Abstract
Intensified efforts to promote protective T cell-based immunity in vaccines and immunotherapies have created a compelling need to expand our understanding of human T cell function and maintenance beyond its characterization in peripheral blood. Mouse studies of T cell immunity show that in response to infection, T cells migrate to diverse sites and persist as tissue-resident memory T cells (TRM) which mediate rapid in situ protection upon antigen recall. Here we discuss new approaches to probe human T cell immunity including novel sampling that indicate a broad distribution and high frequency of human TRM in multiple sites. These newer findings further implicate anatomic compartmentalization as a generalized mechanism for long term maintenance of human T cells throughout life.
Keywords: immune memory, immune homeostasis, naïve T cells, mucosal immunity, peripheral blood
INTRODUCTION
T cells are the primary elements of cell-based adaptive immunity and are essential for establishing and maintaining protective immunity and immune homeostasis. Recent findings have revealed that while T cell-based immunity has a circulating component in blood, a significant proportion of T cells are present as non-circulating tissue resident populations in diverse sites. Studies in mouse models further indicate that the anatomical localization of T cells may be the critical factor impacting this protective capacity, leading to a paradigm shift in the way we study T cell responses and assess their efficacy in vaccines and immunotherapies.
The tissue distribution of T cells is dependent on their subset and differentiation state. Naïve T cells are generated from precursors in the thymus and are exported to the periphery where they primarily localize to lymph nodes, spleen and other lymphoid tissues. Activation of naïve T cells results in altered expression of homing and chemokine receptors enabling the resultant activated/effector T cells to migrate to diverse tissue sites following infection, including mucosal and non-lymphoid tissues. The population of memory T cells that persists following initial activation consists of circulating subsets, which can migrate between sites through peripheral blood, and those which are non-circulating and take up residence in specific tissue compartments. These “tissue-resident” memory T cells (TRM) have been identified in a number of mouse infection models, comprise the predominant T cell subset in lungs, intestines, vaginal mucosa, and skin, and are also present in liver, brain and lymphoid tissues [1, 2]. Human memory T cells bearing TRM phenotypes exhibit a broader tissue distribution than found in mouse, comprising the majority of T cells throughout the body in lymphoid, mucosal, non-lymphoid, and peripheral tissues [3]. These findings further implicate anatomical location as paramount to human T cell immunity, and suggest that localization may be a central feature of T cell responses in humans, integral to their long-term maintenance over decades.
Aside from memory T cells, naïve T cells may also require residence in tissues for their long-term maintenance. While naïve T cells are generally thought to circulate freely between blood and lymphatics due to expression of the homing receptor CCR7 [4, 5], the extent of naïve T cell migration in vivo is not well understood, particularly whether naïve T cells traffic between lymph nodes. In humans, naïve T cells are present throughout life and can persist due to homeostatic turnover [6]. Whether longterm maintenance of naïve T cells is enhanced by localization in specific lymphoid compartments has not been addressed, although there is emerging evidence that naïve T cells in lymphoid tissue are distinct from those in circulation [3]. Thus, the complement of adaptive T cell populations including those emerging from the thymus and those generated by activation at peripheral sites may be more localized than previously appreciated.
The prominent role of anatomical location and tissue residence in T cell responses poses a formidable challenge in human immunology, where sampling and study are largely limited to peripheral blood. Recent years have seen the increased application of T cell-targeted and cell-based immunotherapies for treating cancer, autoimmunity and inflammatory diseases [7], necessitating a deeper understanding of T cell responses in humans in the sites where they function and are maintained, and how the circulating T cell pool relates to T cells in different tissues. Because it is not possible to follow human T cell activation and memory formation to a pathogen in vivo, addressing questions of origin and migration requires extrapolation of information based on systems-wide approaches. Moreover, human studies must take into consideration genetic variability and age, determining which aspects of the T cell response are subject to these influences.
Here we review the current state of knowledge on circulating and tissue T cell populations in humans, including recent studies aimed at dissecting human immunity and its inherent complexity. Recent quantitative evidence from mouse models indicates that the majority of memory T cells generated following infection in mice are resident in multiple diverse tissues, and are not circulating [8]. We discuss evidence from studies in humans suggesting that tissue localization may be a central mechanism for maintaining stable populations of both memory and naïve T cells, and how circulation may be a repository for transiently activated and cycling populations.
Heterogeneity and distribution of human T cell subsets
The heterogeneity of human T cells in peripheral blood has been defined based on differential expression of CD45 isoforms, integrins, chemokine/cytokine receptors and homing receptors [9–11]. The original finding that expression of the lymph node homing/chemokine receptor CCR7 [12] could delineate major subsets of naïve and memory T cells provided compelling evidence that subsets could differ in migration properties. Four major subsets of circulating human T cells are defined by coordinate expression of CD45RA and CCR7 (Table 1): Naïve (CD45RA+CCR7+), central-memory (TCM, CD45RA−CCR7+), effector-memory (TEM, CD45RA−CCR7−) and a subset exhibiting properties of “terminal effector” cells (TEMRA, CD45RA+CCR7−) [10]. Functionally, naïve T cells produce IL-2 when stimulated, while TCM, TEM and TEMRA can all produce effector cytokines (mainly IFN-γ, TNF-α), and naïve and TCM have higher proliferative capacity compared to TEM and TEMRA (reviewed in [13, 14]. Mechanisms for the generation of TCM, TEM and TEMRA cells following activation of naïve T cells are not fully defined; however, based on their differentiation state, proliferative capacity, telomere length and in vitro activation/conversion assays, a progressive differentiation model from naïve to TCM, TEM and ultimately to differentiated effector cells has been proposed [13–15].
Table 1.
