Abstract
Due to significant viral diversity, vaccines that elicit durable and broad protection against influenza have been elusive. Recent research has focused on the potential of highly conserved regions of the viral hemagglutinin (HA) as targets for broadly neutralizing antibody responses. Antibodies that bind the highly conserved stem or stalk of HA can be elicited by vaccination in humans and animal models and neutralize diverse influenza strains. However, the frequency and phenotype of HA stem-specific B cells in vivo remains unclear. Here we characterize HA stem-specific B cell responses following H5N1 vaccination and describe the re-expansion of a pre-existing population of memory B cells specific for stem epitopes. This population utilizes primarily, but not exclusively, IGHV1-69-based immunoglobulins for HA recognition. However within some subjects, allelic polymorphism at the ighv1-69 locus can limit IGHV1-69 immunodominance and may reduce circulating frequencies of stem-reactive B cells in vivo. The accurate definition of allelic selection, recombination requirements and ontogeny of neutralizing antibody responses to influenza will aid rational influenza vaccine design.
Introduction
Antibodies targeting the viral surface protein hemagglutinin (HA) provide the principal protection against the acquisition of influenza infection. However outbreaks of pathogenic avian influenza and the worldwide spread of H1N1pdm underscore the susceptibility of human populations to novel viral serotypes despite seasonal vaccination efforts. The high genetic diversity of HA and capacity for rapid antigenic change limits the conservation of neutralizing epitopes between multiple viral subtypes and poses a challenge to the development of “universal” influenza vaccines for broad and lasting protection.
The isolation and structural characterisation of numerous human monoclonal antibodies revealed a conserved region within the stem (or stalk) domain of HA as a target for broadly neutralizing antibodies (bNAbs) (1-5). Antibodies binding the stem can neutralize diverse influenza strains by inhibiting conformational changes required for membrane fusion and provide passive protection against influenza in small animal influenza challenge models (2, 4-6). In the general population, stem-reactive serum antibodies with analogous neutralizing activity are widespread, but present at low concentrations thought unlikely to prevent infection (7). However, a marked increase in serum stem-reactivity can be elicited following immunization with immunologically novel influenza strains such as H5N1 (8-10) or pH1N1 (7, 11).
We previously reported the phase I influenza clinical vaccine trial (VRC 310), where subjects were immunized against avian H5N1 (A/Indonesia/05/2005) with plasmid DNA or conventional monovalent influenza vaccine (MIV) as a prime, followed by a H5N1 MIV boost delivered to different groups at staggered intervals from one to six months (10). Serological analysis indicated a polyclonal serum antibody response to H5 targeting a diverse epitope repertoire (12) including but not limited to the HA stem (10). The recent development of novel recombinant HA probes allowed the identification of HA-specific B cells within VRC 310 clinical samples (13) and the ready isolation of stem-specific monoclonal antibodies, which in concordance with previous studies were generally IGHV1-69 derived. We now expand those findings to characterize the B cell receptor repertoire and kinetics of HA-stem specific memory B cell expansion following H5N1 immunization. We report the vaccine-responsive, stem-specific memory B cell population is highly polyclonal, with repertoire bias for IGHV1-69 observed in the majority of subjects. Furthermore, we show that ighv1-69 polymorphism alters the immunodominance hierarchy of B cell responses to the stem epitope.
Materials and Methods
Ethics Statement
The study protocol and associated procedures were approved by the NIAID Institutional Review Board. All participants provided written informed consent in accordance with the Declaration of Helsinki.
VRC-310 Study Design and Clinical Samples
The VRC-310 study (ClinicalTrials.gov identifier NCT01086657) (10, 14) was conducted at the National Institutes of Health (NIH), Bethesda, MD by the Vaccine Research Center, NIAID, NIH, DHHS. This Phase I study examined the safety and immunogenicity of H5N1 prime-boost vaccination. One group received inactivated H5N1 vaccine (A/Indonesia/05/2005) for both prime and boost, whilst five other groups were primed with a DNA vaccine expressing H5 (A/Indonesia/05/2005) followed by boosting with inactivated H5N1 vaccine at increasing intervals ranging from 1 to 6 months. Peripheral blood mononuclear cells (PBMCs) were isolated and stored at regular intervals over the course of the trial and used for flow cytometry and sorting experiments as detailed below.
HA-specific probes and flow cytometry
The design and purification of fluorescently labelled recombinant HA probes with ablated sialic acid binding activity has been previously described (13). HA-specific B cells were identified within cryopreserved PBMC samples by co-staining with H1 (A/New Caledonia/20/1999) and H5 (A/Indonesia/05/2005) probes conjugated to streptavidin-PE or −APC (Life Technologies, New York, NY) respectively. B cells were stained with the following antibodies for sorting or phenotypic analysis: CD3-QD655, CD14-QD800, CD27-QD605 (Invitrogen), CD19-ECD (Beckman Coulter), IgM-Cy5.5-PerCP, IgG-FITC or −BV421, IgD-Cy7PE, CD38-Alexa Fluor 700, CD22-Cy5PE, CD24-Cy7PE, CXCR5-Ax488, CD20-Cy7APC (BD Pharmingen). Cell viability was assessed using Aqua Live/Dead amine-reactive dye (Invitrogen). The IGHV1-69 anti-idiotypic mouse monoclonal G6 was biotinylated and conjugated to Ax488 using standard procedures. 1 - 2 million events were collected on an LSR II instrument (BD Immunocytometry Systems) configured to detect 18 fluorochromes and analysis was performed using FlowJo software version 9.5.2 (TreeStar). Where applicable, HA probes were incubated with scFv derived from influenza bNAb F10, or alternatively with VRC01 or VRC04 HIV-1 Env-specific scFv controls. Probes were incubated at room temperature with 10 μg/ml of scFv inhibitors for 1h prior to the addition of PBMCs.
Sequencing, cloning and expression of B cell immunoglobulins
The sequencing and cloning of BCRs from single B cells was performed as previously described (15, 16). Plasmids expressing heavy and light immunoglobulin chains were transfected into Expi293F cells using ExpiFectamine (Invitrogen). Recombinant monoclonal antibodies were purified from culture supernatants using sepharose Protein-A or G (Pierce) as per the manufacturer’s instructions.
Transfection and cell surface binding to HA
Germline-reverted variants of stem antibodies or VRC-01 were synthesised (GeneArt) and used to express receptor IgM on the surface of Expi293 cells as previously described (17). 24 hours after transfection, cells were stained with AQUA Live/Dead amine reactive dye (Invitrogen) and binding to HA was assessed using PE-conjugated HAdSA probes derived from A/New Caledonia/20/1999 (13). Immunoglobulin surface expression was confirmed by staining transfected cells with anti-huIgM Cy5.5PerCP (BD Biosciences). The ability of IGHV1-69 alleles to differentially bind the anti-idiotypic monoclonal antibody G6 was confirmed by co-staining transfected cells with G6-Alexa Fluor 488 (Ax488).
