Abstract
The activation state of an antitumor effector T cell in a tumor depends on the sum of all stimulatory signals and inhibitory signals that it receives in the tumor microenvironment. Accumulating data address the increasing complexity of these signals produced by a myriad of immune checkpoint molecules, cytokines and metabolites. While reductionist experiments have identified key molecules and their importance in signaling, less clear is the integration of all these signals that allows T cells to guide their responses in health and in disease. Mass spectrometry-based proteomics is well poised to offer such insights, including monitoring emergence of resistance mechanisms to immunotherapeutics during treatments. A major application of this technology is in the discovery and characterization of small molecule agents capable of enhancing the response to immunotherapeutic agents. Such an approach would reinvigorate small molecule drug development aimed not at tumor cells but rather at tumor-resident T cells capable of producing dramatic and durable antitumor responses.
Introduction
Over the past few years it has become clear that immunotherapy, previously thought to be useful in only a few select malignancies, has considerable clinical activity in a variety of cancers including lung, bladder, head and neck, cervical, and others. This clinical response has been accomplished with the blockade of a single immunoinhibitory mechanism in the tumor microenvironment, signaling through the PD-1 immune checkpoint receptor on the surface of T cells. Clinical outcomes have been dramatic with very significant improvements in survival, as demonstrated by “raising the tail of the survival curve”, reflecting durable responses produced by these agents (1). However, depending on the cancer, less than half of the patients benefit with disease control (stable disease or better), so there remains considerable opportunity for improvement (2). There are three general categories of mechanisms whereby tumors evade rejection by the immune system: (i) there may be insufficient numbers of tumor-reactive effector T cells generated within the lymphoid compartment; (ii) T cells may not extravasate into the tumor parenchyma; and (iii) if T cells enter the tumor parenchyma they may be shut down by a number of different immunosuppressive mechanisms operational within the tumor microenvironment (TME). Here we will focus on the latter, the situation in which understanding immune signaling proteomes could lead to the discovery of new immunotherapeutic strategies to improve the overall efficacy of this modality.
The activation state of an antitumor effector T cell in a tumor depends on the sum of all stimulating signals and inhibitory signals that it receives in the TME. Accumulating data, nicely reviewed elsewhere, address the increasing complexity of these signals (3, 4). T-cell response to tumors is guided by the activation of the T-cell receptor (TCR) through antigen recognition and is further regulated by both inhibitory and stimulatory co-signals (5). T cells that infiltrate the tumor are previously activated within the lymphoid compartment, since naïve T cells are incapable of extravasating into extra-nodal sites. In patients with progressive cancer, tumors have co-opted the immunosuppressive mechanisms employed in health to physiologically shut down inflammatory immune responses that are no longer needed after a foreign invasive insult such as infection has been cleared. This probably occurs in a variety of different ways including dysregulated gene expression and feedback inhibition of T-cell responses. For example, T cell-produced cytokine γ-interferon (IFNγ) is a potent inducer of PD-L1 expression which can inhibit T cells when it binds to its immune checkpoint receptor, PD-1, on the T-cell surface. The various effector T-cell inhibitory mechanisms in the TME include other T-cell surface checkpoint proteins (CTLA-4, LAG3, TIM3, BTLA, adenosine A2AR), secreted molecules (TGFβ, IL10, PGE2), metabolic alterations (excess adenosine, indoleamine 2,3-dioxygenase, arginase), and immunosuppressive cells (cancer-associated fibroblasts, regulatory T cells, myeloid-derived suppressor cells, tumor-associated macrophages) (4).
