Abstract
Background
Tremendous progress has been made in the past 20 years in understanding the roles played by immunophilins, and in particular the cyclophilins, in supporting the replication cycles of human viruses. A growing body of genetic and biochemical evidence and data from clinical trials confirm that cyclophilins are essential cofactors that contribute to establishing a permissive environment within the host cell that supports the replication of HIV-1 and HCV. Cyclophilin A regulates HIV-1 replication kinetics and infectivity, modulates sensitivity to host restriction factors, and cooperates in the transit of the pre-integration complex into the nucleus of infected cells. Cyclophilin A is an essential cofactor whose expression supports HCV-specific RNA replication in human hepatocytes.
General Significance
Peptidyl-prolyl isomerase inhibitors have been used in clinical trials to validate cyclophilins as antiviral targets for the treatment of HIV-1 and Chronic Hepatitis C virus infection and as molecular probes to identify the roles played by immunophilins in supporting the replication cycles of human viruses.
Scope of Review
This review summarizes emerging research that defines the functions of immunophilins in supporting the replication cycles of HIV-1, HCV, HBV, coronaviruses, and other viral pathogens and describes new information that suggests a role for immunophilins in regulating innate immune responses against chronic viral infection.
Major Conclusions
The dependence on cyclophilins by evolutionarily distinct viruses for accomplishing various steps in replication such as viral entry, initiation of genomic nucleic acid replication, viral genome uncoating, nuclear import and nuclear entry, emphasizes the potential of cyclophilin inhibitors as therapeutic agents. This article is part of a Special Issue entitled Proline-directed Foldases: Cell Signaling Catalysts and Drug Targets.
Abbreviations: ATP, Adenosine triphosphate; PPIase, peptidyl-prolyl isomerase; HIV-1, human immunodeficiency virus type 1; HCV, hepatitis C virus; HBV, hepatitis B virus; AIDS, acquired immunodeficiency syndrome; N-MLV, N-tropic murine leukemia virus; Ref-1, restriction factor 1; TRIM5, Tripartite motif-containing protein 5; Lv-1, lentivirus susceptibility factor 1; NPC, nuclear pore complex; TRN-SR2, transportin SR2; TNPO3, transportin 3; CPSF6, cleavage and polyadenylation specific factor 6
Keywords: Immunophilins, Cyclophilins, Viruses, HIV-1, HCV, Cyclophilin inhibitors
Highlights
-
•
The role of cyclophilin A in HIV-1 replication
-
•
The importance of cyclophilins in HCV replication
-
•
The broad-spectrum of cyclophilin inhibitors as antiviral agents
-
•
Development of resistance to cyclophilin inhibitors
-
•
Innate Response
1. Introduction
The regulatory approvals of cyclosporine A (Neoral, Sandimmune, Gengraf) in 1983 and FK506 (Tacrolimus, Prograf, Advagraf) in 1994 have unquestionably revolutionized the field of solid organ and tissue transplantation. The elucidation of the biological targets and mechanisms-of-action for cyclosporine A and FK506 led directly to the discovery of two multi-enzyme families that exhibit unique catalytic activities. The cyclophilins and the FK Binding Proteins share little sequence homology; however, they are both capable of catalyzing the interconversion of the two energetically preferred conformers (cis and trans) of the planar peptide bond preceding an internal proline residue. Each family of enzymes, referred to as peptidyl prolyl cis/trans isomerases or PPIases, catalyzes a reversible peptide bond isomerization in a non-covalent reaction that does not require the consumption of ATP but rather depends on energy derived from protein substrates. Collectively the cyclophilins and FK binding proteins are now referred to as immunophilins by virtue of their ability to bind these highly immunosuppressive agents. In contrast, the third and most recently discovered class of human PPIase enzymes, the parvulins, is not sensitive to inhibition by either cyclosporine A or FK506. Parvulins, originally described as a novel PPIase activity isolated from Escherichia coli, were shown to be present in higher organisms by their sequence homology to the yeast protein Ess 1 leading in turn to its own identification as a PPIase. The corresponding human ortholog, Pin 1, was identified in a two-hybrid assay as the Protein Interacting with NIMA and was later confirmed to be a PPIase by its ability to complement yeast Ess 1 deletion mutants. Unlike the cyclophilins and FK binding proteins, which are present in the human genome as multi-gene families and express multiple isoforms, parvulin-type PPIases are expressed from only two human genes. Immunophilins and parvulins are ubiquitously distributed throughout nature and have been identified in all organisms examined to date including bacteria, fungi, animals and plants.
Cyclosporine A and FK506 suppress immune function by binding to their respective intracellular targets, cyclophilin A and FK binding protein (FKBP). In a remarkable example of convergent evolution, the binary complexes formed by cyclosporine A/cyclophilin A and FK506/FKBP form high affinity ternary complexes with calcineurin. These ternary complexes block the intrinsic phosphatase activity of calcineurin, which in turn inhibits the de-phosphorylation driven nuclear translocation of members of the nuclear factor of activated T-cells (NF-AT) family of transcription factors. Inhibiting the nuclear translocation of NF-AT reduces the expression of many lymphokines including interleukin-4 (IL-4), which is required for B-cell help and interleukin-2 (IL-2), which is required for T-cell expansion and proliferation. The profound immunosuppressive activity exerted by cyclosporine A and FK506 has provided the rationale to investigate the use of these agents in a wide variety of conditions which are characterized by uncontrolled hyper-inflammatory responses including autoimmune diseases (muscular dystrophy, Crohn's disease, rheumatoid arthritis), ischemic reperfusion injury following myocardial infarction or stroke, ocular disease (uveitis, xerophthalmia), and dermatologic conditions (atopic dermatitis, severe recalcitrant plaque type psoriasis).
