Abstract
Scope
Tissue concentrations of omega-3 fatty acids may reduce cardiovascular disease risk, and genetic variants are associated with circulating fatty acids concentrations. Whether dietary fatty acids interact with genetic variants to modify circulating omega-3 fatty acids is unclear.
Objective
We evaluated interactions between genetic variants and fatty acid intakes for circulating alpha-linoleic acid (ALA), eicosapentaenoic acid (EPA), docosahexaenoic acid (DHA) and docosapentaenoic acid (DPA).
Methods and Results
We conducted meta-analyses (N to 11,668) evaluating interactions between dietary fatty acids and genetic variants (rs174538 and rs174548 in FADS1 (fatty acid desaturase 1), rs7435 in AGPAT3 (1-acyl-sn-glycerol-3-phosphate), rs4985167 in PDXDC1 (pyridoxal-dependent decarboxylase domain-containing 1), rs780094 in GCKR (glucokinase regulatory protein) and rs3734398 in ELOVL2 (fatty acid elongase 2)). Stratification by measurement compartment (plasma vs. erthyrocyte) revealed compartment-specific interactions between FADS1 rs174538 and rs174548 and dietary ALA and linoleic acid for DHA and DPA.
Conclusion
Our findings reinforce earlier reports that genetically-based differences in circulating fatty acids may be partially due to differences in the conversion of fatty acid precursors. Further, fatty acids measurement compartment may modify gene-diet relationships, and considering compartment may improve the detection of gene-fatty acids interactions for circulating fatty acid outcomes.
Keywords: FADS1, gene-diet interactions, meta-analysis, omega-3 fatty acids
Introduction
Tissue concentrations of omega-3 fatty acids may reduce cardiovascular disease (CVD) risk [1–5]. Therefore, furthering our understanding of the determinants of circulating omega-3 fatty acids concentrations is essential. Variants in genes encoding fatty acid biosynthetic enzymes and additional proteins external to the pathway influence circulating omega-3 fatty acid concentrations (6–7). In a previous genome wide association study (GWAS) meta-analysis (N=8866), we reported that FADS1 (fatty acid desaturase 1) and FADS2 (fatty acid desaturase 2) variants were associated with higher alpha-linolenic acid (ALA) and lower eicosapentaenoic acid (EPA) and docosapentaenoic acid (DPA) concentrations. In addition, ELOVL2 (fatty acid elongase 2) variants were associated with higher EPA and DPA and lower docosahexaenoic acid (DHA) concentrations. These findings support prior evidence that enzymatic genetic variation influences flow through the biosynthetic pathway from ALA to DHA (7). The same study reported that a FADS1 variant altered the association between circulating ALA and EPA, implying that FADS1 variation may influence conversion of ALA to EPA. In addition to the genes encoding pathway enzymes, we reported associations between GCKR (glucokinase regulatory protein) and AGPAT3 (1-acyl-sn-glycerol-3-phosphate) variants with circulating DPA and PDXDC1 (pyridoxal-dependent decarboxylase domain containing 1) variants with circulating ALA. Although all of the named loci encode proteins that participate in lipid metabolism, understanding of how AGPAT3, GCKR, and PDXDC1 variants might determine circulating omega-3 fatty acids is limited. Whether habitual diet influences relationships between variants at these six loci for circulating omega-3 fatty acid outcomes is also unexplored, and investigation of gene-diet interactions could increase understanding of the molecular determinants of circulating ALA, EPA, DPA and DHA.
Biologically, several dietary fatty acids (alpha-linolenic acid (ALA), linoleic acid (LA), eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA) could plausibly modify genetic associations for circulating omega-3 fatty acid (ALA, EPA, DPA, DHA) concentrations. First, ALA is a substrate for longer chain omega-3 fatty acids (EPA, DPA, DHA), and genetic factors that influence its conversion could affect long-chain omega-3 concentrations. ALA conversion to EPA/DPA/DHA is limited in humans and demonstrates inter-individual variability that may be related to genetics [8, 9, 10]. A second plausible candidate is the omega-6 fatty acid LA, which may limit ALA conversion to EPA through competitive inhibition of the desaturase enzymes that are shared by the omega-3 and omega-6 fatty acids biosynthetic pathways [11]. Finally, dietary EPA and DHA may also influence omega-3 fatty acid biosynthesis, as shown by differential conversion of ALA to long chain omega-3 fatty acids in fish consumers and non-consumers [12]. Collectively, these studies suggest that habitual intakes of ALA, LA, EPA and DHA may conceivably modulate the genetic contributions to circulating omega-3 fatty acids concentrations.
In addition to being influenced by dietary fatty acids and genetic factors, levels of circulating omega-3 fatty acids vary with the site of fatty acid measurement. Circulating omega-3 fatty acids in human populations are typically measured in total plasma, plasma fractions, or erythrocyte membranes. Differences in fatty acids incorporation, distribution and metabolism may vary based on the measurement compartment [13–16], but understanding of how genetically-based variability in response to diet may be further modified by measurement compartment is limited [17,18].
The primary objective of the current study was to evaluate interactions between habitual dietary fatty acids (ALA, LA, EPA+DHA) intake and selected single nucleotide polymorphisms (SNPs) for the outcome of circulating omega-3 fatty acids (ALA, EPA, DPA and DHA). We hypothesized that the relationship between SNPs and circulating omega-3 fatty acids would be modified by dietary fatty acids. In a secondary analysis we explored whether these gene-diet interactions differed by the fatty acids measurement compartment (plasma and plasma phospholipids vs. erythrocyte membrane). We performed interaction analyses in 9 independent U.S. and European cohorts, with a total number of samples up to 11,668. The fatty acids were measured in plasma phospholipids in 4 cohorts, in total plasma in one cohort, and in erythrocyte membranes in 4 cohorts.
