SUMMARY
Whereas RNA polymerase II (pol II) transcription start sites (TSSs) occur about 30–35 bp downstream of the TATA box in metazoans, TSSs are located 40–120 bp downstream in S. cerevisiae. Promoter melting begins about 12 bp downstream in all eukaryotes, so pol II is presumed to “scan” further downstream before starting transcription in yeast. Here we report that removal of the kinase complex TFIIK from TFIIH shifts the TSS in a yeast system upstream to the location observed in metazoans. Conversely, moving the normal TSS to an upstream location enables a high level of TFIIK-independent transcription in the yeast system. We distinguish two stages of the transcription initiation process: bubble formation by TFIIH, which fills the pol II active center with single stranded DNA; and subsequent scanning downstream, also driven by TFIIH, which requires displacement of the initial bubble. Omission of TFIIK uncouples the two stages of the process.
Graphical abstract
INTRODUCTION
RNA polymerase II (pol II) assembles with five general transcription factors (GTFs) in a pre-initiation complex (PIC) on promoter DNA (Conaway and Conaway, 1993; Kornberg, 2007). The largest GTF, TFIIH, comprises the helicase Ssl2, a six-subunit core complex, and a three-subunit kinase termed TFIIK (Gibbons et al., 2012; Schaeffer et al., 1993; Schultz et al., 2000; Svejstrup et al., 1995). Ssl2 unwinds the promoter duplex to form a “transcription bubble” (Holstege et al., 1997; Kim et al., 2000; Pal et al., 2005; Fishburn et al., 2015), starting about 12 bp downstream from the TATA box, and in metazoans, pol II initiates transcription about 30 bp downstream; in S. cerevisiae, pol II scans further downstream and initiates 40–120 bp from the TATA box. Start site recognition is due in part to properties of pol II (Li et al., 1994; Yang and Ponticelli, 2012; Zhang et al., 2014), but the basis for scanning is unknown. The initiation of transcription is accompanied by phosphorylation of the C-terminal domain (CTD) of pol II (Feaver et al., 1994; Laybourn and Dahmus, 1990; Roy et al., 1994), due to the action of TFIIK. Kinase activity is, however, dispensable for basal transcription in vitro (Li and Kornberg, 1994; Serizawa et al., 1993). Here we report on a requirement for TFIIK for initiation at downstream sites, and thus a likely role of TFIIK in promoter scanning.
RESULTS
Omission of TFIIK shifts transcription initiation upstream
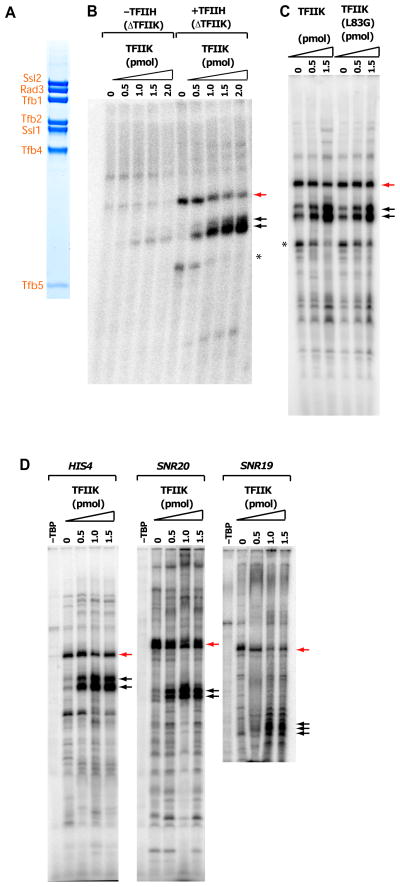
TFIIH bearing a TAP tag on the Tfb4 subunit was purified from a yeast strain lacking the TFB6 gene as described (Murakami et al., 2012). TFIIH bound to IgG-Sepharose through the TAP tag was stripped of TFIIK by washing with high salt (300 mM potassium acetate plus 400 mM ammonium sulfate); following elution and gel filtration, essentially pure TFIIH devoid of TFIIK (TFIIH-ΔTFIIK) was obtained (Fig. 1A). TFIIH-ΔTFIIK in combination with pol II and other general transcription factors proved capable of assembly in a complete pre-initiation complex on a HIS4 promoter fragment as described (Murakami et al., 2013b), and also of transcription initiation (Fig. 1B). In the presence of TFIIH-ΔTFIIK, however, run-off transcripts were longer than expected (indicated by red arrow in Fig. 1B). Transcription required TFIIH-ΔTFIIK (Fig. 1B), TBP (Fig. 1D) and TFIIB (Fig. S1B), confirming promoter-dependence of the reaction. When TFIIK was added back, the expected distal TSS utilization was restored (indicated by black arrows in Fig. 1B) with a concurrent decrease in proximal site use, and a 4-fold increase in total run-off transcription; the same result was obtained when TFIIK bearing a mutation in the Kin28 subunit (TFIIK-L83G) that confers sensitivity to the kinase inhibitor NA-PP1 (Liu et al., 2004) was added to the reaction in the presence of the inhibitor (Figs. 1C, S1A). Evidently TSS determination is dependent on a feature of TFIIK other than the kinase activity. Similar results were obtained with two additional promoters tested (Fig. 1D). Primer extension analysis showed that in the presence of TFIIK, transcription was initiated 63–103 bp downstream of the TATA box, whereas in the absence of TFIIK, transcription was initiated 31–34 bp downstream of the TATA box (summarized in Fig. S2), a location characteristic of transcription initiation in metazoans rather than S. cerevisiae (Ponjavic et al., 2006). TFIIS had a slight effect on TSS utilization, and TFIIA and Mediator had no effect (Fig. S1D–E). All the other general transcription factors were required.