In general the therapeutic effects of cyclosporine A and FK506 for use in their primary indications (the prevention of allograft rejection), secondary indications (atopic dermatitis, xeropthalmia), as well as for investigational uses has been attributed to their ability to bind to their cognate immunophilins and to inhibit calcineurin phosphatase activity. Although a variety of methods have been developed in recent years to measure protein binding to proline rich regions including yeast two hybrid analysis, GST pull down, fluorescence anisotropy, circular dichroism, and nuclear magnetic resonance, these techniques have had limited success in identifying additional binding targets and therefore the roles played by immunophilins in the pathophysiology of other hyper-inflammatory or autoimmune conditions. The natural substrates for PPIases have been difficult if not impossible to identify because they typically exhibit binding constants in the micro-molar range easily escaping detection in protein-based capture techniques. The quantitative characterization of isomerization in native proteins represents a second major difficulty that has been successful only when applied to highly stable proteins such as RNase A and bovine pancreatic trypsin inhibitor. Assay methods have therefore relied on measuring proline isomerization in non-physiological substrates comprised of short chemically modified peptides. Because PPIases catalyze both the forward and reverse isomerization reactions protease coupling techniques have been employed to trap reaction “products” in order to assess unidirectional catalysis.
In contrast to autoimmune diseases and hyper-inflammatory conditions, tremendous progress has been made in the past 20 years in understanding the roles played by immnophilins, and in particular the cyclophilins, in supporting the replication cycles of human viruses. A growing body of genetic and biochemical evidence and data from clinical trials now confirms that cyclophilins are essential host cofactors that contribute to establishing a permissive environment within the host cell that supports the infectivity and replication of HIV-1 and HCV. Pull down experiments have been used to identify specific cyclophilin binding proteins from these viruses including the p24 capsid protein expressed by HIV-1 and the non-structural protein expressed by HCV — NS5A. The relatively greater binding affinity demonstrated by these virally-encoded proteins almost certainly reflects an evolutionary selective advantage on the part of the pathogen allowing it to outcompete the host for the recruitment of critical proteins. To some extent these observations constitute indirect evidence that cyclophilins are indeed essential cofactors necessary to support viral replication. Clinical proof of concept has been demonstrated using second-generation non-immuno suppressive analogs of cyclosporine A confirming that cyclophilins are valid biochemical targets for developing therapeutics for treating patients who are chronically infected with either HIV-1 or HCV. Based in part on these results sensitivity to cyclosporine A in vitro is now used as a surrogate marker to infer that cyclophilins play an obligate role in supporting viral replication. In vitro sensitivity to cyclosporine A has been observed for HBV, coronaviruses, influenza, cytomegalovirus and human papilloma virus.
This review will focus on summarizing the body of research that establishes the roles played by immunophilins in supporting HIV-1 and HCV infectivity and replication. In addition, emerging data describing the potential roles played by immunophilins in supporting the replication of other human viruses will be discussed together with new information suggesting that immunophilins may play a role in regulating innate immune responses against chronic viral infection.
2. HIV-1
During the 1980's when the pathogenesis of HIV-1 infection was poorly understood it was suggested that AIDS was characterized by a phase of disease progression whereby various types of activated lymphocytes participated in the destruction of healthy as well as HIV-1 infected cells drawing analogies between chronic HIV-1 infection and autoimmune cytopenias such as aplastic anemia. This concept of disease progression led many to speculate that the loss of CD4 + lymphocytes in HIV-1 infected individuals could be mitigated by treatment with immunosuppressive therapy. The recent successes of cyclosporine and FK506 in preventing allograft rejection was largely attributed to their ability to inhibit interleukin-2 dependent T-cell activation and proliferation — a step that was also recognized as an important aspect of HIV-1 replication. These observations prompted a pilot evaluation of cyclosporine in patients with advanced HIV-1 related disease [1]. Eight patients, all with evidence of either Pneumocystis carinii pneumonia or Kaposi's sarcoma, received cyclosporine at a total daily dose of 7.5 mg/kg given as a divided dose every 12 h. Doses were adjusted in order to maintain trough plasma concentrations of 100 to 150 ng/mL, which was the typical regimen for transplant recipients. Upon initiation of cyclosporine treatment all patients exhibited clinical signs (nausea, vomiting, fatigue) and laboratory evidence (declines in CD4 + and CD8 + lymphocytes and platelets) of accelerated disease progression. Paradoxically treatment with cyclosporine increased the efficiency with which virus was isolated from all patients. Cessation of treatment led to resolution of all exacerbated symptoms. The researchers concluded that cyclosporine-based immunosuppressive therapy in AIDS patients is not warranted. Nonetheless this work provided the impetus for further evaluations using cyclosporine as a mechanistic probe into the role of cyclophilins in HIV-1 infection and replication.
