Abstract
Objective
Fibroblast-like synoviocytes (FLS) are key players in the synovial pathology of rheumatoid arthritis (RA). Currently, there is no treatment that specifically targets these aggressive cells. By combining 3 different “omics” data sets, i.e., 1) risk genes in RA, 2) differentially expressed genes, and 3) differential DNA methylation in RA versus osteoarthritis (OA) FLS, we identified LBH (limb bud and heart development) as a candidate gene in RA. The present study was undertaken to define the role of this gene in FLS.
Methods
Synovial tissue specimens from RA and OA patients were collected at the time of joint replacement surgery. LBH expression was silenced using small interfering RNA or overexpressed using an LBH expression vector in primary FLS. Gene expression profiles were determined by microarray and assessed using Ingenuity Pathway Analysis. Effects of modified LBH expression were investigated in functional assays.
Results
LBH was expressed in the synovial lining layer in patients with RA. Transforming growth factor β1 significantly increased LBH expression in primary FLS, and platelet-derived growth factor BB decreased it. Pathway analysis of the transcriptome of LBH-deficient FLS compared to control FLS identified “cellular growth and proliferation” as the most significantly enriched pathway. In growth assays, LBH deficiency increased FLS proliferation. Conversely, LBH overexpression significantly inhibited cell growth. Cell cycle analysis demonstrated a marked increase in cells entering the cell cycle in LBH-deficient FLS compared to controls. LBH did not alter apoptosis.
Conclusion
LBH is a candidate gene for synovial pathology in RA. It is regulated by growth factors and modulates cell growth in primary FLS. Our data suggest a novel mechanism for synovial intimal hyperplasia and joint damage in RA.
Rheumatoid arthritis (RA) is a chronic inflammatory disease that predominantly affects diarthrodial joints. Treatment strategies include traditional disease-modifying antirheumatic drugs, novel small molecule kinase inhibitors, and biologic drugs targeting proin-flammatory cytokines, B cells, or the activation of T cells (1). Despite improved outcomes, many patients do not respond to the available therapies, suggesting that yet other factors or cells must be important.
Fibroblast-like synoviocytes (FLS) are key players in the synovial pathology and joint destruction in RA (2). The pathologic changes of the synovial tissue include synovial infiltration of inflammatory cells, hyperplasia of the synovial intimal lining layer, and formation of pannus tissue. RA FLS display an aggressive phenotype that shares many characteristics with transformed cells, such as increased expression of protoon-cogenes, increased production of proinflammatory cytokines and matrix-degrading enzymes, and increased resistance to apoptosis (3,4). In addition, RA FLS can undergo an epithelial-mesenchymal–like transition (5) and potentially spread the disease to distant joints (6). Consequently, they have emerged as important targets for new treatment strategies.
RA, like many other autoimmune diseases, involves both genetic and environmental factors (7). To date, more than 100 risk genes for development of RA have been identified in genome-wide association studies (GWAS) of single-nucleotide polymorphisms (SNPs) (8). How environment affects disease onset and severity is not known, but could be related to epigenetic alterations. We have recently shown that RA FLS display a striking pattern of differential DNA methylation compared with osteoarthritis (OA) and normal FLS (9). Furthermore, the RA-specific FLS methylation signature is stable and includes many genes and pathways involved in RA pathogenesis (10).
To identify and prioritize possible unanticipated RA therapeutic targets, we performed an integrative analysis of methylome, transcriptome, and sequence variation in RA FLS (11). This work implicated several genes that were present in all 3 databases. In the present study, we evaluated one of those genes, LBH (limb bud and heart development), whose function was essentially unknown. This gene encodes a small, highly conserved protein that is a putative transcriptional coactivator and target of Wnt signaling implicated in embryonic development (12). However, its potential role in autoimmune diseases has not been elucidated.
MATERIALS AND METHODS
Biologic samples
Human synovial tissue specimens were obtained during joint replacement surgery. The procedure was approved by the Human Research Protection Program, and all patients provided written informed consent. The RA patients fulfilled the American College of Rheumatology 1987 revised criteria for the disease (13). Homogeneous cultures of primary FLS were established as described earlier (14) and used at passages 4–7.
Cell culture and stimulation
Primary FLS were cultured (at 5% CO2, 37°C) in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with L-glutamine, gentamicin, penicillin/streptomycin, and 10% heat-inactivated fetal bovine serum (15). For stimulation experiments, cells were plated in 6-well plates, serum starved for 24 hours in 0.1% fetal bovine serum, and stimulated with recombinant proteins (R&D Systems) prior to RNA isolation.
Gene silencing and overexpression
Gene silencing was performed using small interfering RNA (siRNA) (Smart Pool On-Target; Thermo Scientific) that was transfected using Amaxa technology according to the instructions of the manufacturer (Lonza). For overexpression studies, control empty vector or pCMVEntry-LBH vector (OriGene) was transfected as previously described (16). Cells were then plated in 6-well plates and incubated for 1–3 days prior to further analysis. In the functional assays, mean LBH gene silencing was 91% (LBHlow) and the mean change in LBH gene expression was 13-fold (LBHhigh), by quantitative polymerase chain reaction (qPCR).
RNA isolation and gene expression assay
Total RNA was extracted from cells using RNA STAT-60 (Tel-Test) (17). Complementary DNA (cDNA) was synthesized from 250 ng of total RNA using TaqMan reverse transcription reagents, and qPCR was performed using TaqMan reagents and primer/ probe sets for GAPDH, LBH, IL6, and MMP3 (Life Technologies). Ct values were normalized to GAPDH expression using a standard curve methodology to obtain relative cell equivalents, as previously described (17). For microarray studies, 4 different RA FLS cultures were transfected, with 1) control siRNA plus control vector, 2) LBH siRNA plus control vector (LBHlow), or 3) control siRNA plus LBH expression vector (LBHhigh). Total RNA from the 12 samples was isolated using an RNeasy Plus Mini kit (Qiagen) and hybridized to an Illumina human HT-12 v4.0 gene expression bead chip.
