Abstract
Antigen presentation by major histocompatibility complex class II molecules (MHC II) to CD4+ T cells plays a key role in the regulation of the adaptive immune response. Loading of antigenic peptides onto MHC II is catalyzed by HLA-DM (DM), a non-classical MHC II molecule. The mechanism of DM-facilitated peptide loading is an outstanding problem in the field of antigen presentation. In this study we systemically explored possible kinetic mechanisms for DM-catalyzed peptide association, by measuring real time peptide association kinetics using fluorescence polarization assays and comparing the experimental data with numerically modeled peptide association reactions. We found that DM does not facilitate peptide association by stabilizing peptide-free MHC II against aggregation. Moreover, DM does not promote transition of an inactive peptide-averse conformation of MHC II to an active peptide-receptive conformation. Instead, DM forms an intermediate with MHC II that binds peptide with faster kinetics than MHC II in the absence of DM. In the absence of peptides, interaction of MHC II with DM leads to inactivation and formation of a peptide-averse form. This study provides novel insights into how DM efficiently catalyzes peptide loading during antigen presentation.
Introduction
Presentation of peptide antigens by major histocompatibility complex class II molecules (MHC II)3 to CD4+ T cells is required for initiation and regulation of adaptive immune responses (1, 2). The intracellular processes leading to peptide loading onto MHC II proteins have been characterized in some detail (3). Newly synthesized MHC II assembles in the endoplasmic reticulum associated with invariant chain and is transported to Golgi apparatus for maturation and subsequently sorting to a specialized endosomal compartment for peptide loading (4). In the MHC II loading compartment the invariant chain is proteolyzed to leave the class II-associated invariant chain peptide (CLIP) fragment occupying the peptide binding groove (5). Antigenic peptides resulting from proteolytic cleavage of exogenous and endogenous proteins are loaded to MHC II (6). Removal of CLIP from MHC II and loading of antigenic peptides to MHC II are catalyzed by DM (HLA-DM in humans and H2-M in mice) (7–10). Peptide-loaded MHC II molecules are transported to the cell surface for presentation to CD4+ T cells and stimulation of an immune response. MHC II recycled from the cell surface can exchange peptide by a similar process (11).
DM plays a key role in MHC II antigen presentation and CD4+ T cell epitope selection (12–17). DM is considered a non-classical MHC II because it is not highly polymorphic and does not have peptide binding capacity (18–22). Instead, DM acts as a peptide editor to catalyze the exchange of CLIP and other peptides onto MHC II during antigen presentation (8–10). We and other researchers have shown that DM-mediated peptide exchange plays a key role in epitope selection by favoring the presentation of peptides with higher kinetic stability (12, 15–17, 23, 24). DM deficiency results in predominant accumulation of MHC II-CLIP complexes at the cell surface and defective peptide loading and exchange, leading to defective negative selection, increased self-reactivity and immunodeficiency (25–27). DM has been shown to promote both peptide binding and release for peptides of varied sequences in vitro, with the combined effect accounting for facilitated peptide exchange (7, 28).
Over the last two decades, much effort has been devoted to elucidation of the mechanism of DM-catalyzed peptide release (18, 21, 29–39). DM binds MHC II near the N-terminus of the peptide binding groove and disrupts peptide binding by inducing or stabilizing a conformation with altered alpha subunit 310 helix and adjacent extended strand (21, 28, 36). The region with altered structure is involved in key MHC II-peptide interactions (21, 32, 35). MHC II-peptide complexes appear to be highly dynamic and adopt various conformations (40, 41). Recently, we and others proposed a model that DM catalyzes peptide release by sensing the dynamic conformation of MHC II-peptide complex constrained by the interactions throughout peptide binding groove (42), and recognizing a conformation involving the rearrangement of MHC II alpha 310 helical and extended region near the N-terminal side of bound peptide in the vicinity of P1 pocket (21).
In contrast to dissociation of bound peptide from MHC II, loading of peptides to MHC II is more complex due to multiple steps and various intermediate species involved in the binding process (43–46). As a result, understanding of the mechanism of DM-catalyzed peptide association is still limited. It has been demonstrated that MHC II undergoes a reversible isomerization between active peptide-receptive and inactive peptide-averse conformations (44, 45). Also proposed is a transient species formed between peptide and MHC II which finally resolves into stable MHC II-peptide complex (43). In different studies, DM has been proposed to catalyze peptide association through different mechanisms. According to one model, DM acts as a molecular chaperone to stabilize empty MHC II against inactivation and aggregation to facilitate peptide loading (47, 48). In another, DM interacts only with MHC II-peptide complex, not empty MHC II, and promotes transition between transient and stable MHC II-peptide complex (49). It has also been suggested that DM catalyzes peptide loading by specifically recognizing unstable conformations of MHC II-peptide complexes and actively converting them into active peptide-receptive MHC II (46). More recently, Grotenbreg et al. proposed a model wherein DM contributes directly to peptide association through formation of a peptide-loading complex between DM and empty MHC II (50). Resolution of this issue requires expression of each model in a testable way and design of experiments that could distinguish each model.
In this study, we tracked real time peptide association kinetics under various experimental conditions using a fluorescence polarization assay, and evaluated several potential kinetic mechanisms for the role of DM in facilitating peptide association. Our data suggest that DM does not stabilize MHC II against aggregation or promote transition of peptide-averse to peptide-receptive conformation. Instead, the experimental data fit with a model wherein DM forms an intermediate with MHC II which binds peptide with faster kinetics but also resolves more quickly into inactive MHC II in the absence of peptides.
Materials and Methods
Peptide synthesis and labeling
For peptide binding assays, N-terminally acetylated influenza hemagglutinin (306–318)-derived HA analog (Ac-PRFVKQNTLRLAT) and CLIP peptide (Ac-VSKMRMATPLLMQ) were synthesized (21st Century Biochemicals, Marlboro, MA) and labeled with Alexa-488 tetrafluorophenyl ester (Invitrogen, Eugene, OR) through primary amine of K5 (HA) and K3 (CLIP). For MHC II stabilization assay, MHC class I HLA-A2 (104–117)-derived W1A peptide (Ac-GSDARFLRGYHQYA) and HA peptide (Ac-PKYVKQNTLKLAT) were synthesized with acetylated N-termini.
Protein expression and purification
Soluble extracellular domains of recombinant HLA-DR1 (DR1) (DRA*0101/DRB1*010101) and DM were expressed in Drosophila S2 cells and purified by immunoaffinity chromatography followed by Superdex200 (GE Healthcare) size exclusion chromatography as described (7, 51). Full-length native DR1 from LG2 lymphoblastoid cells was solubilized using octyl glucoside detergent and isolated by immunoaffinity chromatography as described (52).
Peptide occupancy
To evaluate peptide occupancy, samples of DR1, DM, DR1 preloaded with HA peptide, and full-length DR1 carrying a spectrum of naturally processed peptides were acid-denatured for 1 hour at room temperature with trifluoroacetic acid (final pH ~1.5–2), and characterized by matrix-associated laser desorption ionization (MALDI) mass spectrometry. Mass spectra were obtained using a Micromass MALDI-LR instrument (Waters, Milford, MA). Samples (~4.8 pmol for DR1 and DM, and ~1.0 pmol for DR1 preloaded with HA and DR1 from LG2) and matrix (α-cyano-4-hydroxycinnamic acid, Sigma, St. Louis, MO) were loaded onto the plate and data were acquired in reflectron mode and analyzed using MassLynx software v 4.0 (Waters). For characterization by quantitative amino acid analysis, 2.7 mg samples were denatured in 1ml 1% trifluoracetic acid for 1 hr at room temperature, followed by centrifical ultrafiltration in centricon-10 devices (10 kDa MW cutoff) with filtrate collection and lyophilization. Amino acid analysis was performed by 21st Century Biochemicals (Marlboro MA).
Fluorescence Polarization (FP) assay
Fluorescence polarization (FP) assay was used to measure real time peptide association kinetics as described previously (53). Briefly, 25 nM Alexa488-labeled HA or CLIP was mixed with various concentrations of DR1 (ranging from 0.1 to 1.6 μM) in the presence of different DM concentrations (ranging from 0 to 1.6 μM) in 200 μl pH5.5 binding buffer (100 mM sodium citrate, 50 mM NaCl, 0.1% octylglucoside, 5 mM EDTA, 0.1% NaN3, 1 mM DTT, 1X protease inhibitor cocktail) in 96-well non-binding black polystyrene plates (Corning Incorporated, Corning, NY). Real time peptide binding was monitored by FP using Victor X5 Multilabel plate reader (PerkinElmer, Shelton, CT) at 488nm excitation and 520nm emission.