Materials and Methods
Study Populations
The 9 cohorts included for meta-analysis were the Atherosclerosis Risk in Communities (ARIC) Study, Coronary Artery Risk Development in Young Adults (CARDIA), Cardiovascular Health Study (CHS), Genetics of Lipid Lowering Drugs and Diet Network (GOLDN), Health Professionals Follow-up Study (HPFS), Invecchiare in Chianti (InCHIANTI), Multi-Ethnic Study of Atherosclerosis (MESA), Nurses’ Health Study (NHS) and Women’s Genome Health Study (WGHS) are described in Supplemental Table 1. All studies were approved by local Institutional Review Boards and all participants provided informed consent. These cohorts participate in the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium, which was created in 2008 for the purpose of evaluating genome-wide associations and gene-nutrient interactions for cardiovascular disease and risk factors. Jointly developed plans guide the analysis conducted at each cohort and summary statistics are meta-analyzed centrally.
SNP Selection and Genotyping
Six SNPs from five loci were selected from those highly significantly associated at genome-wide level with plasma phospholipid omega-3 fatty acids in a meta-analysis of GWAS of European origin individuals [7]. The SNPs evaluated were rs174538 and rs174548 in FADS1, rs7435 in AGPAT3, rs4985167 in PDXDC1, rs780094 in GCKR and rs3734398 in ELOVL2. AGPAT3 and PDXDC1 variants were previously identified as determinants of circulating glycerophospholipids and sphingolipids (19), and the AGPAT3 protein participates in the incorporation of DHA into phospholipids. (20). GCKR (protein name glucokinase regulatory protein) is a regulator of glucose phosphorylation that acts through competitive inhibition of glucokinase (21). The GCKR rs780094 variant has been repeatedly associated with metabolic traits including triglycerides and glucose (22) and was suggested to interact with dietary factors (23). Methods for genotyping are described in Supplemental Table 2.
Circulating fatty acids measurement and dietary fatty acids estimates in each study
In ARIC, MESA, CARDIA, and CHS, plasma phospholipids were first isolated by thin layer chromatography and then separated by gas chromatography. In InCHIANTI, total plasma fatty acids were measured using a similar gas chromatography technique. The cohorts ARIC, MESA, CARDIA, CHS and InCHIANTI previously contributed to a meta-analysis of genome-wide associations for circulating omega-3 fatty acids (7). In GOLDN, HPFS, NHS and WGHS fatty acids were measured in erythrocyte membranes. Details of circulating fatty acid measurements for all cohorts are provided in Supplemental Table 3. Dietary fatty acids were estimated from food frequency questionnaires that are described in Supplemental Table 4.
Statistical analyses by each cohort
Each cohort performed linear regression analysis to generate regression coefficients (β) and standard errors for the associations between intake of ALA, LA, EPA+DHA, PUFA and circulating omega-3 fatty acids (ALA, EPA, DPA, DHA) and interactions between genotypes of 6 SNPs and dietary fatty acids for the outcome of circulating fatty acids
Associations between dietary fatty acids and circulating fatty acids were evaluated continuously using a model adjusting for age, gender, total energy intake and population substructure variables as needed. The regression coefficient represents the difference in the circulating fatty acids outcomes (ALA, EPA, DPA, DHA) in association with each 1 gram greater intake of dietary fatty acid.
Fatty acids-SNP interactions were evaluated by cross-product terms using the likelihood ratio test with an additive genetic model. Thus, the interaction regression coefficient represents the difference in the magnitude of association between dietary fatty acids and circulating fatty acids (ALA, EPA, DPA, DHA) per copy of the effect allele. The interaction model was adjusted for age, gender, total energy intake and population substructure variables (as needed).
Meta-analysis and meta-regression
Meta-analysis was performed using an inverse variance-weighted, fixed effects approach. For meta-analysis of dietary fatty acids associations with circulating fatty acids, R software was used. For SNP x dietary fatty acids interaction meta-analysis, METAL software was used (http://umich.edu/csg/abecasis/Metal/). Although the two FADS1 SNPs rs174538 and rs174548 are in moderately strong linkage disequilibrium (r2=0.69 in Europeans) Bonferroni correction was conservatively based on all six SNPs and the four dietary fatty acids evaluated for interaction to establish a significance level with correction for multiple testing (α=0.05/24 tests=0.002). To formally examine a potential source of heterogeneity, we conducted meta-regression analysis with “fatty acids measurement compartment” (plasma phospholipids/plasma vs. erythrocyte membranes) as an independent variable. Meta-regression was performed using Stata software.
Results
Population characteristics, dietary fatty acids and circulating fatty acids in plasma or erythrocytes (expressed as % of total fatty acids) are shown for each cohort (Table 1; Supplemental Table 1). Circulating fatty acids (% total fatty acids) are comparable across the nine cohort studies. Meta-analysis of associations between the selected SNPs and plasma omega-3 fatty acids in five of the nine cohorts were previously described in a meta-analysis of GWAS using plasma phospholipids or total plasma [7]. Similar associations were observed in the current study, in which 4 cohorts that measured erythrocyte fatty acids were included (data not shown).
Table 1.