Western blot analysis
Serum-starved RA FLS cultures were stimulated for 15 minutes with 50 ng/ml tumor necrosis factor (TNF), and Western blot analysis was performed as previously described (18), using anti–phosphorylated p38, anti-phosphorylated ERK, anti-phosphorylated JNK (all from Cell Signaling Technology), or anti–β-actin antibodies (Sigma), and horseradish peroxidase (HRP)–conjugated goat anti-rabbit IgG (Cell Signaling Technology) as secondary antibody. For analysis of LBH expression, total lysates from 3 RA FLS and 1 FLS line transfected with control siRNA or LBH siRNA were collected and subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis and Western blotting. Membranes were probed with rabbit anti-human LBH (1:1,000; Sigma) and anti–β-actin antibodies. Blots were developed using an Immun-Star WesternC Chemiluminescence kit and analyzed using a VersaDoc imaging system and Quantity One software (Bio-Rad).
Immunohistochemistry
Cryosections (5 µm) of frozen human synovial tissue were fixed in acetone for 10 minutes and rehydrated in Tris buffered saline (TBS). Endogenous peroxidase activity was blocked for 10 minutes with Dual endogenous enzyme block (Dako), followed by a quick rinse and blocking for 10 minutes with 5% normal swine serum (Vector). Incubation for 60 minutes with primary rabbit anti-LBH antibody (1:200; Sigma) or normal rabbit Ig (Dako) in phosphate buffered saline (PBS)/1% bovine serum albumin was followed by rinsing (3 times, 10 minutes each) in TBS–Tween and incubation for 30 minutes with biotinylated swine anti-rabbit Ig F(ab′)2. After repeated rinsing, strep-tavidin-HRP was added for 20 minutes, followed by rinsing and incubation with aminoethylcarbazole substrate for 10 minutes. Sections were counterstained with Mayer’s hematoxylin (Dako) for 1 minute, rinsed in water for 10 minutes, and mounted in Faramount (Dako).
Microarray analysis
We analyzed the microarray data using the R library Limma (19,20). We first used the function “neqc” to perform 1) background correction using the control probes, 2) interarray quantile normalization, and 3) log2 transformation. Then any probes that were not found to be above background levels (detection P values ,0.05) in any of the arrays were removed. Next we used the function “lmfit” to fit a linear model to each of the probes. When fitting the linear model, the arrays were weighted to account for the variability in the levels of overexpression that were achieved. All of the control arrays were assigned a weight of 1. As there was little variability in the silencing efficiency (86–91% range), these were also all assigned a weight of 1. The maximum overexpression efficacy in the experiment was an 8.5-fold increase in LBH levels, while the other 3 overexpression efficiencies were 3.8-fold, 2.9-fold, and 2.9-fold, respectively. Thus, we adjusted the weights of the samples with the lower overexpression efficiencies such that they were proportional to the difference in overexpression efficiency; for example, the weight of the array with 3.8-fold overexpression efficiency was calculated as 3.8/8.5 = 0.45.
The linear models were used to perform 3 comparisons: 1) LBHhigh versus control, 2) LBHhigh versus LBHlow, and 3) control versus LBHlow. To ensure that inference was reliable and stable, we applied Empirical Bayes shrinkage (19) of the probewise variances. Then the shrunken standard errors were used to calculate moderated t-statistics, and the resulting P values were adjusted for multiple testing using the Benjamini-Hochberg method (21). Probes that had a multiple testing corrected P value of ,0.05 in any of the 3 comparisons were identified. Z scores rather than actual expression values were used, as they better represent between-condition changes in the levels of individual genes. The Z scores were calculated individually for each probe, using all 12 samples. To perform clustering, Pearson’s correlation coefficient was used to construct a distance matrix that was hierarchically clustered using Ward’s method.
Pathway analysis of gene expression profiles
To identify pathways that are affected by LBH silencing and overexpression, we used Ingenuity Pathway Analysis (IPA; Ingenuity Systems). We performed pathway analysis on the control and LBHlow comparisons, as the knockdown experiment yielded more consistent results than the overexpression experiment, and comparisons of LBHlow and control yield the largest number of significant genes. For the pathway analysis we used 954 differentially expressed genes that had P values of <0.01 in comparisons of LBHlow versus control. These genes and their log fold changes in gene expression were used as input for IPA, which was run using the default options. Networks were ranked according to their degree of relevance to the differentially expressed genes. The score uses a right-tailed Fisher’s exact test to take into account 1) the number of differentially expressed genes in the network, 2) the network size, 3) the total number of differentially expressed genes, and 4) the total number of molecules in the Ingenuity Knowledge Base that could potentially be included in the networks.
Cell migration assay
RA FLS were transfected with control or LBH siRNA, or with control or LBH expression vector. Cells were plated in 6-well plates in complete DMEM and cultured to subconfluence (22). On day 4 posttransfection, 3 linear “wounds” were introduced in each well using a 1-ml pipette tip with subsequent washing 3 times, as previously described (22,23). Images were collected at time 0 and after 24 hours. The diameter of each wound was measured using ImageJ software (National Institutes of Health). The ratio of wound width at 24 hours to wound width at baseline was calculated for each FLS line.
Cell proliferation and apoptosis assay. RA FLS were transfected with control or LBH siRNA, or with control or LBH expression vector. On day 1 posttransfection (silencing) or day 3 posttransfection (overexpression), cells were trypsinized, replated in triplicate in 96-well plates with 1,000 cells/well, and evaluated by MTT assay (22). In parallel with the cell proliferation assay, transfected cells were cultured separately and analyzed for caspase 3 and caspase 7 activity on day 5, using an Apo-One Homogeneous Caspase 3/7 assay (Promega). Fluorescence values were normalized to cell number (measured by MTT).