MHC II stabilization assay
For non peptide-loaded MHC II, 1 μM peptide-free DR1 was incubated alone, or with 1 μM DM, or with 1 μM HA at 37 °C overnight in pH 5.5 binding buffer. For MHC II loaded with a weak binding peptide, 1 μM DR1-W1A was incubated alone, or with 1 μM DM, or with 1 μM DM and 50 μM W1A at 37 °C for 1 hour in pH 5.5 binding buffer. Each sample was injected into Superdex200 size exclusion column and the aggregation of MHC II was analyzed.
Numerical modeling of peptide binding reactions
DM-catalyzed peptide association reactions to MHC II were simulated with KinTek Explorer (39, 54, 55). KinTek Explorer simulates binding reactions by direct numerical integration of coupled rate equations and fits with experimental data to yield estimates for rate constants of microscopic kinetic steps, which has been widely used to determine the kinetic mechanism of enzymatic reactions (54–60). The model described in Scheme V was used to simulate peptide association kinetics and fit with experimental data.
Results
DM catalyzes peptide association to MHC II
We used a FP-based assay to track real time peptide association kinetics to peptide-free DR1 produced in S2 insect cells. Many recombinant MHC II proteins produced in insect cells previously have been reported to be suitable for in vitro peptide binding experiments, including DR1, DR2b, DR3, DR4, DR52, DR2a and DQ (17, 31, 35, 61). Among these proteins DR1 produced in baculovirus-infected Sf9 insect cells cultured in serum-free media in particular appears to be substantially free of associated peptide, as demonstrated by sensitivity to SDS-induced denaturation, increased peptide binding rate and capacity, decreased pH dependence of peptide binding, and most directly, by the absence of associated peptides as quantitated by amino acid analysis (62). To verify that DR1 produced in S2 insect cells was substantially free of associated peptide, we performed quantitative amino acid analysis of acid-eluted material from three samples of DR1 isolated from S2 cells, as originally described for DR1 isolated from baculovirus-infected Sf9 insect cells (62). No significant amino acid content was observed in these samples (1.02, 1.99, and 3.45 nmol amino acid per 50 nmol protein) relative to that present in a control DM sample (3.46 nmol per 50 nmol protein). Amino acid analysis for DR1 preloaded with HA revealed expected stoichiometry (data not shown). Additionally, we examined DR1 acid elutes by MALDI mass spectrometry (Fig. 1). Ions corresponding to peptidic material were not observed in samples of DR1 or peptide-free DM produced in S2 insect cells (Fig. 1A and 1B), whereas peptides were readily observed for DR1 preloaded with HA peptide (Fig. 1C) or for native DR1 isolated from LG2 lymphoblastoid cells (Fig. 1D).
Figure 1. DR1 produced in S2 insect cells is substantially free of associated peptide.
MALDI analysis was performed for (A) DR1 produced in S2 cells, (B) DM, (C) DR1 preloaded with HA, and (D) DR1 isolated from LG2 lymphoblastoid cells.
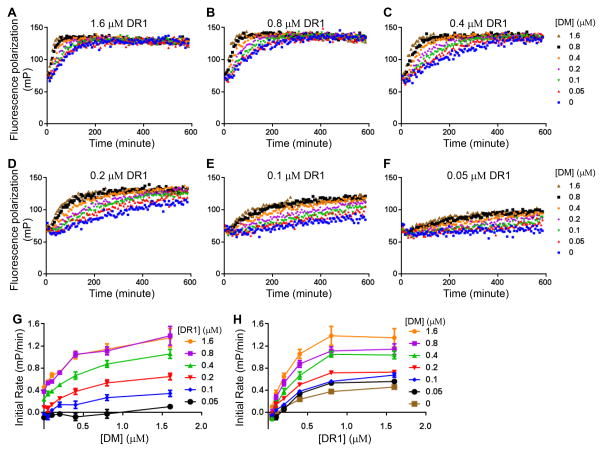
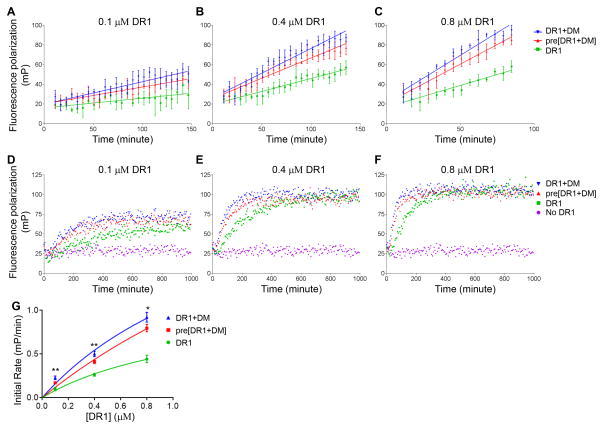
To evaluate the effect of DM on the overall association reaction, we followed CLIP peptide binding to various concentrations of DR1 by the FP assay in the presence of various concentrations of DM (Fig. 2A–F). We observed in each case that DR1 peptide binding proceeded faster in the presence of DM (Fig. 2A–F). Within each set of curves the rate increase depended on the dose of DM. For each curve we calculated the initial rate of peptide binding (the slope of a linear fit to the early part of each association curve), and plotted these against the concentration of DM (Fig. 2G) or DR1 (Fig. 2H). The initial rate of peptide binding increases with the concentration of DM (Fig. 2G). The initial rate of peptide binding increases with DR1 concentration up to a point but then saturates (Fig. 2H), which is an expected behavior for a classic enzyme substrate system as previously observed for DM catalyzed peptide binding to MHC II (7, 28, 29).
Figure 2. DM catalyzes peptide association to MHC II molecules.
Real time association of Alexa488-labeled CLIP to (A) 1.6 μM DR1, (B) 0.8 μM DR1, (C) 0.4 μM DR1, (D) 0.2 μM DR1, (E) 0.1 μM DR1, and (F) 0.05 μM DR1 in the presence of a gradient concentration of DM was tracked by fluorescence polarization. (G) Initial rate versus DM concentration for various DR1 concentrations. Initial rate of peptide binding was calculated as the slope of the linear portion of peptide binding curve. (H) Initial rate versus DR1 concentration for various DM concentrations. These experiments are representative of at least three independent experiments.
Evaluation of models for the role of DM in MHCII- peptide association
We sought to evaluate different kinetic mechanisms for DM-facilitated peptide association. A reversible isomerization between active peptide-receptive (MHCa) and inactive peptide-averse (MHCi) conformations of empty MHC II has been repeatedly observed during intrinsic or DM-catalyzed peptide association (43–45, 47–50). In our evaluation of possible mechanisms for DM-catalyzed peptide association, we always included a step corresponding to transition between MHCa and MHCi. In an earlier study of the non-catalyzed association reaction we observed saturation of the initial peptide-binding rate with increasing peptide or MHC concentration (43). This was explained by a unimolecular conformational change between transient and stable conformations after the initial bimolecular binding (43, 46, 49, 63, 64). We therefore also included in all models conversion of a transient unstable MHC II-peptide complex (MHCpep’) to a stable MHC II-peptide complex (MHCpep). Other aspects of the overall association reaction differ between the various models and these are discussed individually below.
