictyostelium discoideum | gskA | 69 |
| Schizosaccharomyces pombe | Skp1 | 58 |
| Saccharomyces cerevisiae | MCK1/MRK1 | 54–60 |
| Plasmodium flaciparum | GSK-3 | 56 |
| Trifolium repens | GSK-3 | 71 |
| Ricinus communis | shaggy-like | 74 |
| Cicer arietinum | GSK-3 | 73 |
Mammalian GSK-3 was cloned from the rat in 1990 and shown to be the product of two related genes that encoded proteins of 51 and 46 kDa termed GSK-3α and β [46]. The two isoforms have independent activity and do not physically interact although they share similar catalytic and regulatory properties [47]. Both are ubiquitously expressed and the β isoform can be differentially spliced to form a third variant that is expressed in neuronal cells [48].
At the same time the sequences of GSK-3α and β were reported, two Drosophila genetics groups independently published the sequences of a gene implicated in the wingless and notch signaling pathways of the fruitfly termed zeste-white3 and shaggy, respectively [49, 50]. The fly and mammalian proteins shared remarkable identify and were later shown to be functionally replaceable [38, 39]. These findings immediately implicated a role for GSK-3 in the mammalian signaling pathway that is homologous to the fly Wingless pathway, namely the Wnt signaling system.
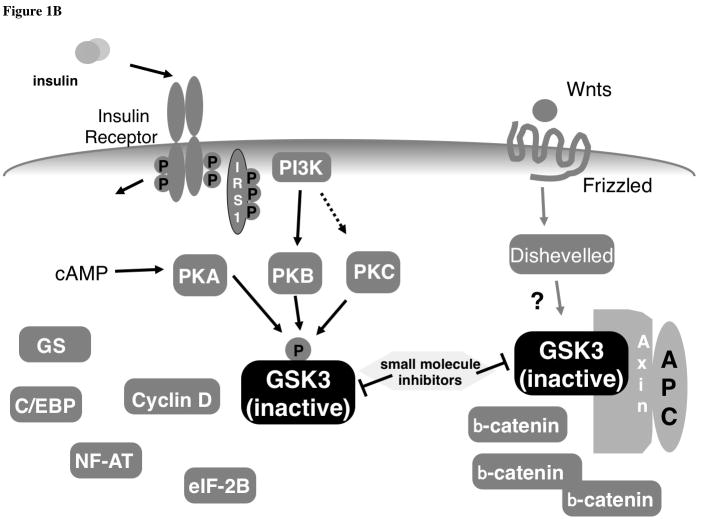
Wnts comprise a family of secreted, glycosylated, protein ligands involved in cell growth, differentiation, migration and fate [51, 52]. Mammals express at least 19 different Wnts and they function in a variety of developmentally regulated processes such as organogenesis and determination of cell fate [51]. The best studied pathway controlled by Wnt molecules is the Wnt/β-catenin system (fig. 3 [53, 54]). Much of the understanding of this pathway originated from genetic analysis of the fly wingless pathway. Binding of Wnt to a serpentine receptor protein of the Frizzled family initiated a cascade of events culminating in the activation of a family of transcriptional regulators termed LEFs or TCFs [55]. The key molecule in this pathway is β-catenin that is tightly controlled at the level of protein degradation. As in the insulin signaling pathway, GSK-3 plays an important inhibitory role in the Wnt pathway. In resting, unstimulated cells, GSK-3 (α or β) phosphorylates β-catenin at several serine residues. When phosphorylated, β-catenin is recognized by an E3 ubiquitin ligase and targeted for ubiquitination and rapid degradation by the 26S proteosome [56]. In the absence of a Wnt signal, cytoplasmic β-catenin levels are therefore kept at very low levels. Binding of Wnt to the Frizzled receptor leads to a reduction in β-catenin phosphorylation by GSK-3 and the subsequent escape of β-catenin from the degradation machinery. The unphosphorylated β-catenin accumulates and interacts with the LEF and TCF DNA binding proteins, promoting transcriptional activation of genes harboring binding sites for these proteins.
Mutations in β-catenin that prevent phosphorylation by GSK-3 resulting in steady-state accumulation have been found in several human cancers of the skin, colon, prostate, liver, endometrium and ovary [54]. The most common mechanism of activation of the pathway involves mutation of another component involved in chaparoning β-catenin between the cytoplasm and nucleus, termed adenomatous polyposis coli (APC). Phosphorylation of β-catenin by GSK-3 only takes place when the two proteins are present in a multi-protein complex, that comprises GSK-3, β-catenin, Axin and APC [58]. APC was first identified as a tumour suppressor gene associated with familial adenomatous polyposis (FAP) and sporadic colorectal cancer [59]. Many patients with colorectal cancer exhibit mutations in APC that result in loss of function. The protein complex is held together through a scaffolding protein termed Axin which harbors several protein-protein interaction domains for GSK-3, APC and β-catenin [58]. Axin and APC are also substrates of GSK-3 with phosphorylation affecting stability and binding affinities [60–62]. The molecular mechanism by which Wnt’s lead to decreased phosphorylation of β-catenin remains unclear although several models have been proposed. Although other proteins are implicated in β-catenin regulation, GSK-3 is a primary player (see below).