Figure 1. Role of TFIIK in transcription initiation.
(A) SDS polyacrylamide gel electrophoresis of purified TFIIH-ΔTFIIK. (B) Run-off transcription with increasing amounts of TFIIK. HIS4 promoter DNA (−96/+112) (Murakami et al., 2013a) was combined with TFIIA, TFIIB, TBP, TFIIE, TFIIH- ΔTFIIK, pol II-TFIIF complex, and the amounts of TFIIK indicated. Transcripts initiated from upstream and downstream TSSs are indicated by red and black arrows. The asterisk indicates a non-specific product that was not observed by primer extension analysis. (C) Run-off transcription with increasing amounts of TFIIK (left panel) or inhibitor-sensitive TFIIK mutant (TFIIK-L83G) (Liu et al., 2004) (right panel). The reactions were performed as in (B) except with the addition of 50 μM NA-PP1. (D) Same as (B) with different promoters. See also Fig. S1.
Transcription from an upstream start site does not require TFIIK
If omission of TFIIK shifts transcription initiation to sites upstream, then it may be dispensable for transcription of a promoter whose TSS is normally located upstream. We created such a promoter by deletion of downstream DNA. We brought the TSS of the SNR20 promoter from the wild type location 91 bp downstream of the TATA box (Fig. 2A, Fig. S2, fragment 91W) to locations 29, 31, and 33 bp downstream (Fig. 2A, Fig. S2, fragments 29D, 31D, 33D); a C+4G mutation was introduced in the TSS to yield only one rather than multiple closely spaced initiation sites. Transcription initiation on fragment 91W was diminished and shifted upstream by the omission of TFIIK (Fig. 2B, lanes 4 and 8), whereas initiation on fragments 31D and 33D was at the same high level (0.25 transcripts per pol II) in the presence or absence of TFIIK (Fig. 2B, lanes 2, 3, 6, 7). It is noteworthy that in the presence of TFIIK, initiation on fragments 31D and 33D was mostly 31 or 33 bp downstream of the TATA box, the distance characteristic of metazoan transcription, and to a lesser extent about 60 bp downstream (Fig. 2B, lanes 6, 7, 10, 11), whereas, in the absence of TFIIK, initiation was exclusively 31 or 33bp downstream (Fig. 2B, lanes 2, 3), confirming a role of TFIIK in initiation at further downstream sites. Initiation on fragment 29D was markedly diminished, further resembling metazoans, in which a 30–31 bp spacing between the TATA box and TSS is most frequent and a spacing shorter than 29 bp is rarely observed (Ponjavic et al., 2006).
Figure 2. Run-off transcription with modified promoters.
(A) Schematic illustration of modified promoters (also see Fig. S2). The wild type SNR20 promoter, the SNR20 promoter with a C+4G mutation introduced in the TSS (SNR20 91W), and the SNR20 promoter with deletions (indicated by dashed lines) bringing the +1 TSS to positions 29, 31, and 33 bp from the TATA box (SNR20 29D, SNR20 31D, SNR20 33D). (B) Run-off transcription with the modified SNR20 promoters. For reactions with the inhibitor-sensitive mutant TFIIK-L83G, 1.1 mM NA-PP1 was added.
The upstream TSS usage in yeast was not previously observed in the nuclear extract system (Lue et al., 1989) or in vivo (Faitar et al., 2001). We determined if the TFIIK effect on TSS utilization might be revealed in vivo by nuclear depletion of Kin28, using the “anchor-away” method (Haruki et al., 2008; Jeronimo and Robert, 2014; Wong et al., 2014). We failed to observe shifting in TSS positions at ADH1 or SNR20 (Fig. S3). These results suggest that TFIIK-subunit activity (Ccl1 or Tfb3) may still be intact when Kin28 is depleted, or that additional factors redundantly enforce downstream TSS usage.