Initial reports of the in vitro antiviral activity of cyclosporine and FK506 described the ability of each compound to decrease the production of infectious HIV-1 by chronically infected T cells; however, no mechanistic data accompanied these reports [2]. Although the inhibition of the PPIase activities of cyclophilin and FKBP could not be excluded as accounting for the observed antiviral effects, it was speculated that T-cell activation through the calcineurin NF–AT pathway could potentially be exploited as a target for the discovery of anti-HIV-1 therapeutics. The application of yeast two-hybrid techniques revealed that two host proteins, cyclophilins A and B, bound specifically to the Gag polyprotein, Pr55gag, and to p24 albeit with differing affinities [3]. These results were confirmed using GST-cyclophilin fusion constructs. The binding of HIV-1 Pr55gag and p24 capsid to cyclophilins A and B was inhibited by cyclosporine suggesting that the PPIase active site of both immunophilins was involved in binding viral proteins. The ability of GST-cyclophilin fusion constructs to capture calcineurin was dependent on the presence of cyclosporine; however, these same constructs were not capable of binding Pr55gag providing the first indirect evidence that the antiviral activity of cyclosporine could be manifested without the involvement of calcineurin through a cyclophilin-dependent pathway. The authors stated that further independence of the antiviral effect from the calcineurin NF-AT pathway could be addressed through the use of MeAla6-CsA, an analog that binds cyclophilin but does not suppress T-cell activation. Using GST-cyp fusion constructs, expression vectors containing only Pr55gag, and the analysis of purified virions it was demonstrated that cyclophilin A is the only immunophilin that HIV-1 specifically incorporates into virions [4], [5]. No evidence was found for the incorporation of cyclophilin B or FKBP12 into virions. Virions produced by cloned SIV or HIV-2 proviruses did not incorporate cyclophilin A, consistent with the specificity of Gag protein species to bind cyclophilin A in vitro. Accordingly cyclosporine does not inhibit the replication of these viruses. Cyclophilin binding within Pr55gag was localized to a proline rich region within which P222 was identified as a critical residue. Viruses harboring the P222A substitution did not incorporate cyclophilin A and exhibited decreased replication indicating that HIV-1 infectivity requires the formation of the Pr55gag-cyclophilin A complex. Reports of the antiviral activity of NIM811, a 4-substituted analog of cyclosporine that retains cyclophilin binding but does not form the ternary immunosuppressive complex with calcineurin, confirmed that immunosuppression through the T-cell activation pathway does not account for the anti-HIV-1 activity of cyclosporine A and its analogs [6]. NIM811 did not inhibit enzymatic activity of non-structural proteins including reverse transcriptase, integrase or protease nor did it inhibit the interaction of Rev with the Rev responsive element. NIM811, and by inference cyclophilin A, was likely involved in an early step in the viral replication cycle occurring immediately post entry. These studies also demonstrated that FK506 did not exert anti-HIV-1 activity confirming that FK binding proteins were not involved in HIV-1 replication. Cyclophilin A binds multiple virally encoded proteins including p17, Nef, Vif, and gp120 [7]. Only the interaction with Pr55gag/p24 is essential for HIV-1 replication — disruption of which accounts for the antiviral activity of cyclosporine. Cyclosporine itself does not interact directly with viral proteins therefore it is not an antiviral drug per se, but it prevents recruitment of host cyclophilin A, which is a critical host cofactor. The authors speculated that cyclophilin A could play a role in the translocation of the pre-integration complex that could directly impact the production of infectious virus. X-ray crystallographic studies identified the solvent exposed loop, comprising residues 85 through 93 of p24, as the primary binding domain for cyclophilin A with proline 90 in the trans conformation [8]. The results suggested that cyclophilin A may facilitate the disassembly of the capsid core of incoming viruses (by structural perturbation of capsid around P90) and transporting the core through the cytoplasm. Additional crystallographic studies confirmed these results and demonstrated that the G89-P90 bond was the primary recognition site for cyclophilin A within the exposed loop with residues 85, 86, 87, 88, and 93 constituting the remainder of the recognition motif [9]. Biochemical studies using well characterized active site mutants of cyclophilin A demonstrated that the exposed loop of p24 interacts with the active site of cyclophilin A [10], [11]. The functional significance of the exposed loop was demonstrated by transferring HIV-1 capsid residues 86–93 to SIVmac thus conferring virion incorporation of cyclophilin A and sensitivity to non-immunosuppressive analogs of cyclosporine A [12]. Transfer of only residues 86–90 conferred cyclophilin A incorporation; however, the presence of cyclosporine A was necessary for efficient viral replication. These results suggested that the presence or absence of a type II tight turn adjacent to the primary cyclophilin A binding site in capsid determines whether cyclophilin A enhances or inhibits viral replication possibly by destabilizing capsid and facilitating uncoating. HIV-1 replication was reduced in cyclophilin A PPIase−/− cells and no further reduction in replication was observed in the presence of cyclosporine A demonstrating that no other cyclophilin was involved in supporting HIV-1 replication [13]. Replication was rescued by re-expression of cyclophilin A. Virions produced from PPIase−/− cells were less infectious than virions produced from wild type cells indicating that cyclophilin A regulates infectivity of HIV-1. Substitution of the cyclophilin A binding region of capsid in an HIV-1 based lentivirus vector with the corresponding region from macrophage tropic HIV-1 Ba-L was resistant to restriction in simian cells and efficiently transduced those cells [14]. The chimeric virus did not require cyclophilin A for early post-entry steps indicating that the cyclophilin A binding region of capsid has a viral specific determinant involved in post entry restriction of HIV-1 based lentivirus vectors. NMR exchange spectroscopy was used to