What mass spectrometry-based signaling proteomics brings to the table
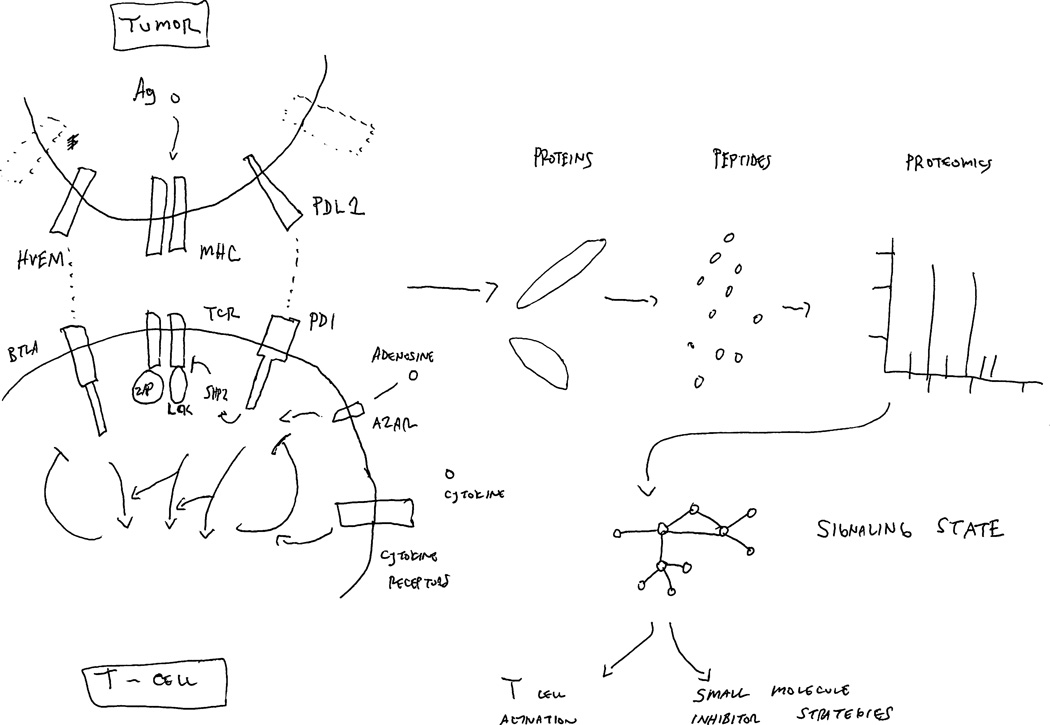
Tumor-infiltrating lymphocytes, depending on the tumor, may be exposed to one or more likely many of these immunosuppressive processes, each delivering a negative signal that is transduced within the T cell through signaling cascades, producing a complex network of interacting signaling pathways. Mapping this immune signaling proteome could provide insight into these networks, which could be a biomarker for resistance to immunotherapeutic agents, and may suggest new immunotherapeutic strategies with agents designed to disrupt critical, converged down-stream signaling pathways. Figure 1 is a cartoon of how mass spectrometry-based signaling proteomics can be used to create a molecular snapshot of T-cell activation circuits to facilitate the design of small molecule inhibitor strategies. Signaling in cells is increasingly recognized as a complex adaptive system produced by contextual expression of cellular gene products and their interaction with environment features and cues. Signaling proteins (e.g. kinases and substrates) act as heterogeneous agents that are decision-makers and evolve over time. Signaling proteins usually do not work in isolation but act in multi-protein complexes and in networks. Emergent behavior results from integrated effects of signaling proteins that affects cell physiology and ultimately drug response. The major challenge and thus opportunity is to understand how components work together and respond to the myriad of on/off molecular switches that affect T-cell recognition of tumor cells.
Figure 1. Proteomic dissection of molecular snapshot of T-cell activation circuits.
The tumor cell:T cell interface initiates a complex signaling environment beginning with MHC/peptide-antigen activation of the T-cell receptor (TCR) along with inputs of other co-stimulatory or inhibitory signaling events, shown as HVEM/BLTA or PD-L1/PD-1 in this cartoon, as well as adenosine signaling through its receptor (A2AR) or cytokines. These along with other immune checkpoints, as well as signals driven by tumor stromal signals such as adenosine, cooperate to drive a signaling network resulting in effector signaling events (ERK, AKT, STAT) within T cells. While shown for simplicity within this cartoon, the system behaves similar to other complex regulatory signaling circuits in normal cells or tumor cells, consisting of many adaptor proteins, signaling kinases and phosphatases, and connected within feedback loops. To comprehend this complexity, mass spectrometry facilitates the dissection of these networks by quantification of peptides and post-translationally modified (phosphorylated, acetylated, ubiquitinated) peptides and can do so in a dynamic fashion. Ultimately, this knowledge can be converted into network maps of T-cell signaling that enables molecular snapshots of T-cell activation circuits and can facilitate a strategy to insert small molecules onto this network.
One major opportunity may lie in utilizing the power of system-level mass spectrometry (MS)-based proteomics to embrace complexity of signaling in T cells. MS-based proteomics allows a system-wide and quantitative view of signaling and drug action in cells of interest, be they tumor cells or immune cells (6–8). Proteomics has the ability to better inform about active pathways and networks governing T-cell responses in a global and unbiased manner and it could inform about combination approaches for targeting complex immunosuppressive mechanisms within the TME. MS-based proteomics could be an important strategy to approach the problem of rapid and adaptive changes in immune cells, such as T cells, induced by the myriad of input signals, including the TCRs and the immunoinhibitory processes described above.
Studies examining system-level changes in phosphorylation in tumor cells have uncovered sometimes paradoxical features. We have observed proteins whose phosphorylation is elevated following kinase inhibition and other groups have started to identify similar compensatory feedback that mediated changes in cancer cells (9–13). In tumor cells, these system-level changes could modulate the effects of kinase inhibitors by activating cell intrinsic compensatory mechanisms sustaining cell survival. Similar events have been observed in yeast in which kinase loss can result in increased levels of phosphorylation in some substrates (9). We suspect similar events occur in immune cells, where therapeutics, such as checkpoint antibodies, may have paradoxical signaling effects that can only be identified through an unbiased and large scale discovery experiment, as offered by MS-based approaches. In addition, therapeutics such as kinase inhibitors or epigenetic agents may impact immune cells in novel ways that are best defined through global proteomic approaches.