Enzyme-linked immunosorbant assay (ELISA)
All incubations were performed at ambient temperature. For the detection of serum IGHV1-69 antibodies, plates were coated overnight with the monoclonal G6 at 2 μg/ml, washed and incubated with serial dilutions of sera. Bound antibodies were detected using 1:30000 goat anti-huIgG-HRP (Bethyl Labs) for 1 hour, developed using one step TMB substrate (Dako) and stopped by the addition of 2M sulphuric acid. Plates were read at 450n using a Spectromax Plus (Molecular Devices).
Generation of fragment antigen-binding (Fab) of monoclonal antibodies
Purified monoclonal antibodies (0.5 mg) were digested with papain-conjugated agarose using a commercial kit (Pierce) for 4 h at 37 °C. After incubation, digestion reactions were separated from papain resin by a spin column. A protein A-conjugated sepharose column was used to separate Fab from undigested antibody and Fc. Purity of Fabs was verified by SDS-PAGE.
Binding kinetics of monoclonal antibodies to HA by biolayer interferometry
A fortéBio Octet HTX instrument (Pall) was used to measure the binding kinetics of soluble HA trimer (A/Indonesia/5/05 H5N1) to Fab from purified monoclonal antibodies. All the assays were performed at 30 °C in tilted bottom 384-well plates (Pall) with agitation set to 1,000 rpm in PBS supplemented with 1% BSA (PBS-BSA) in order to minimize nonspecific interactions. Soluble biotinylated HA trimers (10 μg/ml) in PBS-BSA were immobilized on streptavidin (SA)-coated biosensors (Pall) for 300 s. Typical capture variability within a row of eight tips did not exceed 0.1 nm. Biosensor tips were then equilibrated in PBS-BSA for 180 s prior to binding assessment of Fab of monoclonal antibodies (0 to 200 nM) in PBS-BSA for 300 s. Binding was then allowed to dissociate in PBS-BSA for 300 s. Parallel correction to subtract systematic baseline drift was carried out by subtracting the measurements recorded for a sensor immobilized with biotinylated Epstein-Barr virus gp350 protein. Data were analysed using Octet software, version 8.0 (Pall). Experimental data were fitted with the binding equations describing a 1:1 interaction. Global analyses of the data sets assuming binding was reversible (full dissociation) were carried out using nonlinear least-squares fitting allowing a single set of binding parameters to be obtained simultaneously for all concentrations used in each experiment.
HA pseudovirion neutralization assay
Neutralization of HA-pseudoviruses by purified monoclonal antibodies was assessed as previously described (18). Briefly, HA NA-pseudotyped lentiviral vectors encoding luciferase were incubated with serially diluted sera from H5N1 vaccine recipients prior to addition to 293A cells. Neutralization activity was quantified by the relative decrease in luciferase activity as compared to infection of 293A cells in the absence of sera. A serum dilution yielding a 50% inhibition titer (ID50) was calculated using 5-parameter curve fitting.
Statistical Analyses
Data is generally presented as mean +/− SEM. Statistical significance was assessed by two tailed paired or unpaired Students’ t-test. All statistical analyses were performed using Graphpad Prism ver 5.0 (GraphPad Software Inc.).
Results
H5N1 vaccination elicits the expansion of cross-reactive B cells that bind a conserved epitope within the HA stem
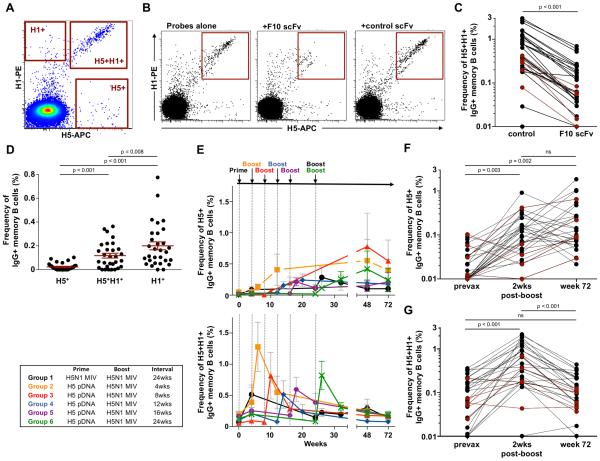
We previously described the development of recombinant HA analogues as flow cytometry probes (13) and how co-staining with H5 (vaccine-strain A/Indonesia/05/2005) and H1 (A/New Caledonia/20/1999) probes could be used to delineate three populations of HA-binding memory B cells within PBMC samples; H1+, vaccine-specific H5+ B cells and H5+H1+ cross-reactive B cells (Fig. 1A; representative gating in Supplementary Fig. 1A and 1B). Non-specific staining associated with HA probes is low (Supplementary Fig. 1C). Based upon the isolation of monoclonal antibodies, B cells within the H5+H1+ cross-reactive population of a single individual were shown to be predominantly specific for the HA stem epitope (13). In order to generalize findings to all H5N1 vaccine recipients, we developed a flow cytometry based competition assay to measure stem specificity. Pre-incubation of HA probes with single chain Fv (scFv) fragments derived from the stem-specific bNAb F10 (4) blocks the HA stem epitope, preventing the binding of cross-reactive H5+H1+, but not H5+ B cells (Fig. 1B). When examined in PBMC samples from H5N1 vaccinees isolated 2 weeks post-MIV boost, F10-scFv inhibition resulted in a ~80-90% reduction in the observed frequencies of H5+H1+ memory B cells relative to a control scFv (Fig. 1C). This suggests a significant majority of the cross-reactive B cell population bound the HA stem, more precisely, epitopes overlapping that of bNAb F10, with the remainder binding currently undefined, alternative cross-reactive epitopes on HA.
Figure 1. Expansion of stem-specific memory B cells following H5N1 vaccination.
(A) Co-staining with H1 (A/New Caledonia/20/1999) and H5 (A/Indonesia/05/2005) probes allows the identification of three distinct populations of memory B cells; H1+, H5+ and a cross-reactive H5+H1+ population. (B) Competition with F10-scFv can selectively block the binding of H5+H1+ memory B cells to HA probes relative to an irrelevant scFv control (VRC01 or VRC04). (C) Change in frequencies of H5+H1+ memory B cells in the presence of F10-scFv relative to an irrelevant scFv control within PBMC samples isolated 2 weeks post-H5N1 MIV boost (N=30, 5 subjects from each vaccine group). (D) Baseline frequencies of HA-specific B cells populations in PBMC samples from a subset of H5N1 vaccine trial participants (N=30, 5 subjects from each vaccine group). (E) Kinetics of H5+ and H5+H1+ memory B cell expansion in each vaccine group following H5N1 immunization assessed in longitudinal PBMC samples. Changes in (F) H5+ and (G) H5+H1+ memory B cell frequencies in subjects (N=30, 5 subjects from each vaccine group) between baseline, two weeks post-H5N1 MIV boost and 72 weeks post-prime. Frequencies of HA specific B cells were compared where indicated using two tailed paired Students’ t-test and p values denoted. ns; non-significant. Subjects subsequently identified as putative L54 homozygotes are indicated by red shading in panels C, F and G.