DM does not facilitate peptide binding by stabilizing MHC II against aggregation
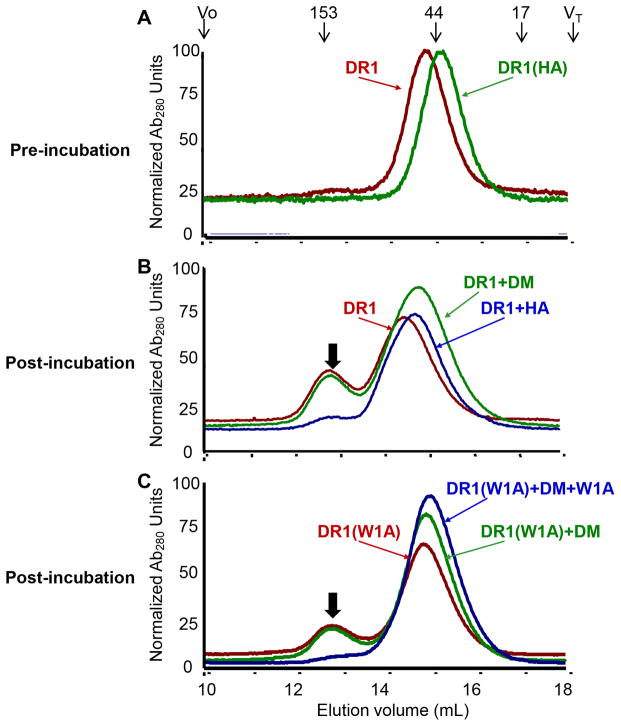
A previous study has indicated that preincubation of DR1 alone leads to the aggregation and functional inactivation of DR1, and preincubation of DR1 and DM together could rescue the activity of DR1 (48). We first evaluated whether DM facilitates peptide binding by stabilizing empty MHC II against aggregation. We incubated purified DR1 monomer alone, or with DM, or with peptide, in each case at 37 °C overnight in buffer with endosomal pH. If DM were able to stabilize DR1 in this manner, we would expect to see that incubation of DR1 alone leads to the aggregation of DR1, whereas incubation of DR1 together with DM would prevent the aggregation of DR1. Before incubation, both purified non peptide-loaded DR1 and pre-loaded DR1-HA eluted as single monomer peaks (Fig. 3A). As previously observed, the elution times of these species differ, with the peptide bound complex adopting a compact conformation and the empty protein adopting a more open conformation (65). Consistent with previous studies (62, 66, 67), empty MHC II aggregated during the incubation period (Fig. 3B, red curve). Inclusion of HA peptide in the incubation mixture stabilized MHC II such that aggregation was not observed (Fig. 3B, blue curve). The extent of aggregation level was similar with DM included in the incubation mixture (Fig. 3B, green curve). The same phenomenon was observed when the incubation time at 37 °C was reduced from overnight to 1 hour (data not shown). Thus we did not observe that DM could directly stabilize MHC II against aggregation. To validate this observation, we loaded DR1 with the weakly binding peptide W1A, which we identified recently as extremely susceptible to DM-mediated peptide release (42), and incubated this complex alone or in the presence of DM or peptide (Fig. 3C). This experiment is similar to the previous one, but with the empty protein generated in situ by release of a weakly bound peptide instead of starting with the non peptide-loaded protein. Consistently, incubation of DR1-W1A at 37 °C for 1 hour in buffer with endosomal pH resulted in the aggregation of MHC II (Fig. 3C, red curve). Excess of peptides but not DM could inhibit the aggregation of MHC II (Fig. 3C, green and blue curves). Therefore, we conclude that DM has no direct effect of stabilizing MHC II against aggregation. We tested whether aggregated MHC II exhibits slower peptide binding kinetics than monomeric MHC II. Surprisingly, aggregated DR1 bound to Alexa488-HA equally well, if not better, compared with monomeric DR1 (supplemental Fig. S1). Thus MHC II aggregation per se is not directly tied to functional inactivation.
Figure 3. DM does not stabilize MHC II by preventing aggregation.
(A) Representative elution profiles of freshly purified DR1 monomer and DR1 loaded with HA (pre-incubation). The elution position and molecular mass of protein standards (unit: kilo Dalton) were shown above traces, with void volume Vo and total included volume VT indicated. Stabilization of (B) non peptide-loaded DR1 or (C) DR1 loaded with a weakly binding peptide W1A by DM or peptides was monitored by gel filtration (post-incubation). Black arrow on each plot indicated the aggregation of DR1. These traces are representative of at least four independent experiments.
DM does not facilitate peptide binding by converting inactive MHC II to active form
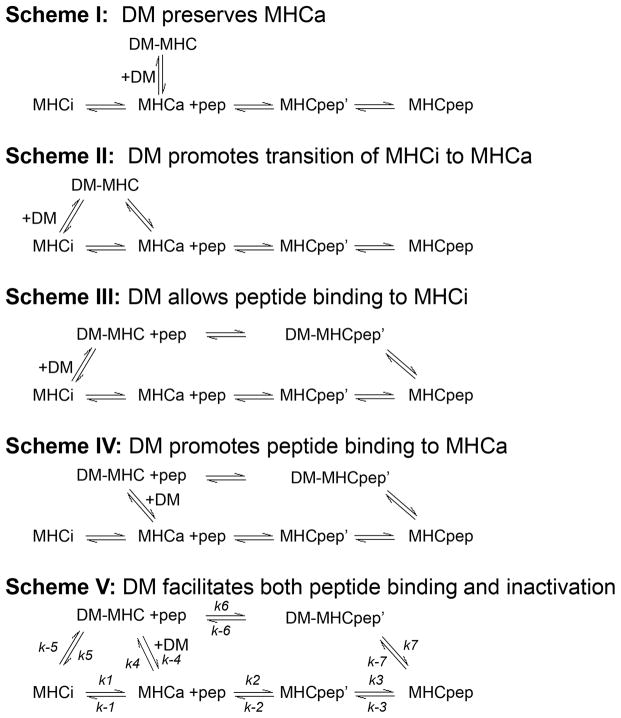
MHC II undergoes reversible isomerization between peptide-receptive and peptide-averse states (44, 45), and up to >90% of MHC II prepared as a recombinant protein can be in the inactive peptide-averse form at equilibrium (43, 44). We evaluated a model wherein DM catalyzes peptide association by promoting the transition of inactive MHC II to active form. We considered four possible mechanisms by which DM could mediate conversion of inactive MHC II to an active form, as shown by Schemes I–IV (Fig. 4). In Scheme I, DM acts as a molecular chaperone for MHCa and prevents the inactivation of MHCa by preserving MHC II in the active form. During peptide binding, MHCa is released from the intermediate with DM and binds peptide. In Scheme II, DM binds to MHCi to promote transition of MHCi to MHCa. In Scheme III, DM directly interacts with MHCi forming an intermediate DM-MHC which allows peptide binding to MHCi. In Scheme IV, DM interacts with MHCa as a molecular chaperone by competing with inactivation and forming an intermediate DM-MHC which binds peptide with a faster rate.
Figure 4. Overview of Scheme I, Scheme II, Scheme III, Scheme IV and Scheme V.
Different models for DM-catalyzed peptide association are described in Schemes I–V. In Scheme I, DM acts a molecular chaperone to stabilize and preserve MHCa against inactivation. In Scheme II, DM binds to MHCi and promotes transition of MHCi to MHCa. In Scheme III, DM allows peptide binding to MHCi by forming an intermediate with MHCi. In Scheme IV, DM promotes peptide binding to MHCa by forming an intermediate with MHCa. In Scheme V, DM facilitates both peptide binding and MHC II inactivation by forming an intermediate with MHCa which binds peptides in a faster kinetics but resolves into MHCi in the absence of peptides.
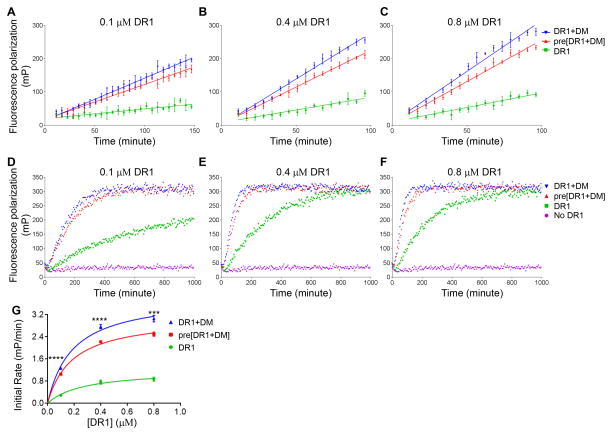
To test Scheme I, Scheme II, Scheme III and Scheme IV, we tracked the binding kinetics of HA and CLIP peptides to DR1 in the absence of DM, in the presence of DM, or in the presence of DM that had been preincubated together with DR1. Our rationale is that if DM catalyzes peptide association by converting inactive MHC II to active form, preincubation of DR1 with DM would largely increase the initial rate of peptide binding due to increased amount of peptide-receptive MHC II. To control for effects of the incubation period on DM activity we separately preincubated DM before adding it to preincubated DR. All these samples were incubated at 37 °C in pH5.5 buffer overnight before adding Alexa488-labeled peptides for binding. We observed faster HA binding to DR1 in the presence of DM, both for DM incubated separately, and for DM preincubated with DR1 (Fig. 5A–C). Surprisingly, preincubation of DR1 with DM decreased initial HA binding compared with adding DM separately (Fig. 5A–C), although they reached the same overall binding level (Fig. 5D–F). The initial rate of binding was 20% lower and significantly different when preincubating DM with DR1 compared with adding DM separately (Fig. 5G). The same experiments were performed for CLIP, which showed the essentially same behavior (Fig. 6). Thus DM does not convert inactive MHC II to active form.