Cell cycle analysis
RA FLS cultures were trans-fected with control or LBH siRNA and plated in duplicate. On day 4 posttransfection, cells were washed and resus-pended in cold PBS, fixed in 70% ethanol, permeabilized, and stained with propidium iodide/0.1% Triton X-100 solution supplemented with RNase A. DNA content was recorded (.65,000 events/sample) using a FACSCalibur and CellQuest Pro Software (Becton Dickinson). Data were analyzed using FlowJo software (Tree Star) and Watson Pragmatic modeling, and percentages of cells in the S+G2/M phases were calculated.
Statistical analysis
The significance of differences between stimulated and unstimulated samples was assessed using Student’s paired t-test, and differences in gene expression in RA versus OA samples were assessed by Student’s unpaired t-test. For cell proliferation experiments, two-way analysis of variance was used, and for cell cycle analysis, t-tests with Holm-Sidak correction for multiple comparisons were used. Data were analyzed with GraphPad Prism version 6.0. P values less than 0.05 were considered significant.
RESULTS
Identification of LBH as a pathogenic gene in RA, by multiple genome-wide assays
We have identified potential RA therapeutic targets by performing an integrative analysis of epigenome, transcriptome, and sequence variation in RA FLS (11). To do this, we first established sets of genes implicated in RA using these 3 genomics approaches separately. For epigenome we used DNA methylation arrays that measure DNA methylation levels at ~450,000 CpG in RA, OA, and normal samples (9,10). For transcriptome we used public microarray data to assess expression of genes in RA, OA, and normal samples (24). For sequence variation we used a catalog of published GWAS that had investigated susceptibility to RA (25,26). Independently, these analyses identified 2,375 differentially methylated genes, 2,947 differentially expressed genes, and 74 risk genes. Determination of where these independent gene sets overlapped identified 357 genes with 2 or 3 forms of evidence, and pathway analysis showed that this set of genes is highly relevant to RA. Six genes that had 3 forms of evidence were identified. One of these triple-evidence genes, LBH, was selected for investigation of its potential role in RA. Figure 1 shows that the significantly hypomethylated loci overlapped the promoter regions of LBH. The SNP identified by GWAS (dbSNP ID rs7579944) was a T-to-C variant that is upstream of the LBH promoter. Microarray transcriptomics analysis indicated that LBH expression was 1.9-fold higher in OA FLS than in RA FLS (24).
Figure 1.
LBH methylation in rheumatoid arthritis (RA) fibroblast-like synoviocytes. The location of the LBH RA single-nucleotide polymorphism (SNP) identified by genome-wide association study (GWAS) and the methylation levels of each of the CpGs on the bead array that are within the LBH promoter region (−2,500 bp to +500 bp from the transcription start site [TSS] [arrow]), and the transcript variant numbers of the RefSeq genes that are transcribed from that TSS are noted. Values are the mean ± SEM. ★= significantly differentially methylated CG (RA versus osteoarthritis [OA] and normal [NL]).
LBH expression and regulation in cultured FLS and RA synovium
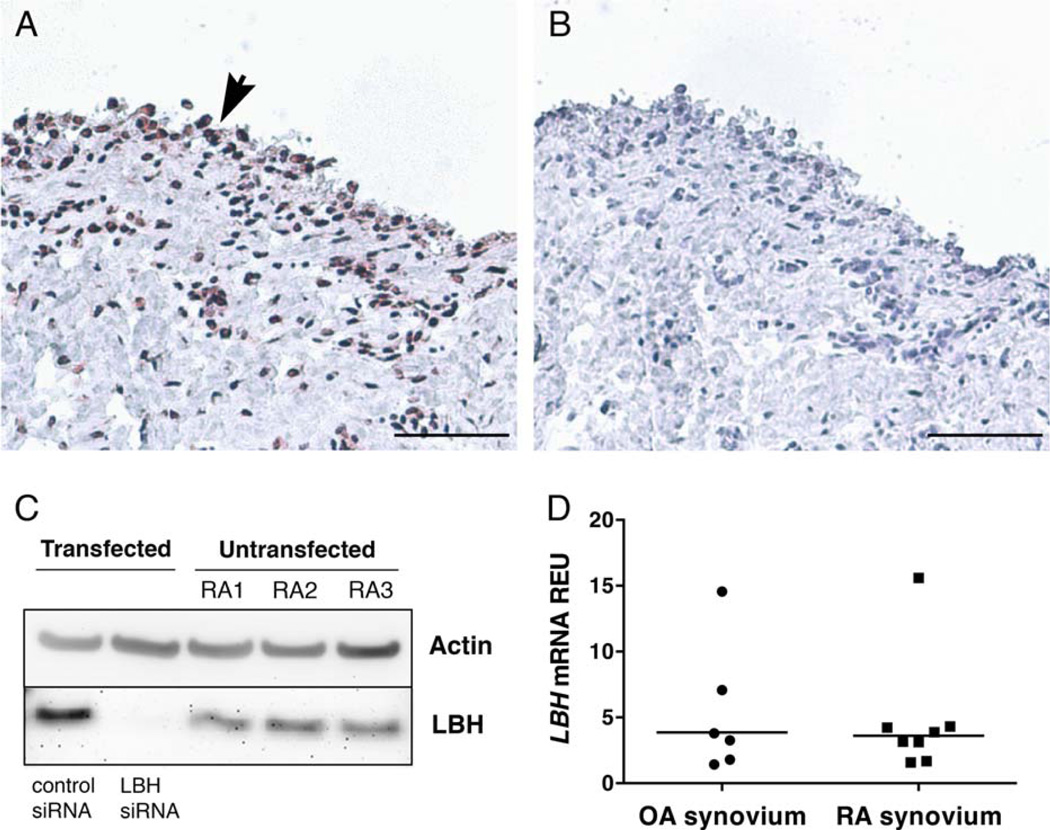
Initial studies were performed to confirm the expression of LBH in cultured FLS. Expression in RA FLS lines was readily detected using qPCR (results not shown) and Western blotting (Figure 2C), confirming constitutive expression in RA FLS. Small interfering RNA–transfected FLS (control and LBH siRNA) were included in the Western blot analysis to confirm antibody specificity. Immunohistochemistry analysis demonstrated that LBH is expressed in the human synovial lining layer, where FLS reside, and in scattered sublining cells in RA (Figures 2A and B). The staining was predominantly nuclear. In situ expression of LBH was also confirmed by qPCR on RA and OA synovial tissue cDNA (Figure 2D). No significant differences between RA and OA synovium were observed, possibly because synovium is a mixture of many cell lineages.