Elicitation of stem-reactive serum antibodies to H5N1 vaccination has been previously reported (8-10). We now assessed the kinetics of stem-specific memory B cell expansion within longitudinal PBMC samples taken from a subset of subjects from VRC 310 (N=30, 5 per group). In most individuals, both H1+ (mean 0.20±0.03%) and H5+H1+ cross-reactive (0.18±0.02%) memory B cell populations were readily detected in baseline samples (Fig. 1D). Memory B cells specific for H5 but not cross-reactive with H1 were comparatively infrequent (0.02±0.01%). Following H5N1 vaccination, irrespective of the prime/boost interval, low baseline frequencies of H5+ B cells underwent a gradual expansion that generally peaked between eight to twenty-four weeks after the final H5N1 MIV boost (Fig. 1E). In comparison, the H5+H1+ cross-reactive memory B cell population underwent rapid re-expansion, peaking at two weeks post-MIV boost before contracting rapidly. In subjects that received H5N1 MIV prime and boost (Group 1), H5+H1+ B cells expanded in response to each vaccine injection. The longer-term effects of H5N1 vaccination were reflected by sustained elevation of circulating H5+ memory B cell frequencies in most individuals, evident over seventy-two weeks after initial immunization (Fig. 1F). In contrast, the frequencies of H5+H1+ cross-reactive memory B cells had generally returned to frequencies comparable to baseline by the seventy-two week sampling (Fig. 1G).
Phenotype of vaccine elicited stem-reactive B cells
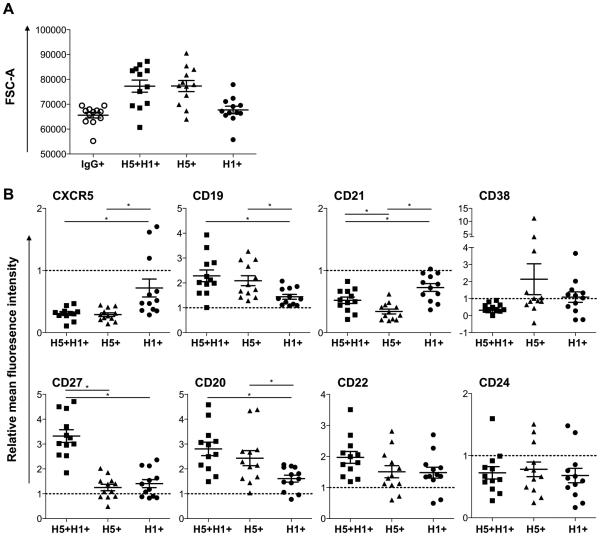
The phenotype of HA-specific B cells at two weeks post H5N1 boost was characterised by flow cytometry in two representative subjects per vaccine group (N=12 total). Using forward scatter (FSC-A) as a marker of cell size, both H5+H1+ and H5+ B cells were generally larger than H1+ B cells and the parental IgG+ memory B cell population (Fig 2A). The mean fluorescent intensity (MFI) for a panel of surface markers was determined for H1+, H5+ and H5+H1+ B populations and normalized relative to the parental IgG+ B cell population in each subject. The intensity of surface marker expression on H1+ B cells (2 – 48 cells/subject, mean = 27) was largely in line with the parental memory B cell population (Fig 2B) consistent with cells mono-specific for an historical H1N1, most likely binding within the variable HA head domain, remaining unresponsive to H5N1 vaccination. In contrast, low relative expression of CXCR5 and CD21 was consistently observed for both H5+H1+ (24 – 1017 cells/subject, mean = 225) and H5+ B cell populations (4 – 178 cells/subject, mean = 46). Increased relative expression of CD19 and CD20 was also noted, although this may merely be reflective of increased cell size. Greater CD27 and CD22 expression was observed only within the double positive H5+H1+ population, potentially reflecting phenotypic differences between a recently class-switched primary response (H5+) and recall responses (H5+H1+). Increasing size, downregulation of CD21 (19) and CXCR5 (20) and upregulation of CD22 (21) and CD27 (22) is broadly consistent with B cell activation and differentiation toward antibody secreting plasmablast/plasma cells. However further experiments, including transcriptional phenotyping, will clarify the extent and nature of activation and/or proliferation within the different HA-specific B cell populations following H5N1 immunization.
Figure 2. Phenotype of vaccine elicited, HA-specific memory B cells.
The phenotype of HA-specific B cell populations was assessed by flow cytometry in PBMC samples collected 2wks post-H5N1 MIV boost (N=12, 2 subjects from each vaccine group). (A) Mean forward scatter area (FSC-A) as a marker of cell size of H5+, H1+ and H5+H1+ memory B cells relative to the parental IgG+ memory B cell population. (B) The relative mean fluorescence intensity (MFI) was calculated for surface markers CD19, CD20, CD21, CD22, CD24, CD27, CD38 and CXCR5 for each HA-specific B cell population (H5+, H1+ and H5+H1+) and normalised to the MFI of the parental IgG+ memory B cell population for each subject (illustrated as a dotted line at 1.0). * denotes p<0.05
The VH repertoire of stem-reactive B cells is dominated by IGHV1-69
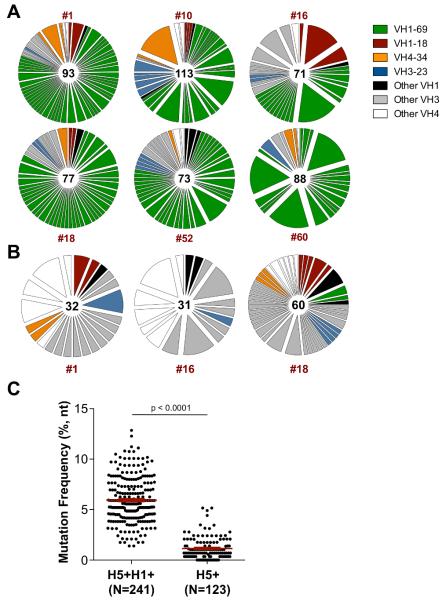
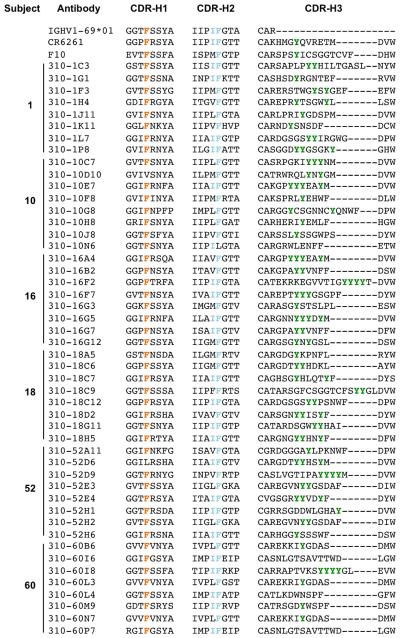
Frequent use of the variable gene segment IGHV1-69 has been reported among isolated HA stem-binding antibodies (3, 6, 11, 23-25). To examine the repertoire of H5+H1+ B cells, single cells were sorted from six representative vaccine recipients and recombined VDJ sequences were recovered as previously described (16). The BCR repertoire was highly polyclonal within each subject, with no single B cell lineage making up more than ~17% of the total response (Fig. 3A). In line with other studies, we observed a marked bias toward IGHV1-69 utilization in all subjects sequenced, making up between 43.6 and 81.5% (mean 68.2%)of recovered sequences. Germline encoded phenylalanine within the CDR-H1 (F29), and a hydrophobic residue at position 53 and phenylalanine at position 54 (F54) within the CDR-H2 were previously shown to be critical for guiding the maturation of IGHV1-69 class stem antibodies (17). More recently, the importance of centrally located tyrosines within the CDR-H3 has been established (26). We similarly observed strong conservation of these critical heavy chain signatures among putative stem-specific IGHV1-69 immunoglobulins sequenced from multiple subjects (Fig. 4). The frequent use of three other germlines, IGHV1-18, IGHV3-23 and IGHV4-34 was also observed in multiple individuals. Paired light chain sequences recovered from 444 H5+H1+ B cells were primarily kappa chain and selected from a wide range of germline families without obvious preference (data not shown).