demonstrate that G89-P90 bond in capsid is bound by cyclophilin A in the trans conformation and that the binding event exerts distal effects on other regions of capsid [15]. Although no evidence for catalysis was found, the catalytically inactive R55A mutant of cyclophilin A bound less tightly to capsid, leading to lower incorporation into virions and less infectivity; indirectly suggesting that isomerization may enhance infectivity by contributing to both of these processes. Evidence that cyclophilin A mediates host cell restriction was demonstrated by showing that the enhancing effects of cyclosporine on HIV-1 infection of Owl Monkey cells was observed only when cyclosporine was incubated with target cells and occurred regardless of whether virions were produced in the presence of cyclosporine [16]. These results clearly indicated that the functionally important interaction between capsid and cyclophilin A occurred in the Owl Monkey target cell and was completely independent of effects in the virus producer cell. The Lv-1 mediated restriction of HIV-1 replication in Owl Monkey cells was relieved by exposure to cyclosporine whereas exposure of human cells to cyclosporine (or mutation of the cyclophilin binding site in the exposed loop of capsid) enhanced recognition and restriction to viral replication by Ref-1. These results suggested that interactions between HIV-1 capsid and cyclophilin A contribute to species-specific restriction of HIV-1 infection and that in humans HIV-1 recruits cyclophilin A in order to evade restriction by Ref-1. Viral capsid was also shown to be an important determinant in human restriction of N-MLV infection by TRIM5a soon after viral entry into the host cell [17]. The discovery that Owl monkeys encode a TRIM5-cyclophilin A fusion protein provided an immediate explanation for the potent restriction of HIV-1 infection in Owl monkeys which could be overcome by the presence of cyclosporine [18], [19]. The retro-transposed cyclophilin A domain in the TRIM5a-cyclophilin fusion protein could represent a type of adaptive molecular targeting whereby functional restriction by TRIM5a was directed specifically to the incoming viral capsids through recognition by the fused cyclophilin domain. Independent retro-transposition of a fused cyclophilin A domain to TRIM5 was observed in some strains of rhesus macaques where the fusion protein restricts infection by HIV-2 providing evidence that the rhesus and owl monkey proteins have been subjected to diverse selective pressures by different pathogenic viruses [20]. For some variants restriction by wild-type TRIM5 depends on the independent expression of cyclophilin A [21]. The anti-HIV-1 activity of DEB025, a 3,4 di-substituted analog of cyclosporine that lacks immunosuppressive properties, was shown to correlate with its PPIase inhibition in vitro [22]. Viral isolates with naturally occurring resistance to DEB025 were found. Resistance was correlated to the presence of multiple substitutions in capsid including V86P, H87Q/P, I91V, and M96I, which are all located in the cyclophilin A binding domain [23]. Although capable of binding cyclophilin A, reverse genetics demonstrated that viruses harboring these changes, were cyclophilin A independent. These viruses were now also capable of infecting normally restrictive Owl monkey cells indicating that these capsid substitutions escaped restriction by the TRIM5-cyclophilin fusion protein.
siRNA screens revealed that expression of karyopherin β transportin-3 (TNPO3) and nucleopore protein complex (NPC) component nucleoporin 153 (NUP153) are important for the infection of non-dividing cells by HIV-1 by allowing passage of the pre-integration complex through the NPC. Depletion of NUP153 did not affect infection by wild-type HIV-1 either in the presence of cyclosporine A or in cells depleted of cyclophilin A. Replication of two well characterized capsid mutations N74D and P90A were also unaffected by NUP153 depletion, suggesting that motifs within capsid determine the utilization of NUP153 and the pathway for importation of the pre-integration complex into the nucleus [24]. Nuclear entry of the pre-integration complex was also shown to utilize NUP358/RanBP2 [25], [26]. Isothermal calorimetry was used to demonstrate the direct binding of capsid to the cytoplasmic C-terminal cyclophilin homology domain of NUP358/RanBP2. Interestingly the complex was not sensitive to inhibition by cyclosporine. Engaging NUP358/RanBP2 leads to nuclear entry through a pathway involving NUP153 that confers optimal HIV-1 replication by targeting preferred regions within the genome for integration. In the presence of cyclosporine or with viruses containing capsid mutants impaired for cyclophilin binding replication proceeds through NUP358/RanBP2 independent pathways and is less efficient. In vitro studies indicate that transportin SR2 (TRN-SR2)/ transportin 3 (TNPO3) facilitates infection through a coordinate action with cyclophilin A [27]. Cyclophilin A acts at an early post-entry point to stabilize incoming capsid possibly in concert with other positive regulators of capsid stability including PDZ Domain Containing Protein 8 (PDZD8) [28]. During transport to the nucleus, the capsid core encounters cytoplasmic TNPO3 that participates in engaging the nuclear pore complex and accelerating uncoating therefore the authors postulate that cyclophilin A and TNPO3 act as positive and negative regulators of the uncoating process. X-ray crystallographic techniques were used to identify a unique binding domain in the N-terminal region of capsid that is recognized by cleavage and polyadenylation specific factor 6 (CPSF6) [29]. Interestingly the N74D mutation in capsid that confers independence from the TNPO3 and NUP358/RanBP2 nuclear entry pathway is no longer capable of binding CPSF6. These findings suggest a model whereby full length CPSF6 recognizes and binds to a region N-terminal to the cyclophilin A binding domain of capsid. This binding event then facilitates the recruitment of TNPO3, NUP358/RanBP2 and NUP153 allowing the efficient nuclear translocation of the pre-integration complex. In this context CPSF6 represents a host factor that facilitates the infection of non-dividing cells. Further evidence for the tight coupling of this process was suggested by the finding that the cyclophilin homology domain of NUP358/RanBP2 is an active isomerase implicating a role for PPIase catalysis and capsid uncoating in efficient nuclear entry [30].