Figure 2.
LBH expression in synovial tissue and fibroblast-like synoviocytes (FLS). A and B, Immunohistochemical staining of human synovial tissue from a rheumatoid arthritis (RA) patient, using anti-LBH antibody (A) and control normal rabbit Ig (B). Arrowhead indicates the lining layer. Bars = 50 µm. C, Expression of LBH in 3 primary RA FLS and in 1 FLS line transfected with control small interfering RNA (siRNA) and LBH siRNA, assessed by Western blotting. The membrane was probed with anti–β-actin antibody and with anti-LBH antibody as indicated. D, Expression of LBH mRNA in whole synovial tissue specimens from osteoarthritis (OA) and RA patients, assessed by quantitative polymerase chain reaction. Ct values were normalized to GAPDH expression. Each symbol represents an individual sample; bars show the mean. REU = relative expression units.
To understand how growth factors implicated in RA might affect LBH gene expression, primary FLS lines were stimulated with various concentrations of transforming growth factor β1 (TGFβ1) or platelet-derived growth factor BB (PDGF-BB), which are growth factors expressed in rheumatoid synovium. LBH messenger RNA (mRNA) expression was significantly increased (mean±SEM 330±26%; P < 0.0001) (n = 8) by stimulation with TGFβ1 at 0.1, 1, and 10 ng/ml (Figure 3A) and significantly decreased (79±6%; P = 0.04) by treatment with 10 ng/ml PDGF-BB (Figure 3C). Time course experiments showed that the peak effects of TGFβ1 and PDGF-BB occurred by 18 hours and 12 hours, respectively (Figures 3B and D). Interleukin-1β (IL-1β) also reduced LBH expression, by 81±5% after 12 hours. Neither Wnt-1, Wnt-3a, Wnt-5a, nor TNF had a significant effect on LBH expression (data not shown).
Figure 3.
Effects of transforming growth factor β1 (TGFβ1) and platelet-derived growth factor (PDGF) on LBH expression in fibroblast-like synoviocytes (FLS). A and C, Expression of LBH mRNA by serum-starved rheumatoid arthritis (RA) FLS without stimulation or after stimulation for 6 hours with recombinant human TGFβ1 at various concentrations (0.1–10 ng/ml) (A) or for 12 hours with PDGF-BB at various concentrations (0.1–10 ng/ml) (C). LBH mRNA expression was measured by quantitative polymerase chain reaction. B and D, Time course of stimulation with TGFβ1 (10 ng/ml) (B) or with PDGF-BB (10 ng/ml) (D). The experiments were repeated 3 times, and Ct values were normalized to GAPDH expression. Values are the mean ± SEM of 3–8 different FLS lines. * = P < 0.05 versus no stimulation. REU = relative expression units.
Lack of a significant effect of LBH on cell migration, cytokine signaling, and gene expression
The aggressive RA FLS phenotype is characterized by increased migration and increased production of proin-flammatory cytokines and matrix-degrading enzymes. To investigate whether LBH is involved in these processes, we performed a variety of functional assays comparing control and LBHlow FLS. There was a trend toward increased migration of LBHlow FLS compared with controls, but this did not reach statistical significance (mean ± SEM ratio of wound width at 24 hours to wound width at baseline 0.82 ± 0.06 versus 0.63 ± 0.02; P = 0.08) (n = 3). A trend toward increased migration when LBHhigh FLS were compared with controls also did not reach significance (0.57 ± 0.06 and 0.48 ± 0.05, respectively; P = 0.06) (n = 3). Next, we stimulated serum-starved control and LBHlow FLS with 50 ng/ml TNF for 9 hours and measured IL6 and MMP3 mRNA levels by qPCR. TNF increased IL6 and MMP3 expression as expected, but no differences were observed with LBH silencing compared to control (data not shown). Furthermore, after stimulation of serum-starved control and LBHlow FLS with 50 ng/ml TNF for 15 minutes to simulate cytokine stimulation in RA synovium, phosphorylation of MAP kinases (p38, ERK, and JNK) did not differ significantly between the groups, as measured by Western blot analysis (data not shown).
Regulation of the FLS transcriptome by LBH
Other studies have indicated that LBH could be a transcriptional coactivator (27,28). However, its regulatory targets in RA remain unknown, and our studies did not identify a clear functional role in traditional measures of pathogenic FLS function. To guide subsequent functional studies, gene expression profiling was performed on RA FLS under 3 conditions: control, LBHlow, and LBHhigh. After correction of P values for multiple testing, we found 47 probes that were significantly differentially expressed and that accurately grouped the samples by LBH status during hierarchical clustering (Figure 4A).
Figure 4.
Genes affected by LBH silencing and overexpression. A, Heatmap showing probes that were significantly differentially expressed between control (C), LBHlow (knockdown [KD]), and LBHhigh (overexpression [O]) fibroblast-like synoviocytes (FLS). Analyses of differential expression included all genes for which the adjusted P value was <0.05 in any of the 3 comparisons (LBHhigh versus control, LBHhigh versus LBHlow, and control versus LBHlow). To make the patterns of differential expression clearer, gene expression levels are represented by a Z score, which was calculated individually for each gene. B, The top-ranked network discovered with Ingenuity Pathway Analysis (IPA) during assessment of LBHlow versus control FLS. The gene expression, cellular development, and cellular growth and proliferation functions were assigned to the network by IPA. Genes that were repressed during knockdown are shown in blue, and those that were induced are shown in red. Genes with P values of <0.01 were used in the analysis. The network contained 43 genes, of which 29 passed the cutoff.