Figure 5. DM does not facilitate HA binding by converting inactive DR1 to active form.
Initial binding of Alexa488-labeled HA to (A) 0.1 μM DR1, (B) 0.4 μM DR1, and (C) 0.8 μM DR1 under conditions incubated DR1 alone (DR1, green), preincubated DR1 with DM (pre[DR1+DM], red) and separately incubated DR1 and DM (DR1+DM, blue) was tracked by fluorescence polarization. DM and Alexa488-HA concentrations were kept at 0.4 μM and 0.025 μM, respectively. The incubation was done at 37 °C for about 16 hours. (D–E) Real time association kinetics for longer time period. Alexa488-labeled HA alone without DR1 (magenta) was also included as a control. Initial rates were calculated for each reaction (A–C), and summarized in (G). These experiments were repeated four times with two replicates for each sample. The two-tailed p-value of unpaired t test for the comparison between DR1 preincubated with DM (red curve) and adding DM together with peptides (blue curve) is shown in (G). *, p<0.05; **, p<0.01; ***, p<0.001; ****, p<0.0001.
Figure 6. DM does not facilitate CLIP binding by converting inactive DR1 to active form.
The experiments described for HA in Figure 5 were repeated for Alexa488-labeled CLIP.
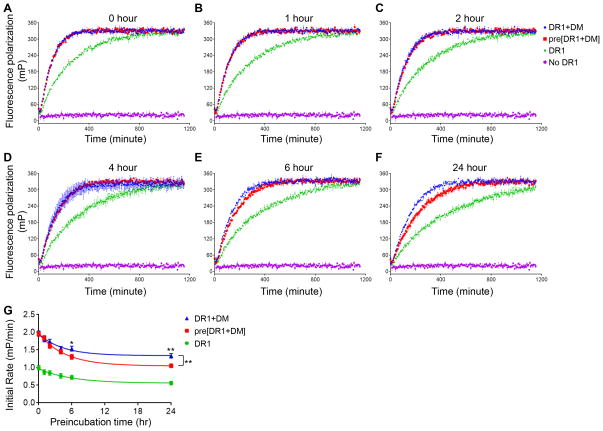
We sought to understand why preincubation of DM and MHC II together led to slower binding than the same reaction where DM and MHCII were preincubated separately. A possibility is that interaction of DM with MHC II in the absence of peptides actually results in inactivation of MHC II. To test the hypothesis that interaction of DM with MHC II in the absence of peptides leads to inactivation not activation of MHC II, we did a time course of preincubation of DR1 with DM and tracked the catalytic effect on HA association (Fig. 7A–F). The initial rate in each condition was plotted (Fig. 7G). We observed that incubation of DR1 alone in the absence of peptide resulted in intrinsic functional inactivation (Fig. 7G, green curve). This intrinsic inactivation was also observed in conditions where DR1 and DM were incubated separately followed by peptide binding as shown by the parallel curves for incubation of DR1 alone (Fig. 7G, green curve) and DR1 separately with DM (Fig. 7G blue curve). However, preincubation of DR1 together with DM leads to facilitated inactivation of DR1, with differences evident at 2 hours and statistically substantial at 6 hours of incubation (Fig. 7G, red curve). Thus, DM does not catalyze peptide association by converting inactive MHC II to active form. Instead, interaction of MHC II with DM in the absence of peptides leads to the inactivation of MHC II.
Figure 7. Preincubation of DR1 with DM in the absence of peptide leads to the inactivation of DR1.
Non peptide-loaded DR1 (0.4 μM) was incubated alone (DR1, green), or together with 0.4 μM DM (pre[DR1+DM], red), or separately with 0.4 μM DM (DR1+DM, blue) for (A) 0 hour, (B) 1hour, (C) 2 hours, (D) 4 hours, (E) 6 hours, and (F) 24 hours 37 °C in pH5.5 buffer. Subsequently, 0.025 μM Alexa488-labeled HA was added to the incubated mixture to track real time peptide binding kinetics by fluorescence polarization. Alexa488-labeled HA alone without DR1 (magenta) was also included as a control. (G) Initial rate was plotted against preincubation time. Each sample has three replicates. The two-tailed p-value of unpaired t test for the comparison between DR1 preincubated with DM (red curve) and adding DM together with peptides (blue curve) is shown in (G). The p-value for the initial rate versus incubation curve fitted to a one-phase decay is also shown for those two groups. Non-significant p-value is not shown. *, p<0.05; **, p<0.01; ***, p<0.001; ****, p<0.0001.
DM catalyzes peptide binding by forming an intermediate with MHC II that binds peptide with faster kinetics but resolves into inactive MHC II in the absence of peptides
Interactions between DM and MHC II have been characterized extensively, both for peptide-free MHC II and for MHC II-peptide complexes (10, 21, 22, 28, 35, 36, 46–48, 68). A transient DM-MHC II complex has been proposed to explain DM catalysis of peptide association or peptide exchange (28, 47, 50, 69). By taking into account the inactivation of MHC II in the absence of peptides and its facilitation by DM (Fig. 5–7), we developed a model that can explain both the observed facilitated peptide binding and also increased inactivation. In this model (Scheme V) DM catalyzes peptide association by forming an intermediate with MHC II, which binds peptides with faster kinetics but resolves into inactive MHC II in the absence of peptides (Fig. 4). In the presence of peptides the transient intermediate DM-MHC binds peptide with faster kinetics as in Scheme IV. In the absence of peptides, DM-MHC resolves into MHCi to facilitate inactivation.
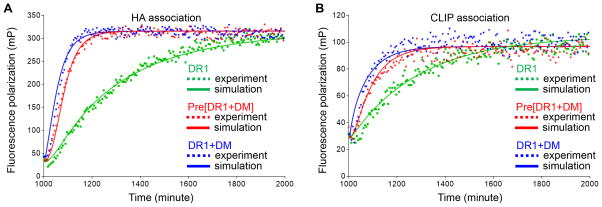
To test Scheme V, we mathematically simulated the binding reactions and fit experimental real time peptide binding kinetic data to the simulated model, using KinTek Explorer software. Peptide binding reactions described in Scheme V were simulated for HA and CLIP binding to DR1 in the absence of DM, in the presence of preincubated DR1 and DM, and in the presence of separately incubated DM, in each case using the 0.4 μM DR1 data in Fig. 5 and Fig. 6. The experimental data and computation simulation fits are shown in Fig. 8 with the experimental set up listed in Supplemental Table I. The model in Scheme II, Scheme III and Scheme IV could not fit the experimental data because it cannot account for the inactivation of DM-interacting MHC II in the absence of peptides (data not shown). We equilibrated the system for 1000 minutes during the preincubation step with or without DM to simulate overnight incubation (Supplemental Table I). The values of k-4 and k-5 were set to 0, because in the model DM only interacts with active MHC II and DM-MHC II complex resolves into inactive MHC II in the absence of peptides as discussed above. The initial starting values for other rate constants were set based on previous literature on MHC II interacting with DM or HA and CLIP peptides (28, 43, 49, 50) (Supplemental Table II). Values for k1, k-1, k4, k-4, k5 and k-5 were kept the same for HA and CLIP, while other rate constants were fitted separately for each peptide. The value for k5 was not previously estimated since it is first developed here. The values for k2CLIP, k-2CLIP, k-3CLIP, k6CLIP, k7CLIP and k-7CLIP have not been previously evaluated experimentally, and were obtained by curve-fitting as described below.
Figure 8. Mathematical modeling of peptide association reactions in Scheme V and fitting with experimental data.