By providing Vpx-mediated relief of restriction of HIV-1 replication it was demonstrated that HIV-1 infection induces dendritic cell maturation, an antiviral type 1 interferon response, and activation of T cells [31]. Although not engaged under normal conditions the innate response, which is dependent on the interaction of newly synthesized capsid with cyclophilin A, may function as a type 1 interferon inducing pathogen associated molecular pattern. Macrophages were shown to be capable of detecting HIV-1 infection and activating an antiviral innate immune signal when specific interactions with HIV-1 cofactors were suppressed or inhibited by mutation, gene silencing, or by pharmacological intervention [32], [33]. These results suggest a mechanism whereby interactions between capsid and CPSF6/cyclophilin A evade detection of viral replication by pathogen associated molecular pattern receptors and promote infection. Human MxB protein associates with cyclophilin A and restricts HIV-1 infection in a cyclophilin A dependent manner [34]. Escape from MxB restriction was associated with an alanine substitution at position 88 in capsid in the cyclophilin A binding domain.
Finally, clinical isolates from various subtypes of HIV-1 exhibit resistance to the non-immunosuppressive cyclosporine analog DEB025 [35]. The majority of these naturally occurring polymorphisms are located within the cyclophilin binding domain of capsid; many of which are also associated with impaired progression of the capsid core through the cytoplasm, incomplete reverse transcription, or lack of transport of the pre-integration complex through the nuclear pore. Although the understanding of the precise mechanisms are yet to be fully understood, it now seems clear that cyclophilin A interacts with the major determinants within capsid for recognition by multiple cellular factors that govern capsid core stability and uncoating, evasion of host restriction factors and masking of pathogen associated molecular pattern receptors, efficient nuclear transport of the pre-integration complex, and optimal integration of viral DNA into the host genome. Irrespective of the exact mechanistic considerations, the binding of capsid represents a highly conserved function for cyclophilin A in the replication cycle of a number of retroviruses and in particular HIV-1.
3. HCV
Over 25 years ago it was reported that the intracellular ligand for cyclosporine A was involved in the replication of non-A non-B hepatitis virus [36]. Those studies demonstrated that 28 days of intravenous administration of cyclosporine A at a dose of 20 mg/kg/day to chronically infected chimpanzees was associated with improvement in liver histometric scores. The authors concluded that cyclosporine A inhibited the proliferation of non-A non-B hepatitis virus albeit through an unknown mechanism of action. The authors speculated that if it were possible to separate the immunosuppressive properties of cyclosporine A from its antiviral activity then “there may be some possibilities of a clinical application of cyclosporine A to certain viral diseases”.
The involvement of immunophilins in the replication cycle of HCV was demonstrated by multiple groups in reports that described the in vitro anti-viral effects of cyclosporine A [37], [38], [39], [40]. The differential effects exerted by cyclosporine A and FK506 on HCV-specific RNA replication implicated cyclophilins, and not FK Binding Proteins, as the only class of immunophilins whose expression was necessary in order to support viral replication [41]. Consistent with this observation, broad knockdown of cyclophilin expression suppressed HCV replication indicating that cyclophilins were the primary target for the anti-viral activity of cyclosporine A. Several groups reported conflicting results concerning the identity of the specific cyclophilin isoform that supported HCV replication; however, these studies clearly established that the anti-HCV activity of cyclosporine A was mediated through its cyclophilin binding activity and not through its P-glycoprotein binding activity or through activation of the NFAT pathway [40], [42], [43]. Using stable knockdown together with specific re-expression and antibody neutralization techniques cyclophilin A was identified as the principal cyclophilin required to support HCV replication [44], [45], [46]. Furthermore, re-expression of active site mutants of cyclophilin A without catalytic activity did not rescue replication in cyclophilin A knock down cells indicating an essential role for PPIase catalysis in HCV replication. These results were confirmed using transgenic mice expressing CD81 and occludin where knockdown of cyclophilin A expression markedly inhibited HCV-specific RNA replication providing the first evidence that cyclophilin A is a genuine cofactor that supports HCV replication in vivo [47].