Network analysis of LBH perturbation
To gain a systems-level understanding of the role of LBH in regulating the transcriptome, we conducted network analysis using IPA. We focused on the comparison of LBHlow versus control cells, as the silencing was more robust than the overexpression and this comparison yielded the largest number of differentially hybridized probes. For the network analysis, we used 954 probes that had unadjusted P values of <0.01. This more relaxed cutoff was used as the small sample size could have resulted in exclusion of many genes that would have been found to be regulated by LBH after P value correction. IPA network analysis identified 25 networks that were affected by LBH silencing. (A complete list of the networks identified is available in Supplementary Table 1, on the Arthritis & Rheumatology web site at http://onlinelibrary.wiley.com/doi/10.1002/art.39060/abstract.) The top-ranked network contained 43 genes, of which 29 were affected in LBHlow samples, and was annotated as being involved in controlling cellular growth and proliferation (Figure 4B). These data suggest that LBH is a transcriptional regulator of the cell cycle.
LBH regulation of cell growth in FLS
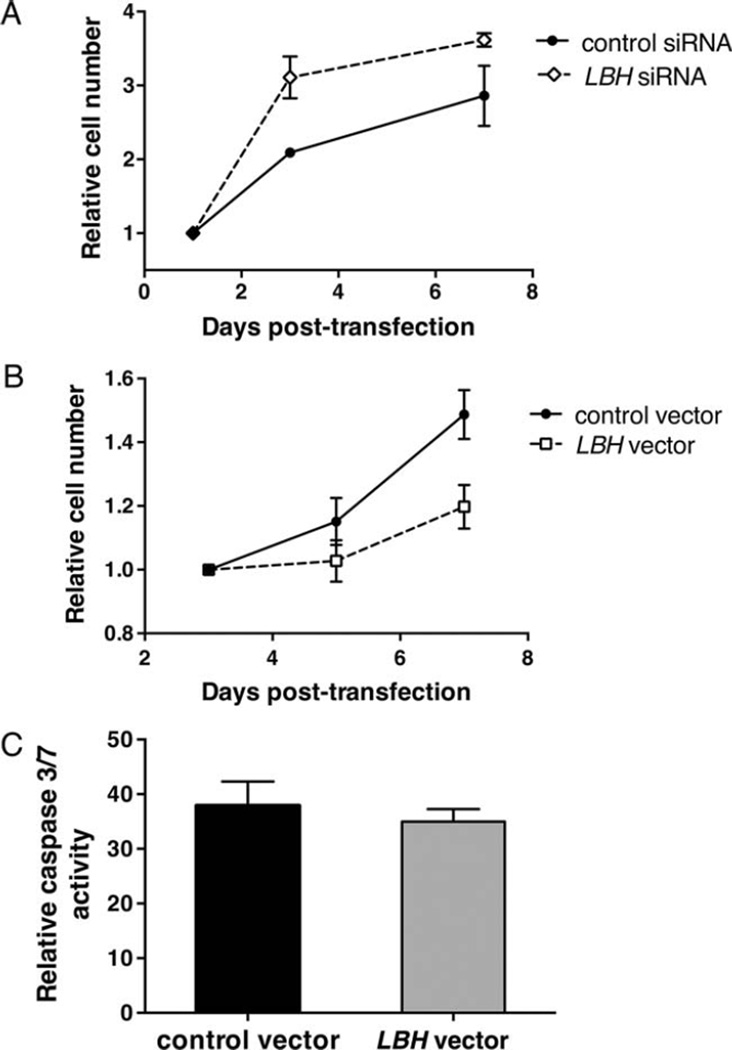
Based on findings of the transcriptome analysis, we then evaluated the role of LBH in cell growth. The gene was silenced in 3 different RA FLS, using siRNA. As shown in Figure 5A, LBH deficiency significantly increased cell growth (by a mean ± SEM of 92 ± 15% on day 3; P = 0.007). These results confirm the micro-array findings suggesting that cell proliferation is a key function of LBH that could be relevant to synovial hyperplasia in RA. To confirm the inhibitory effects of LBH on cell proliferation, additional RA FLS lines were transfected with control or LBH expression vector. A significant decrease in cell growth was readily demonstrated in LBHhigh FLS compared to control FLS (P = 0.005), with 62 ± 10% inhibition on day 7 (Figure 5B). Caspase 3/7 activity assay demonstrated no increased apoptosis in LBHhigh FLS compared to control FLS (Figure 5C), indicating that LBH most likely induces growth arrest rather than apoptosis.
Figure 5.
Effects of modified LBH expression on cell growth in fibroblast-like synoviocytes (FLS). A, Rheumatoid arthritis (RA) FLS lines were transfected with control or LBH small interfering RNA (siRNA) (mean gene silencing 91%). The cells were replated on day 1 posttransfection and subjected to MTT assay on days 1, 3, and 7. Values are the mean ± SEM absorbance of 3 different FLS lines. LBH deficiency significantly increased cell growth (P = 0.007 versus controls). B, RA FLS lines were transfected with control or LBH expression vector (mean overexpression 13-fold). The cells were recultured on day 3 posttransfection and subjected to MTT assay on days 3, 5, and 7. Values are the mean ± SEM absorbance of 6 different FLS lines. LBH overexpression significantly decreased cell growth (P = 0.005 versus controls). C, RA FLS were transfected with control or LBH expression vector. The cells were replated on day 3 posttransfection, and caspase 3/7 activity, as a measure of apoptosis, was determined on day 5. Fluorescence values were normalized to cell number, measured by MTT. Values are the mean±SEM of 3 different RA FLS lines.