Phenotype, function, and tissue distribution of T cell subsets
| Naïvea | TCM | TEM | TEMRA | |
|---|---|---|---|---|
| CD45RA | + | − | − | + |
| CCR7 | + | + | − | − |
| CD69 | −/+ | −/+ | −/++ | −/+ |
| CD103 | − | − | −/++ | − |
| IL-2 | ++ | ++ | + | −/+ |
| IFNγ | − | + | ++ | ++ |
| TNFα | − | + | ++ | ++ |
| Proliferation | − | + | + | + |
| Tissue Localization | Blood and lymphoid tissues | Blood and Lymphoid tissue (CD4+ T cells) | All sites, highest in mucosal tissue | Circulation, lungs, spleen (CD8+ T cells) |
Relative expression levels are indicated as (−), none; (+), low/medium; (++), high, or as ranges (−/+, −/++)
That variations in homing capacity of memory T cells corresponds to anatomical diversity, was initially demonstrated in mice showing persistence of antigen-specific memory T cells in multiple sites distinct from the initial site of infection or immunization [16, 17]. In humans, analysis of tissues has been typically confined to surgical explants or biopsies [18–20]; however, recent analysis of multiple tissues from organ donors [3, 21] has enabled a large scale mapping of T cell subset distribution and heterogeneity throughout the body. Tissue-specific distribution of naïve, TCM, TEM, and TEMRA in blood and 8 different lymphoid (spleen, peripheral and mucosal-draining LN), lungs, and intestines [3] are highly conserved between individuals (Table 1). Specifically, CD4+ T cells in blood, spleen and LN comprise on average, 20–30% naïve T cells, 20–30% TCM, with the remaining 50% being TEM. The complement of CD8+ T cells in these same compartments is different; in blood and spleen, CD8 T cells consist of naïve, TEM and TEMRA in varied proportions, while LN exhibit comparable frequencies of naïve and TEM, with TCM not found in significant frequencies [3, 21]. (This results is in contrast to mice, where TCM-phenotype (CD44hi/CD62Lhi) CD8 T cells comprise between 15–50% of total CD8+ T cells [22, 23]). In mucosal sites, TEM cells predominate for both CD4+ and CD8+ T cells, with some CD4+ TCM found in lungs [3]. The skin is also dominated by memory CD4+ and CD8+ T cells, but in different locations; CD4+ TRM populate the dermis, while the epidermis contains populations of CD4+ and CD8+ TRM cells which exhibit high effector function (a finding not consistent with mouse studies where the epidermis is populated predominantly by CD4+ TRM [18, 24]. Together, these findings show that the organization of T cells in tissues and circulation differs by subset, CD4 or CD8 lineage, and tissue type.
Tissue resident memory T cells
The diverse anatomical distribution of memory T cells could derive from continual surveillance of T cells circulating through tissues, lymphatics, and blood, and/or due to actual residence in tissues. Studies in mouse models of infection have used T cell adoptive transfers [25, 26], parabiosis (surgical conjoining of two mice to create shared circulation) [22], and intravenous infusion of fluorescent antibodies to label T cells in circulation versus those within tissues [27] to distinguish between these possibilities. In mice, tissue T cells comprise both circulating and tissue resident memory T cells (TRM), with TRM found in multiple sites including lungs, intestines, skin, vaginal mucosa, liver, intestines, and to lesser extents in lymph nodes [26–31]. These non-migratory TRM can be generated from site-specific infection in skin, lungs, and vaginal mucosa, and are specifically retained within these sites [26, 32–35]. TRM-associated phenotypic markers include constitutive expression of the early T cell activation marker CD69 for both CD4+ and CD8+ TRM with co-expression of the αE integrin CD103 (which binds e-cadherin on epithelial cells) as a specific feature of CD8+ TRM (for reviews of mouse studies, see [1, 2, 31]). Importantly, TRM in mice can mediate rapid, in situ protective responses to respiratory pathogens such as influenza or M. tuberculosis [26, 28, 36] and to LCMV and herpes virus infection in vaginal mucosa and skin [30, 32, 33, 37–39]. These studies demonstrating enhanced protection by TRM have highlighted the importance of understanding tissue-localized responses for the establishment and maintenance of protective immunity.
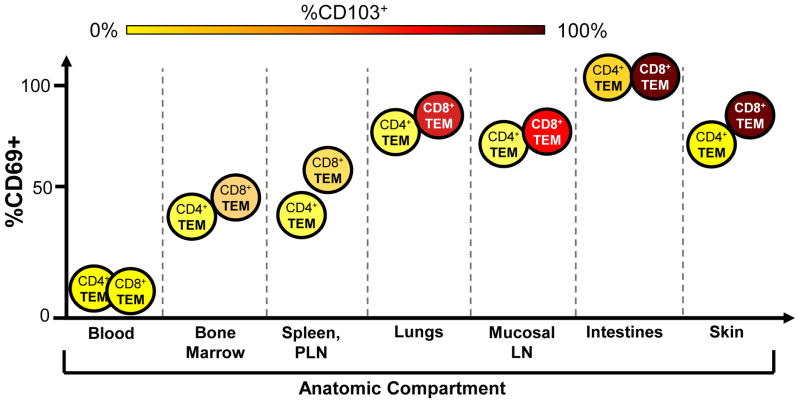
In humans, ex vivo analysis of T cell phenotypes, TCR clonal distribution and studies in patients provide compelling evidence that human TRM exist and are a prominent T cell subset persisting throughout adult life. Phenotypic analysis of the TRM markers CD69 and CD103 on human T cells in circulation and tissues reveal that all T cells in peripheral blood (naïve, TCM, TEM, TEMRA) are uniformly CD69−/CD103−, while tissue T cells express CD69 and CD103 as a function of lineage, subset, and tissue environment. Memory CD4+ and CD8+ T cells in all human tissues including skin, lungs, intestines, stomach, bone marrow, and lymph nodes express CD69, whereas CD103 expression is largely confined to memory CD8 T cells in mucosal sites and skin [3, 18, 20, 21, 40, 41]. Coordinate analysis of CD69/CD103 expression by memory T cells in tissues from organ donors and patients [3, 40, 42] reveals a distinct array of expression based on CD4/CD8 lineage and tissue type (Figure 1). Within an individual, 50–60% TEM in lymph nodes and 70–90% TEM in lung and intestines express CD69, with co-expression of CD103 by CD69+ memory CD8 T cells in small and large intestine and lung, with variable upregulation in mesenteric LN and only a low frequency of CD69+CD103+ memory CD8 T cells in lung-draining and peripheral LN [3] (Figure 1). In human skin, the majority of memory CD4 T cells is CD69+CD103− and are confined to the dermis, while additional populations of CD69+CD103+ memory CD4+ and CD8+ T cells are found in the epidermis [42]. Remarkably, these phenotypic distinctions of memory T cells in different tissues are consistent between diverse individuals [3] and suggest tissue-intrinsic effects on the resident memory T cells.
Figure 1. Variegation of TRM phenotypes in different tissue sites.
Expression of the TRM markers CD69 and CD103 on CD4+ and CD8+ TEM populations in tissue sites (indicated on horizontal axis) is depicted by proportion CD69+ based on position on the vertical axis, and proportion CD103+, indicated by colored shading of each cell type (ranging from yellow=0% to deep red/brown=100%). CD69 is absent on circulating cells and is progressively upregulated on TEM with the highest expression levels seen in mucosal sites. CD103 expression is highest in CD8+ TEM in mucosal tissue sites and mucosal-draining LN with variable expression by CD8+TEM in other tissues, and CD4+ TEM exhibit low or negligible CD103 expression.