Charting Signaling Immunoproteomes
A few studies have reported results using MS-based proteomics related to T-cell activation. Quantitative phosphoproteomics studies in Jurkat T-cell leukemia line identified protein modules activated upon TCR stimulation (14). Pathways of protein phosphorylation have been identified to be pre-organized prior to TCR activation and these pathways can dictate the ability of the T cell to mediate effector function in response to signals activating the TCRs (15). Therefore, organization of signaling cascades, mediated by protein kinases and phosphatases, and written in the language of phosphoproteomic patterns, modulates tumor-specific T cells and their responses both to TCR activation by tumor antigens and to checkpoint inhibitory or stimulatory pathways. Phosphoproteomic studies have also shed light on the manner in which T cells integrate cytokine stimulation into signaling networks (16). While a few studies have reported global signaling events in primary T lymphocytes using MS-based proteomics, there are no studies to date using tumor-infiltrating T cells directly from the tumor. Further opportunities exist to look at post-translational modifications (PTM) beyond phosphorylation. It is well known that the activation state, localization, turnover, and protein-protein interactions are determined by PTMs beyond phosphorylation, ubiquitination and acetylation (17). Multiple PTMs can co-exist on a single protein and this enables cells to fine tune protein functions (18, 19). Results from some studies have suggested crosstalk amongst kinases, acetylase/deacetylase enzymes, and ubiquitin-modifying enzymes, including reports suggesting links between tyrosine kinases and acetylated proteins (p53, Myc) (20–27). Combining studies examining immune signaling events with methods to address PTM crosstalk may provide new opportunities for elucidating complex network-level interactions and opportunities for novel therapeutic approaches. Such studies will be enabled through a new strategy called sequential enrichment of post-translational modifications (SEPTM) that enables quantifying the simultaneous PTM-omes under different stimuli or physiologic conditions from the same set of samples (rather than biological replicates) (18, 28).
Convergent Opportunity Areas for MS-based analysis of signaling proteomes
System-wide signaling states related to immune suppression
There are a myriad of potential immune suppressive mechanisms exploited by tumors to evade rejection. The finding that a single intervention with anti-PD-1 can reverse tumor-mediated evasion of immune-mediated killing suggests that there are “driver” immunosuppressive mechanisms operational in subsets of cancer patients. While some aspects of the CTLA-4 and PD-1 effects of signaling in T cells are known, there are few studies investigated how these affect global signaling networks, especially in tumor-derived T cells (29). Global mechanisms of other checkpoint molecules, such as BTLA and TIM3, remain poorly understood. These results suggest an opportunity to produce an unbiased and system-level analysis of protein phosphorylation in T cells facilitating characterization of both basal signaling networks in T cells and the effects of perturbation by TCR stimulation and checkpoint activation. A well designed MS-based approach can determine the effect of checkpoint inhibitors/stimulators on tumor-derived T-cell signaling pathways, characterize signaling crosstalk, and facilitate future rational combinatorial therapeutic strategies.
Success in these proteomics studies can impact outcome of patients treated with immune checkpoint inhibitors. First, they offer the possibility to examine potential biomarkers in T cells that can help predict which patients may benefit from these types of therapy. Patterns of phosphorylation (or other PTMs) can be related to therapeutic sensitivity and/or resistance. Second, studies on global signaling may guide the clinical development of combinatorial strategies. For example, since kinases drive protein phosphorylation, and numerous clinical-grade kinase inhibitors exist, it is possible to envision therapeutic co-targeting strategies for checkpoint inhibitors and kinase inhibitors, where the underlying target is the T cell, not the tumor cell. There have been few inroads in understanding system-wide mechanisms of checkpoint inhibitors and crosstalk with kinase pathways. Kinases still remain an important target group for cancer therapy yet academic research appears biased toward kinases with well-established roles in signaling - large regions of the kinome remain unexplored and could serve as biomarkers as well as present therapeutic opportunities for immuno-oncology (30). Future studies should aim to characterize these phosphorylation networks induced by tumor antigen and other co-stimulatory/inhibitory molecules employing global and less biased methods that MS offers. One goal could be to better characterize a targetable immuno-suppressive checkpoint on T cells and nominate additional regulatory circuits or signaling proteins that may affect sensitivity or resistance to immune checkpoint therapy.
