SUMMARY
Histone variants are emerging as key regulatory molecules in cancer. Here we report a novel role for the H2A.Z isoform H2A.Z.2 as a driver of malignant melanoma. H2A.Z.2 is highly expressed in metastatic melanoma, correlates with decreased patient survival, and is required for cellular proliferation. Our integrated genomic analyses reveal that H2A.Z.2 controls the transcriptional output of E2F target genes in melanoma cells. These genes are highly expressed and display a distinct signature of H2A.Z occupancy. We identify BRD2 as an H2A.Z interacting protein, whose levels are also elevated in melanoma. We further demonstrate that H2A.Z.2 regulated genes are bound by BRD2 and E2F1 in a H2A.Z.2-dependent manner. Importantly, H2A.Z.2 deficiency sensitizes melanoma cells to chemotherapy and targeted therapies. Collectively, our findings implicate H2A.Z.2 as a mediator of cell proliferation and drug sensitivity in malignant melanoma, holding translational potential for novel therapeutic strategies.
INTRODUCTION
Malignant melanoma is the most lethal form of skin cancer with rising incidence and remains largely incurable. While advances in immune and targeted therapies have made tremendous progress recently (Chapman et al., 2011; Kaufman et al., 2013), they are effective only in distinct subsets of patients or result in the emergence of drug resistance (Lito et al., 2013). Thus, investigation of alternative approaches is essential.
Recent studies have shed light on the importance of epigenetic regulation in melanoma biology. Key roles for BRD4 (Segura et al., 2013), histone methyltransferases SETDB1 (Ceol et al., 2011) and EZH2 (Zingg et al., 2015), and the histone variant macroH2A (Kapoor et al., 2010) have been reported. Relevant to the present study, histone variants and their chaperones are emerging as key regulatory molecules in cancer (Vardabasso et al., 2013).
H2A.Z is a highly conserved H2A variant, with only 60% identity to canonical H2A, and is expressed and incorporated into chromatin throughout the cell cycle (Bonisch and Hake, 2012). While somewhat confounded by species-specific functions and context-dependencies, the role of H2A.Z in transcriptional regulation is well established (Svotelis et al., 2009). H2A.Z is enriched at gene promoters, as well as other regulatory regions, generally exerting a positive role on transcription (Hu et al., 2012; Obri et al., 2014).
Two distinct H2A.Z isoforms, H2A.Z.1 and H2A.Z.2, have been identified in the vertebrate genome as products of two non-allelic genes, H2AFZ and H2AFV, respectively (Dryhurst et al., 2009; Horikoshi et al., 2013; Matsuda et al., 2010). While differing by only three amino acids at the protein level, H2A.Z.1 and H2A.Z.2 are encoded by distinct nucleotide sequences. Isoform-specific functions remain unclear, and H2A.Z.1 mouse knock out studies suggest that the two genes are non-redundant (Faast et al., 2001). In the context of tumorigenesis, H2A.Z is overexpressed in breast, prostate and bladder cancers, where in some cases, it regulates proliferation (reviewed in Vardabasso et al., 2013). However, these studies either focused solely on H2A.Z.1, or did not clearly distinguish between isoforms.
Here we report a distinct role for H2A.Z.2 in melanoma. H2A.Z.2 is highly expressed in melanoma and drives proliferation by promoting expression of E2F target genes. These cell cycle regulatory genes are highly expressed and acquire a unique signature of H2A.Z occupancy – high promoter enrichment and gene body depletion. We further identified the BET (bromodomain and extraterminal domain) protein BRD2 as an H2A.Z interacting protein, whose levels are also elevated in melanoma specimens. Depletion of H2A.Z.2 results in reduced histone acetylation, BRD2 and E2F1 levels, and impairs recruitment of BRD2 and E2F1 to its target genes. Moreover, H2A.Z.2 deficiency cooperates with BET or MEK inhibition to induce melanoma cell death. Hence, our studies suggest that targeting H2A.Z deposition may be effective therapeutically in combination with existing or emerging therapies for melanoma.
RESULTS
H2A.Z isoforms are overexpressed in melanoma
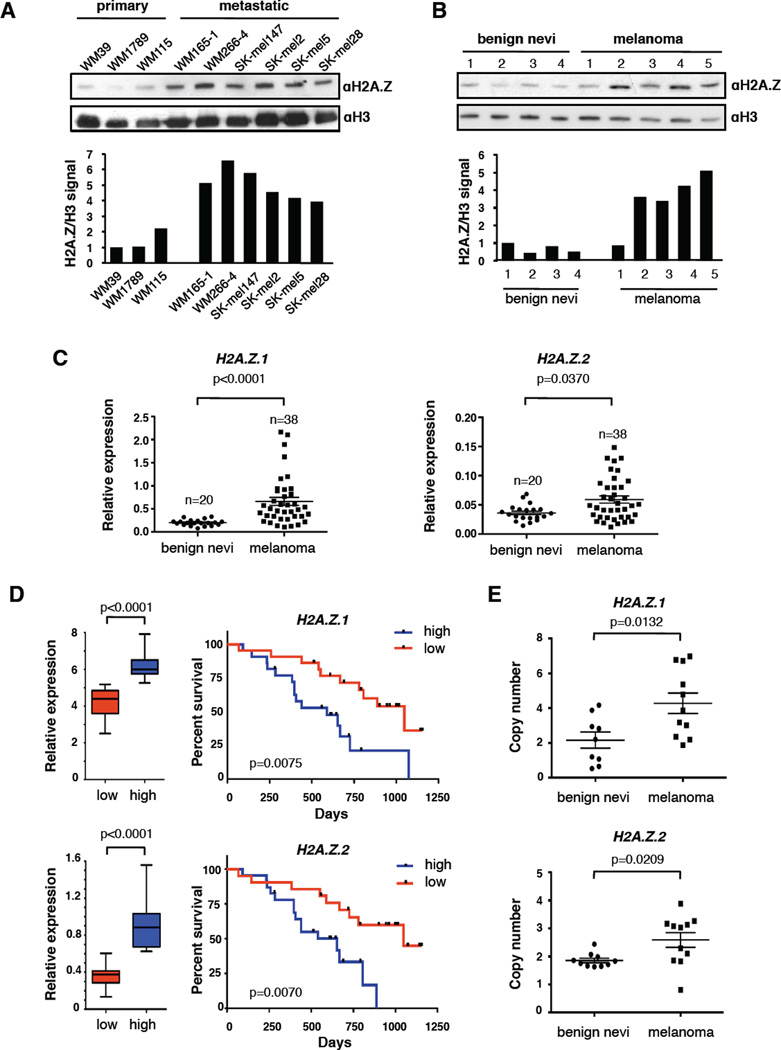
By probing a panel of primary and metastatic melanoma cell lines we detected increased levels of H2A.Z protein in metastatic cells (Figure 1A). Immunoblotting of histones extracted from benign nevi and melanoma specimens revealed increased H2A.Z in melanoma tissues (Figure 1B). We also investigated H2A.Z levels in human primary melanocytes induced to senesce via serial passaging (replicative senescence) and BRAFV600E (oncogene-induced senescence) (Duarte et al., 2014). We observed diminished H2A.Z upon both modes of senescence (Figure S1A). Together, these data link global levels of H2A.Z to cellular proliferation.
Figure 1. H2A.Z.1 and H2A.Z.2 are overexpressed in melanoma.
(A) Chromatin extracted from primary and metastatic cell lines probed with H2A.Z antibody; H3 used for loading. Signals quantified by densitometry. See also Figure S1B for mRNA expression.
(B) H2A.Z immunoblot of acid extracted histones from fresh frozen human benign nevi and melanoma specimens; H3 used for loading. Signals quantified as in (A).
(C) Expression analysis by qRT-PCR of H2A.Z.1 and H2A.Z.2 in benign nevi (n=20) and melanoma (n=38). Values normalized to GAPDH; mean ± SEM. Mann-Whitney test (two-tailed).
(D) Survival of melanoma patients with high and low (above or below the median, respectively) mRNA levels of H2A.Z.1 and H2A.Z.2. Gene expression data of 44 metastatic melanoma tissues (Bogunovic et al., 2009) were used to define high and low expressor groups (boxplots, Mann-Whitney test) and to generate Kaplan-Meier curves (log-rank test).
(E) Analysis of H2A.Z.1 and H2A.Z.2 gene copy number by qPCR of a subset of benign nevi and melanoma in (C), relative to primary melanocytes. Data are mean ± SEM; unpaired Student’s test (two-tailed). See also Figure S1D, E.
To assess whether H2A.Z expression is regulated transcriptionally, as well as to examine the individual H2A.Z isoforms (which is not possible with currently available antibodies), we performed quantitative RT-PCR (qRT-PCR) using isoform-specific primers. H2A.Z isoforms are decreased in human melanocytes induced to senescence (Figure S1A). Conversely, in a panel of benign nevi and melanoma specimens, we observed increased H2A.Z.1 and H2A.Z.2 mRNA in melanoma (Figure 1C). H2A.Z.1 and H2A.Z.2 mRNA levels were also increased in cell lines derived from metastatic versus primary melanoma Figure S1B). Analysis of published transcriptional data is consistent with these findings (Talantov et al., 2005; Riker et al., 2008; Xu et al., 2008) (Figure S1C). Finally, in a cohort of patients followed clinically for 3 years after excision of metastatic lesions (Bogunovic et al., 2009), patients with high H2A.Z.1 and H2A.Z.2 showed significantly lower survival (Figure 1D). Collectively, these findings suggest that H2A.Z isoforms have a functional role in melanoma progression.