Although GSK-3 plays an important role in the Wnt pathway as well as the insulin (and other mitogen) pathway, it is selectively coupled to each agonist such that the consequences of it’s regulation are specific to the agonist. For example, insulin signaling does not impact components of the Wnt signaling pathway. Treatment of cells with insulin does not lead to accumulation of β-catenin. Conversely, Wnt signaling does not affect insulin signaling (e.g. does not activate GS) [63, 64]. The means by which inhibition of the same protein kinase by distinct signaling pathways can allow selective downstream coupling is still not completely understood. In the case of β-catenin regulation, only GSK-3 that is bound to Axin is capable of phosphorylating β-catenin. Axin is present at lower concentrations within cells compared to GSK-3 such that only a fraction of GSK-3 is associated with the scaffold protein. Since only the Axin-associated compartment of GSK-3 is subject to Wnt regulation, this could explain why Wnt doesn’t alter phosphorylation of non-Axin associated substrates of GSK-3. An interesting experiment to test this would be to couple an Axin-binding domain to another GSK-3 substrate and to assess whether this confers Wnt regulation. The reason that insulin does not lead to stabilization of β-catenin is less clear and requires discussion of the molecular mechanism by which GSK-3 is inhibited by insulin.
HOW INSULIN COUPLES TO GSK-3
The catalytic inactivation of GSK-3 induced by insulin and polypeptide mitogens is reversed by treatment with serine/threonine-specific phosphatases indicating that these stimuli cause inhibitory phosphorylation of GSK-3 [65]. The specific residues involved in this inhibition were identified as serines 21 and 9 of GSK-3α and β respectively [66–69]. Both in vitro and in intact cells, these residues are phosphorylated by several protein-serine kinases including protein kinase B/Akt, pp90rsk and cyclic AMP-dependent protein kinase (PKA) [42, 43, 70]. The phosphorylation by PKB is of particular relevance to insulin signaling. Following binding of insulin to its receptor tyrosine kinase, IRS1 becomes highly tyrosine phosphorylated and recruits, among other proteins, phosphatidylinositol 3′ kinase (PI3’K) to the membrane where it encounters is substrate lipids [71]. This results in the membrane-localized generation of 3′ phosphorylated phosphoinositides such as 3,4,5-phosphorylated phosphatidylinositol (PIP3). PIP3 has high affinity for PH domains and its accumulation in the membrane causes translocation of proteins such as PKB and its upstream activating kinase, PDK1, to this cellular location where they interact. The upshot is the activation of PKB by insulin. Once phosphorylated and activated, PKB targets a variety of protein substrates including GSK-3α and β. Polypeptide mitogen-induced inactivation of GSK-3 can be blocked by antagonists of PI3’K such as wortmannin [65] and introduction of activated alleles of PKB into cells stimulates phosphorylation of GSK-3 and its inactivation [72].
The finding that insulin inhibited GSK-3 through phosphorylation of a residue proximal to its N-terminus (i.e. not within its kinase domain) prompted several groups to probe the structural basis for this effect. GSK-3 exhibits and unusual property of preferring to phosphorylate substrates at residues that are just N-terminal to a previously phosphorylate residue [73]. In the case of GS, for example, if completely dephosphorylated, GS is not a substrate for GSK-3 [74]. However, phosphorylation of Ser 657 of human muscle GS by casein kinase-2 facilitates phosphorylation of a serine 4 residues upstream by GSK-3 (Ser 653). This, in turn, allows phosphorylation of Ser 649 that opens up Ser 645 and Ser 641.
637PRPASVPPSPSLSRHSSPHQSEDEED662
The sequence of the inactivating phosphorylation domain of human muscle glycogen synthase. Residues in bold are phosphorylation sites (see text).
Maximal inactivation of GS requires phosphorylation of all four GSK-3 sites (phosphorylation of Ser 657 by casein kinas-2 has little effect on activity). Analysis of other GSK-3 substrates revealed a similar preference for a phosphorylated serine or threonine residue 4 places C-terminal to the GSK-3 target such that the consensus sequence for phosphorylation is Ser-X-X-X-Ser/Thr-P [73]. The intervening residues tend to be enriched for proline but are otherwise quite variable. The protein kinases that provide the “priming” phosphorylation event include casein kinases 1 and 2 and cyclic AMP-dependent protein kinase.
Resolution of the crystal structure of GSK-3β as well as several ingenious biochemical experiments by Frame and Cohen provided insight into how phosphorylation of the N-terminal domain of GSK-3 inhibited its function [75–77]. In resting cells, the N-terminal peptide of GSK-3 is relatively disordered. However, when phosphorylated at Ser 9, this peptide in GSK-3β folds back and forms an electrostatic interaction with Arg 96 which is located in the substrate binding domain of the protein kinase. Normally, this arginine residue plays a key role in allowing substrate entry tot he active site by binding to the phosphate of the priming phosphorylation site residue. Indeed, this binding accounts for the preference of GSK-3 for such pre-phosphorylated proteins. However, if Arg 96 is bound to phospho-Ser 9, the substrate docking site is occluded and GSK-3 cannot bind its substrate protein – resulting in apparent inactivation. At the time of these experiments, phosphorylation of β-catenin was not thought to require priming. This presented a possible explanation for signal-dependent discrimination of GSK-3 targets since if β-catenin could still bind to Ser 9 phosphorylated GSK-3 hence insulin inactivation of the protein kinase would not affect its capacity to phosphorylate β-catenin [78]. Subsequently, however, β-catenin was shown to require priming via phosphorylation of Ser 45 by casein kinase-1 [79,80]. This event allows GSK-3 catalyzed phosphorylation of Thr 41, then Ser 37 and finally Ser 33.
23QQQSYLDSGIHSGATTTAPSLSGK49
Sequence surrounding the phosphorylation domain of human β-catenin. Serine 45 is targeted by casein kinase 1 (priming site). The remaining three bold residues are phosphorylated by GSK-3
Thus, the mechanism that prevents inactivation of Axin-associated GSK-3 in response to insulin treatment is currently unclear. Insulin does increase Ser 9/21 phosphorylation of Axin-bound GSK-3. It is therefore possible that the N-terminal peptide is sterically prevented from interfering with the substrate binding site of GSK-3 through association with another component of the Axin complex – perhaps an Axin mimick of Arg 96 [81]?