Tumor phosphorylation networks and relationships with pY peptide presentation
Another major opportunity relates to understanding how tumor signaling and production of tumor-specific phosphopeptides may relate to tumor rejection and how this knowledge can drive new therapeutic insights. Phosphopeptides can be presented by MHC molecules and these phosphopeptides can serve as strong tumor antigens leading to T-cell activation and tumor rejection (31–34). Recent studies in human leukemia identified tumor-specific phosphopeptides and demonstrated differences across different disease cells (35). These results parallel phosphoproteomic studies in human tumors, which can demonstrate differences in phosphorylation patterns even amongst histologically similar types of tumors (36, 37). One opportunity will be to align patterns of altered tumor phosphoproteomes with phosphopeptides presented by MHC molecules and eliciting recognition by tumor cells. Experiments to conduct these studies would be quite ambitious, since fine-level mapping and quantification of phosphopeptides in tumor would require access to both large numbers of cases and rapid collection and optimized storage of tumors to allow protein phosphorylation to be maintained. Peptide elution from tumor tissues, as opposed to cells, would also require large amounts of starting materials. Challenges aside, the potential exists to connect signaling events in tumors to antigen presentation. As protein phosphorylation is a dynamic process that can be manipulated by drugs (kinase inhibitors), the opportunity exists to perturb systems to alter tumor phosphorylation events facilitating phosphopeptide presentation to antigen-presenting cells (APC) and T-cell recognition. This could usher in new approaches that use small molecule kinase or phosphatase inhibitors to drive production of immunogenic phosphopeptides and combine these with mechanisms to enhance immune response (checkpoint blockade, increasing T-cell influx into tumors, etc) to improve antitumor responses. While discovery approaches may depend on MS-based approaches, it appears now possible to directly examine tumor tissues for peptide:MHC molecule interactions. Proximity ligation assays can be used to directly visualize protein complexes in tumor tissues and a recent report used this technology to demonstrate IDH1R132H presentation in tumor tissues (38, 39). This may then enable biomarkers to identify existence or drug-induced changes in phosphopeptide:MHC interactions on tumor cells.
Signaling proteomes in advanced co-culture models
The major limitation of most in vitro cell-based experiments, especially those used in MS-based proteomics, is the removal of tumor cells or immune cells from their tumor microenvironment. Ideally, it would be best to study immune signaling in models consisting of both tumor cells and T cells. Not only the microenvironment but the true effects of checkpoint engagement, for example, may only be evident after TCR engagement with MHC/peptide and in the presence of other co-receptors or adhesion molecules. The major disadvantage of a co-culture system is the inability to assign contributory peptides/proteins from each cell population. It will be important to overcome this limitation and allow development of a platform to study signaling networks within T cells alongside those of tumor cells. To overcome the inability to assign peptides from each cell population from a co-culture system (for example, T cell/tumor cells), two groups recently reported a novel technology for quantitative MS-based proteomics, named “cell type-specific labeling using amino acid precursors” (CTAP) (40, 41).This technology employs stable isotope-labeling of proteins in analogy to the widely used stable isotope labeling by amino acids in cell culture (SILAC) (42). However, in contrast to SILAC, CTAP uses amino acid precursors that can only be utilized by specifically engineered cells allowing for the simultaneous metabolic labeling of two co-cultured cell types with different abilities to generate labeled amino acids. This enables differential quantitative proteomics analyses of more complex co-culture systems that more accurately incorporate important elements of the tumor microenvironment, e.g. interactions between cancer cells and T cells. This technology still remains complex and it is unclear how immune cells can tolerate culture conditions lacking key amino acid precursors. If technically achievable, further advancements can be made through co-culture models of T cells and primary tumor cells from patients to recapitulate the tumor/immune cell interface. Generating these primary tumor cells directly from tumor tissues along with T cells allows the ability to generate co-culture model systems, for example, for studying influence of tumor cells on T-cell phosphoproteomes and responses of checkpoint inhibitor antibodies (43–45).
Drug:Protein network curation and drug repurposing for immune therapy
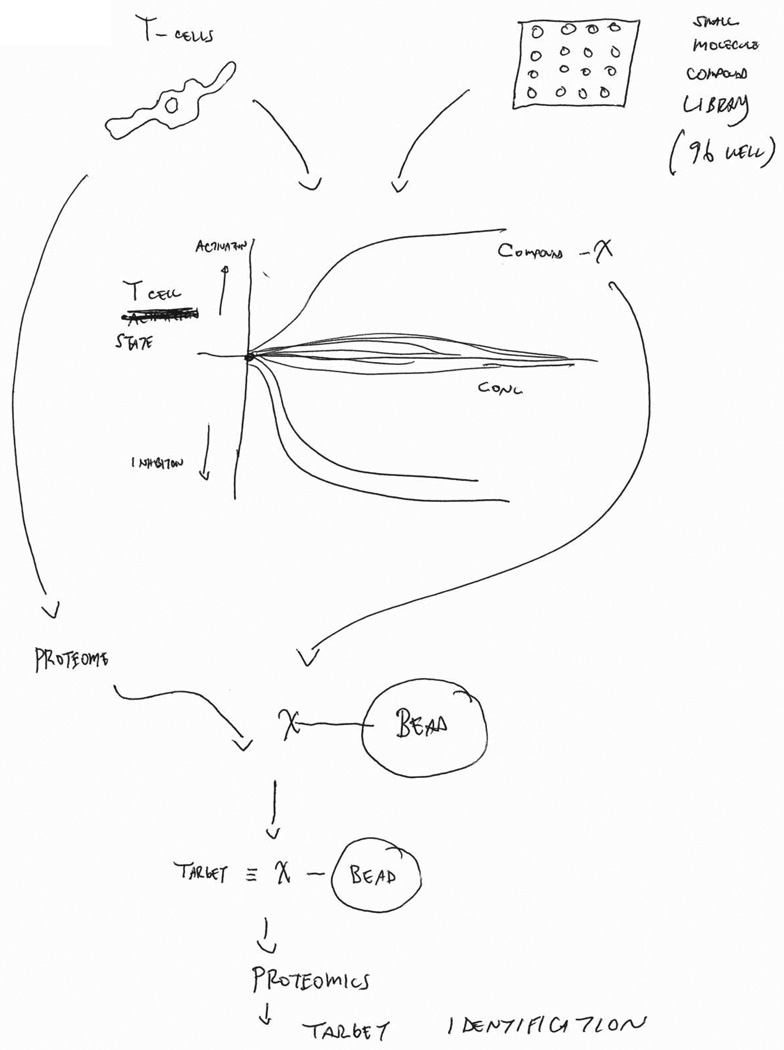