Peptide binding reactions described in Scheme V were simulated and data fitting were done with KinTek Explorer under three conditions (supplemental Table I): non peptide-loaded DR1 was incubated alone for 1000 minutes (Mixing step 1), and Alexa488-labeled HA was added to track real time binding for 1000 minutes (Mixing step 2); non peptide-loaded DR1 was incubated together with DM (Mixing step 1) and Alexa488-labeled HA was added to track real time binding (Mixing step 2); non peptide-loaded DR1 was incubated separately (Mixing step 1), and Alexa488-labeled HA and DM were added to track real time binding (Mixing step 2). Experimental data and simulated values were shown for (A) HA association and (B) CLIP association. The final concentration of total DR1, DM and Alexa488-label peptide was 0.4 μM, 0.4 μM and 0.025 μM respectively.
Fitting the model in Scheme V to experimental data yields excellent fits (Fig. 8A–B). The best fit parameter values are listed in Table I. We found that the fraction of active MHC II preset in the beginning of simulation did not change the data fitting because it was equilibrated out during the preincubation. Multiple combinations of k2 and k3, and k6 and k7 (i.e. k2CLIP=0.1 μM−1min−1, k3CLIP=0.037 min−1, k6CLIP=2.85 μM−1min−1, and k7CLIP=0.028 min−1) are capable of producing the same fits as long as the coupled rates of k2 and k3, and k6 and k7 are similar because the intermediate MHCpep’ or DM-MHCpep’ is not measured in the experiments. Values for k-3HA, k-6HA, k-7HA that are less than 1×10−4 could produce the same fits, which corresponds to the long intrinsic and DM-catalyzed dissociation half-life (~100 hours) for HA compared with short half-life for CLIP (~5 hours) as previously reported (7, 8, 17, 28–31, 33, 34, 37, 43, 49, 70). Notably, k4 must be larger than 1 μM−1min−1, and coupled rate of k6 and k7 must be at least 15 times greater than coupled rate of k2 and k3 in order to capture the fast DM-catalyzed peptide association, which indicated that DM forms an intermediate with active MHC II binding peptides in a faster kinetics. Importantly, k5 must be larger than k-1 in order to capture the facilitated inactivation when incubating DR1 together with DM in the absence of peptides. In summary, we have demonstrated that a model that DM facilitates both peptide binding and MHC II inactivation fits with the experimental data.
Table I.
Summary of best fit values for parameters in Scheme V
| Parameter | HA peptide | CLIP peptide |
|---|---|---|
| k1 (min−1) | 0.022 | 0.022 |
| k-1 (min−1) | 0.0033 | 0.0033 |
| k2 (μM−1min−1) | 0.16 | 0.054 |
| k-2 (min−1) | 0.013 | 0.029 |
| k3 (min−1) | 0.0093 | 0.083 |
| k-3 (min−1) | 10−5 | 0.0058 |
| k4 (μM−1min−1) | 2.23 | 2.23 |
| k-4 (min−1) a | 0 | 0 |
| k5 (min−1) | 0.36 | 0.36 |
| k-5 (μM−1min−1) a | 0 | 0 |
| k6 (μM−1min−1) | 5.42 | 5.21 |
| k-6 (min−1) | 10−5 | 0.022 |
| k7 (min−1) | 0.012 | 0.014 |
| k-7 (μM−1min−1) | 10−5 | 0.020 |
The values of k-4 and k-5 were set to 0, to simulate that DM only interacts with active MHC II and DM-MHC II complex resolves into inactive MHC II in the absence of peptides.
Discussion
The mechanism of DM-facilitated MHC II-peptide exchange is a fundamental problem in antigen presentation. Substantial effort has been devoted to understanding DM-catalyzed peptide dissociation (71, 72), but DM-catalyzed peptide association is not well understood mechanistically (46–50). In this study, we systematically evaluated several potential kinetic mechanisms by which DM could catalyze peptide loading. We tracked real time peptide association kinetics under a wide range of conditions designed to test various kinetic models, and we mathematically simulated peptide binding reactions and fit these to experimental data. Our data demonstrate that DM does not promote peptide loading by stabilizing MHC II against aggregation or by converting inactive MHC II to active forms. Instead, the experimental data are consistent with a model in which DM facilitates peptide loading by forming an intermediate with an active form of MHC II, with the DM-MHC II complex binding peptides with faster kinetics but resolving into inactive MHC II in the absence of peptides.
Several models have been proposed previously to explain DM-facilitated peptide loading (43–50). Most reports agree on a reversible isomerization between active peptide-receptive and inactive peptide-averse MHC II conformations. However, discrepancy exists on whether DM facilitates peptide loading by preventing inactivation (or aggregation) of MHC II (47, 48, 50), or by converting inactive MHC II to active forms (46), or by forming an intermediate with MHC II (50), and whether DM interacts with both non peptide-loaded and peptide-loaded MHC II (47, 48, 50) or only with peptide-loaded MHC II (49). DM has been reported to stabilize MHC II against inactivation assayed by enhanced peptide binding in the presence of DM (47, 48, 50). We did not observe a direct effect of DM on preventing MHC II aggregation. It is possible that the stabilization observed previously is due to the secondary effect of peptide binding, with DM preventing MHC II inactivation indirectly by promoting productive peptide binding. Evidence for this came from the observation that peptide but not DM can prevent MHC II aggregation. An earlier study proposing that DM catalyzes peptide association by converting inactive MHC II to active forms was evidenced by the fact that preincubation of DR1 with DM enhanced peptide binding compared with intrinsic peptide binding (48). This phenomenon was also observed in our study. However, in that study no comparison between preincubating DR1 with DM in the absence of peptides and adding DM together with peptides was made (48), and it is possible that the observed stabilization was indirect. The enhanced peptide binding that we observed for preincubated DR1 and DM be may due to DM forming an intermediate with MHC II which binds peptide in a faster kinetics, as proposed in a recent study (50). In that study the authors also proposed that DM binds MHC II to stabilize it from unfolding and aggregating (50). That study used a photocleavable peptide to generate an active form of empty MHC II after cleavage of bound peptide and release of fragments as a way to start the in vitro peptide binding experiments (50). One caveat of our study is that the purified recombinant DR1 used is not a homogenous population and is a mixture of MHCa and MHCi which may complicate the interactions with DM. For example, DM may interact with both MHCi and MHCa to form different intermediates instead only interacting with MHCa as in Scheme V. We also did not include the possibility that non-productive unstable MHC II-peptide complexes may form with DM working to edit them out as a way to facilitate the overall peptide binding reaction. Due to the fact that the DR1 used is a heterogeneous population of active and inactive MHC II, and some intermediate species of peptide binding reactions are not measured, there are uncertainties in the best fit parameter values of Scheme V. Future studies looking at peptide binding to homogenous MHC II population and capable of capturing intermediate species could add more mechanistic details to the DM-catalyzed peptide association reaction. Nevertheless, in our current study, by directly looking at whether DM could stabilize MHC II or convert inactive MHC II to active forms in the absence of peptides and by including a comparison between preincubated DR1 with DM and adding DM together with peptides, we assessed the direct effect of DM on MHC II stabilization and dissected the kinetic mechanism of DM-catalyzed peptide loading. Thus, this study reconciles discrepancies about the role of DM in peptide association reactions and the model proposed is supported by previous studies on DM-facilitated peptide loading.
A key aspect of the model proposed in this study for DM-catalyzed peptide loading is that DM forms an intermediate with active MHC II, which resolves into inactive conformation in the absence of peptides but can bind peptides in a faster kinetics. This model is consistent with recent structural characterizations. In a structure proposed for the peptide-averse form of empty MHC II based on molecular dynamics simulations, the alpha subunit extended strand region occupies the N-terminal side of the peptide binding groove to inhibit peptide binding (40). Recent crystal structures of DM-DR (21) and DM-DO (22) complexes shows that DM acts to catalyze peptide release by inducing (or stabilizing) conformational changes in the alpha subunit extended strand region which destabilize peptide-MHC interactions at the N-terminal side of the peptide binding groove. Were DM to interact with non peptide-loaded MHC II in the same manner as observed in the DM-DR and DM-DO crystal structures it would have the effect of promoting conversion between active peptide-receptive and inactive peptide-averse forms as observed in the kinetic studies reported here.