YET OTHER FUNCTIONS OF GSK-3
Tau
As shown in Table 1, GSK-3 phosphorylates a panopoly of proteins including transcription factors, metabolic enzymes and structural proteins. The phosphorylation of the microtubule-associated protein Tau is particularly interesting as GSK-3 phosphorylates sites associated with hyperphosphorylated forms of Tau that are found in paired helical filamentous Tau associated with neurofibrilliary tangles in the brains of Alzheimer’s patients [82, reviewed in 83, 84]. A further association with Alzheimer’s disease derives from the finding that Presenilin 1 binds GSK-3 and β-catenin in a similar manner to Axin [85, 86]. Mutations in Presenilin 1 (PS1) are relatively common in patients diagnosed with early-onset, familial Alzheimer’s disease. The protein (along with a related protein, Presenilin 2) plays a role in the processing of the amyloid precursor protein, APP in generating the amyloid plaque-associated protein, β-amyloid.
NF-κB
A novel role for GSK-3β was uncovered following its disruption in the mouse germ-line [87]. Mice lacking both copies of the gene died during embryogenesis due to specific apoptosis of hepatocytes. This phenotype was reminiscent of the effects of disrupting components for the NF-κB signaling pathway, such as the RelA DNA binding protein of NF-κB [88] and I-κB kinase β [89]. Indeed, like these other knockouts, the liver apoptosis could be suppressed by maternal injection of antibodies to tumor necrosis factor-α (TNF-α). NF-κB was originally characterized as an essential component in the regulation of kappa light chain expression in B cells but has subsequently been shown to be a widely expressed protein that is activated by many stress-associated stimuli [reviewed in 90]. In general, the transcriptional targets of NF-κB are associated with survival signals. Mouse embryo fibroblast cells lacking both copies of the GSK-3β gene are insensitive to NF-κB activation in response to inflammatory cytokines such as TNF-α [87]. This cytokine normally induces a mixture of both pro- and anti-apoptotic signals. In the absence of GSK-3β, TNF-α only stimulates the death pathways and so this cytokine promotes apoptosis in GSK-3β null cells. The molecular mechanism by which GSK-3β regulates NF-κB is unclear but likely occurs at the level of the DNA binding subunit since regulation of NF-κB degradation or I-κB degradation is unaffected in GSK-3β mutant cells [87, 91]. The effect on NF-κB presumably reflects a function of GSK-3β that is not shared by GSK-3α. Indeed, these protein kinases share very similar catalytic properties in regulation of substrates such as GS and β-catenin. To date, NF-κB is the only target to demonstrate differential regulation by the two GSK-3 gene products.
Cyclin D1
GSK-3 phosphorylation of β-catenin targets it for ubiquitin-mediated degradation. GSK-3 negatively regulates cyclin D1 by promoting its exclusion from the nucleus [92]. Cyclin D1 positively regulates cyclin-dependent kinase 4 (CDK4) function and expression of the cyclin D1 gene is induced by mitogens and growth factors. By phosphorylating Thr 286 of Cyclin D1, GSK-3 promotes transport of the CDK4 activator from the nucleus to the cytoplasm and thus antagonizes progression of the cell cycle through S-phase [93]. There is also evidence of regulation of the CDK inhibitor, p21CIP, by GSK-3 via phosphorylation of this protein at Thr 57 [94].
NF-AT
Like Cyclin D1, Nuclear Factor of Activated T cells (NF-AT) is also excluded from the nucleus upon phosphorylation by GSK-3 [95, 96]. This protein is an important transcriptional activator and has been most highly characterized with respect to gene inductions occurring following ligation of the T cell receptor (although NF-AT proteins play roles in most tissues) [97]. GSK-3 phosphorylation of this protein is antagonized by calcineurin, a calcium/calmodulin dependent protein phosphatase. The immunosuppressant actions of cyclosporine are largely mediated through inhibition of calcineurin and the subsequent exclusion of NF-AT from the nucleus. Among the target genes for NF-AT is CDK4 [98]. Thus, in resting T cells, GSK-3 plays a double role in suppressing expression of both Cyclin D1 and CDK4.
THE THERAPEUTIC POTENTIAL OF GSK-3 INHIBITORS
The recent availability of small molecule inhibitors to GSK-3 has resulted in several studies that show promising results with respect to activation of glycogen synthesis in models of diabetes [30-34]. These experiments have clearly shown that inhibiting GSK-3 is sufficient to promote glycogen deposition and glucose clearance. The overriding question, however, is whether inhibiting this enzyme will have adverse effects by interfering with other cellular processes controlled by GSK-3. In the case of suppression of Tau phosphorylation, the consequences may be beneficial [99, 100]. However, the remarkable pleiotropism of GSK-3 considerably increases the risk that chronic inhibition may have undesirable effects. The effects of GSK-3β on NF-κB activation could decrease the viability of cells exposed to inflammatory cytokines. This might be avoided by selective inhibition of GSK-3α but this is a tall order for a small molecule inhibitor given the extreme similarity of the catalytic domains of GSK-3α and β. Alternatively, antisense or siRNA approaches may prove useful [101]. The corollary of this approach, of course, there may be specific targets of GSK-3α that, when dysregulated, have deleterious consequences.
Of greater concern, is the effect of inhibiting GSK-3 on growth promotion and oncogenesis [102]. Here, the primary cause for anxiety is β-catenin since mutation of this protein at the GSK-3 phosphorylation sites is sufficient to convert it into an oncogene. Although cells harbor complex mechanisms to effectively insulate the insulin/polypeptide growth factor-dependent regulation of GSK-3 from Wnt regulation, small molecule inhibitors and interfering RNA strategies will not differentiate between the compartments of GSK-3. Hence, all GSK-3 dependent processes will be similarly affected. In addition to β-catenin, the negative effects of GSK-3 on Cyclin D1 and NF-AT may also contribute to anomalous proliferation. GSK-3 has also been implicated in the regulation of the Hedgehog signaling pathway by regulating the degradation of Cubitus interuptus (Ci) [103,104]/ This pathway has been implicated in some human tumours and, like the Wnt pathway, plays an important role in fate determination and organ development during embryogenesis.