Dietary and circulating fatty acids by cohort
| Dietary fatty acids (grams/day) | Circulating fatty acids, % total fatty acids | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | Age (y) |
% Women |
Fatty acids compartment |
ALA | LA | EPA+ DHA |
PUFA | ALA:LA correlation coefficient |
ALA | EPA | DPA | DHA | |
| Atherosclerosis Risk in Communities (ARIC) Study | 3266 | 54±6 | 51 | Plasma phospholipid | 0.7±0.3 | 8.2±4.0 | 0.2±0.2 | 9.4±4.3 | 0.71 | 0.14±0.05 | 0.56±0.3 | 0.9±0.17 | 2.82±0.88 |
| Coronary Artery Risk Development in Young Adults (CARDIA) | 1097 | 46±3 | 55 | Plasma phospholipid | 2.1±2.3 | 17.7±16.9 | 0.2±0.3 | 20.2±19.1 | 0.92 | 0.19±0.09 | 0.85±0.62 | 0.94±0.21 | 3.09±1.12 |
| Cardiovascular Health Study (CHS) | 2305 | 75±5 | 61 | Plasma phospholipid | 1.5±0.6 | 12.6±5.2 | 0.4±0.3 | 14.9±6 | 0.94 | 0.15±0.05 | 0.59±0.37 | 0.83±0.17 | 2.98±0.96 |
| Multi-Ethnic Study of Atherosclerosis (MESA) | 646 | 62±10 | 53 | Plasma phospholipid | 1.1±0.5 | 9.7±5.4 | 0.1±0.1 | 11.1±5.8 | 0.82 | 0.18±0.06 | 0.89±0.59 | 0.96±0.22 | 3.7±1.41 |
| Invecchiare in Chianti (InCHIANTI) | 1059 | 68±15 | 55 | Total Plasma | 1±0.3 | 5.9±2 | NA | 7.4±2.4 | 0.83 | 0.44±0.25 | 0.61±0.22 | NA | 2.29±0.77 |
| Genetics of Lipid Lowering Drugs and Diet Network (GOLDN) | 1038 | 48±16 | 52 | Erythrocyte membranes | 1.5±0.8 | 15.6±7.9 | 0.1±0.1 | 17.3±8.7 | 0.92 | 0.14±0.04 | 0.53±0.28 | 2.11±0.30 | 3.03±0.89 |
| Health Professionals Follow-up Study (HPFS) | 1294 | 64±9 | 0 | Erythrocyte membranes | 1.1±0.4 | 11.2±4.3 | 0.3±0.3 | 12.9±4.7 | 0.82 | 0.21±0.22 | 0.51±0.28 | 1.97±0.39 | 3.67±1.2 |
| Nurses Health Study (NHS) | 597 | 60±6 | 100 | Erythrocyte membranes | 1.1±0.4 | 10.8±3.9 | 0.2±0.2 | 12.5±4.3 | 0.86 | 0.15±0.06 | 0.68±0.67 | 1.67±0.31 | 3.31±1.03 |
| Women’s Genome Health Study (WGHS) | 635 | 54±7 | 100 | Erythrocyte membranes | 1.1±0.4 | 9.7±4.0 | 0.2±0.2 | 11.4±4.6 | 0.86 | 0.16±0.05 | 0.49±0.19 | 1.84±0.39 | 3.27±0.98 |
ALA, alpha linolenic acid; DHA, docosahexaenoic acid; DPA, docosapentaenoic acid; EPA, eicosapentaenoic acid, LA, linoleic acid; NA, not available; PUFA, polyunsaturated fatty acids
Associations between dietary fatty acids and circulating omega-3 fatty acids
Meta-analysis of associations between dietary intake of omega-3 fatty acids (in grams/day) and circulating (plasma or erythrocyte membrane) omega-3 fatty acids are shown in Table 2. Each one gram greater intake of ALA was associated with a higher circulating ALA of 0.006 (95% CI [0.004,0.008]; P<0.001). Each one gram greater intake of LA was associated with a lower circulating EPA of 0.007(95% CI [−0.009,−0.006]), DPA of 0.004(95% CI [−0.005,−0.003]), and DHA of 0.007 (95% CI [−0.011,−0.003]; all P<0.001). Each one gram greater intake of combined EPA+DHA was associated with higher circulating EPA of 0.333 (95% CI [0.298, 0.369] and DHA of 1.49 (95% CI [1.395,1.586]; both P<0.001). The direction of associations between dietary fatty acids and circulating omega-3 fatty acids were generally consistent across individual cohorts.
Table 2.
Meta-analysis of associations between dietary fatty acids and circulating omega-3 fatty acids
| Regression* coefficients (β [95%CI] for dietary ALA (g/d) | P | N | Regression* coefficients (β [95% CI] for dietary LA (g/d) | P | N | Regression* coefficients (β [95% CI] for dietary EPA+DHA (g/d) | P | N | |
|---|---|---|---|---|---|---|---|---|---|
| Circulating fatty acids (%total fatty acids) | |||||||||
| ALA | 0.006 (0.004,0.008) | < 0.001 | 11291 | −0.0002(−0.0004,0.0001) | 0.141 | 10232 | 0.005(0.0003,0.010) | 0.038 | 10656 |
| EPA | 0.011(−0.004,0.026) | 0.143 | 11108 | −0.007(−0.009, −0.006) | < 0.001 | 10049 | 0.333(0.298,0.369) | < 0.001 | 10473 |
| DPA | −0.004(−0.010,0.002) | 0.209 | 10232 | −0.004(−0.005, −0.003) | < 0.001 | 10232 | 0.0004(−0.016,0.017) | 0.964 | 9597 |
| DHA | 0.027(−0.005,0.058) | 0.103 | 11291 | −0.007(−0.011, −0.003) | < 0.001 | 10232 | 1.491(1.395,1.586) | < 0.001 | 10656 |
β represents expected change in circulating fatty acids per one gram greater fatty acid intake
ALA, alpha linolenic acid; DHA, docosahexaenoic acid; DPA, docosapentaenoic acid; EPA, eicosapentaenoic acid, LA, linoleic acid
Interactions between SNPs and dietary fatty acid intake for circulating fatty acids
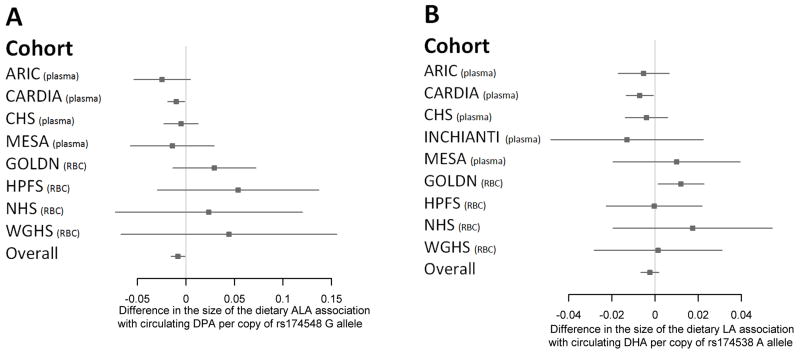
Of the 6 SNPs tested for interaction with dietary fatty acids for the outcomes of circulating omega-3 fatty acids, none reached Bonferroni adjusted statistical significance (data not shown). However, we observed evidence of heterogeneity by the fatty acid measurement compartment for the interactions of FADS1 with dietary ALA and LA for the outcomes of circulating DPA (Figure 1A) and DHA (Figure 1B), respectively. Specifically, for plasma measurements in panel A (ARIC, CARDIA, InCHIANTI, MESA) interaction coefficients are negative and for erythrocyte measurements (GOLDN, HPFS, NHS and WGHS) interaction coefficients are positive.