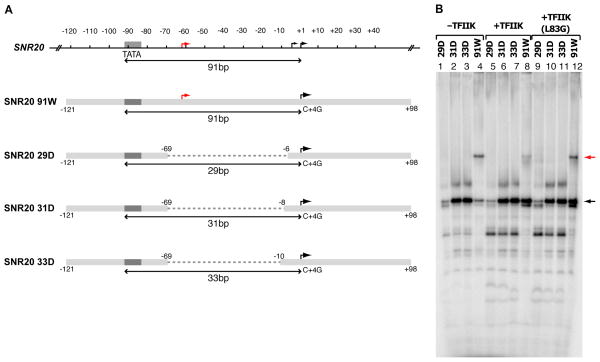
TFIIK affects downstream DNA conformation of PIC as determined by exonuclease III digestion
Exonuclease III mapping of the PIC revealed barriers to digestion at about position -9 relative to the TSS in the presence of TFIIK (Fig. 3, black arrow) and at about position -14 in the absence of TFIIK (Fig. 3, red arrow). No such barriers were observed in complexes formed without TFIIE and TFIIH, showing the barriers are characteristic of a complete PIC. The barriers are located on the opposite (downstream) side of the transcription bubble from sites of interaction with TFIIB (Pardee et al., 1998), TFIIF (Chen et al., 2010; Ghazy et al., 2004) and pol II (Kaplan et al., 2012; Majovski et al., 2005), shown by genetic analysis to be involved in TSS utilization. It is noteworthy that omission of TFIIK had a much greater effect on TSS utilization than a TFIIB mutation (E62K) known to confer a downstream TSS shift (Pinto et al., 1994) (Fig. S1B–C).
Figure 3. Exonuclease III Footprints of the PIC and PIC-ΔTFIIK.
32P-labeled HIS4 (-96/+112) DNA was incubated with TFIIA, TFIIB, TBP, TFIIE, TFIIH-ΔTFIIK, and pol II-TFIIF complex with or without nucleotides as indicated over the lanes, followed by treatment with exonuclease III and gel electrophoresis. Positions of protected fragments are indicated by black (PIC) and red (PIC-ΔTFIIK) arrows.
DISCUSSION
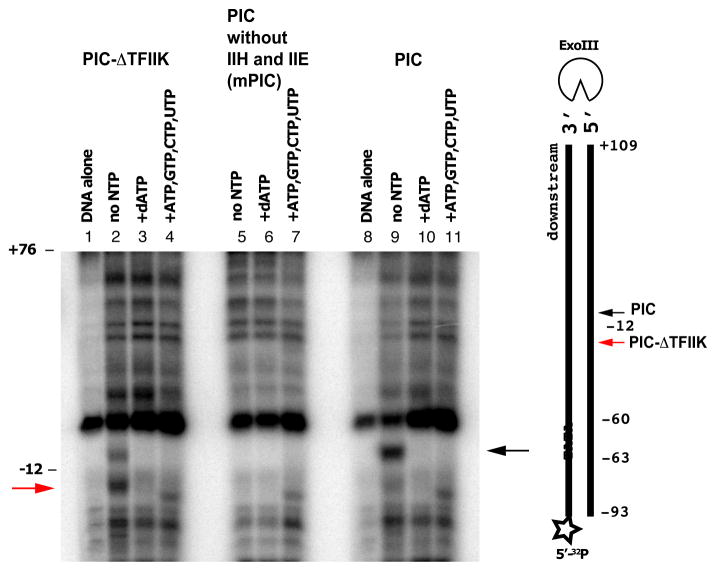
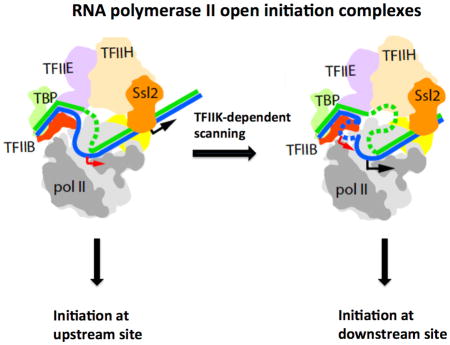
The difference between initiation of pol II transcription about 30 bp downstream of the TATA box in metazoans and initiation further downstream in S. cerevisiae is commonly attributed to scanning downstream by pol II before initiation in S. cerevisiae (Giardina and Lis, 1993). Our findings point to a role of TFIIK in scanning: initiation is shifted upstream in the absence of TFIIK; and movement of a downstream TSS to an upstream location obviates the requirement for TFIIK. Exonuclease protection provides a physical basis for involvement of TFIIK in scanning.
Previous studies are consistent with the participation of TFIIK in TSS determination. A minimal PIC containing TBP, TFIIB, and pol II is sufficient to initiate transcription from artificial bubble templates (Holstege et al., 1995; Parvin and Sharp, 1993), but fails to support proper TSS utilization (Fishburn and Hahn, 2012). TFIIH is required not only for promoter opening but also for subsequent events, including initiation and the transition to RNA chain elongation (Bradsher et al., 2000; Dvir et al., 1997; Spangler et al., 2001). All polypeptides of the closed PIC are retained following promoter opening (Murakami et al., 2013a). The helicase subunit of TFIIH (Ssl2, yeast homolog of XPB, Fig. 4) is an integral component of the PIC and is almost certainly the molecular motor for scanning (Fazal, F. M., Meng, C. A., Block, S. M., personal communication). TFIIK must play an auxiliary role.
Figure 4. Schematic representation of promoter opening and TSS scanning.
Schematic representation of the PIC (left panel) and proposed changes upon promoter opening (middle panel) and TSS scanning (right panel). DNA is blue and green. TFIIK is yellow. Upstream and downstream TSSs are indicated by red and black arrows. See also Fig. S4.