This differential upregulation of CD69 and CD103 by tissue memory T cell populations is likely indicative of distinct signals being perceived in these sites, and may also play functional roles in tissue retention. CD69 expression is induced by TCR signaling, type I IFN and other homeostatic cytokines [43, 44], and CD69 expression by human memory T cells in tissues may therefore reflect in situ activation or responses to cytokines. CD69 can also function in tissue retention via sequestration of the sphingosine-1-phosphate receptor (S1PR1) required for egress from tissues following downregulation of KLF2 [45, 46]. CD103 also functions in retention, through interactions with epithelial cells in mucosal sites [47], and can be upregulated in response to TGF-β [36, 48]. There is evidence that human memory T cells in tissues are responding to cytokine and/or cognate signals based on downregulation of CD28 and IL-7R(CD127) expression, which are indicators of TCR-triggered activation/proliferation and responses to the homostatic cytokine IL-7, respectively [49, 50]. In mucosal sites and bone marrow, CD8 TRM phenotype cells are CD28−CD127+ indicative of TCR activation [3, 51], while in lymphoid sites, TRM are CD28+CD127− suggesting active responses to IL-7 [3, 40]. The origin of these distinct tissue signals could derive from commensal microorganism and/or other environmental/inflammatory signals. For example, analysis of microbial diversity in healthy humans revealed regional compartmentalization of microbial species in diverse sites including oral, genital and intestinal mucosa, and different regions of the skin [52], which could affect the local cytokine and metabolic environment for T cell maintenance [53].
There is increasing evidence that human memory T cells in tissues are maintained distinct from those found in circulation. Skin samples obtained from transplant patients administered the potent T cell-depleting agent alemtuzumab (Anti-CD52) exhibit retention of CD69+CD103+ memory CD8+ and to a lesser extent CD4+ T cells [42, 54], supporting the notion that these populations are tissue-resident. Moreover, the fact that solid organ transplant recipients maintained on peripheral T cell immunosuppression do not succumb to repeat infections provides indirect evidence that protective TRM are preserved [55]. Local challenges to the skin in volunteers also demonstrate involvement of skin-specific T cells in the absence of overt peripheral responses [56]. The repertoire of antigen specificities in tissues has also been shown to be distinct from peripheral blood when examined by tetramer binding and antigenic stimulation assays. Human lung is enriched in influenza-specific T cells compared to blood or spleen and these exhibit a TRM phenotype [20, 31]; bone marrow (BM) contains T cells specific for multiple acute and persistent viruses that are not similarly present in peripheral blood [40] and BM memory CD8 T cells exhibit enhanced effector function compared to blood [51, 57]. Together these results indicate that maintenance of antigen-specific memory T cells is highly compartmentalized.
While assessing antigen specificities provides a measure of how the TCR repertoire is distributed in different sites, TCR sequencing provides a direct measure of individual T cell clone distribution, expansion and diversity (see Box 1). Applying deep sequencing approaches, it is possible to survey all possible TCR rearrangements [58] and quantitatively assess tissue compartmentalization of T cells. TCR analysis of TEM cells sorted from spleen and two peripheral lymph nodes from individual organs donors revealed that the majority of CD4+ TEM in lymphoid tissue are unique to each site, with some overlap between sites, while CD8+ TEM exhibited increased sharing of clonally expanded populations between sites [3]. The increased clonal expansion of CD8+ compared to CD4+ memory T cells was also found by TCR sequencing of peripheral blood subsets [59], and may reflect the continuous activation of CD8 T cells by persistent viruses.
Box 1. TCR clonal analysis of human T cell responses.
It is well established that rearrangement of V, D, and J gene segments results in the generation of millions of unique TCR sequences accounting for the high diversity of naïve T cells and a high capacity to recognize a myriad of new antigens[86]. Different methods have been developed to monitor TCR diversity. Early studies used an array of antibodies specific for the 20 Vβ families identified in humans, for an overall view of TCR usage [87]. However, this did not account for the plethora of individual clones within each family. Spectratyping in which the CDR3 length was measured by DNA sequencing, provided a measure of all distinct CDR3 lengths within a given family and a finer resolution of the number of different types of clones [88, 89]. More recently, next generation deep sequencing has been applied to potentially identify and quantitate each unique TCR clone within a sample. Currently, survey approaches are used in which all CDR3β clones are sequenced from a small number within a given sample. The resultant data provide measure of clonal diversity, clonal expansion, and can also provide a measure of the clonal overlap between samples, replicates and between tissue sites. Because rare clones will be sequenced and may not appear in replicate samples, it is best to analyze parameters of the most frequent clones identified. The sequence information can then be analyzed using information theory approaches to assess diversity and other statistical methods to assess clonal overlap between two or more sites [3]. TCR deep sequencing has been used to estimate the TCR repertoire size in human blood [90, 91], to determine the relative diversity of naïve and memory T cells in circulation and changes with aging [59], has been used to track alloreactive clones in human blood in transplant patients rendered tolerant to the donor alloantigen [92], and for analysis of tumor-infiltrating T cells [93]. The application of TCR analysis is particularly relevant to studies of human immunology where HLA type and antigen-specificity are not usually known.
TCR analysis from tissue samples from patients afflicted with different diseases also reveals distinct repertoires in circulation and in different tissues. T cells infiltrating colorectal tumors contained a distinct, non-overlapping repertoire compared to T cells in the neighboring mucosal tissue [60]. Similarly, T cells in cerebrospinal fluid of patients with multiple sclerosis exhibit minimal overlap of expanded clones compared to peripheral blood [61]. Together, these results suggest localized responses and/or specific recruitment and/or retention of clonally-expanded populations in tissues. Additional studies using deep sequencing and whole transcriptome analyses will provide crucial insights into the tissue-specific influences and compartmentalization of human TRM.