Figure 1.
Interactions between FADS1 rs174548 and rs174538 and dietary fatty acids for circulating omega-3 fatty acids, in each cohort. In ARIC, CARDIA, CHS, InCHIANTI and MESA, fatty acids were measured in plasma/plasma phospholipids; in GOLDN, HPFS, NHS and WGHS, fatty acids were measured in erythrocyte membranes. Panel A illustrates the association of each one gram/day greater intake of dietary alpha linoleic acid (ALA) with circulating docosapentaenoic acid (DPA) per copy of the rs174548 G allele. Panel B illustrates the association of each one gram/day greater intake dietary linoleic acid (LA) with circulating docosahexaenoic (DHA) per copy of the rs174538 A allele. Circulating DPA was not measured in the InCHIANTI study.
Meta-regression
Using meta-regression methods, we tested whether fatty acid measurement compartment explained between-study heterogeneity and determined that compartment was associated (P<0.05) with the magnitude of the regression coefficient for the interaction terms of two FADS1 SNPs (rs174548 and rs174538) and both dietary ALA and LA.
Interaction analyses between two FADS1 SNPs and dietary fatty acids for circulating fatty acids, stratified by fatty acids measurement compartment
Because measurement compartment was associated with differences in meta-analysis of interactions, we stratified the meta-analysis by compartment (Table 3). Several patterns emerged. For diet x SNP interactions involving FADS1 SNPs rs174538 and rs174548, beta coefficient of meta-analysis of interaction consistently differed in direction by compartment (negative in plasma and positive in erythrocytes).
Table 3.
Meta-analysis of interactions between FADS1 SNPs (rs174548 and rs174538) and dietary fatty acids in association with circulating omega-3 fatty acids, stratified by fatty acids measurement compartment
| Interactions between dietary ALA X rs174548 for plasma omega-3 fatty acids | Interactions between dietary ALA X rs174548 for erythrocyte omega-3 fatty acids | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| β | SEM | P | β direction | β | SEM | P | β direction | P* | |
| EPA | −0.0118 | 0.010 | 0.223 | −−+−+ | 0.018 | 0.012 | 0.120 | +−++ | 0.048 |
| DPA | −0.010 | 0.004 | 0.006 | −−−− | 0.034 | 0.017 | 0.048 | ++++ | 0.012 |
| DHA | −0.036 | 0.021 | 0.085 | −−−−+ | 0.13 | 0.05 | 0.005 | ++++ | 0.001 |
| Interactions between dietary LA X rs174548 for plasma omega-3 fatty acids | Interactions between dietary LA X rs174548 for erythrocyte omega-3 fatty acids | ||||||||
| EPA | −0.0008 | 0.001 | 0.408 | +−+−+ | 0.002 | 0.001 | 0.151 | +−++ | 0.103 |
| DPA | −0.001 | 0.001 | 0.057 | +−−− | 0.004 | 0.002 | 0.014 | ++++ | 0.004 |
| DHA | −0.004 | 0.002 | 0.106 | +−−−+ | 0.011 | 0.004 | 0.009 | ++++ | 0.002 |
| Interactions between dietary ALA X rs174538 for plasma omega-3 fatty acids | Interactions between dietary ALA X rs174538 for erythrocyte omega-3 fatty acids | ||||||||
| β | SEM | P | β direction | β | SEM | P | β direction | P* | |
| EPA | −0.0135 | 0.010 | 0.183 | −−+−+ | 0.010 | 0.011 | 0.400 | +−++ | 0.189 |
| DPA | −0.010 | 0.004 | 0.007 | −−−− | 0.024 | 0.017 | 0.146 | ++−+ | 0.043 |
| DHA | −0.046 | 0.022 | 0.033 | −−−−+ | 0.096 | 0.05 | 0.041 | +−++ | 0.006 |
| Interactions between dietary LA X rs174538 for plasma omega-3 fatty acids | Interactions between dietary LA X rs174538 for erythrocyte omega-3 fatty acids | ||||||||
| EPA | −0.0009 | 0.001 | 0.360 | −−+−+ | 0.001 | 0.001 | 0.432 | +−−+ | 0.235 |
| DPA | −0.001 | 0.001 | 0.059 | +−−− | 0.003 | 0.002 | 0.074 | ++−+ | 0.025 |
| DHA | −0.006 | 0.002 | 0.017 | −−−−+ | 0.009 | 0.005 | 0.037 | +−++ | 0.003 |
β represents the difference in the magnitude of the dietary fatty acids association with circulating fatty acids (expressed as difference in circulating fatty acid per 1 gram/day greater intake of fatty acid) per copy of the effect allele
P =P value for meta-analysis of interactions between SNP X dietary fatty acids for the outcome of circulating fatty acids
P* =P value for association between fatty acids compartment for the interaction of dietary fatty acid X SNP on circulating fatty acid.