Effects of LBH on cell cycle progression in FLS
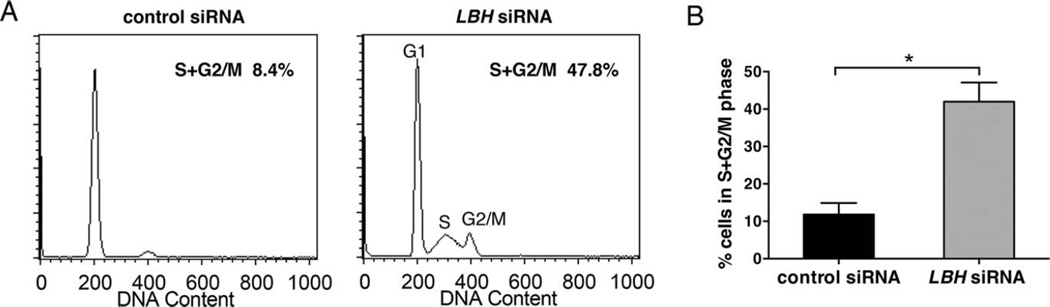
To further explore the effects of perturbed LBH levels on cell growth, we performed cell cycle analysis by flow cytometry. LBH gene expression was silenced in 3 different RA FLS, using siRNA. There was a marked increase in the percentage of cells entering the cell cycle in LBHlow FLS compared to control FLS (Figure 6A). The mean ± SEM percentage of cells (n = 3) in the S + G2/M phases was 42 ± 3% in LBHlow FLS, compared to 12 ± 2% in controls (P = 0.001) (Figure 6B). The percentages of cells in the pre-G1 or post-G2/M phase were negligible. Taken together, these data demonstrate that LBH controls the rate of proliferation in primary FLS.
Figure 6.
Cell cycle progression in fibroblast-like synoviocytes (FLS) after LBH gene silencing. Rheumatoid arthritis (RA) FLS lines were transfected with control or LBH small interfering RNA (siRNA) and cultured until subconfluent. The cells were harvested and fixed on day 4 posttransfection and, after DNA staining, were subjected to flow cytometry. A, Representative histograms of DNA content in RA FLS lines transfected with control siRNA or LBH expression vector. The number of cells in each phase of the cell cycle was estimated using Watson Pragmatic modeling. B, Mean ± SEM percentage of cells in the S + G2/M phases in 3 different RA FLS lines. * = P = 0.001.
DISCUSSION
RA is a chronic synovial disease that leads to joint inflammation and destruction. Despite improved outcomes with targeted therapies, a significant percentage of patients still have active disease after receiving currently available treatments, illustrating the need for identification of additional therapeutic targets. To address this unmet need, we used an unbiased approach integrating “omics” platforms to identify non-obvious candidate genes in RA. Data sets from human primary FLS isolated from the main target tissue of the disease, i.e., the synovium, and robust GWAS data on risk genes in RA were used for this analysis. Based on integrative analysis, LBH was identified as a possible candidate gene. We identified LBH as a candidate gene in RA based on the overlap in results between data sets, including GWAS risk alleles, transcriptomics, and differential DNA methylation. The function of LBH is largely unknown, although forced expression in mouse and chick embryos resulted in cardiovascular anomalies and impaired bone formation (29). In this study, we explored the functions of LBH in FLS and determined how it might contribute to the pathogenesis of RA.
The GWAS-identified RA-associated SNP is located upstream of the LBH transcription start site and might alter gene regulation. Two additional SNPs in the LBH locus have recently been found to be associated with RA and other autoimmune diseases, i.e., celiac disease and systemic lupus erythematosus (8,30,31). The presence of multiple SNPs implicated in autoimmunity suggests that LBH could be a critical checkpoint in immune function. We are currently investigating how risk alleles and differential regulation of CpG loci in critical regulatory regions account for dysregulated LBH expression in RA. In the present study, we focused our attention on how LBH might alter FLS behavior.
We initially investigated how LBH expression is modulated in FLS and found that it is inversely regulated by TGFβ1 and PDGF. These growth factors are expressed in RA synovium and influence fibroblast behavior. TGFβ1 and PDGF regulate cell growth and differentiation of synoviocytes and potentiate TNF and IL-1β-induced production of factors that mediate synovitis and matrix destruction (32). Given the contribution of LBH to synoviocyte proliferation, local production of these growth factors could influence the accumulation of these cells in the intimal lining and the expansion of synovial pannus.
Wnt-responsive elements have been identified in the LBH locus, and expression of LBH in an epithelial cell line was increased by Wnt-3a–conditioned medium (12). Wnt signaling plays a fundamental role in embryonic development as well as cell proliferation and differentiation. Several Wnt types are expressed in RA synovium, and Wnt-5a has been implicated in cytokine production by FLS (33). However, we did not observe an effect of the Wnt types we tested on LBH expression in FLS. This might be due to differences in culture conditions, responsiveness of the different cell types, or other factors that were present in the Wnt-conditioned medium used in previous studies.
A series of experiments was performed to evaluate the role of LBH in various FLS functions that contribute to RA pathogenesis, including migration and cytokine and matrix metalloproteinase gene expression. These experiments were based, in part, on previous studies implicating the gene in activation of signaling pathways, e.g., the MAP kinases (16). Surprisingly, LBH did not significantly influence synoviocyte behavior in these assays. As an alternative strategy to understand the potential role of LBH, we used gene expression arrays and pathway analysis to direct subsequent studies. The array results implicated cell growth and proliferation as an enriched pathway in LBH-deficient FLS. We then experimentally validated this hypothesis in cultured FLS.
Notably, LBH overexpression induced growth arrest, and even more strikingly, LBH deficiency directed cells to enter the cell cycle and increased proliferation. Low LBH expression in RA, as observed in microarray studies, could release FLS from LBH control and contribute to increased proliferation and synovial intimal lining hyperplasia. LBH expression in whole synovial tissues, however, was similar in RA and OA, possibly reflecting the presence of many cell types. Additional studies are needed to determine if the gene regulates proliferation in other cell types, especially since some LBH functions, e.g., MAP kinase and Wnt signaling, are cell-type specific.