We next performed quantitative copy number analysis of H2A.Z.1 and H2A.Z.2 in nevi and metastases by qPCR and detected copy gains for both (Figure 1E). The Cancer Genome Atlas (TCGA) reports increased copy number in 13% and 52% of cutaneous melanomas for H2A.Z.1 and H2A.Z.2, respectively, which correlates with increased mRNA levels (Figure S1D). Fluorescent In Situ Hybridization (FISH) of melanoma cell lines corroborated these findings (Figure S1E).
H2A.Z.2 depletion induces G1/S arrest in melanoma cells
Next we investigated the functional consequences of depleting H2A.Z.1 and H2A.Z.2 in melanoma cell lines. Using sequence-specific shRNAs for H2A.Z isoforms, we established stable SK-mel147 (NRASQ61R), WM266-4 (BRAFV600D), and 501mel (BRAFV600E) cell lines targeting either H2A.Z.1 or H2A.Z.2. Knock down was monitored by qRT-PCR and/or immunoblot (Figure S2A–C). As H2A.Z.1 is the predominant isoform in melanoma (via RNA-sequencing, below), its knock down can be appreciated at the protein level, while H2A.Z.2 knock down is obscured by H2A.Z.1 (Figure S2A, B).
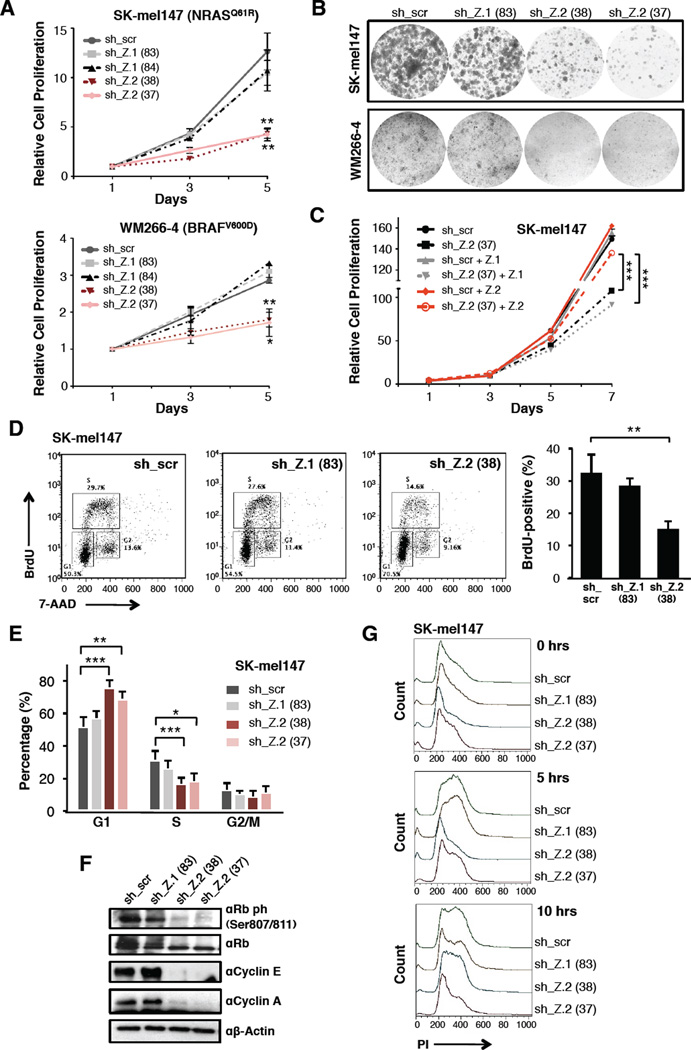
We observed that loss of H2A.Z.2, but not H2A.Z.1, reduced proliferation in all cell lines (Figure 2A, B, S2D). To confirm these variant-specific effects, we generated cells stably expressing H2A.Z.1 or an shRNA-resistant H2A.Z.2 that were infected with sh_Z.2 and a control (sh_scr). Only those cells expressing an shRNA-resistant H2A.Z.2 were able to overcome the proliferation defect induced by sh_Z.2 (Figure 2C). Interestingly, HeLa cells depleted of H2A.Z isoforms did not show proliferation defects (Figure S2E). Thus, H2A.Z isoforms exert distinct and non-redundant functions in melanoma cells.
Figure 2. H2A.Z.2 depletion induces G1/S arrest in melanoma cells.
(A) Proliferation curves of SK-mel147 and WM266-4 cells expressing control and isoform-specific shRNAs as shown. Data are mean ± SEM (n≥3); two-way ANOVA. See also Figure S2D.
(B) Colony formation assays of SK-mel147 and WM266-4 cells expressing shRNAs as in (A).
(C) Proliferation assay of SK-mel147 cells expressing H2A.Z.1, shRNA-resistant H2A.Z.2, or empty vector control. Each line was infected with H2A.Z.2 shRNA (sh_37) and with sh_scr. Data are mean ± SEM (n=2); two-way ANOVA.
(D) BrdU staining of SK-mel147 cells expressing shRNAs as in (A). Profiles from one representative experiment are displayed. BrdU-positive S phase cells are shown as mean values ± SD (n≥3) (right); unpaired Student’s test (two-tailed). See also Figure S2G.
(E) Percentage of SK-mel147 in G1, S or G2/M phases, as revealed by PI incorporation. Values are mean ± SD (n≥3); unpaired Student’s test (two-tailed). Asterisks as follows, in all figures: *p<0.05, **p<0.01, ***p<0.001. See also Figure S2F.
(F) Whole-cell extracts from shRNA-expressing SK-mel147 cells were immunoblotted for unmodified and phosphorylated Rb and cyclins. B-actin used as loading control.
(G) shRNA-expressing SK-mel147 cells were synchronized at early S phase by a double thymidine block and cell synchrony monitored by flow cytometry of PI stained cells at 5 hour intervals. Flow cytometry profiles from a representative experiment are shown.
H2A.Z.2 knock down induced a G1/S cell cycle arrest (Figure 2D, E, S2F, G), accompanied by hypophosphorylation of Rb and decreased levels of Cyclins E and A (Figure 2F). This phenotype was not consistent with cellular senescence, as the expression of cyclin-dependent kinase (CDK) inhibitors (Figure S2H) and β–galactosidase activity (data not shown) were not increased. Moreover, we observed minimal cell death (Figure S2I). Next, SK-mel147 cells were arrested at early S phase via double thymidine block and subsequently released. Both control and H2A.Z.1 knock down cells progressed through S and G2 phases, and at 10 hours ~30% of the cells re-entered G1. However, H2A.Z.2-depleted cells remained largely arrested for the entire duration of the assay (Figure 2G). These findings suggest that H2A.Z.2 loss causes delayed entry into S phase. These data are strikingly similar to htz1 (H2A.Z) mutant budding yeast, which shows delayed DNA replication and cell cycle progression (Dhillon et al., 2006).
H2A.Z.2 regulates E2F target genes
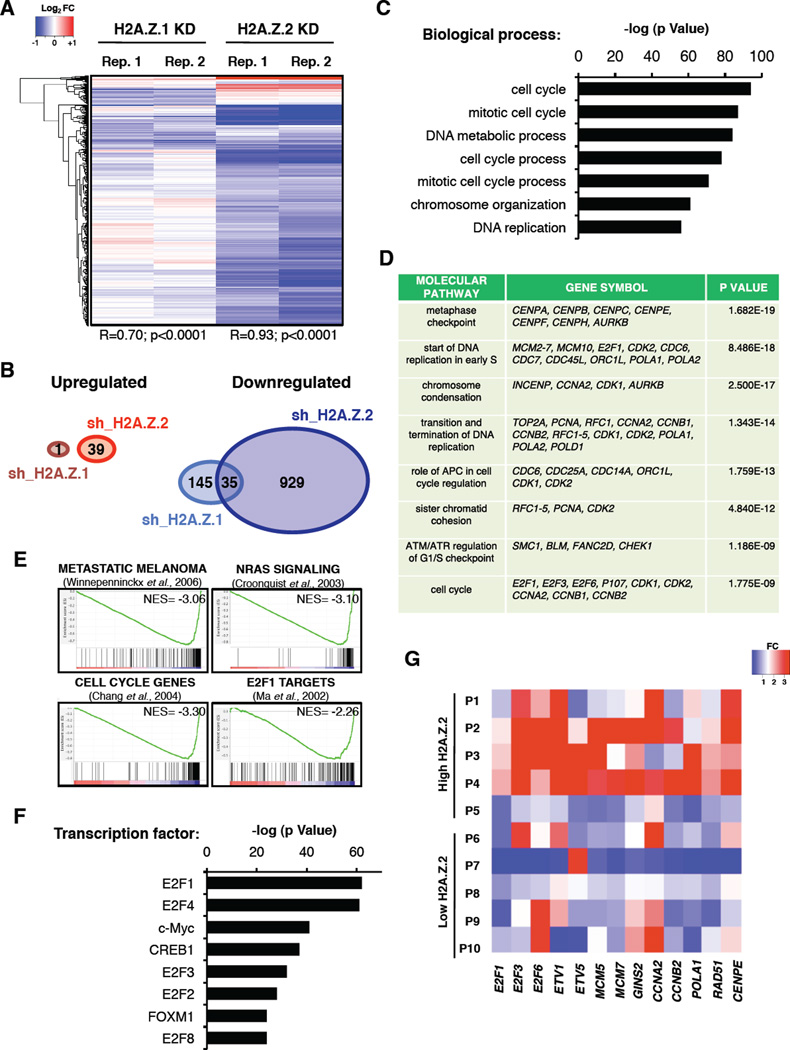
To further understand the observed proliferation defect, we characterized the transcriptional profile of H2A.Z.2-deficient cells. We used Affymetrix microarrays for SK-mel147 and WM266-4 cells depleted of either H2A.Z.1 or H2A.Z.2 (Figure 3A, Table S1). Interestingly, the majority of genes were downregulated (Figure 3A, B), with only 35 overlapping genes between H2A.Z.1 and H2A.Z.2 knock down in SK-mel147 cells (Figure 3B). Similar expression data was observed for WM266-4 cells (Figure S3A, B, Table S1).