Circulating T cells and subset lineages
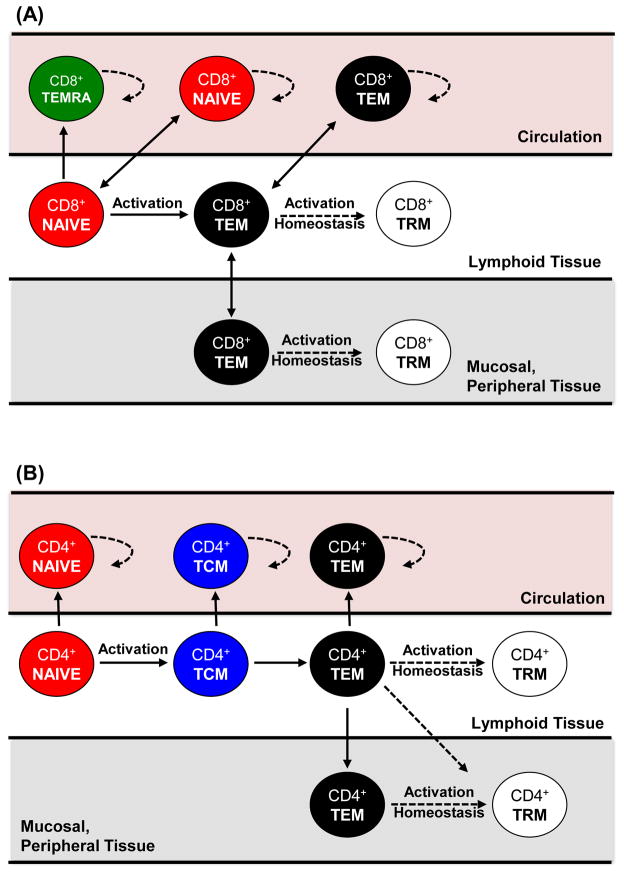
The increasing evidence that tissue localization/residence is integral to T cells responses raises questions concerning the role of circulating T cells and their origin compared to tissue-resident subsets. Relative to tissues, blood appears to be its own compartment, containing specific subsets adapted to circulation. Most strikingly in blood is the presence of CD8+ TEMRA cells that are not found in mucosal sites, skin, or lymph nodes [3]. TEMRA cells exhibit rapid effector function [10], accumulate during aging, and also comprise a major subset specific for persistent viruses such as CMV [62, 63]. For CD4+ T cells, which do not generate significant frequencies of TEMRA cells, TCM appear to be the circulating subset. CD4+ TCM persist in similar frequencies among blood, spleen and lymph nodes and their frequency does not alter as a function of age, suggesting they are freely circulating or constitute an intermediate in differentiation [3]. In addition, TCM cells in skin are depleted in patients with peripheral T cell depletion therapy [42] further suggesting their circulatory nature. The distinct migration and tissue distribution of circulating CD8+ TEMRA and CD4+TCM suggests generation distinct from CD8 and CD4 TEM cells, respectively, with broader migration capabilities (Figure 2). Whether TRM derive from activated precursors/effector cells or from TEM is not clear [64, 65] and both possibilities are illustrated in Figure 2.
Figure 2. Differentiation and homeostasis of circulating and tissue T cell subsets.
Proposed differentiation patterns of CD8+ and CD4+ T cells from naïve (red) to the generation of TRM (white), TEM (black) and TEMRA (green), indicating the tissue-specific maintenance and migration of each subset. (A) Activation of naïve CD8+ T cells may generate TEMRA in blood, or TEM which can migrate to mucosal and other peripheral tissues (e.g., skin). TRM can either be generated from the activated/effector cells which migrate to tissue sites following infection, or could go through a TEM intermediate via further activation or homeostatic turnover. (B) Activation of naïve CD4+T cells can generate TCM as a potential intermediate state migrating to blood and lymphoid tissue, prior to the generation of TEM which migrates to multiple sites. CD4+ TRM could develop from activated/effector CD4+ T cells or CD4+ TEM as in (A). All subsets with access to circulation undergo increased homeostatic proliferation in peripheral blood compared to tissues (see dotted arrows). TEMRA in CD8+ T cells and TCM in CD4+ T cells comprise a circulating subset limited to sites with access to peripheral blood whereas TRM are relegated to tissue sites and are protected from circulation.
There is evidence that circulating T cells in the blood including naïve, TCM, TEM and TEMRA cells are in a more activated state than those in tissues. Higher proportions of TCM and TEM cells in blood express Ki67, a marker of proliferation, compared to their counterparts in lymphoid and mucosal tissues [3], suggesting blood may be enriched in subsets undergoing homeostatic turnover. T cells specific for persistent herpes viruses had more effector-like (CD27−, CD45RA+, CD28−) phenotypes and cytolytic capacity in peripheral blood compared to lymph nodes [66]. During active infection in specific tissues, virus specific CD8 T cells can be detected in blood as seen with Dengue virus [67] influenza virus infection [68], and live yellow fever virus vaccine [69]. Finally, analysis of peripheral blood T cell composition and function in pairs of healthy monozygotic twins revealed high variability and lack of heritable influences [70], contrasting the stable subset distribution observed in similar tissues between unrelated individuals [3]. These results suggest that the blood may be a repository and conduit for T cells responding to antigens and environmental perturbations encountered during the daily human experience.
The more dynamic and transient nature of peripheral blood T cell populations suggest that while sampling blood may be an accurate way to monitor active immune responses, it will not be relevant for assessing the generation and persistence of long-lived memory T cells, including TRM in mucosal and other non-lymphoid sites, and memory T cells in lymphoid tissue. In order to assess the generation of protective T cell responses in vaccines, and to probe how specific immunotherapies may affect long-term memory, it will be necessary to specifically define whether antigen-specific T cells and their associated memory markers are present both in tissues and in circulation. Fine-scale molecular mapping of tissue and blood-borne T cell responses in humans through transcriptome analysis will enable precise assessment of how circulating responses reflect ongoing activation, homeostatic turnover or previous entry into specific tissues.
Naïve T cells and tissues
Naïve T cells mature in the thymus and are then exported through the blood to tissues. In humans, naïve T cells are largely localized to lymph nodes, spleen and blood [3]. The proportion of naïve (CD45RA+ CCR7+) T cells in these sites declines with age due to decreases in thymic volume and output and antigen activation and conversion to memory T cells [6, 71, 72]. Naïve T cell generation and maintenance has been shown to differ between mouse and human [73], with the naïve T cell repertoire in mice maintained almost exclusively by thymic output [74], while in humans naïve T cell maintenance depends more on peripheral expansion [75], with infant thymectomy not adversely affecting T cell-mediated immunity [76, 77]. Through peripheral maintenance, populations of human naïve T cells can persist for decades, are still present in peripheral blood of elderly individuals [6], and comprise up to 20–30% naïve phenotype CD4 and CD8 T cells in lymph nodes derived from individuals in their sixth decade of life [3].