Embryology studies have demonstrated a unique spatiotemporal expression pattern of LBH in primordial limb buds and hearts of mouse embryos (28). Transgenic overexpression (uniform and constitutive as opposed to spatiotemporal) of LBH in the myocardium of the developing heart in mice resulted in severe cardiovascular abnormalities (34). LBH is expressed in chondrocytes, where it appears to regulate cell proliferation and maturation given that overexpression leads to shortened skeletal elements, perturbed bone formation, and reduced osteoclast and endothelial cell invasion in chicken embryos (29). LBH conditional-deficient mice exhibited perturbed mammary gland development at puberty (35). Taken together, these findings also demonstrate the importance of tightly regulated LBH expression in adult tissues and suggest that the gene plays a role in cell differentiation during development. Dysregulation of this gene in synoviocytes could contribute to aberrant proliferation in the intimal lining.
In conclusion, we identified a novel RA-associated risk gene, LBH, by integrating multiple omics technologies. It is regulated by growth factors implicated in RA and controls FLS proliferation. An imbalance in released growth factors, perhaps together with a specific genetic and epigenetic predisposition of FLS with respect to LBH regulation, in the synovium could profoundly influence homeostasis. Aberrant LBH expression in FLS would then contribute to synovial hyperplasia and the pathologic changes in the RA joint. Our data also demonstrate the successful use of a systems biology approach to find novel potential therapeutic targets in a complex disease.
Supplementary Material
ACKNOWLEDGMENT
The authors thank Dennis Young for valuable input on the flow cytometry experiments.
Supported by grants from the Rheumatology Research Foundation, the Arthritis Foundation, the National Institute of Arthritis and Musculoskeletal and Skin Diseases (no. R01-AR-065466), the Swedish Medical Society, the Gothenburg Medical Society, and the Rune and Ulla Amlöv Foundation.
Footnotes
AUTHOR CONTRIBUTIONS
All authors were involved in drafting the article or revising it critically for important intellectual content, and all authors approved the final version to be published. Dr. Firestein had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study conception and design. Ekwall, Whitaker, Hammaker, Wang, Firestein.
Acquisition of data. Ekwall, Whitaker, Hammaker, Bugbee.
Analysis and interpretation of data. Ekwall, Whitaker, Hammaker, Wang, Firestein.
ADDITIONAL DISCLOSURES
Author Whitaker is currently an employee of Janssen Pharmaceuticals, but was employed by the University of California, San Diego during the time the study was conducted.
REFERENCES
- 1.Van Vollenhoven RF. Rheumatoid arthritis in 2012: progress in RA genetics, pathology and therapy. Nat Rev Rheumatol. 2013;9:70–72. doi: 10.1038/nrrheum.2012.232. [DOI] [PubMed] [Google Scholar]
- 2.Bartok B, Firestein GS. Fibroblast-like synoviocytes: key effector cells in rheumatoid arthritis. Immunol Rev. 2010;233:233–255. doi: 10.1111/j.0105-2896.2009.00859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bottini N, Firestein GS. Duality of fibroblast-like synoviocytes in RA: passive responders and imprinted aggressors. Nat Rev Rheumatol. 2013;9:24–33. doi: 10.1038/nrrheum.2012.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ekwall AK, Eisler T, Anderberg C, Jin C, Karlsson N, Brisslert M, et al. The tumour-associated glycoprotein podoplanin is expressed in fibroblast-like synoviocytes of the hyperplastic synovial lining layer in rheumatoid arthritis. Arthritis Res Ther. 2011;13:R40. doi: 10.1186/ar3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steenvoorden MM, Tolboom TC, van der Pluijm G, Lowik C, Visser CP, DeGroot J, et al. Transition of healthy to diseased synovial tissue in rheumatoid arthritis is associated with gain of mesenchymal/fibrotic characteristics. Arthritis Res Ther. 2006;8:R165. doi: 10.1186/ar2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lefevre S, Knedla A, Tennie C, Kampmann A, Wunrau C, Dinser R, et al. Synovial fibroblasts spread rheumatoid arthritis to unaffected joints. Nat Med. 2009;15:1414–1420. doi: 10.1038/nm.2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klareskog L, Catrina AI, Paget S. Rheumatoid arthritis. Lancet. 2009;373:659–672. doi: 10.1016/S0140-6736(09)60008-8. [DOI] [PubMed] [Google Scholar]
- 8.Okada Y, Wu D, Trynka G, Raj T, Terao C, Ikari K, et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506:376–381. doi: 10.1038/nature12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakano K, Whitaker JW, Boyle DL, Wang W, Firestein GS. DNA methylome signature in rheumatoid arthritis. Ann Rheum Dis. 2013;72:110–117. doi: 10.1136/annrheumdis-2012-201526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whitaker JW, Shoemaker R, Boyle DL, Hillman J, Anderson D, Wang W, et al. An imprinted rheumatoid arthritis methylome signature reflects pathogenic phenotype. Genome Med. 2013;5:40. doi: 10.1186/gm444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whitaker JW, Boyle DL, Bartok B, Ball ST, Gay S, Wang W, et al. Integrative omics analysis of rheumatoid arthritis identifies non-obvious therapeutic targets. PloS One. doi: 10.1371/journal.pone.0124254. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rieger ME, Sims AH, Coats ER, Clarke RB, Briegel KJ. The embryonic transcription cofactor LBH is a direct target of the Wnt signaling pathway in epithelial development and in aggressive basal subtype breast cancers. Mol Cell Biol. 2010;30:4267–4279. doi: 10.1128/MCB.01418-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arnett FC, Edworthy SM, Bloch DA, McShane DJ, Fries JF, Cooper NS, et al. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 1988;31:315–324. doi: 10.1002/art.1780310302. [DOI] [PubMed] [Google Scholar]