Intensive structure activity studies for a series of non-immunosuppressive analogs of cyclosporine A established a correlation between inhibition of the PPIase catalytic activity of cyclophilin A and inhibition of HCV-specific RNA replication in a genotype 1b replicon [48]. These studies led to the identification of several analogs including NIM811 [39], [48], DEB025/Alisporivir [49] and SCY-635 [50] as potential candidates for clinical evaluation. Clinical anti-HCV activity was demonstrated for DEB025/Alisporivir as monotherapy in patients who were co-infected with chronic hepatitis C and HIV-1 [51] and for SCY-635 monotherapy in patients with chronic hepatitis C infection [52]. The administration of each drug was associated with rapid and profound reductions in HCV-specific plasma RNA in the absence of dose-limiting toxicities and clearly demonstrated that cyclophilins are valid targets for chemotherapeutic intervention in patients with chronic hepatitis C infection. The clinical development of both compounds has been extensively reviewed [53], [54].
Despite the clinical success of the non-immunosuppressive cyclophilin inhibitors, the exact function of cyclophilin A in the replication cycle of HCV and the antiviral mechanism of cyclosporine A and its non-immunosuppressive analogs is not fully understood. Identifying a discrete role for cyclophilin A has been hampered by the fact that replication is inseparably linked to particle assembly in the HCV replication cycle. As a result, the analysis of mutations associated with decreased sensitivity to cyclosporine A and its non-immunosuppressive analogs has been used to identify biological targets and to infer potential mechanistic roles for cyclophilin A [55], [56], [57], [58]. Several possible functions have been suggested including stabilization of the replication complex [59], modifying the RNA binding capabilities of non-structural protein 5A (NS5A) [60], [61], modifying the catalytic function of the non-structural protein 5B (NS5B) the RNA-dependent RNA polymerase [62], and affecting the processing of the polyprotein [46]. Among these possibilities a growing body of experimental evidence indicates that the NS5A protein is the primary biological target for cyclophilin A. In contrast to HIV-1 where the G89-P90 bond in capsid protein represents the primary binding determinant for cyclophilin A, several groups have shown that multiple proline residues located in Domains II and III of the HCV NS5A protein serve as binding determinants for cyclophilin A and as substrates for its PPIase catalytic activity [63], [64], [65], [66], [67]. Cyclosporine A, its non-immunosuppressive analogs, sanglifehrins and their analogs are all able to disrupt the stable complex formed between NS5A and cyclophilin A; the dissociation of which is associated with the inhibition of HCV replication [52], [68], [69]. Multiple mutations within Domain II of NS5A are required in order to confer resistance to cyclosporine A or DEB025/Alisporivir with D320E being identified as the principal mutation associated with resistance [64], [65], [70], [71]. Mutant NS5A proteins containing the D320E substitution retain their ability to bind cyclophilin A; however, those viruses now replicate in a cyclophilin-independent manner. Introduction of the D320E and Y321N mutations into NS5A restores the ability of mutant viruses harboring these changes in NS5A to replicate in cyclophilin A knockdown cells possibly by introducing conformational flexibility within Domains II and III that reduces the need for cyclophilin A-dependent isomerization within these regions of the protein.
In vitro mechanistic studies with the non-immunosuppressive cyclophilin inhibitor NIM811 indicate that exposure to the drug causes altered trafficking within the cell as evidenced by the accumulation of neutral lipids into lipid droplets and decreased apo B secretion through the VLDL pathway [72]. These studies suggested that cyclophilin A may be important factor for particle secretion since virion assembly and particle release is highly dependent on the integrity of the VLDL pathway. Increasing attention has been focused on the observation that HCV-specific RNA replication occurs in close association with intracellular membranes that form complex membranous replication factories also referred to as the membranous web [73]. Emerging results have now described a coordinate function for the cyclophilin A-NS5A complex in the HCV replication cycle [74], [75]. These reports indicate that the cyclophilin A-NS5A complex does not play a role in RNA synthesis after the formation of the replication complex, but rather at an early step preceding its formation. Cyclophilin inhibitors, including cyclosporine A, DEB025/Alisporivir, and NIM811, suppress the de novo formation of double membrane vesicles, which are highly characteristic structures of hepaciviruses that are induced upon expression of the HCV polyprotein. These structures emanate from the endoplasmic reticulum and contain enzymatically active replicase. Expression of NS5A alone promoted formation of double membrane vesicles; whereas, expression of NS5A in the presence of DEB025/Alisporivir or NIM811 suppressed their formation suggesting that the cyclophilin A-NS5A complex itself governs the formation of these structures. Viruses harboring NS5A containing both the D320E and Y321N mutations were able to generate double membrane vesicles in cyclophilin A knockdown cells consistent with prior observations indicating that these substitutions in NS5A obviate the need for cyclophilin A binding and PPIase catalysis leading to a cyclophilin A independent phenotype. Taken together these studies indicate that cyclophilin A, in concert with NS5A, plays a highly specific and potentially singular role in the replication cycle of HCV by regulating the biogenesis of membranous web formation.