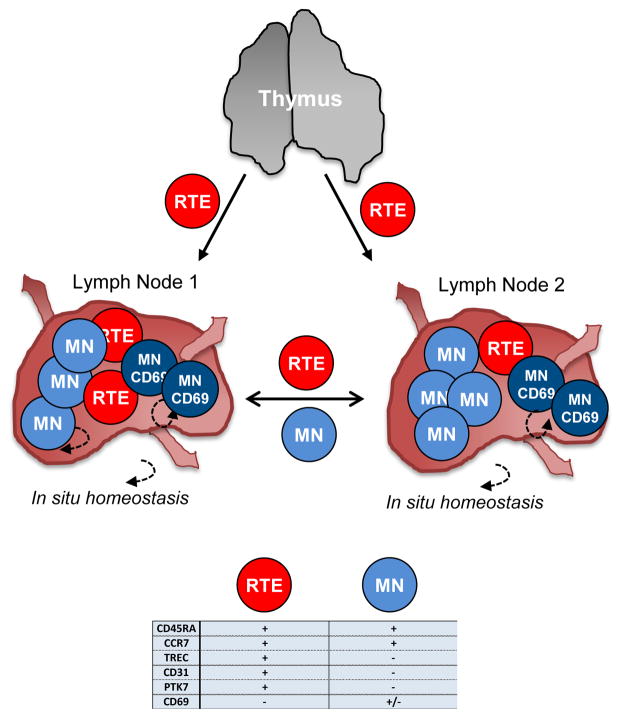
We propose that like memory T cells, tissue localization may promote long-term maintenance of naïve T cells in humans. In mouse models tracking of recent thymic emigrants (RTE), RTE were shown to seed lymphoid tissues and further differentiate into mature naïve T cells [78, 79], indicating a role for tissue localization in naïve T cell maturation (Figure 3). Human CD4+ and CD8+ RTE are most accurately identified by the quantitation of T cell receptor excision circles (TREC) derived from products of TCR rearrangement [80]. TREC+ T cells can be detected at low levels among sorted naïve T cells from peripheral blood of adults [6], suggesting that RTE may differentiate into mature naïve subsets (Figure 3). Human naïve T cells maintain CCR7 expression for trafficking through secondary lymphoid organs [4, 5, 81, 82], although the extent of migration between different LN is not known. We found that a fraction (average 20%) of naïve-phenotype T cells in human LN upregulates CD69 expression (Figure 3), indicating signaling and perhaps transient retention in these sites. TCR repertoire studies of naïve T cells further implicate roles for signaling in their survival. In mice, naïve T cell survival favors low clonal abundance [83], suggesting that maintenance of a diverse naïve T cell repertoire occurs as a result of competition within specific niches. Moreover, human naïve T cells in peripheral blood exhibit a diverse TCR repertoire into old age with some clonal expansion in the elderly [59]. A potential role for tissue retention in naïve T cell clonal competition and homeostasis could be resolved by determining if CD69+ naïve T cells in LN comprise either clonally expanded or more diverse clones. Responses to homeostatic cytokines such as IL-7, enriched in LN as found in mice [84], as well as interactions with LN dendritic and stromal cells [85] could also contribute to naïve T cell survival and/or expansion in situ.
Figure 3. Naïve T cell compartmentalization and migration in lymphoid tissues.
Diagram showing potential compartmentalization of naïve T cells in different lymph nodes (LN1 and LN2) following export from the thymus. Recent thymic emigrants (RTE, red) emerge from the thymus through circulation and traffic to different LN. In tissues, RTE receive signals needed to become mature naïve T cells (MN, blue) and traffic through lymphoid tissues and blood. A fraction of MN upregulate CD69 (darker blue) and are potentially retained in LN sites while RTE and MN may migrate freely throughout lymphoid tissue sites (e.g. LN1 to LN2). Within tissue sites, naïve T cells undergo low levels of proliferation with potential long term maintenance of naïve T cells through homeostatic proliferation in situ. Naïve T cell diversity could also be a function of the interactions of naïve T cells in tissues with dendritic cells or stromal components. Phenotypic markers of RTE and MN are indicated in the table.
Concluding Remarks
The importance of anatomic localization in the development, function and maintenance of T cells underscores the need to advance studies of human immune system development and maintenance in the whole body. The identification of tissue-resident T cell populations in multiple lymphoid, mucosal and peripheral sites underscores the importance of investigating the activation of tissue-specific immune responses for the generation of effective vaccines and immunotherapies. However, many outstanding questions remain (see Box 2) primarily concerning the possibility that tissue residence is perhaps set when naïve T cells seed lymphoid nodes and may be important for their maintenance suggesting that regional adaptations may drive immune responses at their initiation. Additionally, novel markers of tissue residence remain to be identified through high-throughput sequencing of human tissue sites. Information on the identity and function of T cells in tissues can be applied to precisely define how circulating T cell profiles and responses are related, leading to more effective immune monitoring, and new strategies for immune modulation.
Box 2. Outstanding Questions.
Is the accumulation of antigen-specific memory tissue-specific or systemic? Do functional properties of antigen-specific memory populations vary depending on tissue localization?
What are the additional phenotypic and functional markers that distinguish resident T cell populations? Will advances in sequencing and bioinformatics unlock new insights into immune residence?
What are the mechanisms for the maintenance of naïve T cells over life? Is the naïve T cell repertoire preserved through homeostatic proliferation or tissue-specific maintenance?
How compartmentalized is the human naïve T cell immune response? Is there active crosstalk and circulation between lymph node tissue sites?
How important is site-specific vaccination and immunotherapy in the generation of protective memory for the prevention of disease?
Highlights.
Human memory T cells are maintained homeostatically in diverse sites
Localization of human T cells subsets depends on lineage and differentiation state.
Maintenance of human naïve T cells may also depend on tissue localization.
Blood contains distinct subsets and higher levels of turnover and activation.