- 14.Alvaro-Gracia JM, Zvaifler NJ, Brown CB, Kaushansky K, Firestein GS. Cytokines in chronic inflammatory arthritis. VI. Analysis of the synovial cells involved in granulocyte-macrophage colony-stimulating factor production and gene expression in rheumatoid arthritis and its regulation by IL-1 and tumor necrosis factor-α. J Immunol. 1991;146:3365–3371. [PubMed] [Google Scholar]
- 15.Rosengren S, Boyle DL, Firestein GS. Acquisition, culture, and phenotyping of synovial fibroblasts. Methods Mol Med. 2007;135:365–375. doi: 10.1007/978-1-59745-401-8_24. [DOI] [PubMed] [Google Scholar]
- 16.Inoue T, Hammaker D, Boyle DL, Firestein GS. Regulation of p38 MAPK by MAPK kinases 3 and 6 in fibroblast-like synoviocytes. J Immunol. 2005;174:4301–4306. doi: 10.4049/jimmunol.174.7.4301. [DOI] [PubMed] [Google Scholar]
- 17.Boyle DL, Rosengren S, Bugbee W, Kavanaugh A, Firestein GS. Quantitative biomarker analysis of synovial gene expression by real-time PCR. Arthritis Res Ther. 2003;5:R352–R360. doi: 10.1186/ar1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Svensson CI, Inoue T, Hammaker D, Fukushima A, Papa S, Franzoso G, et al. Gadd45b deficiency in rheumatoid arthritis: enhanced synovitis through JNK signaling. Arthritis Rheum. 2009;60:3229–3240. doi: 10.1002/art.24887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:1–25. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 20.Ritchie ME, Silver J, Oshlack A, Holmes M, Diyagama D, Holloway A, et al. A comparison of background correction methods for two-colour microarrays. Bioinformatics. 2007;23:2700–2707. doi: 10.1093/bioinformatics/btm412. [DOI] [PubMed] [Google Scholar]
- 21.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B. 1995;57:289–300. [Google Scholar]
- 22.Bartok B, Boyle DL, Liu Y, Ren P, Ball ST, Bugbee WD, et al. PI3 kinase δ is a key regulator of synoviocyte function in rheumatoid arthritis. Am J Pathol. 2012;180:1906–1916. doi: 10.1016/j.ajpath.2012.01.030. [DOI] [PubMed] [Google Scholar]
- 23.Bartok B, Hammaker D, Firestein GS. Phosphoinositide 3-kinase δ regulates migration and invasion of synoviocytes in rheumatoid arthritis. J Immunol. 2014;192:2063–2070. doi: 10.4049/jimmunol.1300950. [DOI] [PubMed] [Google Scholar]
- 24.Del Rey MJ, Usategui A, Izquierdo E, Canete JD, Blanco FJ, Criado G, et al. Transcriptome analysis reveals specific changes in osteoarthritis synovial fibroblasts. Ann Rheum Dis. 2012;71:275–280. doi: 10.1136/annrheumdis-2011-200281. [DOI] [PubMed] [Google Scholar]
- 25.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hindorff LA, MacArthur J, Morales J, Junkins HA, Hall PN, Klemm AK, et al. A catalog of published genome-wide association studies. URL: http://www.genome.gov/gwastudies.
- 27.Ai J, Wang Y, Tan K, Deng Y, Luo N, Yuan W, et al. A human homolog of mouse Lbh gene, hLBH, expresses in heart and activates SRE and AP-1 mediated MAPK signaling pathway. Mol Biol Rep. 2008;35:179–187. doi: 10.1007/s11033-007-9068-4. [DOI] [PubMed] [Google Scholar]
- 28.Briegel KJ, Joyner AL. Identification and characterization of Lbh, a novel conserved nuclear protein expressed during early limb and heart development. Dev Biol. 2001;233:291–304. doi: 10.1006/dbio.2001.0225. [DOI] [PubMed] [Google Scholar]
- 29.Conen KL, Nishimori S, Provot S, Kronenberg HM. The transcriptional cofactor Lbh regulates angiogenesis and endochondral bone formation during fetal bone development. Dev Biol. 2009;333:348–358. doi: 10.1016/j.ydbio.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhernakova A, Stahl EA, Trynka G, Raychaudhuri S, Festen EA, Franke L, et al. Meta-analysis of genome-wide association studies in celiac disease and rheumatoid arthritis identifies fourteen non-HLA shared loci. PLoS Genet. 2011;7:e1002004. doi: 10.1371/journal.pgen.1002004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu ZY, Lu WS, Zuo XB, Hu J, Yao S, Li Y, et al. One novel susceptibility locus associate with systemic lupus erythematosus in Chinese Han population. Rheumatol Int. 2013;33:2079–2083. doi: 10.1007/s00296-013-2697-0. [DOI] [PubMed] [Google Scholar]
- 32.Rosengren S, Corr M, Boyle DL. Platelet-derived growth factor and transforming growth factor β synergistically potentiate inflammatory mediator synthesis by fibroblast-like synoviocytes. Arthritis Res Ther. 2010;12:R65. doi: 10.1186/ar2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sen M, Lauterbach K, El-Gabalawy H, Firestein GS, Corr M, Carson DA. Expression and function of wingless and frizzled homologs in rheumatoid arthritis. Proc Natl Acad Sci U S A. 2000;97:2791–2796. doi: 10.1073/pnas.050574297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Briegel KJ, Baldwin HS, Epstein JA, Joyner AL. Congenital heart disease reminiscent of partial trisomy 2p syndrome in mice transgenic for the transcription factor Lbh. Development. 2005;132:3305–3316. doi: 10.1242/dev.01887. [DOI] [PubMed] [Google Scholar]
- 35.Lindley LE, Briegel KJ. Generation of mice with a conditional Lbh null allele. Genesis. 2013;51:491–497. doi: 10.1002/dvg.22390. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.