For EPA and DHA in the plasma compartment the cohort order =ARIC, CARDIA, CHS, InCHIANTI, MESA
For DPA the plasma compartment the cohort order =ARIC, CARDIA, CHS, MESA
For EPA, DPA and DHA in the erythrocyte compartment the cohort order =GOLDN, HPFS, NHS, WGHS
ALA, alpha linolenic acid; DHA, docosahexaenoic acid; DPA, docosapentaenoic acid; EPA, eicosapentaenoic acid, LA, linoleic acid
Specifically, FADS1 rs174548 (minor allele G) interacted with dietary ALA for the outcome of plasma DPA with negative betas in all cohorts using plasma (beta= −0.010; P=0.006) and positive betas in all cohorts using erythrocytes (beta= 0.034; P=0.048). Similar cohort-level consistency of beta coefficient direction was also observed for rs174548 interaction with dietary ALA for DHA (beta=0.13; P=0.005) and rs174548 interaction with dietary LA for DHA (beta=0.011; P=0.009) in erythrocytes. For the second FADS1 variant, rs174538 (minor allele A), in interactions with dietary ALA and LA, similar but weaker patterns were observed (Table 3). In particular, dietary ALA interacted with rs174538 for DPA (beta= −0.010; P=0.007) in plasma phospholipids. Interactions between FADS1 variants and dietary ALA for circulating EPA did not reach significance; however, the difference by rs174548 genotype for circulating EPA in erythrocytes was significant at an uncorrected threshold (P=0.048, Table 3), and the regression coefficient was in the same direction as for DPA and DHA. The association of both SNPs with circulating DPA and DHA (main effects) did not differ by compartment (data not shown).
Discussion
In this large study of up to 11,668 individuals, we observed interactions between dietary fatty acids and selected genetic variants for the outcomes of circulating long chain omega-3 fatty acids only when the data were stratified by measurement compartment. Specifically, dietary intake of the precursor fatty acids LA and ALA modulated associations of FADS1 variants for the outcomes of the long chain fatty acids DPA and DHA, in a compartment-specific manner. Our study further showed that these relationships may differ by the site of fatty acid measurement.
FADS1/2 is a well-established genetic determinant of circulating omega -3 fatty acids (ALA, EPA, DPA), but studies that evaluate dietary modulation of these genetic associations are relatively few (6,7). In one previous study, investigation of FADS1 variant rs174561 (r2= 0.86 with FADS1 rs174538 and r2= 0.84 with FADS1 rs174548 in the current study) demonstrated interaction with dietary fatty acids to modulate DPA and EPA in the plasma compartment [10]. In that study (n=36), homozygous minor allele carriers consuming a flaxseed diet (rich in ALA) had lower DPA and EPA compared to major allele carriers, implying that minor allele carriers may exhibit reduced conversion of ALA to longer chain omega-3 fatty acids. Results from the current meta-analysis, suggest that habitual dietary ALA and LA interacted with FADS1 SNPs to modulate circulating DPA and DHA.
Whether fatty acid measurement compartment represents an additional modulator of potential FADS1 gene-diet interactions is unclear. Most large-scale population studies rely on measurements from either the plasma compartment or the erythrocyte compartment, and the current study benefited from the availability of data from both compartments. In general, plasma fatty acids reflect short term fatty acids intakes whereas erythrocyte membranes reflect longer term intakes, so that compartment may be of particular relevance to genetic studies that incorporate dietary data. Further, the consequences of compartment on gene-diet analyses may vary depending on the fatty acid of interest. For example, EPA and DPA are incorporated into the outer erythrocyte leaflet, which readily equilibrates with plasma, whereas DHA is incorporated into the inner erythrocyte leaflet at the time of erythrocyte formation in the bone marrow (16,24). Of potential relevance, data from two feeding trials, both using EPA+DHA, reported differential finding in the two compartments. In one of these trials (n=12 for 12 weeks duration), erythrocyte (but not serum) EPA differed by FADS1/2 genotype [18]. In the second trial (n=310 for 6 months duration), FADS1 genotype appeared to modify delta5 desaturase activity, with some differences between the two compartments [17]. Neither of these studies statistically evaluated the role of compartment. However, a series of previous, non-genetic studies suggest that compartment-based differences in the distribution, metabolism and incorporation of omega-3 fatty acids may be particularly relevant to dietary studies [13–16, 24–25]. In the current study, we statistically evaluated the role of measurement compartment as a source of heterogeneity in FADS1 x diet interaction analyses, and the different patterns in stratified analyses suggest that compartment could be relevant to interaction analyses.