Figure 3. H2A.Z.2 regulates cell cycle promoting genes.
(A) Gene expression profiles of SK-mel147 cells upon H2A.Z.1 and H2A.Z.2 knock down (day 8 post infection). Two biological replicates (with Pearson correlation), and genes displaying a significant (lfdr<0.2) change in each replicate are shown.
(B) Venn diagrams exhibiting the numbers of genes that are significantly up- and downregulated upon H2A.Z.1 and H2A.Z.2 knock down in SK-mel147 cells. See also Figure S3A, B.
(C) Functional annotation (biological process) of genes downregulated upon H2A.Z.2 knock down in SK-mel147 cells. Enriched groups are ranked by the most significant p value.
(D) Functional annotation (molecular pathways) of genes as described in (C). Selected genes belonging to each pathway are shown. p value indicated.
(E) GSEA plots of genes altered upon H2A.Z.2 knock down in SK-mel147 show negative correlation gene signatures as shown. FDR=0.0; NES (Normalized Enrichment Score) as indicated.
(F) TF regulation analysis of genes as described in (C). Enriched groups are ranked by the most significant p value. Analyses for panels (C, D, F) were performed with MetaCore. See also Figure S3C–E.
(G) Heatmap generated by qRT-PCR values of the indicated genes in a subset of melanoma specimens (P1-P10) from Figure 1C. P1–P5 = high H2A.Z.2 and P6–P10 = low H2A.Z.2 expression levels (above and below the median, respectively). Expression levels of each gene are shown as fold change (FC) relative to one patient (not shown).
Consistent with the observed phenotype, functional annotation revealed that H2A.Z.2-regulated genes are enriched for cell cycle regulators (Figure 3C, D, Figure S3C). This is in contrast to H2A.Z.1-regulated genes, which are enriched for immunological pathways (Figure S3D). This is in line with the lack of cell cycle defects observed upon H2A.Z.1 knock down, and implicates a distinct role for H2A.Z.1 in melanoma.
Gene Set Enrichment Analysis (GSEA) and transcription factor (TF) analysis further demonstrated that the H2A.Z.2-regulated genes are associated with transcriptional hallmarks of advanced melanoma and are targets of the E2F family, including E2F1 and E2F4 (Figure 3E, F, S3E). Furthermore, qRT-PCR analysis revealed that E2F target gene expression correlates with H2A.Z.2 levels in human melanoma (Figure 3G). Given that E2F1 and E2F4 promote melanoma progression and metastasis (Alla et al., 2009; Ma et al., 2008), these results implicate concerted H2A.Z.2-E2F function in melanoma progression.
H2A.Z.2-regulated genes show a unique signature of H2A.Z occupancy
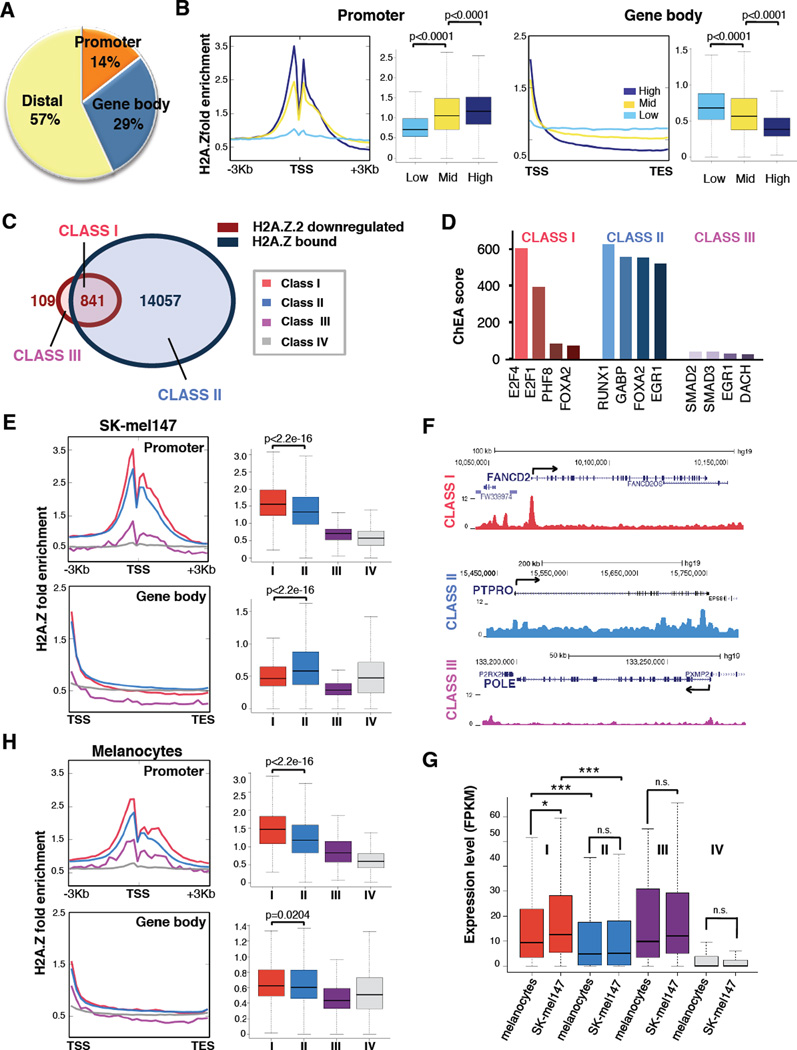
We next performed native chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq) of H2A.Z to determine its genomic occupancy in melanoma cells (Figure 4). ChIP-seq of SK-mel147 cells stably expressing N-terminally eGFP-tagged H2A.Z.1 or H2A.Z.2 (along with eGFP-H2A as a control) was also carried out (Figure S4A), and exhibited highly overlapping genome-wide occupancy patterns with endogenous H2A.Z (Figure S4B, C) as well as with each other (Figure 4SD). Therefore, we utilized endogenous H2A.Z ChIP-seq for further analyses.
Figure 4. A unique signature of H2A.Z occupancy at H2A.Z.2-regulated genes.
(A) Pie chart displaying the percentages of H2A.Z peaks occupying promoters, gene bodies and distal regions. Promoters: −3Kb < TSS < +1Kb; Gene bodies: from +1Kb > TSS to TES; all other regions defined as distal. (TSS: Transcription Start Site; TES: Transcription End Site).
(B) Correlation of H2A.Z signals at the promoter or gene body with mRNA expression levels. Genes were divided by expression level into high (top 25%), medium (middle 50%) and low (bottom 25%) from RNA-sequencing data. Fold enrichment profiles (sliding 100 bp window) and boxplots were calculated around the TSS (−3kb, +3Kb) and over the gene body (TSS to TES) for each group; Mann Whitney test (two-tailed).
(C) Venn diagrams displaying H2A.Z.2 downregulated genes and H2A.Z bound genes by ChIP-seq in SK-mel147. Class I (downregulated in H2A.Z.2 knock down and bound by H2A.Z, red); Class II (bound by H2A.Z but unaffected by H2A.Z.2 knock down, light blue); Class III (downregulated by H2A.Z.2 knock down but not bound by H2A.Z, purple); Class IV (11,003 genes that are not downregulated by H2A.Z.2 knock down and not bound, grey).
(D) ChIP Enrichment Analysis tool (ChEA2) analysis of Class I, Class II and Class III genes (as defined and color coded in panel A). The ChEA2 database contains ChIP-seq data of 200 transcription factors from 221 publications for a total of 458,471 TF-target interactions (Kou et al., 2013). Transcription factors are ranked by ChEA combined score.
(E) H2A.Z occupancy at the promoter and gene body of the four classes of genes defined in (C), in SK-mel147 cells. Profiles and boxplots represent fold enrichment over input. Mann Whitney test (two-tailed).
(F) Captures of the UCSC genome browser (GRCh37/hg19) showing the ChIP-seq profiles for H2A.Z for genes belonging to each of the classes defined in (C). RefSeq annotated genes are displayed on top.
(G) Boxplot representing expression levels (FPKM) for each gene class as in (C), in primary melanocytes and in SK-mel147. Mann Whitney test (two-tailed). Two-way ANOVA.
(H) H2A.Z occupancy at the promoter and gene body of the four classes of genes defined in (C) in primary melanocytes (as in (C)).