Figure 3. H5/H1 cross-reactive B cells are clonally diverse and predominantly utilise IGHV1-69.
Immunoglobulin VH gene family repertoire of (A) H5+H1+ cross-reactive and (B) H5+ memory B cells isolated 2 weeks post-H5N1 MIV boost. The VH repertoire from each subject is shown as a pie chart, with each slice representative of a unique clone or clonally-related family, the width of the slice proportional to the frequency and the colour indicating germline. The number of sequences recovered for each subject is indicated in the centre of the circle. Subjects were drawn from a cross-section of VRC 310 vaccine groups: #1 (Group 3), #10 (Group 6), #16 (Group 4), #18 (Group 2), #52 (Group 6), #60 (Group 1). (C) Frequency of VH gene somatic mutation in H5+H1+ cross-reactive and H5+ memory B populations isolated from three representative vaccine recipients.
Figure 4. Conserved heavy chain signatures putative of IGHV1-69 stem antibody sequences recovered from H5+H1+ B cells from six individuals.
Ten unique representative antibody lineages from each subject were aligned to the IGHV1-69*01 gene and the prototypic bNAbs F10 and CR6261. In line with features known to guide the development of broadly neutralizing stem antibodies, strict conservation of F29 and F54, a preference for hydrophobicity at position 53 and a central tyrosine within the CDR-H3 were all observed.
In contrast to the H5+H1+ population, the BCR repertoire of isolated H5+ cells showed no enrichment for IGHV1-69 but was again highly polyclonal (Fig. 3B). The degree of somatic hypermutation within recovered VDJ sequences from H5+H1+ B cells ranged from 1.4 – 12.9%, (mean 5.9%)(Fig. 3C), broadly in line with previous reports of HA specific antibodies raised to experimental infection or vaccination against seasonal influenza (27, 28) but significantly higher compared with the H5+ B cell population (mean 1.1%). This observation is consistent with both the de novo elicitation of H5+ memory B cells and the re-expansion of pre-existing H5+H1+ memory B cells in response to H5 immunization. Indeed IGHV1-69 bias and extensive immunoglobulin maturation of H5+H1+ B cells was evident prior to H5N1 vaccination (Fig. 5). Expansion of stem reactive antibodies has been previously reported in response to either A/New Jersey/1976 (NJ/76) vaccine (29) or exposure to H2N2 between 1957-1967 (30). Though these findings might suggest cohort-specific differences in HA stem responses, no correlation was observed between age and baseline frequencies of H5+H1+ B cell frequencies (Fig. 6). Therefore exposure to these early strains is not prerequisite and exposure to recently circulating seasonal influenza strains through infection or immunization seems sufficient to seed a cross-reactive stem-specific memory B cell population and establish IGHV1-69 immunodominance.
Figure 5. Repertoire of BCR sequences from H5+H1+ cross-reactive memory B cells seen prior to vaccination.
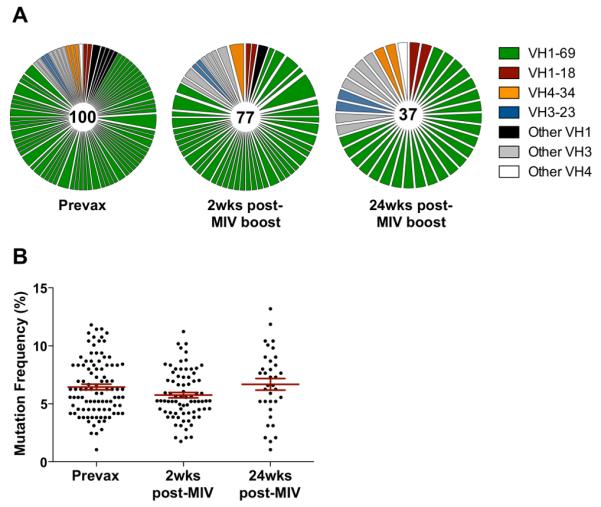
(A) VH gene repertoire of H5+H1+ cross-reactive memory B cells compared at baseline, 2wks and 24wks post-H5N1 MIV boost in a single subject. (B) Mutation frequencies of immunoglobulins recovered from H5+H1+ cross-reactive memory B cells.
Figure 6.