Several limitations must be considered. First, measurement errors in the assessment of dietary intake might have reduced our ability to detect interactions in the overall sample. Food frequency questionnaires that are used to estimate dietary intakes may have limited ability to capture specific fatty acids such as ALA and LA that are used in food preparation. In the current study, since ALA is a substrate for conversion to EPA whereas LA is not converted to EPA, our finding that ALA and LA appear to interact similarly with FADS1 genotypes is unexpected and might represent a statistical artifact. In other words, the high correlations between the dietary estimates of ALA and LA, rather than biology, may account for statistically similar findings for these two fatty acids in the stratified analyses. In addition, we detected significant ALA x SNP interactions for DPA and DHA outcomes (Table 3), but interactions did not reach significance for EPA, the first long-chain PUFA product of the biosynthetic pathway of ALA to EPA/DPA/DHA. The directions of regression coefficients were less consistent across cohorts for EPA compared to DPA and DHA, which could be related to dietary or other unmeasured confounders across the nine cohorts. Finally, we cannot establish functionality for the FADS1 SNPs rs174548 and rs174538, but examination in HapReg v2 software (Broad Institute) showed altered regulatory motifs, promoter or enhancer histone marks, DNAase 1 hypersensitivity, and/or protein binding that support evidence of functionality [26].
Results from the current study improve understanding of the multiple determinants of circulating fatty acids, and may have potential clinical implications. First, they support existing evidence that genetically-based differences in circulating longer chain fatty acids may be due, in part, to differences in the conversion of dietary fatty acids precursors [10]. Accumulating evidence suggests that carriers of FADS1 variants have reduced capacity to synthesize longer chain omega-3 fatty acids, which may be relevant to dietary recommendations. Replication of genetic findings is essential to establishing scientific credibility, and the level of evidence is improved through verification in our large, multi-cohort sample, and under conditions of habitual intake. Second, our findings suggest that fatty acids measurement compartment may modify gene-diet interaction and therefore add heterogeneity to meta-analyses that focus on the simultaneous investigations of gene and diet. Awareness of the extent and circumstances under which this heterogeneity is relevant to analyses may improve the detection of gene-diet interactions for circulating fatty acid outcomes.
Supplementary Material
Acknowledgments
C. Smith is supported by K08 HL112845. Infrastructure for the CHARGE Consortium is supported in part by the National Heart, Lung, and Blood Institute grant HL105756. Funding for the individual cohorts is identified below.
ARIC: The Atherosclerosis Risk in Communities (ARIC) Study is carried out as a collaborative study supported by NHLBI contracts HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C, HHSN268201100012C, R01HL087641, R01HL59367, and R01HL086694; National Human Genome Research Institute (NHGRI) contract U01HG004402; and NIH contract HSN268200625226C. Infrastructure was partly supported by Grant Number UL1RR025005, a component of the NIH Roadmap for Medical Research. CARDIA: The Coronary Artery Risk Development in Young Adults Study (CARDIA) is conducted and supported by the National Heart, Lung, and Blood Institute in collaboration with the University of Alabama at Birmingham (HHSN268201300025C & HHSN268201300026C), Northwestern University (HHSN268201300027C), University of Minnesota (HHSN268201300028C), Kaiser Foundation Research Institute (HHSN268201300029C), and Johns Hopkins University School of Medicine (HHSN268200900041C). CARDIA is also partially supported by the Intramural Research Program of the National Institute on Aging. Genotyping of the CARDIA participants was supported by grants U01-HG-004729, U01-HG-004446, and U01-HG-004424 from the National Human Genome Research Institute. Statistical analyses and fatty acid measures were funded by R01-HL-084099 from the NHLBI to Dr Fornage. CHS: This CHS research was supported by NHLBI contracts HHSN268201200036C, HHSN268200800007C, N01HC55222, N01HC85079, N01HC85080, N01HC85081, N01HC85082, N01HC85083, N01HC85086; and NHLBI grants HL080295, HL087652, HL105756 with additional contribution from the National Institute of Neurological Disorders and Stroke (NINDS). Additional support was provided through AG023629 from the National Institute on Aging (NIA). The provision of genotyping data was supported in part by the National Center for Advancing Translational Sciences, CTSI grant UL1TR000124, and the National Institute of Diabetes and Digestive and Kidney Disease Diabetes Research Center (DRC) grant DK063491 to the Southern California Diabetes Endocrinology Research Center. The fatty acid measurements were supported by grant HL085710 from NHLBI. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. GOLDN: This study was supported by National Heart, Lung, and Blood Institute (NHLBI) grant no. U01HL072524 (Genetic and Environmental Determinants of Triglycerides), NHLBI R01 HL091357 (Genomewide Association Study of Lipid Response to Fenofibrate and Dietary Fat), NHLBI grant number HL54776 and HL078885; and by contracts 53-K06-5-10 and 58-1950-9-001 from the US Department of Agriculture, Agriculture Research Service. Dr CE Smith is supported by K08 HL112845. HPFS and NHS: the research grant P01 CA87969, R01 HL034594, UM1 CA167552, R01 HL35464, HL60712 and CA055075 from National Institutes of Health, a career development award R00HL098459 from the National Heart, Lung, and Blood Institute, and research grant 1-12-JF-13 from American Diabetes Association, and 11SDG7380016 from American Heart Association InCHIANTI: Invecchiare in Chianti (aging in the Chianti area, InCHIANTI) study investigators thank the Intramural Research Program of the NIH, National Institute on Aging who are responsible for the InCHIANTI samples. Investigators also thank the InCHIANTI participants. The InCHIANTI study baseline (1998–2000) was supported as a “targeted project” (ICS110.1/RF97.71) by the Italian Ministry of Health and in part by the U.S. National Institute on Aging (Contracts: 263 MD 9164 and 263 MD 821336).