The inhibitor studies on the diabetic animals did not assess the effects of long-term exposure or incidence of tumors. It is possible that relatively mild inhibition of GSK-3 that is nonetheless sufficient to re-sensitize liver and muscle tissues to insulin will not invoke growth promoting effects. For example, patients with bipolar disorder are frequently prescribed lithium to suppress episodes of mania and depression, yet are not recognized as having an elevated risk of cancer [105]. However, the levels of lithium in the blood of treated patients are only in the order of 1–1.5 mM. In vitro, this results in less than 5% inhibition of GSK-3 [23]. Mice heterozygous for GSK-3β (that is, expressing 75% of the total GSK-3 activity of wild type animals) are fertile and appear quite normal (at least for the first year of life). Indeed, even embryos lacking both copies of GSK-3β appear grossly normal apart from the hepatocyte defect [87]. This is perhaps surprising given that insulin and Wnt signals result in a transient 50–60% inhibition of GSK-3 and this degree of inhibition is sufficient to trigger the biological effects. The knockout animal data suggest that cells reset their thresholds for GSK-3 activity or that total GSK-3 activity is not limiting until more than 50% is inactivated. In the case of the Wnt pathway, fibroblasts completely lacking GSK-3β show no elevation of β-catenin. However, GSK-3α fully compensates for the GSK-3β void since it also binds Axin and is in molar excess over this scaffolding protein. Therefore, GSK-3β null cells experience no change in their capacity to phosphorylate and degrade β-catenin.
With all of these caveats, the safety profile of GSK-3 inhibitors will likely require exhaustive assessment to convince regulatory agencies that chronic treatment is well tolerated and does not lead to increased chance of tumors. Perhaps a less risky, albeit complicated, strategy for an insulin-sensitizer would be to target the GS/GSK-3 molecular interface since this blocking this interaction appears sufficient for promoting glucose clearance. For the immediate future, the availability of potent reagents that selectively antagonize GSK-3 should result in better understanding of the complex functions of this enigmatic protein kinase – regardless of its ultimate suitability as a therapeutic target.
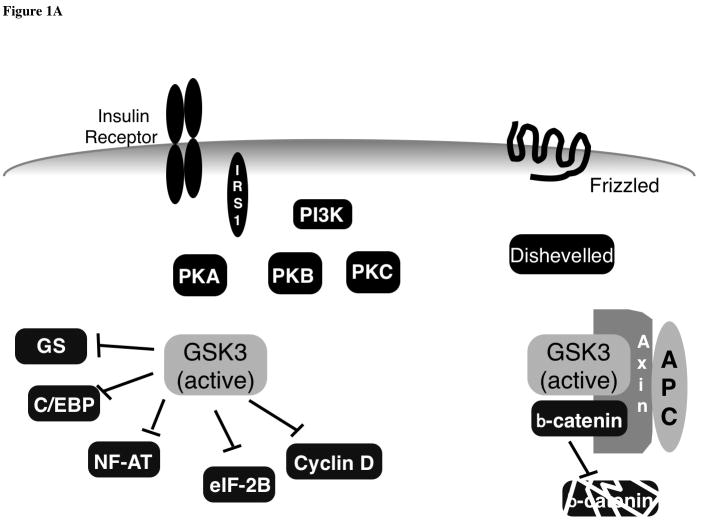
Figure 1.
Acknowledgments
Work in the author’s laboratory is supported by grants from the Canadian Institutes of Health Research, National Cancer Institute of Canada and the Terry Fox Foundation. JRW is a CIHR Senior Scientist and International Scholar of the Howard Hughes Medical Institute.
References
- 1.Cohen P, Yellowlees D, Aitken A, Donella-Deana A, Hemmings BA, Parker PJ. Eur J Biochem. 1982;124(1):21–35. doi: 10.1111/j.1432-1033.1982.tb05902.x. [DOI] [PubMed] [Google Scholar]
- 2.Poulter L, Ang SG, Gibson BW, Williams DH, Holmes CF, Caudwell FB, Pitcher J, Cohen P. Eur J Biochem. 1988;175(3):497–510. doi: 10.1111/j.1432-1033.1988.tb14222.x. [DOI] [PubMed] [Google Scholar]
- 3.Eldar-Finkelman H, Argast GM, Foord O, Fischer EH, Krebs EG. Proc Natl Acad Sci USA. 1996;93(19):10228–10233. doi: 10.1073/pnas.93.19.10228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parker PJ, Embi N, Caudwell FB, Cohen P. Eur J Biochem. 1982;124(1):47–55. doi: 10.1111/j.1432-1033.1982.tb05904.x. [DOI] [PubMed] [Google Scholar]
- 5.Parker PJ, Caudwell FB, Cohen P. Eur J Biochem. 1983;130(1):227–234. doi: 10.1111/j.1432-1033.1983.tb07140.x. [DOI] [PubMed] [Google Scholar]
- 6.Dent P, Lavoinne A, Nakielny S, Caudwell FB, Watt P, Cohen P. Nature. 1990;348(6299):302–308. doi: 10.1038/348302a0. [DOI] [PubMed] [Google Scholar]
- 7.Welsh GI, Proud CG. Biochem J. 1993;316(3):241–246. doi: 10.1016/0014-5793(93)81300-o. [DOI] [PubMed] [Google Scholar]
- 8.Moule SK, Welsh GI, Edgell NJ, Foulstone EJ, Proud CG, Denton RM. J Biol Chem. 1997;272(12):7713–7719. doi: 10.1074/jbc.272.12.7713. [DOI] [PubMed] [Google Scholar]