Amongst H2A.Z-bound sites, 14% lie within promoters and 29% in gene bodies (Figure 4A). By integrating RNA-sequencing and ChIP analyses in SK-mel147 cells, we found that H2A.Z promoter levels positively correlate with expression (Figure 4B), as previously reported (Barski et al., 2007; Hu et al., 2012). Intriguingly, gene body occupancy shows a striking negative correlation with gene expression (Figure 4B). Finally, in line with previous reports (Barski et al., 2007; Hu et al., 2012; Obri et al., 2014), our results show that half of the H2A.Z-bound sites lie within intergenic regions (Figure 4A).
Next we integrated H2A.Z ChIP-seq with H2A.Z.2 downregulated genes. Taking into account both promoter and gene body occupancy, we defined four classes of genes: H2A.Z-bound and H2A.Z.2 downregulated (Class I), H2A.Z-bound but not H2A.Z.2 downregulated (Class II), H2A.Z.2 downregulated but not H2A.Z-bound (Class III), and neither downregulated nor bound (Class IV) (Figure 4C, Table S2). Gene ontology analyses revealed that Class I is enriched for cell cycle genes, while Class II is enriched for metabolic processes (Figure S4E). Accordingly, ChIP Enrichment Analysis (ChEA2) (Kou et al, 2013) uncovered distinct TF binding profiles for each class, with only Class I showing enrichment for E2Fs (Figure 4D). By examining H2A.Z distribution, we found that Class I genes were significanly enriched at the promoter, and depleted within the gene body (Figure 4E, F). Conversely, many Class II genes lie within broader H2A.Z domains (Figure 4F). Expression of Class I genes is significantly higher than Class II genes in melanoma (Figure 4G), consistent with our findings in Figure 4B.
H2A.Z ChIP-seq in normal human melanocytes revealed that the Class I signature is not detectable, as H2A.Z is not depleted in the gene body (Figure 4H). Consistent with this, Class I genes are expressed at significant lower levels in melanocytes than melanoma cells, while expression of all other classes remains largely unchanged (Figure 4G).
Collectively, our analyses revealed that H2A.Z.2 regulated/H2A.Z bound genes (Class I) have unique features in melanoma cells. They are E2F targets, highly expressed, and enriched for H2A.Z at the promoter and depleted in the gene body. These features do not apply to H2A.Z.1-downregulated genes (Figure S4F, G, Table S3), suggesting a unique chromatin signature at genes that regulate melanoma cell proliferation.
BRD2 interacts with H2A.Z-containing nucleosomes and is overexpressed in melanoma
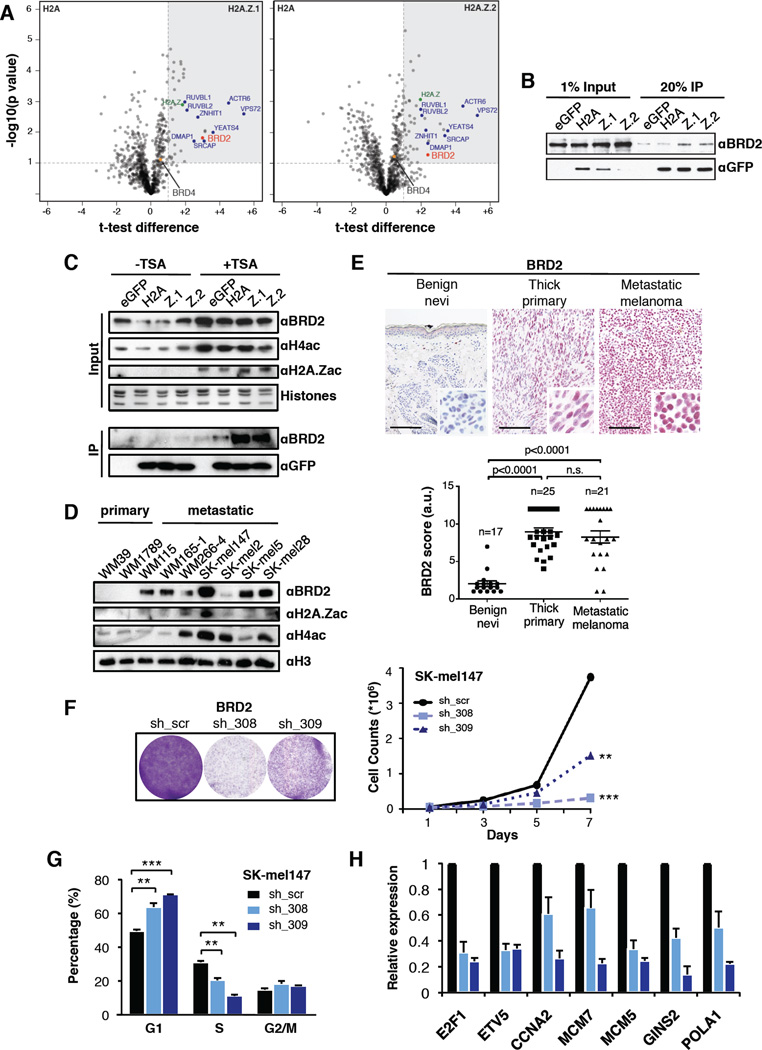
To further decipher H2A.Z function, we investigated the factors that interact with H2A.Z-containing nucleosomes in melanoma cells. To this aim, we utilized unbiased label-free quantitative mass spectrometry (MS) (Eberl et al., 2013). Chromatin isolated from SK-mel147 cells stably expressing eGFP, eGFP-H2A, eGFP-H2A.Z.1 or eGFP-H2A.Z.2 was digested to mononucleosomes (Figure S5A), immunoprecipitated (Figure S5B) and LC-MS/MS analyzed. We found significant enrichment of ~45 H2A.Z interactors as compared to H2A-containing nucleosomes (Figure 5A, S5C). The majority of these proteins were found in both H2A.Z variant IPs, including members of the H2A.Z histone chaperone complex, SRCAP (Billon and Cote, 2012).
Figure 5. BRD2 interacts with H2A.Z-containing nucleosomes and is overexpressed in melanoma.
(A) Volcano plots of label-free interactions of eGFP-H2A.Z.1- or eGFP-H2A.Z.2-containing nucleosomes. Significantly enriched proteins over eGFP-H2A containing nucleosomes are shown in the upper right box (grey shading). Members of the H2A.Z-specific chaperone/remodeling complex SRCAP are highlighted in blue, H2A.Z in green, BRD2 and BRD4 as red and orange dots, respectively. See also Figure S5A–C.
(B) Immunoblots for BRD2 and GFP upon immunoprecipitation of mononucleosomes generated from SK-mel147 cells expressing eGFP, eGFP-H2A, -H2A.Z.1 or -H2A.Z.2.
(C) SK-mel147 cells as in (B) were treated with DMSO or TSA (200nM for 2 hours), and chromatin was probed for BRD2, H4ac, and H2A.Zac (upper panels). Histones used for loading. (Bottom) Immunoblots for BRD2 and GFP upon immunoprecipitation are shown.
(D) Chromatin extracted from primary and metastatic cell lines probed with BRD2, H2A.Zac and H4ac antibodies; H3 used for loading. See also Figure 1A.
(E) IHC for BRD2 in representative intradermal nevi, thick primary, and metastatic melanoma tissue. Images at 20× magnification; insets at 40× magnification. Scale bar represents 100 µm. Scores derived by multplying the number of positively stained cells (1–4) by intensity of stain (1–3); Mann-Whitney (two-tailed).
(F) Colony formation and proliferation assays of SK-mel147 cells expressing control or BRD2 shRNAs as shown. Data are mean ± SEM (n≥2); two-way ANOVA.
(G) Percentage of SK-mel147 cells in G1, S or G2 phases, as shown by PI incorporation. Values are mean ± SD (n≥3); unpaired Student’s test (two-tailed).
(H) Expression of a handful of Class I genes was analyzed by qRT-PCR upon BRD2 knock down. Expression is shown normalized to GAPDH and relative to scrambled shRNA. Mean ± SD is shown (n≥3).
We identified BRD2 to be enriched in H2A.Z.1- and H2A.Z.2-containing nucleosomes (Figure 5A, B). BET proteins (BRD2, BRD3, BRD4 and BRDT) bind to acetylated lysine residues in histones (LeRoy et al., 2008; LeRoy et al., 2012), and function as scaffolds to recruit chromatin modifying enzymes and TFs, thereby coupling histone acetylation to transcription (reviewed in (Belkina and Denis, 2012)). While BRD2 and BRD4 are both overexpressed in melanoma (Segura et al., 2013), only BRD2 interacts with H2A.Z-containing nucleosomes (Figure 5A and data not shown). We next tested whether hyperacetylation of histones would enhance the interaction between BRD2 and H2A.Z isoforms in melanoma cells. Treatment with the HDAC inhibitor trichostatin A (TSA) resulted in increased histone H4 and H2A.Z acetylation and increased BRD2 chromatin association (Figure 5C). Furthermore, the BRD2-H2A.Z interaction was enhanced (Figure 5C). By probing primary and metastatic melanoma cell lines (as in Figure 1A), we observed hyperacetylation of H4 and H2A.Z, and high levels of BRD2 in metastatic cells (Figure 5D). Collectively, these results are consistent with the fact that BRD2 is recruited to chromatin by a combination of acetylated H4 (H4ac) and H2A.Z (Draker et al., 2012).