From the combination of our functional studies with previous structural information, we arrive at a picture of the initiation process (Fig. 4). Initial melting through the action of TFIIH draws some 24 bp of DNA into the polymerase cleft, filling the single stranded DNA-binding region of the active center. If a TSS is recognized in the single stranded bubble, transcription begins, an RNA-DNA hybrid helix develops, TFIIB is ejected as proposed (Bushnell et al., 2004; Liu et al., 2010), and transcription elongation ensues. Alternatively, if no TSS resides within the initial single stranded bubble, then further action of TFIIH drives scanning downstream. Additional single stranded DNA entering the pol II cleft must displace that which was bound before, through sliding and the formation of a larger bubble. Evidently TFIIK is necessary for this process, interacting with the DNA either directly (as suggested by exonuclease III mapping) or indirectly (for example, by bridging between TFIIH and pol II). In the absence of TFIIK, the DNA may disengage from the entrance of the pol II channel and its unwinding by the TFIIH helicase may not be coupled to the delivery of a TSS to the active center of pol II (Fig. S4).
EXPERIMENTAL PROCEDURES
Protein purification
TFIIA, TFIIB, TBP, and TFIIS were available in recombinant form, and TFIIF, TFIIH-ΔTFIIK, and pol II were isolated from yeast as previously published(Murakami et al., 2013a; Murakami et al., 2012).
For the isolation of TFIIK or TFIIK-L83G, yeast strain CB010 (Murakami et al., 2012) or SHY532 (Liu et al., 2004) harboring TAP-tagged Tfb3 was grown in 100L YPAD. The yeast culture was harvested at OD 7.0–8.0 and lysed by disruption with glass beads in buffer (400) (50 mM HEPES (pH7.6), 1 mM EDTA, 5% glycerol, 0.1% 3-(decyldimethyl-ammonio) propanesulfonate (Sigma), with the mM concentration of potassium acetate in parentheses) supplemented with 1 mM benzamidine, 100 μM leupeptin, 10 μM pepstatin A, and 1 mM PMSF. The cell extract was stirred with 100 mM ammonium sulfate and 0.2 % polyethyleneimine (PEI) for 1 hour, centrifuged, loaded onto an IgG column, and washed with buffer (300). A mixture of holo-TFIIH and TFIIK was eluted using buffer (300) after incubating with TEV protease for 15 hours at 4°C, and loaded onto UnoQ column (Bio-Rad). The flow-through fraction containing TFIIK was concentrated, and further purified by using Superdex 200 16/60 (GE healthcare) in a buffer containing 20 mM HEPES (pH7.6), 300 mM potassium acetate, 5 mM DTT, 10% glycerol, and 0.1% 3-(decyldimethyl-ammonio) propanesulfonate.
For the isolation of TFIIE, S. cerevisiae harboring the TAP-tagged Tfa2 (Borggrefe et al., 2001) was grown in 100L YPAD. The yeast culture was harvested at OD 8.0–10.0 and lysed by disruption with glass beads in a lysis buffer (50 mM HEPES (pH7.6), 250 mM ammonium sulfate, 1 mM EDTA, 5% glycerol, 3 mM DTT, 1 mM benzamidine, 100 μM leupeptin, 10 μM pepstatin A, and 1 mM PMSF). The cell extract was stirred with 0.2 % PEI for 30 min, centrifuged, and was loaded onto an IgG column. The column was washed with buffer 300 (50 mM HEPES (pH7.6), 300 mM potassium acetate, 1 mM EDTA, 5% glycerol, 2.5 mM DTT, 1 mM benzamidine, 100 μM leupeptin, 10 μM pepstatin A) containing 400 mM ammonium sulfate, washed with buffer 300, and eluted after incubating with TEV protease for 15 hours at 4°C. The IgG elution was further purified by using Superose 6 XK 16/70 (GE healthcare) in a buffer containing 20 mM HEPES (pH7.6), 300 mM potassium acetate, 5 mM DTT, and 2 mM magnesium acetate.
For the purification of Mediator, yeast strain CB010 harboring TAP-tagged Med7 was grown in 16L YPAD. The yeast culture was harvested at OD 10–12 and lysed by disruption with zirconium beads in HGE(200) (50 mM HEPES (pH7.6), 5% glycerol, 1 mM EDTA with the mM concentration of ammonium sulfate in parentheses supplemented with 5 mM dithiothreitol, 1 mM benzamidine, 100 μM leupeptin, 10 μM pepstatin A, and 1 mM PMSF. The cell lysate was centrifuged in a JA-14 rotor at 8,000 RPM for 20 minutes. The soluble lysate was extracted with 0.25% PEI for 1 hour then centrifuged at 14,000 RPM for 45 minutes. The PEI soluble extract was further extracted with a 65% ammonium sulfate cut. After the ammonium sulfate pellet was dissolved in 15% the original cell slurry volume of HGE(0), it was loaded onto an 8 ml IgG Fast Flow 4 column at ten column volumes per hour. After washing the IgG column with 20 column volumes of HGE(250), Mediator was eluted overnight (4°C) in 25 mM HEPES, 5% glycerol, 75 mM ammonium sulfate, and 2 mM dithiothreitol with 15 μg/ml of TEV protease. Eluted Mediator was applied to a 1ml HiTrap Q column (GE healthcare) and was eluted in a linear gradient from 75 mM to 1000 mM ammonium sulfate gradient in 25 mM HEPES, 5% glycerol, and 2 mM dithiothreitol. Peak fractions containing Meidator between 150 and 200 mM ammonium sulfate were pooled and concentrated to 3.1 mg/ml.