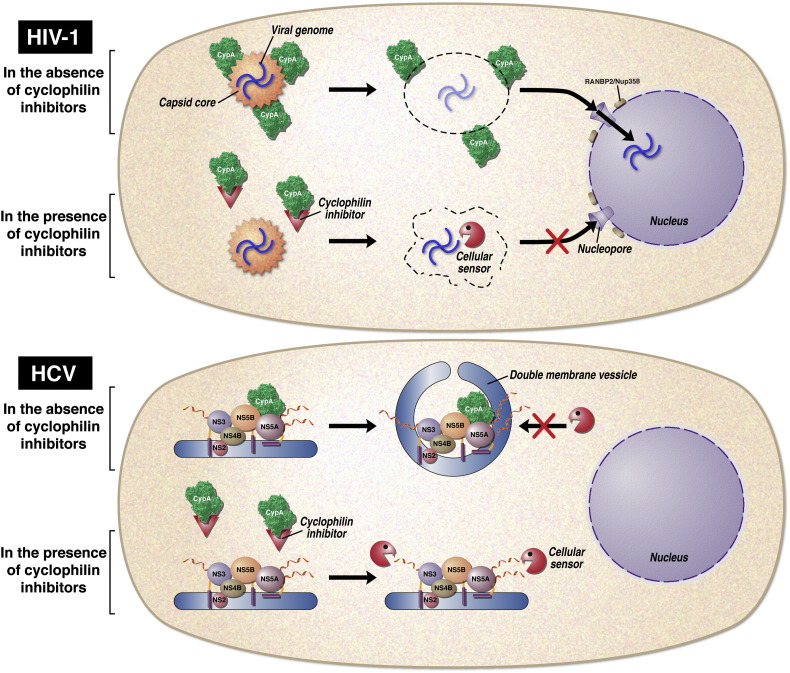
There is therefore a close resemblance between the role played by cyclophilin A in HIV-1 and HCV infection. Cyclophilin A apparently plays a protective role in both virus life cycles against innate cell sensors. For HIV-1, cyclophilin A, by binding to capsid, stabilizes the integrity of the capsid core and protects the viral genome from innate cell sensor recognition during its transport to the nucleus where it migrates through the nucleopore and ultimately integrates into host chromosomes (Fig. 1 , top model). For HCV, cyclophilin A, by binding to NS5A, triggers the formation of a double membrane vesicle, which serves as a compartment where viral RNA replication occurs sheltered from innate cell sensors.
Fig. 1.
Models for the respective roles of cyclophilin A in HIV-1 (top model) and HCV infection (bottom model). HIV-1: Immediately after viral entry and delivery of the viral capsid core into the cytosol of the target cell, cyclophilin A binds to the capsid core that surrounds the viral genome and preserves the stability of the core during its transport to the nucleus. After docking to the nucleopore via specific components of the nucleopore complex such as Nup358/RANBP2 and Nup153, the viral genome undergoes the so-called core uncoating allowing the passage of the viral genome through the nucleopore and its integration into the host DNA. Cyclophilin inhibitors by binding to the enzymatic pocket of cyclophilin A prevent its binding to the incoming capsid core. In the absence of cyclophilin A, the capsid core is destabilized and brakes apart allowing the recognition of the viral genome by cellular sensors resulting in its degradation. HCV: After entry, the viral RNA is decapsidated and used both for polyprotein translation and replication in the cytoplasm. Translation occurs at rough endoplasmic reticulum (ER) and produces single polyprotein, which is then cleaved by cellular and viral proteases, to produce structural and nonstructural proteins (NS). Replication and post-translational processing take place in a membranous web made of NS and host proteins located at the perinuclear membrane. Cyclophilin A by binding to the membrane-anchored NS5A protein triggers the creation of double membrane vesicles (DMVs). In this new membranous compartment, HCV RNA occurs in a protected manner. In the presence of cyclophilin inhibitors, cyclophilin A is unable to bind to NS5A and to mediate DMVs. Unprotected viral RNA and proteins are now exposed to cellular sensors and degradation.