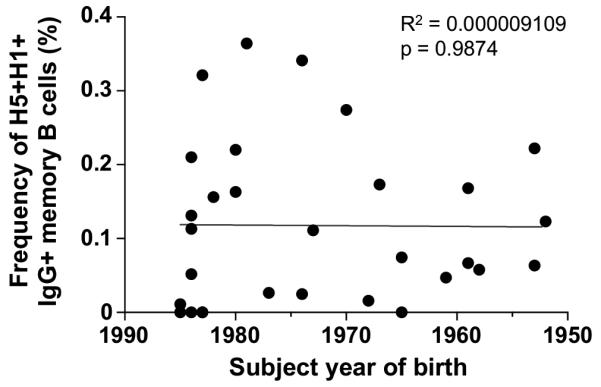
No relationship between a subject’s year of birth and baseline frequencies of H5+H1+ memory B cells
IGHV1-69 polymorphism alters the immunodominance of stem reactive antibody responses
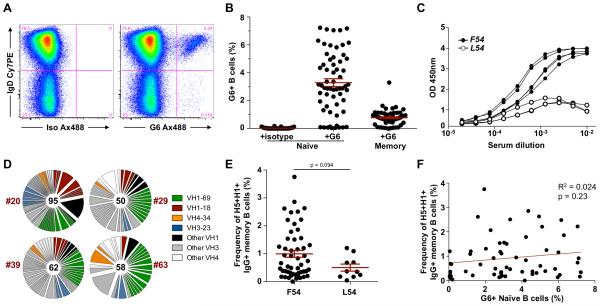
The mechanisms that drive IGHV1-69 bias within stem antibody responses remain unclear. Thirteen distinct ighv1-69 alleles have been described encompassing F54L, T56I and G49R polymorphisms within and proximal to the CDR-H2 (31). Notably, seven alleles encode F54 and six alleles instead encode a leucine (L54), with 11% of the general population reported to be homozygous for L54-encoding alleles (32). Almost exclusively, IGHV1-69 derived stem bNAbs isolated to date have been derived from F54-encoding alleles *01, *03, *06 and *12 (6, 13, 23, 25, 26). Recently, allelic polymorphism was shown to limit the serological titres of stem antibodies, with levels significantly lower in L54 homozygotes (25). We therefore examined the effect of allelic polymorphism at the ighv1-69 locus upon the immunodominance of IGHV1-69 among stem-specific antibodies elicited in vivo. The mAb G6 binds selectively to F54-encoding idiotypes of IGHV1-69 (33) and can be used to identify B cells expressing such immunoglobulins (Fig. 7A). Ten of 61 VRC 310 subjects screened lacked G6+ B cells (Fig. 7B) and for three representative subjects tested, also lacked G6-reactive serum antibodies (Fig. 7C). Therefore subjects lacking G6+ B cells were putatively identified as L54 homozygotes and from four representative individuals single H5+H1+ B cells taken 2wks post-MIV boost were isolated and sequenced (Fig. 7D). The repertoires of H5+H1+ B cells were again highly polyclonal and conserved stem-reactive germlines IGHV1-18, 3-23 and 4-34 were detectable in all subjects. However, IGHV1-69 was evidently less immunodominant; 21.1-34.0% (mean 26.9%) of recovered sequences and derived primarily from ighv1-69*02 or ighv1-69*09 alleles. Reversion to F54 was rare. The inability to utilise a dominant IGHV1-69-based pathway for stem antibody elicitation may contribute to a reduction in the frequencies of H5+H1+ B cells observed in L54 homozygotes following H5N1 immunization (Fig. 7E). However, this difference failed to reach statistical significance and confirmation would be required using larger cohorts of L54 homozygotes. No correlation was observed between the frequencies of naïve G6+ B cells and H5+H1+ B cells elicted post-vaccination (Fig. 7F), suggesting a single F54-encoding allele is sufficient to populate the pool of IGHV1-69 B cells targeting the HA stem.
Figure 7. Homozygosity for L54-encoding alleles of ighv1-69 alters the immunodominance of stem-specific B cell responses.
(A) The mouse monoclonal G6 specifically binds immunoglobulins derived from ighv1-69 alleles that encode F54. Flow cytometric staining of CD19+ B cells with G6 allows identification of surface expressed of F54-encoding IGHV1-69, compared to an isotype control. (B) Frequencies of G6+ naïve and memory B cells from VRC 310 subjects (N=61) with the anti-idiotypic mAb G6. (C) F54-encoding IGHV1-69 serum antibodies were measured by G6-capture ELISA from representative subjects with G6+ B cells (closed circles) and without G6+ B cells (open circles). (D) VH gene repertoire from four subjects lacking F54-encoding IGHV1-69 alleles. (E) Frequencies of H5+H1+ B cells at 2wks post-MIV boost observed within subjects with F54-encoding IGHV1-69 alleles (left) and L54-encoding homozygotes (right). (F) Lack of correlation between the frequencies H5+H1+ B cells at 2wks post-MIV boost and the proportion of naïve B cells that stain positive for G6.
Germline binding to HA by IGHV1-69*09 does not account for subdominant antibody responses to the stem
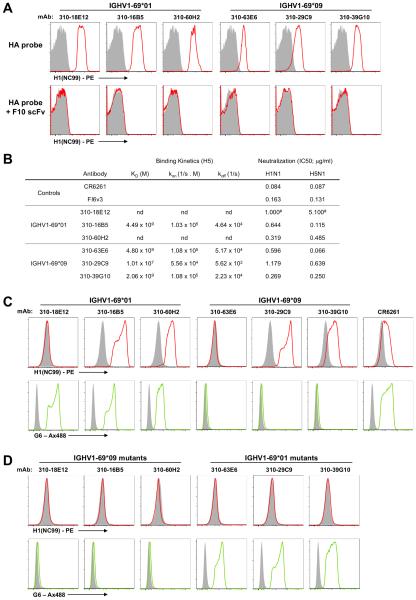
Sequences for three canonical ighv1-69*01-derived stem antibodies and three ighv1-69*09-derived stem antibodies were recovered from subjects with and lacking F54 alleles respectively. Each antibody was expressed as membrane associated IgM on the surface of 293 cells as previously described (17) and binding to the HA stem epitope was confirmed based upon F10-scFv inhibition (Fig. 8A). Affinity measurements were obtained by biolayer interferometry using purified Fab fragments in solution binding to biotinylated H5 (A/Indonesia/5/05) (Fig. 8B). Antibodies derived from both IGHV1-69 alleles were capable of binding HA at nanomolar affinities, similar to values previously reported for CR6261 (4.1 nM) (2). Moreover, the neutralization potency against H1N1 and H5N1 pseudoviruses was broadly comparable for antibodies derived from both IGHV1-69 alleles (Fig. 8B). These data suggest that IGHV1-69*09 derived stem antibodies maintain a level of functional equivalence to the canonical IGHV1-69*01 derived population.
Figure 8. HA engagement by germline reverted immunoglobulins of different IGHV1-69 alleles.
(A) ighv1-69*01-derived and ighv1-69*09-derived stem-specific immunoglobulins were expressed as surface IgM by transfection into Expi293 cells. Relative to a mock transfected control (VRC-01;grey shaded), the binding of PE-labelled HAdSA probes (A/New Caledonia/20/1999) (red line) is shown alone or in the presence of F10 scFv. (B) The binding kinetics and affinity of stem antibodies from different IGHV1-69 alleles was assessed by biolayer interferometry. Neutralization of H1N1 (A/New Caledonia/20/1999) and H5N1 (A/Vietnam/1203/2004) pseudoviruses was assessed and is reported as the 50% inhibitory concentration (IC50) in μg/ml. #Denotes neutralization titres previously reported in (13). (C) Unmutated germline-reverted variants of ighv1-69*01-derived and ighv1-69*09-derived stem-specific immunoglobulins were synthesized and binding to PE-labelled HAdSA probes was measured using transfected Expi293 cells as before. The expression of specific IGHV1-69 allelic variants was confirmed by costaining with the anti-idiotypic monoclonal G6. (D) The effect of allelic context on HA binding was assessed by incorporation of G49R, F54L and T56I amino acid substitutions into IGHV1-69*01-derived antibodies. Reciprocal changes were also made in IGHV1-69*09-derived antibodies and binding to HA measured as before.