DNA templates
For a series of SNR20 spacing promoters, SNR20 (−122/+297) was subcloned into the pDrive Cloning Vector using forward (5′-CCCGGGCTGCAGGCCGTTTCCGATGGG-3′) and reverse (5′-CCCGGGTAATGAGCCTCATTGAG-3′) primers. A C+4G point mutation and a series of deletions were generated by the QuikChange Site Directed Mutagenesis. DNA templates were amplified by PCR, purified using Superose 6 10/300 column (GE healthcare) in a buffer containing 20 mM HEPES (pH7.6), 300 mM potassium acetate, 5 mM DTT, and 2 mM magnesium acetate, and concentrated up to 5–10 μM using Vivacon 500 3K MWCO (Vivaproducts), yielding ~0.3–0.5 nmol from 2 mL PCR reaction.
HIS4 DNA fragment was prepared as previously published (Murakami et al., 2013a). SNR19 DNA fragment was amplified from yeast genomic DNA using forward (5′-GATATCCTGCAGCTAAGGCGACGAGTTTTCC-3′) and reverse (5′-GATATCGCTTAACGTCCTTCTACTATTGG-3′) primers, and purified using Superose 6 10/300 (GE healthcare) in a buffer containing 20 mM HEPES (pH7.6), 300 mM potassium acetate, 5 mM DTT, and 2 mM magnesium acetate.
Run-off transcription assay
All transcription assays were performed in an in vitro reconstituted system including TFIIA, TFIIB, TBP, TFIIE, TFIIF, TFIIH, and pol II unless otherwise indicated. 1.5 pmol of DNA fragment was combined with 3.7 pmol of TFIIA, 3.7 pmol of TFIIB, 1.5 pmol of TBP, 3.7 pmol of TFIIE, 1.5 pmol of pol II-TFIIF complex, and 1.5 pmol of TFIIH-ΔTfb6 with or without TFIIK in 5 μl of buffer 300 (50 mM HEPES (pH7.6), 300 mM potassium acetate, 5 mM DTT, and 5% glycerol), diluted with 5 μl of buffer 10 (20 mM HEPES (pH7.6), 10 mM potassium acetate, 5 mM magnesium sulfate, and 5 mM DTT), and incubated for more than 1hr on ice. The transcription was initiated by adding an equal volume of 2xNTP buffer containing 20 mM HEPES (pH7.6), 10 mM potassium acetate, 5 mM magnesium sulfate, 10 mM magnesium acetate, 1 unit of RNaseOut, 5 mM DTT, 1.6 mM ATP, 1.6 mM GTP, 1.6 mM CTP, 40 μM UTP, and 0.83 μM [α-32P] UTP (2.5 μCi). The reaction was stopped in 15 min by adding 185 μl of stop buffer (300 mM sodium acetate (pH 5.5), 5 mM EDTA, 0.7% SDS, 0.1 mg/ml glycogen, 0.013 mg/ml of proteinase K (Sigma)). Transcripts were precipitated by adding 700 μL of ethanol, dried, and analysed by a denaturing 6% acrylamide gel.
In vitro kinase assay of PIC
HIS4 (−96/+112) DNA (1.5 pmol) was incubated with 3.7 pmol of TFIIB, 3.7 pmol of TFIIA, 1.8 pmol of TBP, 3.7 pmol of TFIIE, 1.0 pmol of RNA polymerase II-TFIIF complex, and 1.5 pmol of TFIIH-ΔTFIIK with TFIIK or inhibitor-sensitive TFIIK mutant (TFIIK-L83G) (2.0 pmol). Phosphorylation was initiated by adding an equal volume of 2xATP buffer (20 mM HEPES (pH7.6), 10 mM potassium acetate, 5 mM magnesium sulfate, 10 mM magnesium acetate, 1 mM ATP, and 1.0 μCi/μl of [γ-32P]). The reaction was stopped in 6 min by adding 4x SDS-PAGE loading buffer and was analyzed by SDS-PAGE.