Acknowledgments
DLF is supported by NIH grants AI100119, AI106697, and HL116136. JJCT is supported by an NIH F31 pre-doctoral fellowship AG047003 and a BD Bioscience research grant.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mueller SN, et al. Memory T cell subsets, migration patterns, and tissue residence. Annu Rev Immunol. 2013;31:137–161. doi: 10.1146/annurev-immunol-032712-095954. [DOI] [PubMed] [Google Scholar]
- 2.Schenkel JM, Masopust D. Tissue-Resident Memory T Cells. Immunity. 2014;41:886–897. doi: 10.1016/j.immuni.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thome JJC, et al. Spatial Map of Human T Cell Compartmentalization and Maintenance over Decades of Life. Cell. 2014;159:814–828. doi: 10.1016/j.cell.2014.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Debes GF, et al. CC chemokine receptor 7 expression by effector/memory CD4+ T cells depends on antigen specificity and tissue localization during influenza A virus infection. J Virol. 2004;78:7528–7535. doi: 10.1128/JVI.78.14.7528-7535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bromley SK, et al. Chemokine receptor CCR7 guides T cell exit from peripheral tissues and entry into afferent lymphatics. Nat Immunol. 2005;6:895–901. doi: 10.1038/ni1240. [DOI] [PubMed] [Google Scholar]
- 6.Goronzy JJ, et al. Naive T Cell Maintenance and Function in Human Aging. J Immunol. 2015;194:4073–4080. doi: 10.4049/jimmunol.1500046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis MM. A prescription for human immunology. Immunity. 2008;29:835–838. doi: 10.1016/j.immuni.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steinert EM, et al. Quantifying Memory CD8 T Cells Reveals Regionalization of Immunosurveillance. Cell. 2015;161:737–749. doi: 10.1016/j.cell.2015.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang HH, et al. CCR2 identifies a stable population of human effector memory CD4+ T cells equipped for rapid recall response. J Immunol. 2010;185:6646–6663. doi: 10.4049/jimmunol.0904156. [DOI] [PubMed] [Google Scholar]
- 10.Sallusto F, et al. Central memory and effector memory T cell subsets: function, generation, and maintenance. Annu Rev Immunol. 2004;22:745–763. doi: 10.1146/annurev.immunol.22.012703.104702. [DOI] [PubMed] [Google Scholar]
- 11.Gattinoni L, et al. A human memory T cell subset with stem cell-like properties. Nat Med. 2011;17:1290–1297. doi: 10.1038/nm.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sallusto F, et al. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions [see comments] Nature. 1999;401:708–712. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- 13.Farber DL, et al. Human memory T cells: generation, compartmentalization and homeostasis. Nat Rev Immunol. 2014;14:24–35. doi: 10.1038/nri3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Restifo NP, Gattinoni L. Lineage relationship of effector and memory T cells. Curr Opin Immunol. 2013;25:556–563. doi: 10.1016/j.coi.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lanzavecchia A, Sallusto F. Progressive differentiation and selection of the fittest in the immune response. Nat Rev Immunol. 2002;2:982–987. doi: 10.1038/nri959. [DOI] [PubMed] [Google Scholar]
- 16.Masopust D, et al. Preferential localization of effector memory cells in nonlymphoid tissue. Science. 2001;291:2413–2417. doi: 10.1126/science.1058867. [DOI] [PubMed] [Google Scholar]
- 17.Reinhardt RL, et al. Visualizing the generation of memory CD4 T cells in the whole body. Nature. 2001;410:101–105. doi: 10.1038/35065111. [DOI] [PubMed] [Google Scholar]
- 18.Clark RA. Resident memory T cells in human health and disease. Sci Transl Med. 2015;7:269rv261. doi: 10.1126/scitranslmed.3010641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clark RA, et al. A novel method for the isolation of skin resident T cells from normal and diseased human skin. J Invest Dermatol. 2006;126:1059–1070. doi: 10.1038/sj.jid.5700199. [DOI] [PubMed] [Google Scholar]
- 20.Purwar R, et al. Resident memory T cells (T(RM)) are abundant in human lung: diversity, function, and antigen specificity. PLoS One. 2011;6:e16245. doi: 10.1371/journal.pone.0016245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sathaliyawala T, et al. Distribution and compartmentalization of human circulating and tissue-resident memory T cell subsets. Immunity. 2013;38:187–197. doi: 10.1016/j.immuni.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klonowski KD, et al. Dynamics of blood-borne CD8 memory T cell migration in vivo. Immunity. 2004;20:551–562. doi: 10.1016/s1074-7613(04)00103-7. [DOI] [PubMed] [Google Scholar]
- 23.Chiu BC, et al. Cutting edge: Central memory CD8 T cells in aged mice are virtual memory cells. J Immunol. 2013;191:5793–5796. doi: 10.4049/jimmunol.1302509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clark RA. Skin-resident T cells: the ups and downs of on site immunity. J Invest Dermatol. 2010;130:362–370. doi: 10.1038/jid.2009.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Masopust D, et al. Activated primary and memory CD8 T cells migrate to nonlymphoid tissues regardless of site of activation or tissue of origin. J Immunol. 2004;172:4875–4882. doi: 10.4049/jimmunol.172.8.4875. [DOI] [PubMed] [Google Scholar]
- 26.Teijaro JR, et al. Cutting edge: tissue-retentive lung memory CD4 T cells mediate optimal protection to respiratory virus infection. J Immunol. 2011;187:5510–5514. doi: 10.4049/jimmunol.1102243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anderson KG, et al. Intravascular staining for discrimination of vascular and tissue leukocytes. Nat Protoc. 2014;9:209–222. doi: 10.1038/nprot.2014.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sakai S, et al. Cutting edge: control of Mycobacterium tuberculosis infection by a subset of lung parenchyma-homing CD4 T cells. J Immunol. 2014;192:2965–2969. doi: 10.4049/jimmunol.1400019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schenkel JM, et al. Cutting edge: resident memory CD8 T cells occupy frontline niches in secondary lymphoid organs. J Immunol. 2014;192:2961–2964. doi: 10.4049/jimmunol.1400003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schenkel JM, et al. Sensing and alarm function of resident memory CD8 T cells. Nat Immunol. 2013 doi: 10.1038/ni.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turner DL, Farber DL. Mucosal resident memory CD4 T cells in protection and immunopathology. Front Immunol. 2014;5:331. doi: 10.3389/fimmu.2014.00331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang X, et al. Skin infection generates non-migratory memory CD8+ TRM cells providing global skin immunity. Nature. 2012;483:227–231. doi: 10.1038/nature10851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shin H, Iwasaki A. A vaccine strategy that protects against genital herpes by establishing local memory T cells. Nature. 2012;491:463–467. doi: 10.1038/nature11522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turner DL, et al. Lung niches for the generation and maintenance of tissue-resident memory T cells. Mucosal Immunol. 2014;7:501–510. doi: 10.1038/mi.2013.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gebhardt T, et al. Memory T cells in nonlymphoid tissue that provide enhanced local immunity during infection with herpes simplex virus. Nat Immunol. 2009;10:524–530. doi: 10.1038/ni.1718. [DOI] [PubMed] [Google Scholar]