- 9.Hurel SJ, Rochford JJ, Borthwick AC, Wells AM, Vandenheede JR, Turnbull DM, Yeaman SJ. Biochem J. 1996;320(Pt 3):871–877. doi: 10.1042/bj3200871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Resink TJ, Hemmings BA, Tung HY, Cohen P. Eur J Biochem. 1983;133(2):455–461. doi: 10.1111/j.1432-1033.1983.tb07485.x. [DOI] [PubMed] [Google Scholar]
- 11.Hughes K, Ramakrishna S, Benjamin WB, Woodgett JR. Biochem J. 1992;288(Pt 1):309–314. doi: 10.1042/bj2880309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eldar-Finkelman H, Krebs EG. Proc Natl Acad Sci USA. 1997;94(18):9660–9664. doi: 10.1073/pnas.94.18.9660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lochhead PA, Coghlan M, Rice SQ, Sutherland C. Diabetes. 2001;50(5):937–946. doi: 10.2337/diabetes.50.5.937. [DOI] [PubMed] [Google Scholar]
- 14.Ross SE, Erickson RL, Hemati N, MacDougald OA. Mol Cell Biol. 1999;19(12):8433–8441. doi: 10.1128/mcb.19.12.8433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beck-Nielsen H, Vaag A, Damsbo P, Handberg A, Nielsen OH, Henriksen JE, Thye-Ronn P. Diabetes Care. 1992;15(3):418–429. doi: 10.2337/diacare.15.3.418. [DOI] [PubMed] [Google Scholar]
- 16.Thorburn A, Andrikopoulos S, Proietto J. Metabolism. 1995;44(10):1298–1302. doi: 10.1016/0026-0495(95)90033-0. [DOI] [PubMed] [Google Scholar]
- 17.Henry RR, Ciaraldi TP, Abrams-Carter L, Mudaliar S, Park KS, Nikoulina SE. J Clin Invest. 1996;98(5):1231–1236. doi: 10.1172/JCI118906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petersen KF, Shulman GI. Am J Cardiol. 2002;90(5A):11G–18G. doi: 10.1016/s0002-9149(02)02554-7. [DOI] [PubMed] [Google Scholar]
- 19.Eldar-Finkelman H, Schreyer SA, Shinohara MM, LeBoeuf RC, Krebs EG. Diabetes. 1999;48(8):1662–1666. doi: 10.2337/diabetes.48.8.1662. [DOI] [PubMed] [Google Scholar]
- 20.Kaidanovich O, Eldar-Finkelman H. Expert Opin Ther Targets. 2002;6(5):555–561. doi: 10.1517/14728222.6.5.555. [DOI] [PubMed] [Google Scholar]
- 21.Nikoulina SE, Ciaraldi TP, Mudaliar S, Mohideen P, Carter L, Henry RR. Diabetes. 2000;49(2):263–271. doi: 10.2337/diabetes.49.2.263. [DOI] [PubMed] [Google Scholar]
- 22.Ciaraldi TP, Nikoulina SE, Henry RR. J Diabetes Complications. 2002;16 (1):69–71. doi: 10.1016/s1056-8727(01)00193-3. [DOI] [PubMed] [Google Scholar]
- 23.Klein PS, Melton DA. Proc Natl Acad Sci USA. 1996;93(16):8455–8459. doi: 10.1073/pnas.93.16.8455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He X, Saint-Jeannet JP, Woodgett JR, Varmus HE, Dawid IB. Nature. 1995;374(6523):617–622. doi: 10.1038/374617a0. [DOI] [PubMed] [Google Scholar]
- 25.Stambolic V, Ruel L, Woodgett JR. Curr Biol. 1996;6(12):1664–1668. doi: 10.1016/s0960-9822(02)70790-2. [DOI] [PubMed] [Google Scholar]
- 26.Cheng K, Creacy S, Larner J. Mol Cell Biochem. 1983;56(2):177–182. doi: 10.1007/BF00227218. [DOI] [PubMed] [Google Scholar]
- 27.Orena SJ, Torchia AJ, Garofalo RS. J Biol Chem. 2000;275(21):15765–15772. doi: 10.1074/jbc.M910002199. [DOI] [PubMed] [Google Scholar]
- 28.Nyfeler F, Walter P. FEBS Lett. 1979;108(1):197–199. doi: 10.1016/0014-5793(79)81209-0. [DOI] [PubMed] [Google Scholar]
- 29.Choi WS, Sung CK. Biochim Biophys Acta. 2000;1475(3):225–230. doi: 10.1016/s0304-4165(00)00068-4. [DOI] [PubMed] [Google Scholar]
- 30.Coghlan MP, Culbert AA, Cross DA, Corcoran SL, Yates JW, Pearce NJ, Rausch OL, Murphy GJ, Carter PS, Roxbee Cox L, Mills D, Brown MJ, Haigh D, Ward RW, Smith DG, Murray KJ, Reith AD, Holder JC. Chem Biol. 2000;7(10):793–803. doi: 10.1016/s1074-5521(00)00025-9. [DOI] [PubMed] [Google Scholar]
- 31.Cline GW, Johnson K, Regittnig W, Perret P, Tozzo E, Xiao L, Damico C, Shulman GI. Diabetes. 2002;51(10):2903–2910. doi: 10.2337/diabetes.51.10.2903. [DOI] [PubMed] [Google Scholar]
- 32.Nikoulina SE, Ciaraldi TP, Mudaliar S, Carter L, Johnson K, Henry RR. Diabetes. 2002;51(7):2190–2198. doi: 10.2337/diabetes.51.7.2190. [DOI] [PubMed] [Google Scholar]