MESA: The Multi-Ethnic Study of Atherosclerosis (MESA) and MESA SHARe were supported by contracts N01-HC-95159 through N01-HC-95169 and RR-024156 from the National Heart, Lung, and Blood Institute. Funding for MESA SHARe genotyping was provided by NHLBI Contract N02HL64278. The provision of genotyping data was supported in part by the National Center for Advancing Translational Sciences, CTSI grant UL1TR000124, and the National Institute of Diabetes and Digestive and Kidney Disease Diabetes Research Center (DRC) grant DK063491 to the Southern California Diabetes Endocrinology Research Center. The authors thank the other investigators, the staff, and the participants of the MESA study for their valuable contributions. A full list of participating MESA investigators and institutions can be found at http://www.mesa-nhlbi.org.
WGHS: The WGHS is supported by HL043851 and HL080467 from the National Heart, Lung, and Blood Institute (NHLBI) and CA047988 from the National Cancer Institute (NCI), the Donald W. Reynolds Foundation and the Fondation Leducq, with collaborative scientific support and funding for genotyping provided by Amgen. The WHS is supported by HL043851, HL080467 and HL099355 from the NHLBI and CA047988 from NCI. The measurement of erythrocyte fatty acid is supported by 0735390N from the American Heart Association (AHA). Dr. Wang is supported by R00-HL095649 from the NHLBI.
Abbreviations used
- AGPAT3
1-acyl-sn-glycerol-3-phosphate
- ALA
alpha-linolenic acid
- ARIC
Atherosclerosis Risk in Communities
- CARDIA
Coronary Artery Risk Development in Young Adults
- CHARGE
Cohorts for Heart and Aging Research in Genomic Epidemiology
- CHS
Cardiovascular Health Study
- DPA
docosapentaenoic acid
- DHA
docosahexaenoic acid
- ELOVL2
fatty acid elongase 2
- EPA
eicosapentaenoic acid
- FADS1
fatty acid desaturase 1
- FADS2
fatty acid desaturase 2
- GCKR
glucokinase regulatory protein
- GOLDN
Genetics of Lipid Lowering Drugs and Diet Network
- GWAS
genome wide association study
- HPFS
Health Professionals Follow-up Study
- InCHIANTI
Invecchiare in Chianti
- LA
linoleic acid
- MESA
Multi-Ethnic Study of Atherosclerosis
- NHS
Nurses Health Study
- PDXDC1
pyridoxal-dependent decarboxylase domain
- SNP
single nucleotide polymorphism
- WGHS
Women’s Genome Health Study
Footnotes
Author Contributions:
The authors’ responsibilities were as follows- CES, RL: designed the study; MF, LF, DKA, DSS, FBH, QS, SB, MYT, JMO, JIR, DIC, PMR, YIC, IBK: provided essential materials, CES, JLF, JAN, MF, JHYW, YM, TT, AWM, HW, AYC, SEC: contributed to the statistical analyses; CES, DM, RL, BM, LD, DSS, TL, WT: interpretation of data; CES, RL, BMP, BM, LW, DSS writing of the manuscript; EKK, SSR, BMP, YM, RL, BMcK, LMS, DSS, JAN: critical reading of the manuscript; CES had primary responsibility for the final content and all authors read and approved the final version of the manuscript.
Conflict of interest statement: B. Psaty serves on the DSMB of a clinical trial for the device manufacturer funded by Zoll LifeCor and on the Steering Committee of the Yale Open Data Access Project funded by Johnson & Johnson. Other authors declare no conflict of interest.
References
- 1.Kaur G, Cameron-Smith D, Garg M, Sinclair AJ. Docosapentaenoic acid (22:5n-3): a review of its biological effects. Prog Lip Res. 2011;50(1):28–34. doi: 10.1016/j.plipres.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 2.Mozaffarian D, Lemaitre RN, King IB, Song X, et al. Circulating long-chain -3 fatty acids and incidence of congestive heart failure in older adults: the cardiovascular health study: a cohort study. Ann Int Med. 2011;155(3):160–70. doi: 10.1059/0003-4819-155-3-201108020-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khaw KT, Friesen MD, Riboli E, Luben R, et al. Plasma phospholipid fatty acid concentration and incident coronary heart disease in men and women: the EPIC-Norfolk prospective study. PLoS Medicine. 2012;9(7):e1001255. doi: 10.1371/journal.pmed.1001255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilk JB, Tsai MY, Hanson NQ, Gaziano JM, et al. Plasma and dietary omega-3 fatty acids, fish intake, and heart failure risk in the Physicians’ Health Study. Am J Clin Nutr. 2012;96(4):882–8. doi: 10.3945/ajcn.112.042671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mozaffarian D, Lemaitre RN, King IB, Song X, et al. Plasma phospholipid long-chain omega-3 fatty acids and total and cause-specific mortality in older adults: a cohort study. Ann Int Med. 2013;158(7):515–525. doi: 10.7326/0003-4819-158-7-201304020-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tanaka T, Shen J, Abecasis GR, Kisialiou A, et al. Genome-wide association study of plasma polyunsaturated fatty acids in the InCHIANTI Study. PLoS Genetics. 2009;5(1):e1000338. doi: 10.1371/journal.pgen.1000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lemaitre RN, Tanaka T, Tang W, Manichaikul A, et al. Genetic loci associated with plasma phospholipid n-3 fatty acids: a meta-analysis of genome-wide association studies from the CHARGE Consortium. PLoS Genetics. 2011;7(7):e1002193. doi: 10.1371/journal.pgen.1002193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burdge GC, Jones AE, Wootton SA. Eicosapentaenoic and docosapentaenoic acids are the principal products of alpha-linolenic acid metabolism in young men. Brit J Nutr. 2002;88(4):355–63. doi: 10.1079/BJN2002662. [DOI] [PubMed] [Google Scholar]