- 36.Wu T, et al. Lung-resident memory CD8 T cells (TRM) are indispensable for optimal cross-protection against pulmonary virus infection. J Leukoc Biol. 2014;95:215–224. doi: 10.1189/jlb.0313180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schenkel JM, et al. T cell memory. Resident memory CD8 T cells trigger protective innate and adaptive immune responses. Science. 2014;346:98–101. doi: 10.1126/science.1254536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ariotti S, et al. T cell memory. Skin-resident memory CD8(+) T cells trigger a state of tissue-wide pathogen alert. Science. 2014;346:101–105. doi: 10.1126/science.1254803. [DOI] [PubMed] [Google Scholar]
- 39.Mackay LK, et al. Long-lived epithelial immunity by tissue-resident memory T (TRM) cells in the absence of persisting local antigen presentation. Proc Natl Acad Sci U S A. 2012;109:7037–7042. doi: 10.1073/pnas.1202288109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Okhrimenko A, et al. Human memory T cells from the bone marrow are resting and maintain long-lasting systemic memory. Proc Natl Acad Sci U S A. 2014;111:9229–9234. doi: 10.1073/pnas.1318731111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Booth JS, et al. Characterization and functional properties of gastric tissue-resident memory T cells from children, adults, and the elderly. Front Immunol. 2014;5:294. doi: 10.3389/fimmu.2014.00294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Watanabe R, et al. Human skin is protected by four functionally and phenotypically discrete populations of resident and recirculating memory T cells. Sci Transl Med. 2015;7:279ra239. doi: 10.1126/scitranslmed.3010302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ziegler SF, et al. The activation antigen CD69. Stem Cells. 1994;12:456–465. doi: 10.1002/stem.5530120502. [DOI] [PubMed] [Google Scholar]
- 44.Shiow LR, et al. CD69 acts downstream of interferon-alpha/beta to inhibit S1P1 and lymphocyte egress from lymphoid organs. Nature. 2006;440:540–544. doi: 10.1038/nature04606. [DOI] [PubMed] [Google Scholar]
- 45.Matloubian M, et al. Lymphocyte egress from thymus and peripheral lymphoid organs is dependent on S1P receptor 1. Nature. 2004;427:355–360. doi: 10.1038/nature02284. [DOI] [PubMed] [Google Scholar]
- 46.Skon CN, et al. Transcriptional downregulation of S1pr1 is required for the establishment of resident memory CD8+ T cells. Nat Immunol. 2013;14:1285–1293. doi: 10.1038/ni.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schon MP, et al. Mucosal T lymphocyte numbers are selectively reduced in integrin alpha E (CD103)-deficient mice. J Immunol. 1999;162:6641–6649. [PubMed] [Google Scholar]
- 48.Zhang N, Bevan MJ. Transforming growth factor-beta signaling controls the formation and maintenance of gut-resident memory T cells by regulating migration and retention. Immunity. 2013;39:687–696. doi: 10.1016/j.immuni.2013.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lo DJ, et al. Selective targeting of human alloresponsive CD8+ effector memory T cells based on CD2 expression. Am J Transplant. 2011;11:22–33. doi: 10.1111/j.1600-6143.2010.03317.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mazzucchelli R, Durum SK. Interleukin-7 receptor expression: intelligent design. Nat Rev Immunol. 2007;7:144–154. doi: 10.1038/nri2023. [DOI] [PubMed] [Google Scholar]
- 51.Zhang X, et al. Human bone marrow: a reservoir for “enhanced effector memory” CD8+ T cells with potent recall function. J Immunol. 2006;177:6730–6737. doi: 10.4049/jimmunol.177.10.6730. [DOI] [PubMed] [Google Scholar]
- 52.Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Belkaid Y, Naik S. Compartmentalized and systemic control of tissue immunity by commensals. Nat Immunol. 2013;14:646–653. doi: 10.1038/ni.2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Clark RA, et al. Skin effector memory T cells do not recirculate and provide immune protection in alemtuzumab-treated CTCL patients. Sci Transl Med. 2012;4:117ra117. doi: 10.1126/scitranslmed.3003008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Turner DL, et al. Tissue-resident T cells, in situ immunity and transplantation. Immunol Rev. 2014;258:150–166. doi: 10.1111/imr.12149. [DOI] [PubMed] [Google Scholar]
- 56.Agius E, et al. Decreased TNF-alpha synthesis by macrophages restricts cutaneous immunosurveillance by memory CD4+ T cells during aging. J Exp Med. 2009;206:1929–1940. doi: 10.1084/jem.20090896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Racanelli V, et al. Bone marrow of persistently hepatitis C virus-infected individuals accumulates memory CD8+ T cells specific for current and historical viral antigens: a study in patients with benign hematological disorders. J Immunol. 2007;179:5387–5398. doi: 10.4049/jimmunol.179.8.5387. [DOI] [PubMed] [Google Scholar]
- 58.Robins H. Immunosequencing: applications of immune repertoire deep sequencing. Curr Opin Immunol. 2013;25:646–652. doi: 10.1016/j.coi.2013.09.017. [DOI] [PubMed] [Google Scholar]
- 59.Qi Q, et al. Diversity and clonal selection in the human T-cell repertoire. Proc Natl Acad Sci U S A. 2014;111:13139–13144. doi: 10.1073/pnas.1409155111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sherwood AM, et al. Tumor-infiltrating lymphocytes in colorectal tumors display a diversity of T cell receptor sequences that differ from the T cells in adjacent mucosal tissue. Cancer Immunol Immunother. 2013;62:1453–1461. doi: 10.1007/s00262-013-1446-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lossius A, et al. High-throughput sequencing of TCR repertoires in multiple sclerosis reveals intrathecal enrichment of EBV-reactive CD8+ T cells. Eur J Immunol. 2014;44:3439–3452. doi: 10.1002/eji.201444662. [DOI] [PubMed] [Google Scholar]
- 62.Wertheimer AM, et al. Aging and cytomegalovirus infection differentially and jointly affect distinct circulating T cell subsets in humans. J Immunol. 2014;192:2143–2155. doi: 10.4049/jimmunol.1301721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lilleri D, et al. Human cytomegalovirus-specific memory CD8+ and CD4+ T cell differentiation after primary infection. J Infect Dis. 2008;198:536–543. doi: 10.1086/590118. [DOI] [PubMed] [Google Scholar]