Exonuclease footprinting
Exonuclease footprinting was performed as described before (Murakami et al., 2013a). Briefly, 32P-labled HIS4 (−96/+112) DNA (1.5 pmol) was incubated with 3.7 pmol of TFIIB, 3.7 pmol of TFIIA, 2.0 pmol of TBP, 3.7 pmol of TFIIE, 2.0 pmol of RNA polymerase II-TFIIF complex, and 2.0 pmol of TFIIH-ΔTFIIK with or without 3.0 pmol of TFIIK in 5 μl of buffer 300 (50 mM HEPES (pH7.6), 300 mM potassium acetate, 5 mM DTT, and 5% glycerol), then combined with 5 μl of buffer 30 (50 mM HEPES (pH7.6), 5 mM magnesium sulfate, 30 mM potassium acetate, and 5 mM DTT), and incubated for more than 1 hr at 4°C. The reconstituted PIC was combined with an equal volume of 2xNTP buffer (1.6 mM NTP(s) or 1.6 mM dATP, 50 mM HEPES (pH7.6), 5 mM magnesium sulfate, 30 mM potassium acetate, 5 mM DTT, 10 mM magnesium acetate, and 5 units of RNaseOUT) and incubated for 4 min at 30°C. Exonuclease III digestion was performed with 5–10 units (NEB) for 9 min at 30°C, and was stopped by adding 185 μl of stop buffer (300 mM sodium acetate (pH 5.5), 5 mM EDTA, 0.7% SDS, 0.1 mg/ml glycogen, 0.013 mg/ml of proteinase K (Sigma), 0.5 mg/ml Salmon Sperm DNA (Invitrogen)). DNAs were precipitated by adding 700 μL of ethanol, dried, and analysed by a denaturing 6% acrylamide gel.
In vivo analysis of Kin28 function in TSS selection
kin28-FRB or control “anchor-away” yeast strains (Jeronimo and Robert, 2014) were treated with 1 μg/ml Rapamycin (Sigma) for up to 1 hour followed by total RNA isolation and primer extension analysis as previously described (Kaplan et al., 2012). Primer extension products were resolved on denaturing 8% polyacrylamide (19:1 bisacrylamide:acrylamide) gels in 1× TBE. Primer sequences available upon request.
Supplementary Material
Highlights.
Omission of TFIIK shifts the TSS in a yeast system to a metazoan location
Moving the TSS to a metazoan location enables transcription in the yeast system
Omission of TFIIK uncouples promoter opening from start site scanning
Acknowledgments
We thank Dr. Peter Geiduschek, Furqan Fazal, and Cong Meng for critical reading of the manuscript and discussions, and thank Kyle Eagen and Igor Eliseev for help with DNA manipulation and protein purification. We thank Dr. Steve Hahn (Fred Hutchinson Cancer Center, Seattle) for supplying yeast strain carrying KIN28 L83G. We thank François Robert (IRCM, Montreal, Canada) for the gift of kin28 and control “anchor-away” strains and plasmids for constructing kin28-FRB strains. We thank Dr. Xin Liu (UT Southwestern, Texas) for providing TFIIB E62K. This research was supported by NIH grants GM36659 and AI21144 to R.D.K and by GM097260 to C.D.K..
Footnotes
Author Contributions
K.M. and R.D.K designed the experiments. K.M. P-J.M., R.E.D. purified proteins. K.M., R.E.D, H.J., and C.D.K. performed the experiments. K.M. and R.D.K. wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Borggrefe T, Davis R, Bareket-Samish A, Kornberg RD. Quantitation of the RNA polymerase II transcription machinery in yeast. The Journal of biological chemistry. 2001;276:47150–47153. doi: 10.1074/jbc.M109581200. [DOI] [PubMed] [Google Scholar]
- Bradsher J, Coin F, Egly JM. Distinct roles for the helicases of TFIIH in transcript initiation and promoter escape. The Journal of biological chemistry. 2000;275:2532–2538. doi: 10.1074/jbc.275.4.2532. [DOI] [PubMed] [Google Scholar]
- Bushnell DA, Westover KD, Davis RE, Kornberg RD. Structural basis of transcription: an RNA polymerase II-TFIIB cocrystal at 4.5 Angstroms. Science. 2004;303:983–988. doi: 10.1126/science.1090838. [DOI] [PubMed] [Google Scholar]
- Chen ZA, Jawhari A, Fischer L, Buchen C, Tahir S, Kamenski T, Rasmussen M, Lariviere L, Bukowski-Wills JC, Nilges M, et al. Architecture of the RNA polymerase II-TFIIF complex revealed by cross-linking and mass spectrometry. EMBO J. 2010;29:717–726. doi: 10.1038/emboj.2009.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaway RC, Conaway JW. General initiation factors for RNA polymerase II. Annu Rev Biochem. 1993;62:161–190. doi: 10.1146/annurev.bi.62.070193.001113. [DOI] [PubMed] [Google Scholar]
- Dvir A, Conaway RC, Conaway JW. A role for TFIIH in controlling the activity of early RNA polymerase II elongation complexes. Proc Natl Acad Sci U S A. 1997;94:9006–9010. doi: 10.1073/pnas.94.17.9006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faitar SL, Brodie SA, Ponticelli AS. Promoter-specific shifts in transcription initiation conferred by yeast TFIIB mutations are determined by the sequence in the immediate vicinity of the start sites. Molecular and cellular biology. 2001;21:4427–4440. doi: 10.1128/MCB.21.14.4427-4440.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feaver WJ, Svejstrup JQ, Henry NL, Kornberg RD. Relationship of CDK-activating kinase and RNA polymerase II CTD kinase TFIIH/TFIIK. Cell. 1994;79:1103–1109. doi: 10.1016/0092-8674(94)90040-x. [DOI] [PubMed] [Google Scholar]