There is currently a high interest and demand for identifying small molecules that amplify antitumor immune therapy and broaden the patient base that responds to immune checkpoint targeting. As a consequence, various cell-based highthroughput drug screens are being conducted. However, in addition to the aforementioned challenge to distinguish target proteins from tumor and microenvironment cells, small molecule drugs display wide ranges of target selectivity. This is well recognized for kinase inhibitors (46 47), but has also been reported for other drug classes, such as histone deacetylase (HDAC) (48, 49) and poly ADP ribose polymerase (PARP) inhibitors (50). There is mounting evidence that targeting multiple proteins simultaneously does not necessarily imply a safety liability, but that such polypharmacology also can be advantageous by enhancing a compound’s cellular activity. Accordingly, the cellular mechanism of action of polypharmacology drugs can be complex and either involve one or more targets in addition to the intended target (51) or can be entirely unrelated to it. As this may similarly apply for small molecules that modulate the efficacy of anticancer immune therapy, proteome-wide characterization of drug target profiles in cancer and microenvironment cells may be necessary to dissect the molecular mechanisms underlying immunostimulatory drug activities of interest. Chemical proteomics is a post-genomic drug affinity purification approach that is powered by modern high resolution MS and bioinformatics and that is excellently suited to this task (52). For instance, applying a chemical proteomics approach we recently described several protein targets to be engaged by the multi-targeted kinase inhibitors dasatinib and sunitinib in primary lung tumors (53). Interestingly, comparison with the target profiles of lung cancer cell lines and mouse xenografts suggested kinase targets not only in the lung cancer cells themselves, but also in the tumor microenvironment, particularly cells of hematopoietic origin. Combining chemical proteomics with sophisticated labeling technologies, such as CTAP, can be a powerful approach to dissect target profiles and mechanisms of action of immune stimulatory drug effects in co-cultures of cancer cells with for instance T cells, ultimately leading to novel actionable targets, drug combinations and therapeutic strategies. Figure 2 shows a possible chemical proteomics workflow using T-cell activation screen data to drive identification of targets that mediate immune stimulatory drug activities.
Figure 2. Identification of T-cell activators through chemical proteomics.
Compound libraries can be screened for effects on T cells (or other immune cells) that are taken directly from tumors. Compounds showing desired properties, such as activation of T cells, can be developed via chemical proteomics and used to probe the underlying biology of the tumor. Chemical proteomics is a post-genomic drug-affinity purification approach that is powered by modern high-resolution mass spectrometry and bioinformatics. In this illustrated case, active compounds are immobilized on beads and used to probe T-cell proteomes. Targets bound to the compound:bead matrix are eluted and identified using mass spectrometry and bioinformatics.
Conclusion
Regulation of tumor cell survival by receptor tyrosine kinase (RTK) signaling has parallels between T-cell activation driven by the engaged T-cell receptor. Both systems require cooperation of signaling molecules including kinases, phosphatases, and key adaptor molecules. These molecules, acting in a coordinated fashion and interconnected through networks, are able to fine tune responses. Similarly, deregulation of both systems can lead to disease pathology, either transformation of cells by RTK leading to cancer, or deregulation of T-cell signaling leading to immune suppression in cancer or autoimmunity diseases. While reductionist experiments have identified key molecules and their importance in signaling, the time has come to produce insights that arise from integration of all signaling inputs. MS-based proteomics is well poised to offer such insights. Through this approach insights will be realized related to monitoring emergence of resistance mechanisms to immunotherapeutics during treatments (e.g. by detection of phosphorylation or dephosphorylation events associated with a resistance pathway) that can also contribute to biomarker development. In addition, we believe that a major application of this technology is in the discovery and characterization of small molecules capable of enhancing the response to immunotherapeutic agents. Global studies can also inform new directions for development of immunotherapeutic approaches gearing towards neutralizing resistance to specific immunotherapeutics analogous to studies that are identifying global mechanism of resistance to small molecule agents used in cancer therapy. Combining these landscape approaches with functional experiments and chemical proteomics allow assessment of drug effects on tumor and key non-tumor immune cells, and therefore provide rationale for specific combinatory approaches. Achieving the promise of immune proteomics will require collaborative teams of immune biologists, immune therapy physicians, experts in cellular signaling, and experts in mass spectrometry.