The major conclusion of this work is that DM promotes peptide binding by forming an intermediate with MHC II which binds peptides with faster kinetics compared with MHC II alone. Efficient loading of antigenic peptides to MHC II is required for the initiation and regulation of adaptive immune responses (73). In response to a variety of stimuli, professional antigen-presenting cells, such as dendritic cells transform from an immature state specialized for protein antigen uptake into a mature state specialized for presentation of antigenic peptides for T cell activation (74). Dendritic cell maturation is accompanied by enhanced lysosomal acidification which results in increased antigen proteolysis (74, 75). The proteolytic activity is tightly controlled in antigen-presenting cells to prevent complete destruction of protein antigens and to ensure efficient presentation of immunodominant epitopes (74, 76). DM resides in endosomal peptide-loading compartments together with MHC II (10, 47, 77). By forming an intermediate with MHC II which binds peptides in a faster kinetics, DM could efficiently facilitate peptide loading during infection to initiate adaptive immunity. This idea is supported by the observation that cells from DM-deficient mice poorly present both whole proteins and short peptides (26, 27). The mechanism we describe for DM-catalyzed peptide association also involves facilitated MHC II inactivation in the absence of peptides (Scheme V). Although this phenomenon happens in the physiological time frame of MHC II and DM residence half-life in peptide loading compartment (78, 79), the physiological relevance is still unknown because conditions under which there are absence or limited amount of peptides are not well illustrated.
In summary, by tracking real time peptide association kinetics, mathematically modeling peptide binding reactions, and fitting with experimental data, we systematically established that DM catalyzes peptide loading by forming an intermediate with MHC II which binds peptides with faster kinetics but resolves into inactive MHC II in the absence of peptides. Our study provides novel insight into how DM efficiently catalyzes antigenic peptides loading during antigen presentation.
Supplementary Material
Acknowledgments
We would like to thank Liying Lu and Loretta Lee for expression and purification of soluble DR1 and DM, and Zachary Maben for assistance with aggregation studies.
Footnotes
This work was supported by National Institute of Health grant AI-38996
Abbreviations used in this paper: MHC II, major histocompatibility complex class II molecules; CLIP, class II-associated invariant chain peptide; DM, HLA-DM; MALDI, matrix-associated laser desorption ionization; FP, fluorescence polarization; HA, hemagglutinin; DR1, HLA-DR1
References
- 1.Unanue ER. Antigen-presenting function of the macrophage. Annual review of immunology. 1984;2:395–428. doi: 10.1146/annurev.iy.02.040184.002143. [DOI] [PubMed] [Google Scholar]
- 2.Cresswell P. Assembly, transport, and function of MHC class II molecules. Annual review of immunology. 1994;12:259–293. doi: 10.1146/annurev.iy.12.040194.001355. [DOI] [PubMed] [Google Scholar]
- 3.Jensen PE. Recent advances in antigen processing and presentation. Nat Immunol. 2007;8:1041–1048. doi: 10.1038/ni1516. [DOI] [PubMed] [Google Scholar]
- 4.Bakke O, Dobberstein B. MHC class II-associated invariant chain contains a sorting signal for endosomal compartments. Cell. 1990;63:707–716. doi: 10.1016/0092-8674(90)90137-4. [DOI] [PubMed] [Google Scholar]
- 5.Riberdy JM, Newcomb JR, Surman MJ, Barbosa JA, Cresswell P. HLA-DR molecules from an antigen-processing mutant cell line are associated with invariant chain peptides. Nature. 1992;360:474–477. doi: 10.1038/360474a0. [DOI] [PubMed] [Google Scholar]
- 6.West MA, Lucocq JM, Watts C. Antigen processing and class II MHC peptide-loading compartments in human B-lymphoblastoid cells. Nature. 1994;369:147–151. doi: 10.1038/369147a0. [DOI] [PubMed] [Google Scholar]
- 7.Sloan VS, Cameron P, Porter G, Gammon M, Amaya M, Mellins E, Zaller DM. Mediation by HLA-DM of dissociation of peptides from HLA-DR. Nature. 1995;375:802–806. doi: 10.1038/375802a0. [DOI] [PubMed] [Google Scholar]
- 8.Kropshofer H, Vogt AB, Moldenhauer G, Hammer J, Blum JS, Hammerling GJ. Editing of the HLA-DR-peptide repertoire by HLA-DM. EMBO J. 1996;15:6144–6154. [PMC free article] [PubMed] [Google Scholar]
- 9.Morris P, Shaman J, Attaya M, Amaya M, Goodman S, Bergman C, Monaco JJ, Mellins E. An essential role for HLA-DM in antigen presentation by class II major histocompatibility molecules. Nature. 1994;368:551–554. doi: 10.1038/368551a0. [DOI] [PubMed] [Google Scholar]
- 10.Denzin LK, Cresswell P. HLA-DM induces CLIP dissociation from MHC class II alpha beta dimers and facilitates peptide loading. Cell. 1995;82:155–165. doi: 10.1016/0092-8674(95)90061-6. [DOI] [PubMed] [Google Scholar]
- 11.Haque A, Hajiaghamohseni LM, Li P, Toomy K, Blum JS. Invariant chain modulates HLA class II protein recycling and peptide presentation in nonprofessional antigen presenting cells. Cellular immunology. 2007;249:20–29. doi: 10.1016/j.cellimm.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pu Z, Lovitch SB, Bikoff EK, Unanue ER. T cells distinguish MHC-peptide complexes formed in separate vesicles and edited by H2-DM. Immunity. 2004;20:467–476. doi: 10.1016/s1074-7613(04)00073-1. [DOI] [PubMed] [Google Scholar]
- 13.Lazarski CA, Chaves FA, Sant AJ. The impact of DM on MHC class II-restricted antigen presentation can be altered by manipulation of MHC-peptide kinetic stability. J Exp Med. 2006;203:1319–1328. doi: 10.1084/jem.20060058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amria S, Hajiaghamohseni LM, Harbeson C, Zhao D, Goldstein O, Blum JS, Haque A. HLA-DM negatively regulates HLA-DR4-restricted collagen pathogenic peptide presentation and T cell recognition. Eur J Immunol. 2008;38:1961–1970. doi: 10.1002/eji.200738100. [DOI] [PubMed] [Google Scholar]
- 15.Hartman IZ, Kim A, Cotter RJ, Walter K, Dalai SK, Boronina T, Griffith W, Lanar DE, Schwenk R, Krzych U, Cole RN, Sadegh-Nasseri S. A reductionist cell-free major histocompatibility complex class II antigen processing system identifies immunodominant epitopes. Nat Med. 2010;16:1333–1340. doi: 10.1038/nm.2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kremer AN, van der Meijden ED, Honders MW, Goeman JJ, Wiertz EJ, Falkenburg JH, Griffioen M. Endogenous HLA class II epitopes that are immunogenic in vivo show distinct behavior toward HLA-DM and its natural inhibitor HLA-DO. Blood. 2012;120:3246–3255. doi: 10.1182/blood-2011-12-399311. [DOI] [PubMed] [Google Scholar]