- 9.Burdge GC. Metabolism of alpha-linolenic acid in humans. Prostaglandins Leukot Essent Fatty Acids. 2006;75(3):161–8. doi: 10.1016/j.plefa.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 10.Gillingham LG, Harding SV, Rideout TC, Yurkova N, et al. Dietary oils and FADS1-FADS2 genetic variants modulate [13C]-linolenic acid metabolism and plasma fatty acid composition. Am J Clin Nutr. 2013;97(1):195–207. doi: 10.3945/ajcn.112.043117. [DOI] [PubMed] [Google Scholar]
- 11.Goyens PL, Spilker ME, Zock PL, Katan MB, et al. Conversion of alpha-linolenic acid in humans is influenced by the absolute amounts of alpha-linolenic acid and linoleic acid in the diet and not by their ratio. Am J Clin Nutr. 2006;84(1):44–53. doi: 10.1093/ajcn/84.1.44. [DOI] [PubMed] [Google Scholar]
- 12.Welch AA, Shakya-Shrestha S, Lentjes MA, Wareham NJ, et al. Dietary intake and status of n-3 polyunsaturated fatty acids in a population of fish-eating and non-fish-eating meat-eaters, vegetarians, and vegans and the precursor-product ratio of a-linolenic acid to long-chain n-3 polyunsaturated fatty acids: results from the EPIC-Norfolk cohort. Am J Clin Nutr. 2010;92(5):1040–51. doi: 10.3945/ajcn.2010.29457. [DOI] [PubMed] [Google Scholar]
- 13.Cao J, Schwichtenberg KA, Hanson NQ, Tsai MY. Incorporation and clearance of omega-3 fatty acids in erythrocyte membranes and plasma phospholipids. Clin Chem. 2006;52(12):2265–72. doi: 10.1373/clinchem.2006.072322. [DOI] [PubMed] [Google Scholar]
- 14.Patel PS, Sharp SJ, Jansen E, Luben RN, et al. Fatty acids measured in plasma and erythrocyte-membrane phospholipids and derived by food-frequency questionnaire and the risk of new-onset type 2 diabetes: a pilot study in the European Prospective Investigation into Cancer and Nutrition (EPIC)-Norfolk cohort. Am J Clin Nutr. 2010;92(5):1214–22. doi: 10.3945/ajcn.2010.29182. [DOI] [PubMed] [Google Scholar]
- 15.Miller E, Kaur G, Larsen A, Loh SP, et al. A short-term n-3 DPA supplementation study in humans. Eur J Nutr. 2013;52(3):895–904. doi: 10.1007/s00394-012-0396-3. [DOI] [PubMed] [Google Scholar]
- 16.Cartwright IJ, Pockley AG, Galloway JH, Greaves M, et al. The effects of dietary omega-3 polyunsaturated fatty acids on erythrocyte membrane phospholipids, erythrocyte deformability and blood viscosity in healthy volunteers. Atherosclerosis. 1985;55(3):267–81. doi: 10.1016/0021-9150(85)90106-6. [DOI] [PubMed] [Google Scholar]
- 17.Al-Hilal M, Alsaleh A, Maniou Z, Lewis FJ, Hall WL, Sanders TA, O’Dell SD MARINA study team. Genetic variation at the FADS1-FADS2 gene locus influences delta-5 desaturase activity and LC-PUFA proportions after fish oil supplement. J Lipid Res. 2013;54(2):542–51. doi: 10.1194/jlr.P032276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roke K, Mutch DM. The role of FADS1/2 polymorphisms on cardiometabolic markers and fatty acid profiles in young adults consuming fish oil supplements. Nutrients. 2014;6(6):2290–304. doi: 10.3390/nu6062290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Demirkan A, van Duijn CM, Ugocsai P, Isaacs A, et al. Genome-Wide Association Study Identifies Novel Loci Associated with Circulating Phospho- and Sphingolipid Concentrations. PLoS Genet. 2012;8(2):e1002490. doi: 10.1371/journal.pgen.1002490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kitson AP, Stark KD, Duncan RE. Enzymes in brain phospholipid docosahexaenoic acid accretion: a PL-ethora of potential PL-ayers. Prostaglandins Leukot Essent Fatty Acids. 2012;87(1):1–10. doi: 10.1016/j.plefa.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 21.Agius L. Glucokinase and molecular aspects of liver glycogen metabolism. Biochem J. 2008;414:1–18. doi: 10.1042/BJ20080595. [DOI] [PubMed] [Google Scholar]
- 22.Murata-Mori F, Hayashida N, Ando T, Ikeoka T, et al. Association of the GCKR rs780094 polymorphism with metabolic traits including carotid intima-media thickness in Japanese community-dwelling men, but not in women. Clin Chem Lab Med. 2014;52(2):289–95. doi: 10.1515/cclm-2013-0092. [DOI] [PubMed] [Google Scholar]
- 23.Nettleton JA, McKeown NM, Kanoni S, Lemaitre RN, et al. Interactions of dietary whole-grain intake with fasting glucose– and insulin-related genetic loci in individuals of European descent: a meta-analysis of 14 cohort studies. Diabetes Care. 2010;33:2684–2691. doi: 10.2337/dc10-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brown AJ, Pang E, Roberts DC. Persistent changes in the fatty acid composition of erythrocyte membranes after moderate intake of n-3 polyunsaturated fatty acids: study design implications. Am J Clin Nutr. 1991;54:668–673. doi: 10.1093/ajcn/54.4.668. [DOI] [PubMed] [Google Scholar]
- 25.King IB, Lemaitre RN, Kestin M. Effect of a low-fat diet on fatty acid composition in red cells, plasma phospholipids, and cholesterol esters: investigation of a biomarker of total fat intake. Am J Clin Nutr. 2006;83:227–36. doi: 10.1093/ajcn/83.2.227. [DOI] [PubMed] [Google Scholar]
- 26.Ward LD, Kellis HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40(Database issue):D930–4. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.