In order to model the initial engagement of naïve B cells with the HA stem, inferred germline immunoglobulins were constructed for each antibody lineage. Cells transfected with the germline-reverted CR6261 control and two of three ighv1-69*01-derived antibodies efficiently bound soluble HA from A/New Caledonia/20/1999 (Fig. 8C). However, efficient binding was also observed for two of three ighv1-69*09-derived antibodies despite the lack of canonical CDR-H2 residues, confirmed by the absence of G6 staining. The incorporation of G49R, F54L and T56I amino acid substitutions into ighv1-69*01-derived antibodies in order to model presentation in an IGHV1-69*09 context resulted in a loss of HA binding (Fig. 8D), consistent with past observations (17). However this was also the case for the converse, where R49G, L54F and I56T mutations led to a loss of HA binding by ighv1-69*09-derived antibodies. Our observations suggest that immunoglobulins from both L54- and F54-encoding ighv1-69 alleles are capable of engaging HA at the germline level and germline-encoded CDR-H2 contacts are critical. However, the modes of HA recognition for ighv1-69*09-derived antibodies appears distinct from that engendering ighv1-69*01 antibodies.
Discussion
Broadly neutralizing antibodies that target the influenza HA stem have the potential to protect against diverse influenza serotypes and could form the basis of a universal vaccine. We have now shown that increased stem-reactive serum antibody following H5N1 immunization coincides with a rapid re-expansion of stem-specific memory B cells. Moreover, these memory B cells predominantly arise from a maturation pathway reliant upon IGHV1-69 alleles with germline-encoded F54, analogous to that of bNAbs CR6261. We previously demonstrated that all IGHV1-69-derived monoclonal antibodies isolated from a representative H5N1 vaccine recipient had comparable influenza neutralization potency and breadth to the prototypic bNAbs CR6261 and F10 (13). Thus despite maintenance as a highly polyclonal population, IGHV1-69-encoding B cells represent a dominant elicitation pathway for responses targeting the HA stem in vivo and yield antibodies of comparable affinities and neutralizing potencies. The high prevalence at baseline of stem-specific memory B cells among the cohort, independent of age or other factors, suggests that repeated exposure to seasonal influenza strains establishes IGHV1-69 immunodominance and seeds a population of circulating memory B cells. This population is then primed for rapid expansion upon subsequent exposure to immunologically novel HA, such as pH1N1 and H5N1, where recall responses to the stem seem to be favoured (7, 10, 11, 23). The degree to which seasonal tri- and quadrivalent vaccines elicit stem reactivity in naïve recipients remains to be clarified.
The existence of a dominant and conserved molecular pathway for the elicitation of influenza bNAbs is encouraging for stem epitope-based universal influenza vaccine candidates (34-36). However, the reliance upon a subset of IGHV1-69 alleles means genetic differences may lead to vaccine variability between subjects. The maturation pathways for stem bNAbs using IGHV1-69*01 and related alleles have been well defined (17, 25, 26), with germline-encoded F54 thought to be critical for naïve B cell selection in vivo, based upon HA binding to inferred germline-reverted immunoglobulins (17, 25). However we find a lack of germline specificity for the HA stem does not explain why IGHV1-69*09-encoding naïve B cells are not favoured in vivo, which similarly maintain a requirement for germline-encoded residues within the CDR-H2 for HA engagement. The diminished prominence of IGHV1-69 in the stem response of L54 homozygotes, coupled with the lower serological titres of stem antibodies in such individuals (25), suggests antibodies derived from L54-encoding alleles may be intrinsically less productive in vivo. However the precise mechanisms that bias recruitment of ighv1-69*01 and related alleles into the initial stem response requires further elucidation.
Our findings highlight that immunoglobulin gene polymorphism is an important consideration for rational immunogen design not just for influenza, but potentially for other vaccines intended to elicit class-specific bNAbs such as ighv1-2*02 restricted VRC01-like bNAbs targeting the CD4 binding site of HIV-1 Env (37). Encouragingly, immunoglobublins derived from non-IGHV1-69 germlines that similarly bind the HA stem epitope (13), were found in multiple subjects suggesting alternative conserved pathways of stem antibody elicitation exist. Crystallographic characterization of IGHV1-69*09-derived, or other non-IGHV1-69 stem antibodies may clarify the mechanisms driving their elicitation and potentially allow fine-tuning of HA immunogen design to simultaneously elicit subdominant antibody responses for the widest potential applicability. Finally our studies suggest that the selective design and combinatorial use of antigen-specific B cell probes have great utility for clarifying genetic determinants of immunoglobulins specific for a given neutralizing epitope. Analogous studies of other identified broadly neutralizing epitopes of HA such as the receptor-binding site are warranted. The accurate delineation of conserved and pre-existing pathways for bNAb elicitation at a population level will significantly aid the rational design of vaccines that promote neutralizing activity that extends beyond the autologous virus subtype.
Supplementary Material
Acknowledgements
The authors wish to thank all VRC 310 vaccine trial participants and the VRC 310 clinical team for their contribution and commitment to vaccine research. In addition, we thank: members of the R.A.K. laboratory for advice and discussions; Marissa Johnson and Danielle Scheer for technical assistance; Richard Nguyen and David Ambrosak for expert flow cytometry assistance; and Professor Roy Jefferis for providing the G6 monoclonal antibody.
Funding: This work was supported by the Intramural Research Program of the Vaccine Research Center, NIAID, National Institutes of Health.