- 64.Mackay LK, et al. The developmental pathway for CD103(+)CD8+ tissue-resident memory T cells of skin. Nat Immunol. 2013;14:1294–1301. doi: 10.1038/ni.2744. [DOI] [PubMed] [Google Scholar]
- 65.Masopust D, et al. Stimulation history dictates memory CD8 T cell phenotype: implications for prime-boost vaccination. J Immunol. 2006;177:831–839. doi: 10.4049/jimmunol.177.2.831. [DOI] [PubMed] [Google Scholar]
- 66.Remmerswaal EB, et al. Human virus-specific effector-type T cells accumulate in blood but not in lymph nodes. Blood. 2012;119:1702–1712. doi: 10.1182/blood-2011-09-381574. [DOI] [PubMed] [Google Scholar]
- 67.Rivino L, et al. Virus-specific T lymphocytes home to the skin during natural dengue infection. Sci Transl Med. 2015;7:278ra235. doi: 10.1126/scitranslmed.aaa0526. [DOI] [PubMed] [Google Scholar]
- 68.Sridhar S, et al. Cellular immune correlates of protection against symptomatic pandemic influenza. Nat Med. 2013;19:1305–1312. doi: 10.1038/nm.3350. [DOI] [PubMed] [Google Scholar]
- 69.Miller JD, et al. Human effector and memory CD8+ T cell responses to smallpox and yellow fever vaccines. Immunity. 2008;28:710–722. doi: 10.1016/j.immuni.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 70.Brodin P, et al. Variation in the human immune system is largely driven by non-heritable influences. Cell. 2015;160:37–47. doi: 10.1016/j.cell.2014.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bains I, et al. Quantifying thymic export: combining models of naive T cell proliferation and TCR excision circle dynamics gives an explicit measure of thymic output. Journal of immunology. 2009;183:4329–4336. doi: 10.4049/jimmunol.0900743. [DOI] [PubMed] [Google Scholar]
- 72.Palmer DB. The effect of age on thymic function. Frontiers in immunology. 2013;4:316. doi: 10.3389/fimmu.2013.00316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.den Braber I, et al. Maintenance of peripheral naive T cells is sustained by thymus output in mice but not humans. Immunity. 2012;36:288–297. doi: 10.1016/j.immuni.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 74.Hale JS, et al. Thymic output in aged mice. Proc Natl Acad Sci U S A. 2006;103:8447–8452. doi: 10.1073/pnas.0601040103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Qi Q, et al. Mechanisms shaping the naive T cell repertoire in the elderly - thymic involution or peripheral homeostatic proliferation? Experimental gerontology. 2014;54:71–74. doi: 10.1016/j.exger.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wells WJ, et al. Neonatal thymectomy: does it affect immune function? The Journal of thoracic and cardiovascular surgery. 1998;115:1041–1046. doi: 10.1016/S0022-5223(98)70403-9. [DOI] [PubMed] [Google Scholar]
- 77.Eysteinsdottir JH, et al. The influence of partial or total thymectomy during open heart surgery in infants on the immune function later in life. Clin Exp Immunol. 2004;136:349–355. doi: 10.1111/j.1365-2249.2004.02437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Scollay R. Thymus cell migration: cells migrating from thymus to peripheral lymphoid organs have a “mature” phenotype. Journal of immunology. 1982;128:1566–1570. [PubMed] [Google Scholar]
- 79.Boursalian TE, et al. Continued maturation of thymic emigrants in the periphery. Nat Immunol. 2004;5:418–425. doi: 10.1038/ni1049. [DOI] [PubMed] [Google Scholar]
- 80.Hazenberg MD, et al. T cell receptor excision circles as markers for recent thymic emigrants: basic aspects, technical approach, and guidelines for interpretation. J Mol Med. 2001;79:631–640. doi: 10.1007/s001090100271. [DOI] [PubMed] [Google Scholar]
- 81.Forster R, et al. CCR7 coordinates the primary immune response by establishing functional microenvironments in secondary lymphoid organs. Cell. 1999;99:23–33. doi: 10.1016/s0092-8674(00)80059-8. [DOI] [PubMed] [Google Scholar]
- 82.Berzins SP, et al. The role of the thymus and recent thymic migrants in the maintenance of the adult peripheral lymphocyte pool. The Journal of experimental medicine. 1998;187:1839–1848. doi: 10.1084/jem.187.11.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hataye J, et al. Naive and memory CD4+ T cell survival controlled by clonal abundance. Science. 2006;312:114–116. doi: 10.1126/science.1124228. [DOI] [PubMed] [Google Scholar]
- 84.Guimond M, et al. Interleukin 7 signaling in dendritic cells regulates the homeostatic proliferation and niche size of CD4+ T cells. Nat Immunol. 2009;10:149–157. doi: 10.1038/ni.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kumar V, et al. A dendritic-cell-stromal axis maintains immune responses in lymph nodes. Immunity. 2015;42:719–730. doi: 10.1016/j.immuni.2015.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gellert M. V(D)J recombination: RAG proteins, repair factors, and regulation. Annu Rev Biochem. 2002;71:101–132. doi: 10.1146/annurev.biochem.71.090501.150203. [DOI] [PubMed] [Google Scholar]
- 87.Pannetier C, et al. The sizes of the CDR3 hypervariable regions of the murine T-cell receptor beta chains vary as a function of the recombined germ-line segments. Proc Natl Acad Sci U S A. 1993;90:4319–4323. doi: 10.1073/pnas.90.9.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Arstila TP, et al. A direct estimate of the human alphabeta T cell receptor diversity. Science. 1999;286:958–961. doi: 10.1126/science.286.5441.958. [DOI] [PubMed] [Google Scholar]
- 89.Baron V, et al. The repertoires of circulating human CD8(+) central and effector memory T cell subsets are largely distinct. Immunity. 2003;18:193–204. doi: 10.1016/s1074-7613(03)00020-7. [DOI] [PubMed] [Google Scholar]
- 90.Robins HS, et al. Comprehensive assessment of T-cell receptor beta-chain diversity in alphabeta T cells. Blood. 2009;114:4099–4107. doi: 10.1182/blood-2009-04-217604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Robins HS, et al. Overlap and effective size of the human CD8+ T cell receptor repertoire. Sci Transl Med. 2010;2:47ra64. doi: 10.1126/scitranslmed.3001442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Morris H, et al. Tracking donor-reactive T cells: Evidence for clonal deletion in tolerant kidney transplant patients. Sci Transl Med. 2015;7:272ra210. doi: 10.1126/scitranslmed.3010760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Emerson RO, et al. High-throughput sequencing of T-cell receptors reveals a homogeneous repertoire of tumour-infiltrating lymphocytes in ovarian cancer. J Pathol. 2013;231:433–440. doi: 10.1002/path.4260. [DOI] [PMC free article] [PubMed] [Google Scholar]