Through immunohistochemistry (IHC) of BRD2 in a cohort of patient samples including benign nevi, thick primary melanoma and metastatic melanoma, we detected a significant increase of BRD2 in primary and metastatic melanoma specimens as compared to dermal melanocytes of nevi (Figure 5E). Next, we investigated BRD2 knock down in multiple melanoma cell lines and observed proliferation defects via G1/S arrest (Figure 5F, G, S5D–F). BRD2 knock down altered gene expression of selected E2F targets (Figure 5H, S5G). Collectively, BRD2 knock down recapitulated the phenotype observed upon H2A.Z.2 loss, suggesting that H2A.Z.2 and BRD2 work together to promote Class I transcription.
An H2A.Z.2-BRD2-E2F1 axis in melanoma
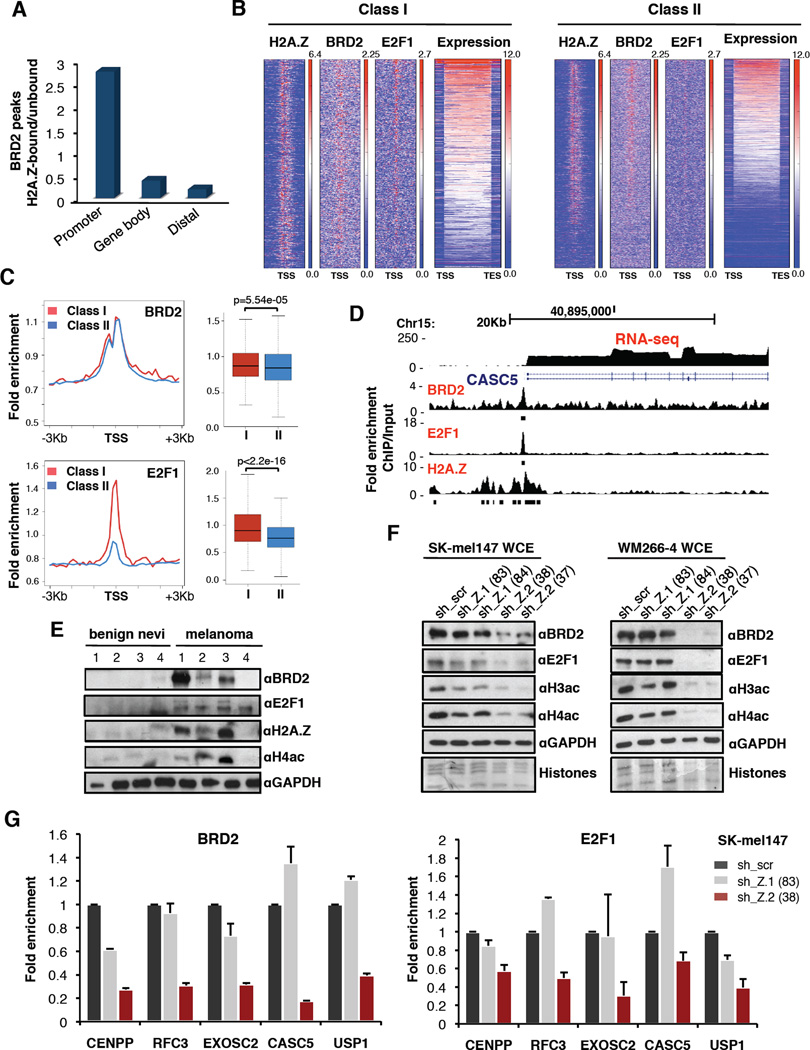
To test this hypothesis, we performed ChIP-seq of BRD2 in SK-mel147 cells, and found a similar genomic distribution as H2A.Z (Figure 4A, S6A). We next determined the extent of co-localization of BRD2 and H2A.Z genome-wide and found that BRD2 peaks overlap with H2A.Z largely at promoters (Figure 6A). Promoters of Classes I and II genes are bound by BRD2 (Figure 6B, S6B), consistent with the fact that BRD2 interacts with both H2A.Z isoforms. However, H2A.Z and BRD2 have the highest enrichment at Class I genes (Figure 4E, 6C).
Figure 6. An H2A.Z.2-BRD2-E2F1 axis in melanoma.
(A) Histograms of the ratio between BRD2 peaks bound by H2A.Z and BRD2 peaks not bound by H2A.Z at promoters, gene bodies and distal regions as defined in Figure 4A.
(B) Heatmaps of promoters (−3Kb, +3Kb) of Class I and Class II genes based on H2A.Z, BRD2 and E2F1 fold enrichment over input, and ranked by expression level. Expression is indicated as log2 RNA-seq signal. See also Figure S6B.
(C) BRD2 and E2F1 occupancy at the promoter Class I and Class II genes in SK-mel147 cells. Profiles and boxplots represent fold enrichment over input. Mann-Whitney test (two-tailed).
(D) UCSC genome browser (GRCh37/hg19) capture of 30Kb region of human chromosome 15 depicting a Class I gene. Read counts (normalized fold enrichment of ChIP over input DNA) for BRD2, E2F1 and H2A.Z and FPKM for RNA-seq are shown. RefSeq annotated genes are displayed above.
(E) Whole-cell extracts from fresh frozen benign nevi and metastatic specimens probed with BRD2, E2F1, H2A.Z and H4ac antibodies; GAPDH used for loading.
(F) Whole-cell extracts from control and isoform-depleted SK-mel147 and WM266-4 cells were immunoblotted for BRD2, E2F1, H3ac and H4ac. GAPDH served as loading control. See also Figure S6C, E.
(G) ChIP-qPCR for BRD2 (left) and E2F1 (right) at Class I genes in SK-mel147 expressing control or isoform-specific shRNAs as indicated. Fold enrichment ChIP/input is plotted, relative to scrambled shRNA. One representative experiment shown; values are mean ± SD (n≥2).
Because our in silico analyses predicted that Class I genes are E2F targets (Figure 4D), and BRD2 interacts with E2F1 to mediate its recruitment to chromatin (Denis et al., 2006; Sinha et al., 2005), we next queried whether Class I genes are bound by E2F1. ChIP-seq of E2F1 in SK-mel147 cells showed this was indeed the case (Figure 6B, C, S6B). Taken together, these data suggest that H2A.Z.2 works cooperatively with BRD2 and E2F1 (Figure 6D) to promote high levels of transcription at Class I genes in melanoma. Next, we probed a panel of benign nevi and melanoma tissues for H2A.Z, BRD2, E2F1 and H4ac, and found evidence of the H2A.Z-BRD2-E2F axis in melanoma specimens (Figure 6E). This reinforces the relevance of our findings for melanoma disease.
H2A.Z.2 depletion impairs BRD2 and E2F1 function
The findings above prompted us to investigate BRD2 and E2F1 levels upon H2A.Z silencing. We observed marked reduction of BRD2 and E2F1 levels upon H2A.Z.2, but not H2A.Z.1, knock down across melanoma cell lines (Figure 6F, S6C). BET family members are not transcriptionally regulated by H2A.Z.2 (Figure S6D), suggesting that H2A.Z.2 stabilizes these factors. This was paralleled by a dramatic loss of H4 and H3 acetylation (Figure 6F and S6C, E). These data suggest that BRD2 and E2F1 chromatin recruitment to Class I genes, mediated by histone acetylation, is impaired in H2A.Z.2-deficient cells. Thus, we performed ChIP-qPCR for BRD2 and E2F1 in either control or H2A.Z isoform-depleted cells, and found that BRD2 and E2F1 recruitment to Class I promoters is dependent on H2A.Z.2 (Figure 6G). Overall, H2A.Z.2 deficiency results in dramatic alterations in chromatin structure, thereby clearly distinguishing it from H2A.Z.1. Overall, our ChIP studies demonstrate that H2A.Z.2, BRD2 and E2F1 work cooperatively to promote high levels of transcription at cell cycle promoting genes in melanoma.
H2A.Z.2 deficiency sensitizes melanoma cells to therapy
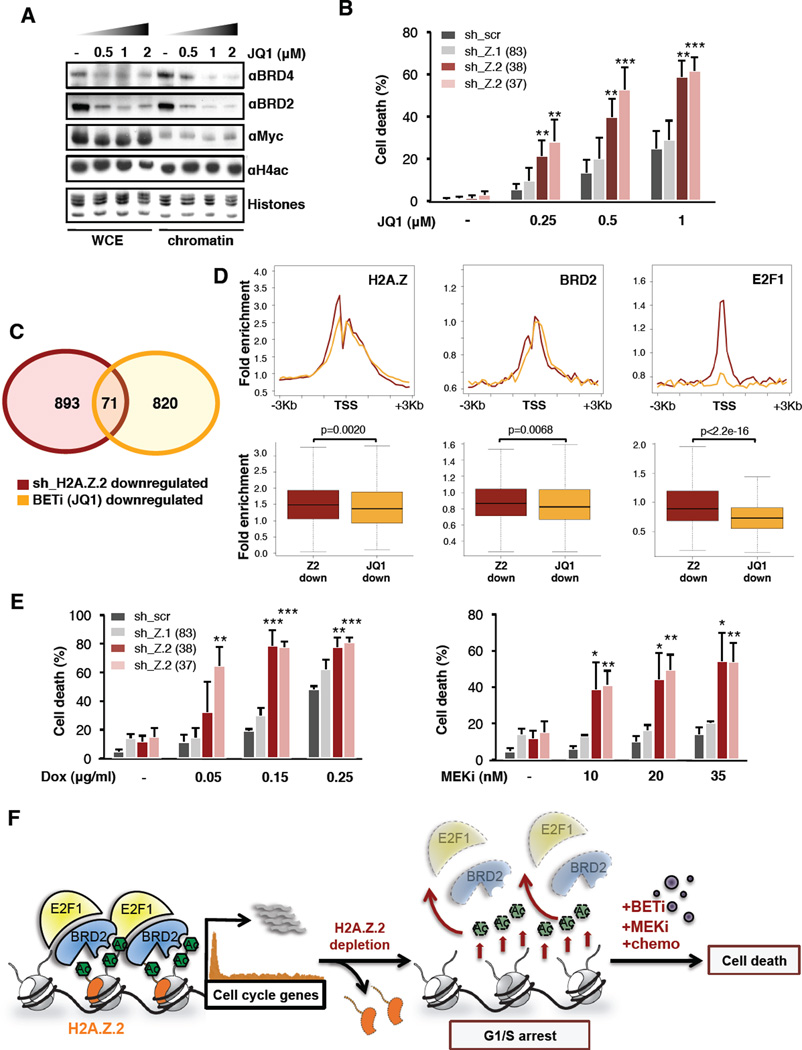
Because we observed loss of histone acetylation upon H2A.Z.2 knock down, we queried whether H2A.Z.2 depletion might potentiate the effects of BET inhibitors (BETi). BETi prevent the acetyllysine binding of bromodomains with high affinity, and are effective agents in a number of tumors (Dawson et al., 2012; Segura et al., 2013). We first assessed the sensitivity of melanoma cells to JQ1 (Filippakopoulos et al., 2010) (Figure 7A), and found a dose-dependent growth inhibitiory effect in the majority of cell lines tested (Figure S7A–C). Cells treated with JQ1 for 4 days accumulated in G2/M (Figure S7B), with minor induction of apoptosis (data not shown).