ACKNOWLEDGEMENTS
Research in our labs has been funded by the National Cancer Institute, the American Lung Association, the V Foundation, the LUNGevity Foundation, the Moffitt Cancer Center, and the Moffitt Lung Cancer Center of Excellence.
LITERATURE CITED
- 1.Brahmer JR, Tykodi SS, Chow LQ, Hwu WJ, Topalian SL, Hwu P, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med. 2012;366:2455–2465. doi: 10.1056/NEJMoa1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antonia SJ, Larkin J, Ascierto PA. Immuno-oncology combinations: a review of clinical experience and future prospects. Clin Cancer Res. 2014;20:6258–6268. doi: 10.1158/1078-0432.CCR-14-1457. [DOI] [PubMed] [Google Scholar]
- 3.Brownlie RJ, Zamoyska R. T cell receptor signalling networks: branched, diversified and bounded. Nat Rev Immunol. 2013;13:257–269. doi: 10.1038/nri3403. [DOI] [PubMed] [Google Scholar]
- 4.Chen L, Flies DB. Molecular mechanisms of T cell co-stimulation and co-inhibition. Nat Rev Immunol. 2013;13:227–242. doi: 10.1038/nri3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen DS, Mellman I. Oncology meets immunology: the cancer-immunity cycle. Immunity. 2013;39(1):1–10. doi: 10.1016/j.immuni.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 6.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 7.Meissner F, Mann M. Quantitative shotgun proteomics: considerations for a high-quality workflow in immunology. Nat Immunol. 2014;15:112–117. doi: 10.1038/ni.2781. [DOI] [PubMed] [Google Scholar]
- 8.Mann M. Can proteomics retire the western blot? J Proteome Res. 2008;7:3065. doi: 10.1021/pr800463v. [DOI] [PubMed] [Google Scholar]
- 9.Bodenmiller B, Wanka S, Kraft C, Urban J, Campbell D, Pedrioli PG, et al. Phosphoproteomic analysis reveals interconnected system-wide responses to perturbations of kinases and phosphatases in yeast. Science signaling. 2010;3:s4. doi: 10.1126/scisignal.2001182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duncan JS, Whittle MC, Nakamura K, Abell AN, Midland AA, Zawistowski JS, et al. Dynamic Reprogramming of the Kinome in Response to Targeted MEK Inhibition in Triple-Negative Breast Cancer. Cell. 2012;149:307–321. doi: 10.1016/j.cell.2012.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chandarlapaty S, Sawai A, Scaltriti M, Rodrik-Outmezguine V, Grbovic-Huezo O, Serra V, et al. AKT inhibition relieves feedback suppression of receptor tyrosine kinase expression and activity. Cancer Cell. 2011;19:58–71. doi: 10.1016/j.ccr.2010.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodrik-Outmezguine VS, Chandarlapaty S, Pagano NC, Poulikakos PI, Scaltriti M, Moskatel E, et al. mTOR kinase inhibition causes feedback-dependent biphasic regulation of AKT signaling. Cancer Discov. 2011;1:248–259. doi: 10.1158/2159-8290.CD-11-0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bai Y, Kim JY, Watters JM, Fang B, Kinose F, Song L, et al. Adaptive Responses to Dasatinib-Treated Lung Squamous Cell Cancer Cells Harboring DDR2 Mutations. Cancer Res. 2014;74:7217–7228. doi: 10.1158/0008-5472.CAN-14-0505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mayya V, Lundgren DH, Hwang SI, Rezaul K, Wu L, Eng JK, et al. Quantitative phosphoproteomic analysis of T cell receptor signaling reveals system-wide modulation of protein-protein interactions. Science signaling. 2009;2:ra46. doi: 10.1126/scisignal.2000007. [DOI] [PubMed] [Google Scholar]
- 15.Navarro MN, Goebel J, Feijoo-Carnero C, Morrice N, Cantrell DA. Phosphoproteomic analysis reveals an intrinsic pathway for the regulation of histone deacetylase 7 that controls the function of cytotoxic T lymphocytes. Nat Immunol. 2011;12:352–361. doi: 10.1038/ni.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arneja A, Johnson H, Gabrovsek L, Lauffenburger DA, White FM. Qualitatively different T cell phenotypic responses to IL-2 versus IL-15 are unified by identical dependences on receptor signal strength and duration. J Immunol. 2014;192:123–135. doi: 10.4049/jimmunol.1302291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deribe YL, Pawson T, Dikic I. Post-translational modifications in signal integration. Nat Struct Mol Biol. 2010;17:666–672. doi: 10.1038/nsmb.1842. [DOI] [PubMed] [Google Scholar]