- Fishburn J, Hahn S. Architecture of the yeast RNA polymerase II open complex and regulation of activity by TFIIF. Molecular and cellular biology. 2012;32:12–25. doi: 10.1128/MCB.06242-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishburn J, Tomko E, Galburt E, Hahn S. Double-stranded DNA translocase activity of transcription factor TFIIH and the mechanism of RNA polymerase II open complex formation. Proc Natl Acad Sci U S A. 2015;2015;112:3961–3966. doi: 10.1073/pnas.1417709112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazy MA, Brodie SA, Ammerman ML, Ziegler LM, Ponticelli AS. Amino acid substitutions in yeast TFIIF confer upstream shifts in transcription initiation and altered interaction with RNA polymerase II. Molecular and cellular biology. 2004;24:10975–10985. doi: 10.1128/MCB.24.24.10975-10985.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardina C, Lis JT. DNA melting on yeast RNA polymerase II promoters. Science. 1993;261:759–762. doi: 10.1126/science.8342041. [DOI] [PubMed] [Google Scholar]
- Gibbons BJ, Brignole EJ, Azubel M, Murakami K, Voss NR, Bushnell DA, Asturias FJ, Kornberg RD. Subunit architecture of general transcription factor TFIIH. Proc Natl Acad Sci U S A. 2012;109:1949–1954. doi: 10.1073/pnas.1105266109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruki H, Nishikawa J, Laemmli UK. The anchor-away technique: rapid, conditional establishment of yeast mutant phenotypes. Molecular cell. 2008;31:925–932. doi: 10.1016/j.molcel.2008.07.020. [DOI] [PubMed] [Google Scholar]
- Holstege FC, Fiedler U, Timmers HT. Three transitions in the RNA polymerase II transcription complex during initiation. EMBO J. 1997;16:7468–7480. doi: 10.1093/emboj/16.24.7468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstege FC, Tantin D, Carey M, van der Vliet PC, Timmers HT. The requirement for the basal transcription factor IIE is determined by the helical stability of promoter DNA. EMBO J. 1995;14:810–819. doi: 10.1002/j.1460-2075.1995.tb07059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeronimo C, Robert F. Kin28 regulates the transient association of Mediator with core promoters. Nature structural & molecular biology. 2014;21:449–455. doi: 10.1038/nsmb.2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan CD, Jin H, Zhang IL, Belyanin A. Dissection of Pol II trigger loop function and Pol II activity-dependent control of start site selection in vivo. PLoS Genet. 2012;8:e1002627. doi: 10.1371/journal.pgen.1002627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Ebright RH, Reinberg D. Mechanism of ATP-dependent promoter melting by transcription factor IIH. Science. 2000;288:1418–1422. doi: 10.1126/science.288.5470.1418. [DOI] [PubMed] [Google Scholar]
- Kornberg RD. The molecular basis of eukaryotic transcription. Proc Natl Acad Sci U S A. 2007;104:12955–12961. doi: 10.1073/pnas.0704138104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laybourn PJ, Dahmus ME. Phosphorylation of RNA polymerase IIA occurs subsequent to interaction with the promoter and before the initiation of transcription. The Journal of biological chemistry. 1990;265:13165–13173. [PubMed] [Google Scholar]
- Li Y, Flanagan PM, Tschochner H, Kornberg RD. RNA polymerase II initiation factor interactions and transcription start site selection. Science. 1994;263:805–807. doi: 10.1126/science.8303296. [DOI] [PubMed] [Google Scholar]
- Li Y, Kornberg RD. Interplay of positive and negative effectors in function of the C-terminal repeat domain of RNA polymerase II. Proc Natl Acad Sci U S A. 1994;91:2362–2366. doi: 10.1073/pnas.91.6.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Bushnell DA, Wang D, Calero G, Kornberg RD. Structure of an RNA polymerase II-TFIIB complex and the transcription initiation mechanism. Science. 2010;327:206–209. doi: 10.1126/science.1182015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Kung C, Fishburn J, Ansari AZ, Shokat KM, Hahn S. Two cyclin-dependent kinases promote RNA polymerase II transcription and formation of the scaffold complex. Molecular and cellular biology. 2004;24:1721–1735. doi: 10.1128/MCB.24.4.1721-1735.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lue NF, Flanagan PM, Sugimoto K, Kornberg RD. Initiation by yeast RNA polymerase II at the adenoviral major late promoter in vitro. Science. 1989;246:661–664. doi: 10.1126/science.2510298. [DOI] [PubMed] [Google Scholar]
- Majovski RC, Khaperskyy DA, Ghazy MA, Ponticelli AS. A functional role for the switch 2 region of yeast RNA polymerase II in transcription start site utilization and abortive initiation. The Journal of biological chemistry. 2005;280:34917–34923. doi: 10.1074/jbc.M502932200. [DOI] [PubMed] [Google Scholar]