Figure 7. H2A.Z.2 deficiency sensitizes melanoma cells to chemotherapy and targeted therapies.
Next we investigated whether H2A.Z.2 knock down cooperates with JQ1 to enhance the antiproliferative effect of melanoma cells. Whereas JQ1 treatment or H2A.Z.2 knock down alone induced growth arrest (Figure S7B, 2D–G), the combination resulted in cell death, in both BRAF and NRAS mutant lines (Figure 7B, S7D). Functional annotation of the SK-mel147 transcriptome upon JQ1 treatment (Table S4) revealed enrichment in developmental processes, distinguishing it from cell cycle annotation associated with H2A.Z.2 silencing (see Figure 3C, S7E). Consistent with this synergy, we observed that the transcriptional profiles of H2A.Z.2 knock down and BETi show minimal overlap (Figure 7C, Table S4), suggesting that while H2A.Z.2-regulated genes are BRD2 and E2F1 targets, the mode of action of JQ1 is largely distinct from H2A.Z.2. While H2A.Z and BRD2 are enriched on promoters of JQ1 regulated genes (although significantly less so than on H2A.Z.2-regulated genes), E2F1 binding is absent (Figure 7D). JQ1 regulated genes are instead targets of distinct TFs (Figure S7F).
H2A.Z.2 loss not only enhances sensitivity of melanoma cells to BETi, but also to chemo- and targeted therapies (MEKi) used clinically for melanoma (Figure 7E). Collectively, these data suggest that H2A.Z.2 is a critical mediator of melanoma drug sensitivity and regulating its deposition may serve as an important target for novel therapeutic strategies.
DISCUSSION
H2A.Z.2 is a novel driver of malignant melanoma
While histone variants and their chaperones have emerged as critical players in cancer biology (Vardabasso et al., 2013), our mechanistic understanding remains limited. Here we report a unique role for H2A.Z.2 in driving melanoma cell proliferation and drug sensitivity. To our knowledge this is the first study to identify a specific role for H2A.Z.2 in any tumor type. This may suggest a melanoma-specific role for H2A.Z.2 in driving proliferation, and it will be of interest to learn if H2A.Z.2 plays a similar role in other tumors. Importantly, we do not exclude a role for H2A.Z.1 in melanoma, as it is also upregulated and correlates with shorter patient survival.
Because H2A.Z isoforms have distinct roles in melanoma, we hypothesized they may have unique interaction partners and genomic occupancy. Our studies indicate that H2A.Z.1 and H2A.Z.2 share genomic occupancy patterns and interact with similar histone chaperone complexes. However, it is clear that H2A.Z.2 is critical for promoting cell cycle progression in melanoma, and acts distinctly from H2A.Z.1. Our data strongly suggest that a unique property of H2A.Z.2 is to promote and/or maintain BRD2, E2F1 and histone acetylation levels. While the exact mechanism remains unclear, H2A.Z.2 likely acts together with histone acetylation to recruit co-activators and TFs, such as BRD2 and E2F1 respectively, to promote expression of cell cycle regulators (see below).
A unique signature of H2A.Z occupancy at E2F target genes
Our analyses revealed that H2A.Z.2 promotes the expression of E2F target genes. In melanoma cells, these genes are characterized by a unique signature of H2A.Z occupancy - highly enriched at the TSS and depleted within the gene body - and this pattern associates with high gene expression levels. Our findings are in line with previous observations in plants and yeast (Coleman-Derr and Zilberman, 2012; Sadeghi et al., 2011; Zilberman et al., 2008); for example, H2A.Z is excluded from the bodies of actively transcribed genes in Arabidopsis (Coleman-Derr and Zilberman, 2012; Zilberman et al., 2008). Intriguingly, the DREAM complex was recently reported to promote H2A.Z gene body incorporation to repress cell cycle progression genes in C. elegans (Latorre et al., 2015). Together, these studies suggest that H2A.Z is differentially distributed across promoters and gene bodies at distinct subsets of genes to regulate their expression levels.
An H2A.Z.2-BRD2-E2F1 axis in melanoma
Our study has identified BRD2 as an H2A.Z-interacting protein in malignant melanoma. Work by Denis and colleagues initially demonstrated that BRD2 has oncogenic potential: BRD2 transforms mouse fibroblasts in the context of oncogenic Ras (Denis et al., 2000), and Eμ-BRD2 transgenic mice develop B-cell lymphoma and leukemia (Greenwald et al., 2004). In fact, BRD2 has a crucial role in cell cycle control, and by interaction with E2F1, it regulates the expression of cyclins and other cell cycle regulatory genes (Denis et al., 2000; Denis et al, 2006; Sinha et al., 2005).
Our loss-of-function approach revealed that the chromatin association and total levels of BRD2, E2F1 and histone acetylation are H2A.Z.2- dependent. This is in line with the fact that BRD2's preference for H2A.Z-containing nucleosomes is mediated by a combination of hyperacetylated H4, and features on H2A.Z itself (Draker et al., 2012), and that histone acetyltransferase (HAT) activity is contained within BRD2 nuclear complexes (Sinha et al., 2005). Furthermore, we find evidence of an H2A.Z-BRD2-E2F axis in melanoma tissues. Accordingly, our ChIP analyses show that BRD2 and E2F1 are enriched at promoters of Class I genes and that H2A.Z.2 is required for recruitment of these factors to these E2F targets. Interestingly, Draker at al., found that recruitment of BRD2 to androgen receptor (AR)-regulated genes in prostate cancer cells is dependent on H2A.Z.1. Thus, BRD2 may associate with distinct TFs and H2A.Z isoforms to achieve oncogenic gene transcription in different tumor types.
Collectively, we envision that H2A.Z.2 recruits BRD2 and E2Fs, along with HAT activity, to E2F target genes in melanoma cells. This in turn results in increased expression of cell cycle genes, and ultimately promotes proliferation (Figure 7F). Our findings implicate the H2A.Z.2-BRD2-E2F1 axis as a driver of melanoma progression. Of these molecules, BRD2 represents a key target for therapy.
Novel epigenetic therapeutic strategies to treat melanoma
Metastatic melanoma is notoriously refractory to conventional cancer therapies, and remains largely resistant to current targeted therapies (Lito et al., 2013). Here we show that in combination with BET inhibition, H2A.Z.2 depletion is effective in inducing cell death. Because a tool to disrupt H2A.Z deposition is currently lacking, it is plausible that combining BETi with a potent inhibitor of HAT activity will potentiate melanoma cell death (Figure 7F). This combination may not only evict BET proteins from chromatin, but cause additional destabilization of BET proteins and their associated TFs due to loss of acetylation (Figure 7F). It will be of interest to create BRD-specific inhibitors, if achievable, as our study suggests that BRDs function distinctly in disease. Finally, our findings implicate H2A.Z.2 as a mediator of cell proliferation and drug sensitivity in malignant melanoma. Owing to the fact that histone modification and deposition are reversible processes, our study holds therapeutic potential for this highly intractable neoplasm.
EXPERIMENTAL PROCEDURES
Cell culture, plasmids and infections
Primary (WM115, WM1789, WM39), metastatic (SK-mel147, WM266-4, 501mel, A375, SK-mel2, SK-mel28, SK-mel239, SK-mel5, M14, WM165-1) melanoma cell lines, HeLa cells, and human melanocytes were cultured as described in Extended Experimental Procedures. Lentiviral vectors and shRNAs used for the generation of stable cell lines are described in Extended Experimental Procedures. Infections were performed according to standard procedures (Kapoor et al., 2010).
Chromatin fractionation, acid extraction of histones and immunoblotting
Chromatin fractionation and acid extraction of histones were performed as described (Kapoor et al., 2010) and in Extended Experimental Procedures. Antibodies used in this study are listed in Extended Experimental Procedures.