- 17.Yin L, Calvo-Calle JM, Dominguez-Amorocho O, Stern LJ. HLA-DM constrains epitope selection in the human CD4 T cell response to vaccinia virus by favoring the presentation of peptides with longer HLA-DM-mediated half-lives. J Immunol. 2012;189:3983–3994. doi: 10.4049/jimmunol.1200626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mohan JF, Unanue ER. A novel pathway of presentation by class II-MHC molecules involving peptides or denatured proteins important in autoimmunity. Mol Immunol. 2013;55:166–168. doi: 10.1016/j.molimm.2012.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim CY, Quarsten H, Bergseng E, Khosla C, Sollid LM. Structural basis for HLA-DQ2-mediated presentation of gluten epitopes in celiac disease. Proc Natl Acad Sci U S A. 2004;101:4175–4179. doi: 10.1073/pnas.0306885101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nicholson MJ, Moradi B, Seth NP, Xing X, Cuny GD, Stein RL, Wucherpfennig KW. Small molecules that enhance the catalytic efficiency of HLA-DM. J Immunol. 2006;176:4208–4220. doi: 10.4049/jimmunol.176.7.4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pos W, Sethi DK, Call MJ, Schulze MS, Anders AK, Pyrdol J, Wucherpfennig KW. Crystal structure of the HLA-DM-HLA-DR1 complex defines mechanisms for rapid peptide selection. Cell. 2012;151:1557–1568. doi: 10.1016/j.cell.2012.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guce AI, Mortimer SE, Yoon T, Painter CA, Jiang W, Mellins ED, Stern LJ. HLA-DO acts as a substrate mimic to inhibit HLA-DM by a competitive mechanism. Nat Struct Mol Biol. 2013;20:90–98. doi: 10.1038/nsmb.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lazarski CA, Chaves FA, Jenks SA, Wu S, Richards KA, Weaver JM, Sant AJ. The kinetic stability of MHC class II:peptide complexes is a key parameter that dictates immunodominance. Immunity. 2005;23:29–40. doi: 10.1016/j.immuni.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 24.Sant AJ, Chaves FA, Jenks SA, Richards KA, Menges P, Weaver JM, Lazarski CA. The relationship between immunodominance, DM editing, and the kinetic stability of MHC class II:peptide complexes. Immunol Rev. 2005;207:261–278. doi: 10.1111/j.0105-2896.2005.00307.x. [DOI] [PubMed] [Google Scholar]
- 25.Fung-Leung WP, Surh CD, Liljedahl M, Pang J, Leturcq D, Peterson PA, Webb SR, Karlsson L. Antigen presentation and T cell development in H2-M-deficient mice. Science. 1996;271:1278–1281. doi: 10.1126/science.271.5253.1278. [DOI] [PubMed] [Google Scholar]
- 26.Martin WD, Hicks GG, Mendiratta SK, Leva HI, Ruley HE, Van Kaer L. H2-M mutant mice are defective in the peptide loading of class II molecules, antigen presentation, and T cell repertoire selection. Cell. 1996;84:543–550. doi: 10.1016/s0092-8674(00)81030-2. [DOI] [PubMed] [Google Scholar]
- 27.Miyazaki T, Wolf P, Tourne S, Waltzinger C, Dierich A, Barois N, Ploegh H, Benoist C, Mathis D. Mice lacking H2-M complexes, enigmatic elements of the MHC class II peptide-loading pathway. Cell. 1996;84:531–541. doi: 10.1016/s0092-8674(00)81029-6. [DOI] [PubMed] [Google Scholar]
- 28.Pashine A, Busch R, Belmares MP, Munning JN, Doebele RC, Buckingham M, Nolan GP, Mellins ED. Interaction of HLA-DR with an acidic face of HLA-DM disrupts sequence-dependent interactions with peptides. Immunity. 2003;19:183–192. doi: 10.1016/s1074-7613(03)00200-0. [DOI] [PubMed] [Google Scholar]
- 29.Weber DA, Evavold BD, Jensen PE. Enhanced dissociation of HLA-DR-bound peptides in the presence of HLA-DM. Science. 1996;274:618–620. doi: 10.1126/science.274.5287.618. [DOI] [PubMed] [Google Scholar]
- 30.Stratikos E, Wiley DC, Stern LJ. Enhanced catalytic action of HLA-DM on the exchange of peptides lacking backbone hydrogen bonds between their N-terminal region and the MHC class II alpha-chain. J Immunol. 2004;172:1109–1117. doi: 10.4049/jimmunol.172.2.1109. [DOI] [PubMed] [Google Scholar]
- 31.Narayan K, Chou CL, Kim A, Hartman IZ, Dalai S, Khoruzhenko S, Sadegh-Nasseri S. HLA-DM targets the hydrogen bond between the histidine at position beta81 and peptide to dissociate HLA-DR-peptide complexes. Nat Immunol. 2007;8:92–100. doi: 10.1038/ni1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Narayan K, Su KW, Chou CL, Khoruzhenko S, Sadegh-Nasseri S. HLA-DM mediates peptide exchange by interacting transiently and repeatedly with HLA-DR1. Mol Immunol. 2009;46:3157–3162. doi: 10.1016/j.molimm.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou Z, Callaway KA, Weber DA, Jensen PE. Cutting edge: HLA-DM functions through a mechanism that does not require specific conserved hydrogen bonds in class II MHC-peptide complexes. J Immunol. 2009;183:4187–4191. doi: 10.4049/jimmunol.0901663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferrante A, Gorski J. Cutting edge: HLA-DM-mediated peptide exchange functions normally on MHC class II-peptide complexes that have been weakened by elimination of a conserved hydrogen bond. J Immunol. 2010;184:1153–1158. doi: 10.4049/jimmunol.0902878. [DOI] [PubMed] [Google Scholar]
- 35.Anders AK, Call MJ, Schulze MS, Fowler KD, Schubert DA, Seth NP, Sundberg EJ, Wucherpfennig KW. HLA-DM captures partially empty HLA-DR molecules for catalyzed removal of peptide. Nat Immunol. 2011;12:54–61. doi: 10.1038/ni.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Painter CA, Negroni MP, Kellersberger KA, Zavala-Ruiz Z, Evans JE, Stern LJ. Conformational lability in the class II MHC 310 helix and adjacent extended strand dictate HLA-DM susceptibility and peptide exchange. Proc Natl Acad Sci U S A. 2011;108:19329–19334. doi: 10.1073/pnas.1108074108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ferrante A, Anderson MW, Klug CS, Gorski J. HLA-DM mediates epitope selection by a “compare-exchange” mechanism when a potential peptide pool is available. PLoS One. 2008;3:e3722. doi: 10.1371/journal.pone.0003722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yin L, Stern LJ. HLA-DM Focuses on Conformational Flexibility Around P1 Pocket to Catalyze Peptide Exchange. Frontiers in immunology. 2013;4:336. doi: 10.3389/fimmu.2013.00336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yin L, Stern LJ. A novel method to measure HLA-DM-susceptibility of peptides bound to MHC class II molecules based on peptide binding competition assay and differential IC(50) determination. J Immunol Methods. 2014;406:21–33. doi: 10.1016/j.jim.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Painter CA, Cruz A, Lopez GE, Stern LJ, Zavala-Ruiz Z. Model for the peptide-free conformation of class II MHC proteins. PLoS One. 2008;3:e2403. doi: 10.1371/journal.pone.0002403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Painter CA, Stern LJ. Conformational variation in structures of classical and non-classical MHCII proteins and functional implications. Immunol Rev. 2012;250:144–157. doi: 10.1111/imr.12003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yin L, Trenh P, Guce A, Wieczorek M, Lange S, Sticht J, Jiang W, Bylsma M, Mellins ED, Freund C, Stern LJ. Susceptibility to HLA-DM protein is determined by a dynamic conformation of major histocompatibility complex class II molecule bound with peptide. J Biol Chem. 2014;289:23449–23464. doi: 10.1074/jbc.M114.585539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Joshi RV, Zarutskie JA, Stern LJ. A three-step kinetic mechanism for peptide binding to MHC class II proteins. Biochemistry. 2000;39:3751–3762. doi: 10.1021/bi9923656. [DOI] [PubMed] [Google Scholar]
- 44.Rabinowitz JD, Vrljic M, Kasson PM, Liang MN, Busch R, Boniface JJ, Davis MM, McConnell HM. Formation of a highly peptide-receptive state of class II MHC. Immunity. 1998;9:699–709. doi: 10.1016/s1074-7613(00)80667-6. [DOI] [PubMed] [Google Scholar]
- 45.Natarajan SK, Assadi M, Sadegh-Nasseri S. Stable peptide binding to MHC class II molecule is rapid and is determined by a receptive conformation shaped by prior association with low affinity peptides. J Immunol. 1999;162:4030–4036. [PubMed] [Google Scholar]
- 46.Chou CL, Sadegh-Nasseri S. HLA-DM recognizes the flexible conformation of major histocompatibility complex class II. J Exp Med. 2000;192:1697–1706. doi: 10.1084/jem.192.12.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Denzin LK, Hammond C, Cresswell P. HLA-DM interactions with intermediates in HLA-DR maturation and a role for HLA-DM in stabilizing empty HLA-DR molecules. J Exp Med. 1996;184:2153–2165. doi: 10.1084/jem.184.6.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kropshofer H, Arndt SO, Moldenhauer G, Hammerling GJ, Vogt AB. HLA-DM acts as a molecular chaperone and rescues empty HLA-DR molecules at lysosomal pH. Immunity. 1997;6:293–302. doi: 10.1016/s1074-7613(00)80332-5. [DOI] [PubMed] [Google Scholar]