- Murakami K, Calero G, Brown CR, Liu X, Davis RE, Boeger H, Kornberg RD. Formation and Fate of a Complete, 31-Protein, RNA polymerase II Transcription Initiation Complex. The Journal of biological chemistry. 2013a doi: 10.1074/jbc.M112.433623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami K, Elmlund H, Kalisman N, Bushnell DA, Adams CM, Azubel M, Elmlund D, Levi-Kalisman Y, Liu X, Gibbons BJ, et al. Architecture of an RNA polymerase II transcription preinitiation complex. Science. 2013b;342:1238724. doi: 10.1126/science.1238724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami K, Gibbons BJ, Davis RE, Nagai S, Liu X, Robinson PJ, Wu T, Kaplan CD, Kornberg RD. Tfb6, a previously unidentified subunit of the general transcription factor TFIIH, facilitates dissociation of Ssl2 helicase after transcription initiation. Proc Natl Acad Sci U S A. 2012;109:4816–4821. doi: 10.1073/pnas.1201448109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal M, Ponticelli AS, Luse DS. The role of the transcription bubble and TFIIB in promoter clearance by RNA polymerase II. Molecular cell. 2005;19:101–110. doi: 10.1016/j.molcel.2005.05.024. [DOI] [PubMed] [Google Scholar]
- Pardee TS, Bangur CS, Ponticelli AS. The N-terminal region of yeast TFIIB contains two adjacent functional domains involved in stable RNA polymerase II binding and transcription start site selection. The Journal of biological chemistry. 1998;273:17859–17864. doi: 10.1074/jbc.273.28.17859. [DOI] [PubMed] [Google Scholar]
- Parvin JD, Sharp PA. DNA topology and a minimal set of basal factors for transcription by RNA polymerase II. Cell. 1993;73:533–540. doi: 10.1016/0092-8674(93)90140-l. [DOI] [PubMed] [Google Scholar]
- Pinto I, Wu WH, Na JG, Hampsey M. Characterization of sua7 mutations defines a domain of TFIIB involved in transcription start site selection in yeast. The Journal of biological chemistry. 1994;269:30569–30573. [PubMed] [Google Scholar]
- Ponjavic J, Lenhard B, Kai C, Kawai J, Carninci P, Hayashizaki Y, Sandelin A. Transcriptional and structural impact of TATA-initiation site spacing in mammalian core promoters. Genome Biol. 2006;7:R78. doi: 10.1186/gb-2006-7-8-r78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy R, Adamczewski JP, Seroz T, Vermeulen W, Tassan JP, Schaeffer L, Nigg EA, Hoeijmakers JH, Egly JM. The MO15 cell cycle kinase is associated with the TFIIH transcription-DNA repair factor. Cell. 1994;79:1093–1101. doi: 10.1016/0092-8674(94)90039-6. [DOI] [PubMed] [Google Scholar]
- Schaeffer L, Roy R, Humbert S, Moncollin V, Vermeulen W, Hoeijmakers JH, Chambon P, Egly JM. DNA repair helicase: a component of BTF2 (TFIIH) basic transcription factor. Science. 1993;260:58–63. doi: 10.1126/science.8465201. [DOI] [PubMed] [Google Scholar]
- Schultz P, Fribourg S, Poterszman A, Mallouh V, Moras D, Egly JM. Molecular structure of human TFIIH. Cell. 2000;102:599–607. doi: 10.1016/s0092-8674(00)00082-9. [DOI] [PubMed] [Google Scholar]
- Serizawa H, Conaway JW, Conaway RC. Phosphorylation of C-terminal domain of RNA polymerase II is not required in basal transcription. Nature. 1993;363:371–374. doi: 10.1038/363371a0. [DOI] [PubMed] [Google Scholar]
- Spangler L, Wang X, Conaway JW, Conaway RC, Dvir A. TFIIH action in transcription initiation and promoter escape requires distinct regions of downstream promoter DNA. Proc Natl Acad Sci U S A. 2001;98:5544–5549. doi: 10.1073/pnas.101004498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svejstrup JQ, Wang Z, Feaver WJ, Wu X, Bushnell DA, Donahue TF, Friedberg EC, Kornberg RD. Different forms of TFIIH for transcription and DNA repair: holo-TFIIH and a nucleotide excision repairosome. Cell. 1995;80:21–28. doi: 10.1016/0092-8674(95)90447-6. [DOI] [PubMed] [Google Scholar]
- Wong KH, Jin Y, Struhl K. TFIIH phosphorylation of the Pol II CTD stimulates mediator dissociation from the preinitiation complex and promoter escape. Molecular cell. 2014;54:601–612. doi: 10.1016/j.molcel.2014.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Ponticelli AS. Evidence that RNA polymerase II and not TFIIB is responsible for the difference in transcription initiation patterns between Saccharomyces cerevisiae and Schizosaccharomyces pombe. Nucleic Acids Res. 2012;40:6495–6507. doi: 10.1093/nar/gks323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Degen D, Ho MX, Sineva E, Ebright KY, Ebright YW, Mekler V, Vahedian-Movahed H, Feng Y, Yin R, et al. GE23077 binds to the RNA polymerase ‘i’ and ‘i+1’ sites and prevents the binding of initiating nucleotides. eLife. 2014;3:e02450. doi: 10.7554/eLife.02450. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.