Clinical specimens
Approval to collect melanoma specimens was granted by Mount Sinai Biorepository Cooperative and the New York University Interdisciplinary Melanoma Cooperative Group (project number HSD08-00565 and IRB number 10362, respectively). Approval to collect benign nevi was granted by ISMMS (Icahn School of Medicine at Mount Sinai) Division of Dermatopathology (project number 08-0964).
RNA extraction, qRT-PCR and microarray hybridization
For RNA extraction, qRT-PCR, and primers, see Extended Experimental Procedures. RNA amplification, labeling and hybridization to Human Gene 1.0 ST Arrays (Affymetrix) were performed as described previously (Wiedemann et al., 2010), and data processed in R/bioconductor (www.bioconductor.org). For data analysis, see Extended Experimental Procedures.
Cell proliferation, colony formation and flow cytometry
For proliferation curves, cells were counted up to 7 days and normalized to cell counts at day 1. Colony formation assay was performed by seeding cells at low density and allowing growth for 2 weeks. Cells were washed with phosphate buffered saline, fixed in 10% methanol/acetic acid solution and stained with 1% crystal violet. Flow cytometry experiments were performed as described in Extended Experimental Procedures.
Native and cross-linked ChIP and next-generation sequencing
Chromatin from SK-mel147 cells was digested with micrococcal nuclease (MNase) and used for ChIP with H2A.Z (Abcam ab4174) and GFP Trap Beads (Chromotek), essentially as described (Hasson et al., 2013). SK-mel147 cells stably expressing control or isoform-specific shRNAs were cross-linked for 10’ with 1% formaldehyde and immunoprecipitated with BRD2 and E2F1 antibodies (Bethyl Laboratories A302-583A and Santa Cruz sc-193, respectively) as described in Extended Experimental Procedures. Sequencing libraries were generated and barcoded for multiplexing as described (Hasson et al., 2013) and libraries were submitted for 100-bp, single-end Illumina sequencing on a HiSeq 2500. For data processing and analysis, see Extended Experimental Procedures.
RNA-sequencing
Total RNA samples were isolated from human melanocytes and enantiomer- or JQ1-treated SK-mel147 using miRNeasy mini kit (Qiagen) following manufacturer's protocol. Sequencing libraries were prepared and data analysis performed as described in Extended Experimental Procedures.
Mononucleosome Immunoprecipitation
Mononucleosomes were generated according to (Sansoni et al., 2014) and described in Extended Experimental Procedures.
LC-MS/MS Analysis and MS data analysis
See Extended Experimental Procedures for details.
Statistical methodologies
Statistical tests were applied as indicated in figure legends. Asterisks as follow: *p<0.05; ** p<0.01; ***p<0.001. Boxplots represent Tukey boxplots with outliers omitted.
Supplementary Material
HIGHLIGHTS.
High levels of H2A.Z isoforms in metastatic melanoma correlate with poor survival
H2A.Z.2 promotes expression of E2F targets which display unique H2A.Z occupancy
BRD2 and E2F1 bind E2F targets in a H2A.Z.2-dependent manner
H2A.Z.2 silencing sensitizes melanoma cells to chemo- and targeted therapies
ACKNOWLEDGEMENTS
The authors are grateful for assistance and reagents provided by Luis F. Duarte, Avnish Kapoor, Nicholas Mills, Clemens Bönisch, Pauline Rimmele, Danielle Martinez (Baylor), and the laboratories of Robert Fisher, Stuart Aaronson, Ramon Parsons and Jay Bradner. We thank Cristina Montagna and Jidong Shan at Molecular Cytogenetic Core at Albert Einstein College of Medicine, Genomics core facility at Mount Sinai, Avi Ma’ayan for statistical support, Robert Phelps, Madelaine Haddican, Shelbi Jim On, and Giselle Singer for assistance with benign nevi collection, and Mark Lebwohl for his support. Funding was supported by a Melanoma Research Development Award (Mount Sinai) to C.V., DOD BCRP Postdoctoral fellowship (W81XWH-11-1-0018) and NYSCF Druckenmiller Fellowship to A.G.-M., Graduate fellowship from CONACyT (239663) to D.V-G, International Max Planck Research School for Life Science (IMPRS-LS) to S.P., Bundesministerium für Bildung und Forschung (FKZ01GS0861, DiGtoP consortium) to M.M., DOD Collaborative Award CA093471 (W81XWH-10-1-080), 1R01CA155234, and 1R01CA163891 to E.H., Deutsche Forschungsgemeinschaft (DFG) through the Collaborative Research Center SFB 1064 (project A10 to S.B.H. and project Z04 to T.S.), HA 5437/4-1 and the Center for Integrated Protein Science Munich (CIPSM) to S.B.H, and Worldwide Cancer Research, Hirschl/Weill-CaulierResearch Award and NCI/NIH R01CA154683 to E.B. We dedicate this study in memory of Estela Medrano.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers
All microarray, RNA-seq and ChIP-seq data sets have been deposited to NCBI’s Gene Expression Omnibus (GSE59060).
AUTHOR CONTRIBUTIONS
C.V. and E.B. conceived this study with guidance from S.B.H. C.V. performed all qPCR, shRNA studies and generated eGFP melanoma cell lines. C.V. and T.P. performed immunoblots and cell cycle analyses. S.B.H. and T. Straub performed and analyzed microarrays, respectively. C.V., D.H. and S.P. performed ChIP-seq experiments and data analyses was lead by C.V., A.G.-M., D.V.-G, D.H. and T. Straub. S.P. performed IPs for MS, and E.C.K. performed MS with the support of M.M. C.V. and T. Strub performed drug treatments and S.P., C.V. and T. Strub performed BRD2 immunoblots. J.D. performed BRD2 IHC and J.L.Y. and R.S. scored tissues. C.-Y.C. assisted with network analyses and A.V. provided copy number analysis support. M.F.S., B.F.-C. and E.H. provided assistance with patient data and with BETi experiments. B.F.-C. prepared samples for RNA-seq, and D.H. analyzed RNA-seq data. C.V., A.G.-M., D.V.-G., D.H., S.P, E.C.K., S.B.H. and E.B. designed experiments and interpreted results. C.V., E.B. and S.B.H. wrote the manuscript with contributions from all other coauthors.
REFERENCES
- Alla V, Engelmann D, Niemetz A, Pahnke J, Schmidt A, Kunz M, Emmrich S, Steder M, Koczan D, Putzer BM. E2F1 in melanoma progression and metastasis. J Natl Cancer Inst. 2009;102:127–133. doi: 10.1093/jnci/djp458. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Belkina AC, Denis GV. BET domain co-regulators in obesity, inflammation and cancer. Nat Rev Cancer. 2012;12:465–477. doi: 10.1038/nrc3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billon P, Cote J. Precise deposition of histone H2A.Z in chromatin for genome expression and maintenance. Biochim Biophys Acta. 2012;1819:290–302. doi: 10.1016/j.bbagrm.2011.10.004. [DOI] [PubMed] [Google Scholar]
- Bogunovic D, O'Neill DW, Belitskaya-Levy I, Vacic V, Yu YL, Adams S, Darvishian F, Berman R, Shapiro R, Pavlick AC, et al. Immune profile and mitotic index of metastatic melanoma lesions enhance clinical staging in predicting patient survival. Proc Natl Acad Sci U S A. 2009;106:20429–20434. doi: 10.1073/pnas.0905139106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonisch C, Hake SB. Histone H2A variants in nucleosomes and chromatin: more or less stable? Nucleic Acids Res. 2012;40:10719–10741. doi: 10.1093/nar/gks865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceol CJ, Houvras Y, Jane-Valbuena J, Bilodeau S, Orlando DA, Battisti V, Fritsch L, Lin WM, Hollmann TJ, Ferre F, et al. The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset. Nature. 2011;471:513–517. doi: 10.1038/nature09806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, Dummer R, Garbe C, Testori A, Maio M, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman-Derr D, Zilberman D. Deposition of histone variant H2A.Z within gene bodies regulates responsive genes. PLoS Genet. 2012;8:e1002988. doi: 10.1371/journal.pgen.1002988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson MA, Kouzarides T, Huntly BJ. Targeting epigenetic readers in cancer. N Engl J Med. 2012;367:647–657. doi: 10.1056/NEJMra1112635. [DOI] [PubMed] [Google Scholar]
- Denis GV, McComb ME, Faller DV, Sinha A, Romesser PB, Costello CE. Identification of transcription complexes that contain the double bromodomain protein Brd2 and chromatin remodeling machines. J Proteome Res. 2006;5:502–511. doi: 10.1021/pr050430u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis GV, Vaziri C, Guo N, Faller DV. RING3 kinase transactivates promoters of cell cycle regulatory genes through E2F. Cell Growth Differ. 2000;11:417–424. [PMC free article] [PubMed] [Google Scholar]