- 33.Henriksen EJ, Kinnick TR, Teachey MK, O’Keefe MP, Ring D, Johnson KW, Harrison SD. Am J Physiol Endocrinol Metab. 2003 doi: 10.1152/ajpendo.00346.2002. epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 34.Ring DB, Johnson KW, Henriksen EJ, Nuss JM, Goff D, Kinnick TR, Ma ST, Reeder JW, Samuels I, Slabiak T, Wagman AS, Hammond ME, Harrison SD. Diabetes. 2003;52(3):588–595. doi: 10.2337/diabetes.52.3.588. [DOI] [PubMed] [Google Scholar]
- 35.Martinez A, Castro A, Dorronsoro I, Alonso M. Med Res Rev. 2002;22 (4):373–384. doi: 10.1002/med.10011. [DOI] [PubMed] [Google Scholar]
- 36.Eldar-Finkelman H. Trends Mol Med. 2002;8(3):126–132. doi: 10.1016/s1471-4914(01)02266-3. [DOI] [PubMed] [Google Scholar]
- 37.Ali A, Hoeflich KP, Woodgett JR. Chem Rev. 2001;101(8):2527–2540. doi: 10.1021/cr000110o. [DOI] [PubMed] [Google Scholar]
- 38.Siegfried E, Chou TB, Perrimon N. Cell. 1992;71(7):1167–1179. doi: 10.1016/s0092-8674(05)80065-0. [DOI] [PubMed] [Google Scholar]
- 39.Ruel L, Bourouis M, Heitzler P, Pantesco V, Simpson P. Nature. 1993;362(6420):557–560. doi: 10.1038/362557a0. [DOI] [PubMed] [Google Scholar]
- 40.Harwood AJ, Plyte SE, Woodgett J, Strutt H, Kay RR. Cell. 1995;80 (1):139–148. doi: 10.1016/0092-8674(95)90458-1. [DOI] [PubMed] [Google Scholar]
- 41.Plyte SE, O’Donovan E, Woodgett JR, Harwood AJ. Development. 1999;126(2):325–333. doi: 10.1242/dev.126.2.325. [DOI] [PubMed] [Google Scholar]
- 42.Fang X, Yu SX, Lu Y, Bast RC, Jr, Woodgett JR, Mills GB. Proc Natl Acad Sci USA. 2000;97(22):11960–11965. doi: 10.1073/pnas.220413597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li M, Wang X, Meintzer MK, Laessig T, Birnbaum MJ, Heidenreich KA. Mol Cell Biol. 2000;20(24):9356–9363. doi: 10.1128/mcb.20.24.9356-9363.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim L, Liu J, Kimmel AR. Cell. 1999;99(4):399–408. doi: 10.1016/s0092-8674(00)81526-3. [DOI] [PubMed] [Google Scholar]
- 45.Kim L, Harwood A, Kimmel AR. Dev Cell. 2002;3(4):523–532. doi: 10.1016/s1534-5807(02)00269-1. [DOI] [PubMed] [Google Scholar]
- 46.Woodgett JR. EMBO J. 1990;9(8):2431–2438. doi: 10.1002/j.1460-2075.1990.tb07419.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hughes K, Pulverer BJ, Theocharous P, Woodgett JR. Eur J Biochem. 1992;203(1–2):305–311. doi: 10.1111/j.1432-1033.1992.tb19860.x. [DOI] [PubMed] [Google Scholar]
- 48.Mukai F, Ishiguro K, Sano Y, Fujita SC. J Neurochem. 2002;81(5):1073–1083. doi: 10.1046/j.1471-4159.2002.00918.x. [DOI] [PubMed] [Google Scholar]
- 49.Siegfried E, Perkins LA, Capaci TM, Perrimon N. Nature. 1990;345(6278):825–829. doi: 10.1038/345825a0. [DOI] [PubMed] [Google Scholar]
- 50.Bourouis M, Moore P, Ruel L, Grau Y, Heitzler P, Simpson P. EMBO J. 1990;9(9):2877–2884. doi: 10.1002/j.1460-2075.1990.tb07477.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miller JR. Genome Biol. 2002;3(1):REVIEWS 3001. doi: 10.1186/gb-2001-3-1-reviews3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smalley MJ, Dale TC. J Mammary Gland Biol Neoplasia. 2001;6(1):37–52. doi: 10.1023/a:1009564431268. [DOI] [PubMed] [Google Scholar]
- 53.Seidensticker MJ, Behrens J. Biochim Biophys Acta. 2000;1495(2):168–182. doi: 10.1016/s0167-4889(99)00158-5. [DOI] [PubMed] [Google Scholar]
- 54.Polakis P. Genes Dev. 2000;14(15):1837–1851. [PubMed] [Google Scholar]
- 55.Barker N, Morin PJ, Clevers H. Adv Cancer Res. 2000;77:1–24. doi: 10.1016/s0065-230x(08)60783-6. [DOI] [PubMed] [Google Scholar]
- 56.Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. EMBO J. 1997;16 (13):3797–3804. doi: 10.1093/emboj/16.13.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chan EF, Gat U, McNiff JM, Fuchs E. Nat Genet. 1999;21(4):410–413. doi: 10.1038/7747. [DOI] [PubMed] [Google Scholar]
- 58.Hinoi T, Yamamoto H, Kishida M, Takada S, Kishida S, Kikuchi A. J Biol Chem. 2000;275(44):34399–34406. doi: 10.1074/jbc.M003997200. [DOI] [PubMed] [Google Scholar]
- 59.Polakis P. Biochim Biophys Acta. 1997;1332(1):F127–147. doi: 10.1016/s0304-419x(97)00008-5. [DOI] [PubMed] [Google Scholar]
- 60.Ikeda S, Kishida S, Yamamoto H, Murai H, Koyama S, Kikuchi A. ) EMBO J. 1998;17(5):1371–1384. doi: 10.1093/emboj/17.5.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jho E, Lomvardas S, Costantini F. Biochem Biophys Res Commun. 1999;266(1):28–35. doi: 10.1006/bbrc.1999.1760. [DOI] [PubMed] [Google Scholar]