- 18.Swaney DL, Beltrao P, Starita L, Guo A, Rush J, Fields S, et al. Global analysis of phosphorylation and ubiquitylation cross-talk in protein degradation. Nat Methods. 2013;10:676–682. doi: 10.1038/nmeth.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Guan S, Acharya P, Liu Y, Thirumaran RK, Brandman R, et al. Multisite phosphorylation of human liver cytochrome P450 3A4 enhances Its gp78- and CHIP-mediated ubiquitination: a pivotal role of its Ser-478 residue in the gp78-catalyzed reaction. Mol Cell Proteomics. 2012;11:M111 010132. doi: 10.1074/mcp.M111.010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lopez-Otin C, Hunter T. The regulatory crosstalk between kinases and proteases in cancer. Nat Rev Cancer. 2010;10:278–292. doi: 10.1038/nrc2823. [DOI] [PubMed] [Google Scholar]
- 21.Zhang D, Zaugg K, Mak TW, Elledge SJ. A role for the deubiquitinating enzyme USP28 in control of the DNA-damage response. Cell. 2006;126:529–542. doi: 10.1016/j.cell.2006.06.039. [DOI] [PubMed] [Google Scholar]
- 22.Murphy G. The ADAMs: signalling scissors in the tumour microenvironment. Nat Rev Cancer. 2008;8:929–941. doi: 10.1038/nrc2459. [DOI] [PubMed] [Google Scholar]
- 23.Duex JE, Sorkin A. RNA interference screen identifies Usp18 as a regulator of epidermal growth factor receptor synthesis. Mol Biol Cell. 2009;20:1833–1844. doi: 10.1091/mbc.E08-08-0880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bengoechea-Alonso MT, Ericsson J. A phosphorylation cascade controls the degradation of active SREBP1. J Biol Chem. 2009;284:5885–5895. doi: 10.1074/jbc.M807906200. [DOI] [PubMed] [Google Scholar]
- 25.Lee SM, Bae JH, Kim MJ, Lee HS, Lee MK, Chung BS, et al. Bcr-Abl-independent imatinib-resistant K562 cells show aberrant protein acetylation and increased sensitivity to histone deacetylase inhibitors. J Pharmacol Exp Ther. 2007;322:1084–1092. doi: 10.1124/jpet.107.124461. [DOI] [PubMed] [Google Scholar]
- 26.Goel A, Janknecht R. Acetylation-mediated transcriptional activation of the ETS protein ER81 by p300, P/CAF, and HER2/Neu. Mol Cell Biol. 2003;23:6243–6254. doi: 10.1128/MCB.23.17.6243-6254.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanchez-Arevalo Lobo VJ, Doni M, Verrecchia A, Sanulli S, Faga G, Piontini A, et al. Dual regulation of Myc by Abl. Oncogene. 2013;32:5261–5271. doi: 10.1038/onc.2012.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mertins P, Qiao JW, Patel J, Udeshi ND, Clauser KR, Mani DR, et al. Integrated proteomic analysis of post-translational modifications by serial enrichment. Nat Methods. 2013;10:634–637. doi: 10.1038/nmeth.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yokosuka T, Takamatsu M, Kobayashi-Imanishi W, Hashimoto-Tane A, Azuma M, Saito T. Programmed cell death 1 forms negative costimulatory microclusters that directly inhibit T cell receptor signaling by recruiting phosphatase SHP2. J Exp Med. 2012;209:1201–1217. doi: 10.1084/jem.20112741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fedorov O, Muller S, Knapp S. The (un)targeted cancer kinome. Nature chemical biology. 2010;6:166–169. doi: 10.1038/nchembio.297. [DOI] [PubMed] [Google Scholar]
- 31.Depontieu FR, Qian J, Zarling AL, McMiller TL, Salay TM, Norris A, et al. Identification of tumor-associated, MHC class II-restricted phosphopeptides as targets for immunotherapy. Proc Natl Acad Sci U S A. 2009;106:12073–12078. doi: 10.1073/pnas.0903852106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mohammed F, Cobbold M, Zarling AL, Salim M, Barrett-Wilt GA, Shabanowitz J, et al. Phosphorylation-dependent interaction between antigenic peptides and MHC class I: a molecular basis for the presentation of transformed self. Nat Immunol. 2008;9:1236–1243. doi: 10.1038/ni.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zarling AL, Polefrone JM, Evans AM, Mikesh LM, Shabanowitz J, Lewis ST, et al. Identification of class I MHC-associated phosphopeptides as targets for cancer immunotherapy. Proc Natl Acad Sci U S A. 2006;103:14889–14894. doi: 10.1073/pnas.0604045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zarling AL, Ficarro SB, White FM, Shabanowitz J, Hunt DF, Engelhard VH. Phosphorylated peptides are naturally processed and presented by major histocompatibility complex class I molecules in vivo. J Exp Med. 2000;192:1755–1762. doi: 10.1084/jem.192.12.