References
- 1.Corti D, Voss J, Gamblin SJ, Codoni G, Macagno A, Jarrossay D, Vachieri SG, Pinna D, Minola A, Vanzetta F, Silacci C, Fernandez-Rodriguez BM, Agatic G, Bianchi S, Giacchetto-Sasselli I, Calder L, Sallusto F, Collins P, Haire LF, Temperton N, Langedijk JP, Skehel JJ, Lanzavecchia A. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science (New York, N.Y.) 2011;333:850–856. doi: 10.1126/science.1205669. [DOI] [PubMed] [Google Scholar]
- 2.Throsby M, van den Brink E, Jongeneelen M, Poon L, Alard P, Cornelissen L, Bakker A, Cox F, van Deventer E, Guan Y, Cinatl J, ter Meulen J, Lasters I, Carsetti R, Peiris M, de Kruif J, Goudsmit J. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PloS one. 2008:3. doi: 10.1371/journal.pone.0003942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ekiert DC, Bhabha G, Elsliger MA, Friesen RH, Jongeneelen M, Throsby M, Goudsmit J, Wilson IA. Antibody recognition of a highly conserved influenza virus epitope. Science. 2009;324:246–251. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sui J, Hwang WC, Perez S, Wei G, Aird D, Chen L.-m. M., Santelli E, Stec B, Cadwell G, Ali M, Wan H, Murakami A, Yammanuru A, Han T, Cox NJ, Bankston LA, Donis RO, Liddington RC, Marasco WA. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nature structural & molecular biology. 2009;16:265–273. doi: 10.1038/nsmb.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dreyfus C, Laursen NS, Kwaks T, Zuijdgeest D, Khayat R, Ekiert DC, Lee JH, Metlagel Z, Bujny MV, Jongeneelen M, van der Vlugt R, Lamrani M, Korse HJ, Geelen E, Sahin O, Sieuwerts M, Brakenhoff JP, Vogels R, Li OT, Poon LL, Peiris M, Koudstaal W, Ward AB, Wilson IA, Goudsmit J, Friesen RH. Highly conserved protective epitopes on influenza B viruses. Science. 2012;337:1343–1348. doi: 10.1126/science.1222908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corti D, Suguitan AL, Pinna D, Silacci C, Fernandez-Rodriguez BM, Vanzetta F, Santos C, Luke CJ, Torres-Velez FJ, Temperton NJ, Weiss RA, Sallusto F, Subbarao K, Lanzavecchia A. Heterosubtypic neutralizing antibodies are produced by individuals immunized with a seasonal influenza vaccine. The Journal of clinical investigation. 2010;120:1663–1673. doi: 10.1172/JCI41902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sui J, Sheehan J, Hwang WC, Bankston LA, Burchett SK, Huang C-YY, Liddington RC, Beigel JH, Marasco WA. Wide prevalence of heterosubtypic broadly neutralizing human anti-influenza A antibodies. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2011;52:1003–1009. doi: 10.1093/cid/cir121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ellebedy AH, Krammer F, Li G-MM, Miller MS, Chiu C, Wrammert J, Chang CY, Davis CW, McCausland M, Elbein R, Edupuganti S, Spearman P, Andrews SF, Wilson PC, Garcïa-Sastre A, Mulligan MJ, Mehta AK, Palese P, Ahmed R. Induction of broadly cross-reactive antibody responses to the influenza HA stem region following H5N1 vaccination in humans. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:13133–13138. doi: 10.1073/pnas.1414070111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nachbagauer R, Wohlbold TJ, Hirsh A, Hai R, Sjursen H, Palese P, Cox RJ, Krammer F. Induction of broadly reactive anti-hemagglutinin stalk antibodies by an H5N1 vaccine in humans. Journal of virology. 2014;88:13260–13268. doi: 10.1128/JVI.02133-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ledgerwood JE, Zephir K, Hu Z, Wei C-JJ, Chang L, Enama ME, Hendel CS, Sitar S, Bailer RT, Koup RA, Mascola JR, Nabel GJ, Graham BS, V. R. C. S. Team Prime-boost interval matters: a randomized phase 1 study to identify the minimum interval necessary to observe the H5 DNA influenza vaccine priming effect. The Journal of infectious diseases. 2013;208:418–422. doi: 10.1093/infdis/jit180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li GM, Chiu C, Wrammert J, McCausland M, Andrews SF, Zheng NY, Lee JH, Huang M, Qu X, Edupuganti S, Mulligan M, Das SR, Yewdell JW, Mehta AK, Wilson PC, Ahmed R. Pandemic H1N1 influenza vaccine induces a recall response in humans that favors broadly cross-reactive memory B cells. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:9047–9052. doi: 10.1073/pnas.1118979109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khurana S, Wu J, Dimitrova M, King L, Manischewitz J, Graham B, Ledgerwood J, Golding H. DNA priming prior to inactivated influenza A(H5N1) vaccination expands the antibody epitope repertoire and increases affinity maturation in a boost-interval-dependent manner in adults. The Journal of infectious diseases. 2013;208:413–417. doi: 10.1093/infdis/jit178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Whittle JRR, Wheatley AK, Wu L, Lingwood D, Kanekiyo M, Ma SS, Narpala SR, Yassine HM, Frank GM, Yewdell JW, Ledgerwood JE, Wei C-JJ, McDermott AB, Graham BS, Koup RA, Nabel GJ. Flow cytometry reveals that H5N1 vaccination elicits cross-reactive stem-directed antibodies from multiple Ig heavy-chain lineages. Journal of virology. 2014;88:4047–4057. doi: 10.1128/JVI.03422-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ledgerwood JE, Wei C-JJ, Hu Z, Gordon IJ, Enama ME, Hendel CS, McTamney PM, Pearce MB, Yassine HM, Boyington JC, Bailer R, Tumpey TM, Koup RA, Mascola JR, Nabel GJ, Graham BS, V. R. C. S. Team DNA priming and influenza vaccine immunogenicity: two phase 1 open label randomised clinical trials. The Lancet. Infectious diseases. 2011;11:916–924. doi: 10.1016/S1473-3099(11)70240-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scheid JF, Mouquet H, Feldhahn N, Walker BD, Pereyra F, Cutrell E, Seaman MS, Mascola JR, Wyatt RT, Wardemann H, Nussenzweig MC. A method for identification of HIV gp140 binding memory B cells in human blood. Journal of immunological methods. 2009;343:65–67. doi: 10.1016/j.jim.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tiller T, Meffre E, Yurasov S, Tsuiji M, Nussenzweig MC, Wardemann H. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. Journal of immunological methods. 2008;329:112–124. doi: 10.1016/j.jim.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lingwood D, McTamney PM, Yassine HM, Whittle JRR, Guo X, Boyington JC, Wei C-JJ, Nabel GJ. Structural and genetic basis for development of broadly neutralizing influenza antibodies. Nature. 2012;489:566–570. doi: 10.1038/nature11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei C-JJ, Boyington JC, McTamney PM, Kong W-PP, Pearce MB, Xu L, Andersen H, Rao S, Tumpey TM, Yang Z-YY, Nabel GJ. Induction of broadly neutralizing H1N1 influenza antibodies by vaccination. Science (New York, N.Y.) 2010;329:1060–1064. doi: 10.1126/science.1192517. [DOI] [PubMed] [Google Scholar]