- 62.Yamamoto H, Kishida S, Kishida M, Ikeda S, Takada S, Kikuchi A. J Biol Chem. 1999;274(16):10681–10684. doi: 10.1074/jbc.274.16.10681. [DOI] [PubMed] [Google Scholar]
- 63.Ding VW, Chen RH, McCormick F. J Biol Chem. 2000;275(42):32475–32481. doi: 10.1074/jbc.M005342200. [DOI] [PubMed] [Google Scholar]
- 64.Yuan H, Mao J, Li L, Wu D. J Biol Chem. 1999;274(43):30419–30423. doi: 10.1074/jbc.274.43.30419. [DOI] [PubMed] [Google Scholar]
- 65.Cross DA, Alessi DR, Vandenheede JR, McDowell HE, Hundal HS, Cohen P. Biochem J. 1994;303(Pt 1):21–26. doi: 10.1042/bj3030021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sutherland C, Leighton IA, Cohen P. Biochem J. 1993;296(Pt 1):15–19. doi: 10.1042/bj2960015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sutherland C, Cohen P. FEBS Lett. 1994;338(1):37–42. doi: 10.1016/0014-5793(94)80112-6. [DOI] [PubMed] [Google Scholar]
- 68.Stambolic V, Woodgett JR. Biochem J. 1994;303(Pt 3):701–704. doi: 10.1042/bj3030701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shaw M, Cohen P, Alessi DR. FEBS Lett. 1997;416(3):307–311. doi: 10.1016/s0014-5793(97)01235-0. [DOI] [PubMed] [Google Scholar]
- 70.Cross DA, Alessi DR, Cohen P, Andjelkovich M, Hemmings BA. Nature. 1995;378(6559):785–789. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- 71.Cantley LC. Science. 2002;296(5573):1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 72.Hajduch E, Alessi DR, Hemmings BA, Hundal HS. Diabetes. 1998;47 (7):1006–1013. doi: 10.2337/diabetes.47.7.1006. [DOI] [PubMed] [Google Scholar]
- 73.Roach PJ. J Biol Chem. 1991;266(22):14139–14142. [PubMed] [Google Scholar]
- 74.Picton C, Woodgett J, Hemmings B, Cohen P. FEBS Lett. 1982;150(1):191–196. doi: 10.1016/0014-5793(82)81332-x. [DOI] [PubMed] [Google Scholar]
- 75.Dajani R, Fraser E, Roe SM, Young N, Good V, Dale TC, Pearl LH. Cell. 2001;105(6):721–732. doi: 10.1016/s0092-8674(01)00374-9. [DOI] [PubMed] [Google Scholar]
- 76.ter Haar E, Coll JT, Austen DA, Hsiao HM, Swenson L, Jain J. Nat Struct Biol. 2001;8(7):593–596. doi: 10.1038/89624. [DOI] [PubMed] [Google Scholar]
- 77.Frame S, Cohen P, Biondi RM. Mol Cell. 2001;7(6):1321–1327. doi: 10.1016/s1097-2765(01)00253-2. [DOI] [PubMed] [Google Scholar]
- 78.Cohen P, Frame S. Nat Rev Mol Cell Biol. 2001;2(10):769–776. doi: 10.1038/35096075. [DOI] [PubMed] [Google Scholar]
- 79.Liu C, Li Y, Semenov M, Han C, Baeg GH, Tan Y, Zhang Z, Lin X, He X. Cell. 2002;108(6):837–847. doi: 10.1016/s0092-8674(02)00685-2. [DOI] [PubMed] [Google Scholar]
- 80.Gao ZH, Seeling JM, Hill V, Yochum A, Virshup DM. Proc Natl Acad Sci USA. 2002;99(3):1182–1187. doi: 10.1073/pnas.032468199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dajani R, Fraser E, Roe SM, Yeo M, Good VM, Thompson V, Dale TC, Pearl LH. EMBO J. 2003;22(3):494–501. doi: 10.1093/emboj/cdg068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hanger DP, Hughes K, ;Woodgett JR, Brion JP, Anderton BH. Neurosci Lett. 1992;147(1):58–62. doi: 10.1016/0304-3940(92)90774-2. [DOI] [PubMed] [Google Scholar]
- 83.Mattson MP. Trends Neurosci. 2001;24(5):255–256. doi: 10.1016/s0166-2236(00)01838-5. [DOI] [PubMed] [Google Scholar]
- 84.Maccioni RB, Munoz JP, Barbeito L. Arch Med Res. 2001;32(5):367–381. doi: 10.1016/s0188-4409(01)00316-2. [DOI] [PubMed] [Google Scholar]
- 85.Nishimura M, Yu G, Levesque G, Zhang DM, Ruel L, Chen F, Milman P, Holmes E, Liang Y, Kawarai T, Jo E, Supala A, Rogaeva E, Xu DM, Janus C, Levesque L, Bi Q, Duthie M, Rozmahel R, Mattila K, Lannfelt L, Westaway D, Mount HT, Woodgett J, St George-Hyslop P. Nat Med. 1999;5(2):164–169. doi: 10.1038/5526. [DOI] [PubMed] [Google Scholar]
- 86.Palacino JJ, Murphy MP, Murayama O, Iwasaki K, Fujiwara M, Takashima A, Golde TE, Wolozin B. J Biol Chem. 2001;276(42):38563–38569. doi: 10.1074/jbc.M105376200. [DOI] [PubMed] [Google Scholar]