- 49.Zarutskie JA, Busch R, Zavala-Ruiz Z, Rushe M, Mellins ED, Stern LJ. The kinetic basis of peptide exchange catalysis by HLA-DM. Proc Natl Acad Sci U S A. 2001;98:12450–12455. doi: 10.1073/pnas.211439398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grotenbreg GM, Nicholson MJ, Fowler KD, Wilbuer K, Octavio L, Yang M, Chakraborty AK, Ploegh HL, Wucherpfennig KW. Empty class II major histocompatibility complex created by peptide photolysis establishes the role of DM in peptide association. J Biol Chem. 2007;282:21425–21436. doi: 10.1074/jbc.M702844200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Busch R, Doebele RC, von Scheven E, Fahrni J, Mellins ED. Aberrant intermolecular disulfide bonding in a mutant HLA-DM molecule: implications for assembly, maturation, and function. J Immunol. 1998;160:734–743. [PubMed] [Google Scholar]
- 52.Gorga JC, Horejsi V, Johnson DR, Raghupathy R, Strominger JL. Purification and characterization of class II histocompatibility antigens from a homozygous human B cell line. J Biol Chem. 1987;262:16087–16094. [PubMed] [Google Scholar]
- 53.Yin L, Stern LJ. Measurement of Peptide Binding to MHC Class II Molecules by Fluorescence Polarization. In: Coligan John E, et al., editors. Current protocols in immunology. Vol. 106. 2014. pp. 5 10 11–15 10 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Johnson KA. Fitting enzyme kinetic data with KinTek Global Kinetic Explorer. Methods in enzymology. 2009;467:601–626. doi: 10.1016/S0076-6879(09)67023-3. [DOI] [PubMed] [Google Scholar]
- 55.Johnson KA, Simpson ZB, Blom T. Global kinetic explorer: a new computer program for dynamic simulation and fitting of kinetic data. Analytical biochemistry. 2009;387:20–29. doi: 10.1016/j.ab.2008.12.024. [DOI] [PubMed] [Google Scholar]
- 56.Johnson KA, Simpson ZB, Blom T. FitSpace explorer: an algorithm to evaluate multidimensional parameter space in fitting kinetic data. Analytical biochemistry. 2009;387:30–41. doi: 10.1016/j.ab.2008.12.025. [DOI] [PubMed] [Google Scholar]
- 57.Farfan-Arribas DJ, Stern LJ, Rock KL. Using intein catalysis to probe the origin of major histocompatibility complex class I-presented peptides. Proc Natl Acad Sci U S A. 2012;109:16998–17003. doi: 10.1073/pnas.1210271109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hsieh FK, Kulaeva OI, Patel SS, Dyer PN, Luger K, Reinberg D, Studitsky VM. Histone chaperone FACT action during transcription through chromatin by RNA polymerase II. Proc Natl Acad Sci U S A. 2013;110:7654–7659. doi: 10.1073/pnas.1222198110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Roberts KM, Pavon JA, Fitzpatrick PF. Kinetic mechanism of phenylalanine hydroxylase: intrinsic binding and rate constants from single-turnover experiments. Biochemistry. 2013;52:1062–1073. doi: 10.1021/bi301675e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Colabroy KL, Smith IR, Vlahos AH, Markham AJ, Jakubik ME. Defining a kinetic mechanism for l-DOPA 2,3 dioxygenase, a single-domain type I extradiol dioxygenase from Streptomyces lincolnensis. Biochimica et biophysica acta. 2014;1844:607–614. doi: 10.1016/j.bbapap.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 61.Hall FC, Rabinowitz JD, Busch R, Visconti KC, Belmares M, Patil NS, Cope AP, Patel S, McConnell HM, Mellins ED, Sonderstrup G. Relationship between kinetic stability and immunogenicity of HLA-DR4/peptide complexes. Eur J Immunol. 2002;32:662–670. doi: 10.1002/1521-4141(200203)32:3<662::AID-IMMU662>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 62.Stern LJ, Wiley DC. The human class II MHC protein HLA-DR1 assembles as empty alpha beta heterodimers in the absence of antigenic peptide. Cell. 1992;68:465–477. doi: 10.1016/0092-8674(92)90184-e. [DOI] [PubMed] [Google Scholar]
- 63.Natarajan SK, Stern LJ, Sadegh-Nasseri S. Sodium dodecyl sulfate stability of HLA-DR1 complexes correlates with burial of hydrophobic residues in pocket 1. J Immunol. 1999;162:3463–3470. [PubMed] [Google Scholar]
- 64.Ferrante A, Gorski J. A Peptide/MHCII conformer generated in the presence of exchange peptide is substrate for HLA-DM editing. Sci Rep. 2012;2:386. doi: 10.1038/srep00386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zarutskie JA, Sato AK, Rushe MM, Chan IC, Lomakin A, Benedek GB, Stern LJ. A conformational change in the human major histocompatibility complex protein HLA-DR1 induced by peptide binding. Biochemistry. 1999;38:5878–5887. doi: 10.1021/bi983048m. [DOI] [PubMed] [Google Scholar]
- 66.Germain RN, Rinker AG., Jr Peptide binding inhibits protein aggregation of invariant-chain free class II dimers and promotes surface expression of occupied molecules. Nature. 1993;363:725–728. doi: 10.1038/363725a0. [DOI] [PubMed] [Google Scholar]
- 67.Sadegh-Nasseri S, Stern LJ, Wiley DC, Germain RN. MHC class II function preserved by low-affinity peptide interactions preceding stable binding. Nature. 1994;370:647–650. doi: 10.1038/370647a0. [DOI] [PubMed] [Google Scholar]
- 68.Doebele RC, Busch R, Scott HM, Pashine A, Mellins ED. Determination of the HLA-DM interaction site on HLA-DR molecules. Immunity. 2000;13:517–527. doi: 10.1016/s1074-7613(00)00051-0. [DOI] [PubMed] [Google Scholar]
- 69.Sanderson F, Thomas C, Neefjes J, Trowsdale J. Association between HLA-DM and HLA-DR in vivo. Immunity. 1996;4:87–96. doi: 10.1016/s1074-7613(00)80301-5. [DOI] [PubMed] [Google Scholar]
- 70.Belmares MP, Busch R, Wucherpfennig KW, McConnell HM, Mellins ED. Structural factors contributing to DM susceptibility of MHC class II/peptide complexes. J Immunol. 2002;169:5109–5117. doi: 10.4049/jimmunol.169.9.5109. [DOI] [PubMed] [Google Scholar]
- 71.Pos W, Sethi DK, Wucherpfennig KW. Mechanisms of peptide repertoire selection by HLA-DM. Trends in immunology. 2013;34:495–501. doi: 10.1016/j.it.2013.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mellins ED, Stern LJ. HLA-DM and HLA-DO, key regulators of MHC-II processing and presentation. Curr Opin Immunol. 2014;26:115–122. doi: 10.1016/j.coi.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Germain RN. MHC-dependent antigen processing and peptide presentation: providing ligands for T lymphocyte activation. Cell. 1994;76:287–299. doi: 10.1016/0092-8674(94)90336-0. [DOI] [PubMed] [Google Scholar]
- 74.Trombetta ES, Ebersold M, Garrett W, Pypaert M, Mellman I. Activation of lysosomal function during dendritic cell maturation. Science. 2003;299:1400–1403. doi: 10.1126/science.1080106. [DOI] [PubMed] [Google Scholar]
- 75.McCurley N, Mellman I. Monocyte-derived dendritic cells exhibit increased levels of lysosomal proteolysis as compared to other human dendritic cell populations. PLoS One. 2010;5:e11949. doi: 10.1371/journal.pone.0011949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lennon-Dumenil AM, Bakker AH, Maehr R, Fiebiger E, Overkleeft HS, Rosemblatt M, Ploegh HL, Lagaudriere-Gesbert C. Analysis of protease activity in live antigen-presenting cells shows regulation of the phagosomal proteolytic contents during dendritic cell activation. J Exp Med. 2002;196:529–540. doi: 10.1084/jem.20020327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fernandez-Borja M, Verwoerd D, Sanderson F, Aerts H, Trowsdale J, Tulp A, Neefjes J. HLA-DM and MHC class II molecules co-distribute with peptidase-containing lysosomal subcompartments. International immunology. 1996;8:625–640. doi: 10.1093/intimm/8.5.625. [DOI] [PubMed] [Google Scholar]
- 78.Xu X, Song W, Cho H, Qiu Y, Pierce SK. Intracellular transport of invariant chain-MHC class II complexes to the peptide-loading compartment. J Immunol. 1995;155:2984–2992. [PubMed] [Google Scholar]
- 79.Schafer PH, Green JM, Malapati S, Gu L, Pierce SK. HLA-DM is present in one-fifth the amount of HLA-DR in the class II peptide-loading compartment where it associates with leupeptin-induced peptide (LIP)-HLA-DR complexes. J Immunol. 1996;157:5487–5495. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.