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cobbold M, De La Pena H, Norris A, Polefrone JM, Qian J, English AM, et al. MHC class I-associated phosphopeptides are the targets of memory-like immunity in leukemia. Sci Transl Med. 2013;5:203ra125. doi: 10.1126/scitranslmed.3006061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rikova K, Guo A, Zeng Q, Possemato A, Yu J, Haack H, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 37.Machida K, Eschrich S, Li J, Bai Y, Koomen J, Mayer BJ, et al. Characterizing tyrosine phosphorylation signaling in lung cancer using SH2 profiling. PLoS One. 2010;5:e13470. doi: 10.1371/journal.pone.0013470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith MA, Hall R, Fisher K, Haake SM, Khalil F, Schabath MB, et al. Annotation of human cancers with EGFR signaling-associated protein complexes using proximity ligation assays. Sci Signal. 2015;8:ra4. doi: 10.1126/scisignal.2005906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bunse L, Schumacher T, Sahm F, Pusch S, Oezen I, Rauschenbach K, et al. Proximity ligation assay evaluates IDH1R132H presentation in gliomas. J Clin Invest. 2015;125:593–606. doi: 10.1172/JCI77780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tape CJ, Norrie IC, Worboys JD, Lim L, Lauffenburger DA, Jorgensen C. Cell-specific labeling enzymes for analysis of cell-cell communication in continuous co-culture. Mol Cell Proteomics. 2014;13:1866–1876. doi: 10.1074/mcp.O113.037119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gauthier NP, Soufi B, Walkowicz WE, Pedicord VA, Mavrakis KJ, Macek B, et al. Cell-selective labeling using amino acid precursors for proteomic studies of multicellular environments. Nat Methods. 2013;10:768–773. doi: 10.1038/nmeth.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 43.Liu X, Ory V, Chapman S, Yuan H, Albanese C, Kallakury B, et al. ROCK inhibitor and feeder cells induce the conditional reprogramming of epithelial cells. The American journal of pathology. 2012;180:599–607. doi: 10.1016/j.ajpath.2011.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suprynowicz FA, Upadhyay G, Krawczyk E, Kramer SC, Hebert JD, Liu X, et al. Conditionally reprogrammed cells represent a stem-like state of adult epithelial cells. Proc Natl Acad Sci U S A. 2012;109:20035–20040. doi: 10.1073/pnas.1213241109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yuan H, Myers S, Wang J, Zhou D, Woo JA, Kallakury B, et al. Use of reprogrammed cells to identify therapy for respiratory papillomatosis. N Engl J Med. 2012;367:1220–1227. doi: 10.1056/NEJMoa1203055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Winter GE, Rix U, Carlson SM, Gleixner KV, Grebien F, Gridling M, et al. Systems-pharmacology dissection of a drug synergy in imatinib-resistant CML. Nat Chem Biol. 2012;8:905–912. doi: 10.1038/nchembio.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Davis MI, Hunt JP, Herrgard S, Ciceri P, Wodicka LM, Pallares G, et al. Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol. 2011;29:1046–1051. doi: 10.1038/nbt.1990. [DOI] [PubMed] [Google Scholar]
- 48.Khan N, Jeffers M, Kumar S, Hackett C, Boldog F, Khramtsov N, et al. Determination of the class and isoform selectivity of small-molecule histone deacetylase inhibitors. Biochem J. 2008;409:581–589. doi: 10.1042/BJ20070779. [DOI] [PubMed] [Google Scholar]
- 49.Bradner JE, West N, Grachan ML, Greenberg EF, Haggarty SJ, Warnow T, et al. Chemical phylogenetics of histone deacetylases. Nat Chem Biol. 2010;6:238–243. doi: 10.1038/nchembio.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wahlberg E, Karlberg T, Kouznetsova E, Markova N, Macchiarulo A, Thorsell AG, et al. Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors. Nat Biotechnol. 2012;30:283–288. doi: 10.1038/nbt.2121. [DOI] [PubMed] [Google Scholar]
- 51.Li J, Rix U, Fang B, Bai Y, Edwards A, Colinge J, et al. A chemical and phosphoproteomic characterization of dasatinib action in lung cancer. Nat Chem Biol. 2010;6:291–299. doi: 10.1038/nchembio.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rix U, Superti-Furga G. Target profiling of small molecules by chemical proteomics. Nat Chem Biol. 2009;5:616–624. doi: 10.1038/nchembio.216. [DOI] [PubMed] [Google Scholar]
- 53.Gridling M, Ficarro S, Breitwieser FP, Song L, Parapatics K, Colinge J, et al. Identification of kinase inhibitor targets in the lung cancer microenvironment by chemical and phosphoproteomics. Mol Cancer Ther. 2014;13:2751–2762. doi: 10.1158/1535-7163.MCT-14-0152. [DOI] [PMC free article] [PubMed] [Google Scholar]