- 87.Hoeflich KP, Luo J, Rubie EA, Tsao MS, Jin O, Woodgett JR. Nature. 2000;406(6791):86–89. doi: 10.1038/35017574. [DOI] [PubMed] [Google Scholar]
- 88.Beg AA, Baltimore D. Science. 1996;274(5288):782–784. doi: 10.1126/science.274.5288.782. [DOI] [PubMed] [Google Scholar]
- 89.Li Q, Van Antwerp D, Mercurio F, Lee KF, Verma IM. Science. 1999;284(5412):321–325. doi: 10.1126/science.284.5412.321. [DOI] [PubMed] [Google Scholar]
- 90.Li Q, Verma IM. Nat Rev Immunol. 2002;2(10):725–734. doi: 10.1038/nri910. [DOI] [PubMed] [Google Scholar]
- 91.Schwabe RF, Brenner DA. Am J Physiol Gastrointest Liver Physiol. 2002;283(1):G204–211. doi: 10.1152/ajpgi.00016.2002. [DOI] [PubMed] [Google Scholar]
- 92.Diehl JA, Cheng M, Roussel MF, Sherr CJ. Genes Dev. 1998;12(22):3499–3511. doi: 10.1101/gad.12.22.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Alt JR, Cleveland JL, Hannink M, Diehl JA. Genes Dev. 2000;14(24):3102–3114. doi: 10.1101/gad.854900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rossig L, Badorff C, Holzmann Y, Zeiher AM, Dimmeler S. J Biol Chem. 2002;277(12):9684–9689. doi: 10.1074/jbc.M106157200. [DOI] [PubMed] [Google Scholar]
- 95.Beals CR, Sheridan CM, Turck CW, Gardner P, Crabtree GR. Science. 1997;275(5308):1930–1934. doi: 10.1126/science.275.5308.1930. [DOI] [PubMed] [Google Scholar]
- 96.Sheridan CM, Heist EK, Beals CR, Crabtree GR, Gardner P. J Biol Chem. 2002;277(50):48664–48976. doi: 10.1074/jbc.M207029200. [DOI] [PubMed] [Google Scholar]
- 97.Crabtree GR, Olson EN. Cell. 2002;109(Suppl):S67–79. doi: 10.1016/s0092-8674(02)00699-2. [DOI] [PubMed] [Google Scholar]
- 98.Baksh S, Widlund HR, Frazer-Abel AA, Du J, Fosmire S, Fisher DE, DeCaprio JA, Modiano JF, Burakoff SJ. Mol Cell. 2002;10(5):1071–1081. doi: 10.1016/s1097-2765(02)00701-3. [DOI] [PubMed] [Google Scholar]
- 99.Bhat RV, Budd SL. Neurosignals. 2002;11(5):251–261. doi: 10.1159/000067423. [DOI] [PubMed] [Google Scholar]
- 100.Lahiri DK, Farlow MR, Sambamurti K, Greig NH, Giacobini E, Schneider LS. Curr Drug Targets. 2003;4(2):97–112. doi: 10.2174/1389450033346957. [DOI] [PubMed] [Google Scholar]
- 101.Yu JY, Taylor J, DeRuiter SL, Vojtek AB, Turner DL. Mol Ther. 2003;7(2):228–236. doi: 10.1016/s1525-0016(02)00037-0. [DOI] [PubMed] [Google Scholar]
- 102.Manoukian AS, Woodgett JR. Adv Cancer Res. 2002;84:203–29. doi: 10.1016/s0065-230x(02)84007-6. [DOI] [PubMed] [Google Scholar]
- 103.Price MA, Kalderon D. Cell. 2002;108(6):823–835. doi: 10.1016/s0092-8674(02)00664-5. [DOI] [PubMed] [Google Scholar]
- 104.Jia J, Amanai K, Wang G, Tang J, Wang B, Jiang J. Nature. 2002;416(6880):548–552. doi: 10.1038/nature733. [DOI] [PubMed] [Google Scholar]
- 105.Vestergaard P, Licht RW. World J Biol Psychiatry. 2001;2(1):18–26. doi: 10.3109/15622970109039980. [DOI] [PubMed] [Google Scholar]
- 106.Sasaki T, Taoka M, Ishiguro K, Uchida A, Saito T, Isobe T, Hisanaga S. J Biol Chem. 2002;277(39):36032–36039. doi: 10.1074/jbc.M206674200. [DOI] [PubMed] [Google Scholar]
- 107.Kirschenbaum F, Hsu SC, Cordell B, McCarthy JV. J Biol Chem. 2001;276(33):30701–30707. doi: 10.1074/jbc.M102849200. [DOI] [PubMed] [Google Scholar]
- 108.Yost C, Torres M, Miller JR, Huang E, Kimelman D, Moon RT. Genes Dev. 1996;10(12):1443–1454. doi: 10.1101/gad.10.12.1443. [DOI] [PubMed] [Google Scholar]
- 109.Rubinfeld B, Albert I, Porfiri E, Fiol C, Munemitsu S, Polakis P. Science. 1996;272(5264):1023–1026. doi: 10.1126/science.272.5264.1023. [DOI] [PubMed] [Google Scholar]
- 110.Xavier IJ, Mercier PA, McLoughlin CM, Ali A, Woodgett JR, Ovsenek N. J Biol Chem. 2000;275(37):29147–29152. doi: 10.1074/jbc.M002169200. [DOI] [PubMed] [Google Scholar]
- 111.Boyle WJ, Smeal T, Defize LH, Angel P, Woodgett JR, Karin M, Hunter T. Cell. 1991;64(3):573–584. doi: 10.1016/0092-8674(91)90241-p. [DOI] [PubMed] [Google Scholar]
- 112.Sears R, Nuckolls F, Haura E, Taya Y, Tamai K, Nevins JR. Genes Dev. 2000;14(19):2501–2514. doi: 10.1101/gad.836800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Fiol CJ, Williams JS, Chou CH, Wang QM, Roach PJ, Andrisani OM. J Biol Chem. 1994;269(51):32187–32193. [PubMed] [Google Scholar]