- Dhillon N, Oki M, Szyjka SJ, Aparicio OM, Kamakaka RT. H2A.Z functions to regulate progression through the cell cycle. Mol Cell Biol. 2006;26:489–501. doi: 10.1128/MCB.26.2.489-501.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draker R, Ng MK, Sarcinella E, Ignatchenko V, Kislinger T, Cheung P. A combination of H2A.Z and H4 acetylation recruits Brd2 to chromatin during transcriptional activation. PLoS Genet. 2012;8:e1003047. doi: 10.1371/journal.pgen.1003047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte LF, Young ARJ, Wang Z, Wu H-A, Panda T, Kou Y, Kapoor A, Hasson D, Mills NR, Ma'ayan A, Narita M, Bernstein E. Histone H3.3 and its proteolitically processed form drive a cellular senescence program. Nat Commun. 2014;5:5210. doi: 10.1038/ncomms6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dryhurst D, Ishibashi T, Rose KL, Eirin-Lopez JM, McDonald D, Silva-Moreno B, Veldhoen N, Helbing CC, Hendzel MJ, Shabanowitz J, et al. Characterization of the histone H2A.Z-1 and H2A.Z-2 isoforms in vertebrates. BMC Biol. 2009;7:86. doi: 10.1186/1741-7007-7-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberl HC, Spruijt CG, Kelstrup CD, Vermeulen M, Mann M. A map of general and specialized chromatin readers in mouse tissues generated by label-free interaction proteomics. Mol Cell. 2013;49:368–378. doi: 10.1016/j.molcel.2012.10.026. [DOI] [PubMed] [Google Scholar]
- Faast R, Thonglairoam V, Schulz TC, Beall J, Wells JR, Taylor H, Matthaei K, Rathjen PD, Tremethick DJ, Lyons I. Histone variant H2A.Z is required for early mammalian development. Curr Biol. 2001;11:1183–1187. doi: 10.1016/s0960-9822(01)00329-3. [DOI] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwald RJ, Tumang JR, Sinha A, Currier N, Cardiff RD, Rothstein TL, Faller DV, Denis GV. E mu-BRD2 transgenic mice develop B-cell lymphoma and leukemia. Blood. 2004;103:1475–1484. doi: 10.1182/blood-2003-06-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasson D, Panchenko T, Salimian KJ, Salman MU, Sekulic N, Alonso A, Warburton PE, Black BE. The octamer is the major form of CENP-A nucleosomes at human centromeres. Nat Struct Mol Biol. 2013;20:687–695. doi: 10.1038/nsmb.2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horikoshi N, Sato K, Shimada K, Arimura Y, Osakabe A, Tachiwana H, Hayashi-Takanaka Y, Iwasaki W, Kagawa W, Harata M, et al. Structural polymorphism in the L1 loop regions of human H2A.Z.1 and H2A.Z.2. Acta Crystallogr D Biol Crystallogr. 2013;69:2431–2439. doi: 10.1107/S090744491302252X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Cui K, Northrup D, Liu C, Wang C, Tang Q, Ge K, Levens D, Crane-Robinson C, Zhao K. H2A.Z facilitates access of active and repressive complexes to chromatin in embryonic stem cell self-renewal and differentiation. Cell Stem Cell. 2012;12:180–192. doi: 10.1016/j.stem.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor A, Goldberg MS, Cumberland LK, Ratnakumar K, Segura MF, Emanuel PO, Menendez S, Vardabasso C, Leroy G, Vidal CI, et al. The histone variant macroH2A suppresses melanoma progression through regulation of CDK8. Nature. 2010:1105–1109. doi: 10.1038/nature09590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman HL, Kirkwood JM, Hodi FS, Agarwala S, Amatruda T, Bines SD, Clark JI, Curti B, Ernstoff MS, Gajewski T, et al. The Society for Immunotherapy of Cancer consensus statement on tumour immunotherapy for the treatment of cutaneous melanoma. Nat Rev Clin Oncol. 2013;10:588–598. doi: 10.1038/nrclinonc.2013.153. [DOI] [PubMed] [Google Scholar]
- Kou Y, Chen EY, Clark NR, Tan CM, Ma‘ayan A. ChEA2: Gene-Set Libraries from ChIP-X Experiments to De 13code the Transcription Regulome. (2013). Multidisciplinary Research and Practice for Information Systems. CD-ARES 2013. Lecture Notes in Computer Science. 2013;8127:416–430. [Google Scholar]
- Latorre I, Chesney MA, Garrigues JM, Stempor AA, Francesconi M, Strome s, Ahringer J. The DREAM complex promotes gene body H2A.Z for target repression. Genes Dev. 2015;29(5):495–500. doi: 10.1101/gad.255810.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeRoy G, Rickards B, Flint SJ. The double bromodomain proteins Brd2 and Brd3 couple histone acetylation to transcription. Mol Cell. 2008;30:51–60. doi: 10.1016/j.molcel.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeRoy G, Chepelev I, DiMaggio PA, Blanco MA, Zee BM, Zhao K, Garcia BA. Proteogenomic characterization and mapping of nucleosomes decoded by Brd and HP1 proteins. Genome Biol. 2012;13(8):R68. doi: 10.1186/gb-2012-13-8-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lito P, Rosen N, Solit DB. Tumor adaptation and resistance to RAF inhibitors. Nat Med. 2013;19:1401–1409. doi: 10.1038/nm.3392. [DOI] [PubMed] [Google Scholar]
- Ma Y, Kurtyka CA, Boyapalle S, Sung SS, Lawrence H, Guida W, Cress WD. A small-molecule E2F inhibitor blocks growth in a melanoma culture model. Cancer Res. 2008;68:6292–6299. doi: 10.1158/0008-5472.CAN-08-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda R, Hori T, Kitamura H, Takeuchi K, Fukagawa T, Harata M. Identification and characterization of the two isoforms of the vertebrate H2A.Z histone variant. Nucleic Acids Res. 2010;38:4263–4273. doi: 10.1093/nar/gkq171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obri A, Ouararhni K, Papin C, Diebold ML, Padmanabhan K, Marek M, Stoll I, Roy L, Reilly PT, Mak TW, et al. ANP32E is a histone chaperone that removes H2A.Z from chromatin. Nature. 2014;505:648–653. doi: 10.1038/nature12922. [DOI] [PubMed] [Google Scholar]
- Riker AI, Enkemann SA, Fodstad O, Liu S, Ren S, Morris C, Xi Y, Howell P, Metge B, Samant RS, et al. The gene expression profiles of primary and metastatic melanoma yields a transition point of tumor progression and metastasis. BMC Med Genomics. 2008;1:13. doi: 10.1186/1755-8794-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadeghi L, Bonilla C, Stralfors A, Ekwall K, Svensson JP. Podbat: a novel genomic tool reveals Swr1-independent H2A.Z incorporation at gene coding sequences through epigenetic meta-analysis. PLoS Comput Biol. 2011;7:e1002163. doi: 10.1371/journal.pcbi.1002163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansoni V, Casas-Delucchi CS, Rajan M, Schmidt A, Bonisch C, Thomae AW, Staege MS, Hake SB, Cardoso MC, Imhof A. The histone variant H2A.Bbd is enriched at sites of DNA synthesis. Nucleic Acids Res. 2014 doi: 10.1093/nar/gku303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura MF, Fontanals-Cirera B, Gaziel-Sovran A, Guijarro MV, Hanniford D, Zhang G, Gonzalez-Gomez P, Morante M, Jubierre L, Zhang W, et al. BRD4 Sustains Melanoma Proliferation and Represents a New Target for Epigenetic Therapy. Cancer Res. 2013;73:6264–6276. doi: 10.1158/0008-5472.CAN-13-0122-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha A, Faller DV, Denis GV. Bromodomain analysis of Brd2-dependent transcriptional activation of cyclin A. Biochem J. 2005;387:257–269. doi: 10.1042/BJ20041793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svotelis A, Gevry N, Gaudreau L. Regulation of gene expression and cellular proliferation by histone H2A.Z. Biochem Cell Biol. 2009;87:179–188. doi: 10.1139/O08-138. [DOI] [PubMed] [Google Scholar]
- Talantov D, Mazumder A, Yu JX, Briggs T, Jiang Y, Backus J, Atkins D, Wang Y. Novel genes associated with malignant melanoma but not benign melanocytic lesions. Clin Cancer Res. 2005;11:7234–7242. doi: 10.1158/1078-0432.CCR-05-0683. [DOI] [PubMed] [Google Scholar]
- Vardabasso C, Hasson D, Ratnakumar K, Chung CY, Duarte LF, Bernstein E. Histone variants: emerging players in cancer biology. Cell Mol Life Sci. 2013 doi: 10.1007/s00018-013-1343-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemann SM, Mildner SN, Bonisch C, Israel L, Maiser A, Matheisl S, Straub T, Merkl R, Leonhardt H, Kremmer E, et al. Identification and characterization of two novel primate-specific histone H3 variants, H3.X and H3.Y. J Cell Biol. 2010;190:777–791. doi: 10.1083/jcb.201002043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Shen SS, Hoshida Y, Subramanian A, Ross K, Brunet JP, Wagner SN, Ramaswamy S, Mesirov JP, Hynes RO. Gene expression changes in an animal melanoma model correlate with aggressiveness of human melanoma metastases. Mol Cancer Res. 2008;6:760–769. doi: 10.1158/1541-7786.MCR-07-0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D, Coleman-Derr D, Ballinger T, Henikoff S. Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature. 2008;456:125–129. doi: 10.1038/nature07324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zingg D, Debbache J, Schaefer SM, Tuncer E, Frommel SC, Cheng P, Arenas-Ramirez N, Haeusel J, Zhang Y, Bonalli M, et al. The epigenetic modifier EZH2 controls melanoma growth and metastasis through silencing of distinct tumour suppressors. Nat Commun. 2015;6:6051. doi: 10.1038/ncomms7051. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.