- 19.Tedder TF, Clement LT, Cooper MD. Expression of C3d receptors during human B cell differentiation: immunofluorescence analysis with the HB-5 monoclonal antibody. Journal of immunology (Baltimore, Md. : 1950) 1984;133:678–683. [PubMed] [Google Scholar]
- 20.Chan TD, Gatto D, Wood K, Camidge T, Basten A, Brink R. Antigen affinity controls rapid T-dependent antibody production by driving the expansion rather than the differentiation or extrafollicular migration of early plasmablasts. Journal of immunology (Baltimore, Md. : 1950) 2009;183:3139–3149. doi: 10.4049/jimmunol.0901690. [DOI] [PubMed] [Google Scholar]
- 21.Wakabayashi C, Adachi T, Wienands J, Tsubata T. A distinct signaling pathway used by the IgG-containing B cell antigen receptor. Science (New York, N.Y.) 2002;298:2392–2395. doi: 10.1126/science.1076963. [DOI] [PubMed] [Google Scholar]
- 22.Avery DT, Ellyard JI, Mackay F, Corcoran LM, Hodgkin PD, Tangye SG. Increased expression of CD27 on activated human memory B cells correlates with their commitment to the plasma cell lineage. Journal of immunology (Baltimore, Md. : 1950) 2005;174:4034–4042. doi: 10.4049/jimmunol.174.7.4034. [DOI] [PubMed] [Google Scholar]
- 23.Thomson CA, Wang Y, Jackson LM, Olson M, Wang W, Liavonchanka A, Keleta L, Silva V, Diederich S, Jones RB, Gubbay J, Pasick J, Petric M, Jean F, Allen VG, Brown EG, Rini JM, Schrader JW. Pandemic H1N1 Influenza Infection and Vaccination in Humans Induces Cross-Protective Antibodies that Target the Hemagglutinin Stem. Frontiers in immunology. 2012;3:87. doi: 10.3389/fimmu.2012.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wrammert J, Koutsonanos D, Li GM, Edupuganti S, Sui J, Morrissey M, McCausland M, Skountzou I, Hornig M, Lipkin WI, Mehta A, Razavi B, Del Rio C, Zheng NY, Lee JH, Huang M, Ali Z, Kaur K, Andrews S, Amara RR, Wang Y, Das SR, O'Donnell CD, Yewdell JW, Subbarao K, Marasco WA, Mulligan MJ, Compans R, Ahmed R, Wilson PC. Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. The Journal of experimental medicine. 2011;208:181–193. doi: 10.1084/jem.20101352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pappas L, Foglierini M, Piccoli L, Kallewaard NL, Turrini F, Silacci C, Fernandez-Rodriguez B, Agatic G, Giacchetto-Sasselli I, Pellicciotta G, Sallusto F, Zhu Q, Vicenzi E, Corti D, Lanzavecchia A. Rapid development of broadly influenza neutralizing antibodies through redundant mutations. Nature. 2014 doi: 10.1038/nature13764. [DOI] [PubMed] [Google Scholar]
- 26.Avnir Y, Tallarico AS, Zhu Q, Bennett AS, Connelly G, Sheehan J, Sui J, Fahmy A, Huang CY, Cadwell G, Bankston LA, McGuire AT, Stamatatos L, Wagner G, Liddington RC, Marasco WA. Molecular signatures of hemagglutinin stem-directed heterosubtypic human neutralizing antibodies against influenza A viruses. PLoS pathogens. 2014;10:e1004103. doi: 10.1371/journal.ppat.1004103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moody MA, Zhang R, Walter EB, Woods CW, Ginsburg GS, McClain MT, Denny TN, Chen X, Munshaw S, Marshall DJ, Whitesides JF, Drinker MS, Amos JD, Gurley TC, Eudailey JA, Foulger A, DeRosa KR, Parks R, Meyerhoff RR, Yu J-SS, Kozink DM, Barefoot BE, Ramsburg EA, Khurana S, Golding H, Vandergrift NA, Alam SM, Tomaras GD, Kepler TB, Kelsoe G, Liao H-XX, Haynes BF. H3N2 influenza infection elicits more cross-reactive and less clonally expanded anti-hemagglutinin antibodies than influenza vaccination. PloS one. 2011:6. doi: 10.1371/journal.pone.0025797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tan YC, Blum LK, Kongpachith S, Ju CH, Cai X, Lindstrom TM, Sokolove J, Robinson WH. High-throughput sequencing of natively paired antibody chains provides evidence for original antigenic sin shaping the antibody response to influenza vaccination. Clinical immunology. 2014;151:55–65. doi: 10.1016/j.clim.2013.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller MS, Tsibane T, Krammer F, Hai R, Rahmat S, Basler CF, Palese P. 1976 and 2009 H1N1 influenza virus vaccines boost anti-hemagglutinin stalk antibodies in humans. The Journal of infectious diseases. 2013;207:98–105. doi: 10.1093/infdis/jis652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller MS, Gardner TJ, Krammer F, Aguado LC, Tortorella D, Basler CF, Palese P. Neutralizing antibodies against previously encountered influenza virus strains increase over time: a longitudinal analysis. Science translational medicine. 2013:5. doi: 10.1126/scitranslmed.3006637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sasso EH, Willems van Dijk K, Bull AP, Milner EC. A fetally expressed immunoglobulin VH1 gene belongs to a complex set of alleles. The Journal of clinical investigation. 1993;91:2358–2367. doi: 10.1172/JCI116468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sasso EH, Johnson T, Kipps TJ. Expression of the immunoglobulin VH gene 51p1 is proportional to its germline gene copy number. The Journal of clinical investigation. 1996;97:2074–2080. doi: 10.1172/JCI118644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Potter KN, Li Y, Mageed RA, Jefferis R, Capra JD. Molecular characterization of the VH1-specific variable region determinants recognized by anti-idiotypic monoclonal antibodies G6 and G8. Scandinavian journal of immunology. 1999;50:14–20. doi: 10.1046/j.1365-3083.1999.00524.x. [DOI] [PubMed] [Google Scholar]
- 34.Krammer F, Pica N, Hai R, Margine I, Palese P. Chimeric hemagglutinin influenza virus vaccine constructs elicit broadly protective stalk-specific antibodies. Journal of virology. 2013;87:6542–6550. doi: 10.1128/JVI.00641-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kanekiyo M, Wei C-JJ, Yassine HM, McTamney PM, Boyington JC, Whittle JRR, Rao SS, Kong W-PP, Wang L, Nabel GJ. Self-assembling influenza nanoparticle vaccines elicit broadly neutralizing H1N1 antibodies. Nature. 2013;499:102–106. doi: 10.1038/nature12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goff PH, Eggink D, Seibert CW, Hai R, Martinez-Gil L, Krammer F, Palese P. Adjuvants and immunization strategies to induce influenza virus hemagglutinin stalk antibodies. PloS one. 2013;8:e79194. doi: 10.1371/journal.pone.0079194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu X, Zhou T, Zhu J, Zhang B, Georgiev I, Wang C, Chen X, Longo NS, Louder M, McKee K, O'Dell S, Perfetto S, Schmidt SD, Shi W, Wu L, Yang Y, Yang ZY, Yang Z, Zhang Z, Bonsignori M, Crump JA, Kapiga SH, Sam NE, Haynes BF, Simek M, Burton DR, Koff WC, Doria-Rose NA, Connors M, Mullikin JC, Nabel GJ, Roederer M, Shapiro L, Kwong PD, Mascola JR. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science. 2011;333:1593–1602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.