4. Other viruses
Although the role of cyclophilins has been extensively studies for HIV-1 and HCV, they have been shown to participate in the life cycle of other viruses. Table 1 summarizes the participation of cyclophilin members in various virus life cycles. It has been reported that the neutralization of cyclophilins decreases HBV replication. Specifically, either the addition of cyclophilin inhibitors – cyclosporine A, alisporivir and the sanglifehrin derivate NVP018 – to hepatocytes or knocking down the expression of cyclophilin members – cyclophilin A, C and D – significantly inhibit HBV replication [76], [77], [78]. How cyclophilins assist HBV remains to be understood, although a study proposed an interaction between cyclophilin A and the HBV surface antigen [76]. Recent reports suggest that cyclophilins also play an important role in the replication of coronaviruses. Specifically, the presence of cyclophilin inhibitors – cyclosporin D, alisporivir and NIM811 – as well as the suppression of cyclophilin A expression inhibit human coronavirus HCoV-NL63 replication [79]. Moreover, low non-cytotoxic concentrations of cyclosporin A reduced the replication of the severe acute respiratory syndrome coronavirus (SARS-CoV) as well the human coronavirus 229E [80]. As for HBV, the role of cyclophilin A in coronavirus infection remains to be identified. Similarly, silencing expression of cyclophilin A or adding cyclosporine A blocks the replication of human cytomegalovirus [81], [82], whereas silencing expression of cyclophilin B or adding cyclosporine A blocks Japanese encephalitis virus (JEV) replication [83]. Cyclophilin B has also been shown to enhance human papillomavirus (HPV) infectivity at the viral internalization step as well as at another step downstream [84]. The neutralization of cyclophilin A with cyclosporine A or NIM811 inhibits the replication of a serotype of vesicular stomatitis virus (VSV) [85]. Conflicting results have been described for a role of cyclophilin A in influenza A virus infection. Indeed, in one study, Liu et al. showed that knocking down cyclophilin A expression enhances influenza virus replication through protecting the M1 protein from degradation [86], and in another study, they reported that cyclosporine A blocks influenza virus replication [87]. Similar conflicting results have been reported for another flavivirus, the Dengue virus. One study showed that silencing the expression of cyclophilin A in hepatoma cells or the addition of cyclosporine A inhibits Dengue replication [88]. However, two independent studies demonstrated that Dengue infects equally parental and cyclophilin A knockdown hepatoma cells, suggesting that Dengue does not rely on cyclophilin A to replicate [45], [46].
Table 1.
Cyclophilin members participating in the replication of divergent viruses.
| Cyclophilins | Viruses | Virus families |
|---|---|---|
| Cyclophilin A | HIV-1 | Retroviridae |
| Cyclophilin A, B, D, H and 40 | HCV | Flaviviridae |
| Cyclophilin A, B, D, G and H | CoV | Coronaviridae |
| Cyclophilin A, C and D | HBV | Hepadnaviridae |
| Cyclophilin B | HPV-16 | Papillomaviridae |
| Cyclophilin A | CMV | Herpesviridae |
| Cyclophilin A and B | Influenza A | Orthomyxoviridae |
| Cyclophilin B | JEV | Flaviviridae |
| Cyclophilin A, B and C | Dengue | Flaviviridae |
| Cyclophilin A and B | YFV | Flaviviridae |
| Cyclophilin A and E | VSV | Rhabdoviridae |
Altogether these studies demonstrate that members of the cyclophilin family can control the infection and replication of highly divergent viruses including current prime threats such as HIV-1, HCV and HBV. The dependence on cyclophilins of distinct viruses for various steps of replication such as viral entry, formation of protective membranous compartments, RNA synthesis and multiplication, viral genome uncoating, nuclear import and nuclear entry, emphasizes the potential of cyclophilin inhibitors as therapeutic agents. The apparent broad-spectrum of cyclophilin inhibitors may suggest that they could be exploited as a first line of protection against new and emerging viruses.
Acknowledgements
We acknowledge financial support from the U.S. Public Health Service grant no. AI087746 (P.A.G.). This is publication no. 28055 from the Department of Immunology & Microbial Science, The Scripps Research Institute, La Jolla, CA.
Footnotes
This article is part of a Special Issue entitled Proline-directed Foldases: Cell Signaling Catalysts and Drug Targets.
Contributor Information
Sam Hopkins, Email: s.hopkins@autoimmune.com.
Philippe A. Gallay, Email: gallay@scripps.edu.
References
- 1.Phillips A., Wainberg M.A., Coates R., Klein M., Rachlis A., Read S., Shepherd F., Vellend H., Walmsley S., Halloran P. Cyclosporine-induced deterioration in patients with AIDS. CMAJ. 1989;140:1456–1460. [PMC free article] [PubMed] [Google Scholar]
- 2.Karpas A., Lowdell M., Jacobson S.K., Hill F. Inhibition of human immunodeficiency virus and growth of infected T cells by the immunosuppressive drugs cyclosporin A and FK 506. Proc. Natl. Acad. Sci. U. S. A. 1992;89:8351–8355. doi: 10.1073/pnas.89.17.8351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luban J., Bossolt K.L., Franke E.K., Kalpana G.V., Goff S.P. Human immunodeficiency virus type 1 Gag protein binds to cyclophilins A and B. Cell. 1993;73:1067–1078. doi: 10.1016/0092-8674(93)90637-6. [DOI] [PubMed] [Google Scholar]
- 4.Franke E.K., Yuan H.E., Luban J. Specific incorporation of cyclophilin A into HIV-1 virions. Nature. 1994;372:359–362. doi: 10.1038/372359a0. [DOI] [PubMed] [Google Scholar]
- 5.Thali M., Bukovsky A., Kondo E., Rosenwirth B., Walsh C.T., Sodroski J., Göttlinger H.G. Functional association of cyclophilin A with HIV-1 virions. Nature. 1994;372:363–365. doi: 10.1038/372363a0. [DOI] [PubMed] [Google Scholar]
- 6.Rosenwirth B., Billich A., Datema R., Donatsch P., Hammerschmid F., Harrison R., Hiestand P., Jaksche H., Mayer P., Peichl P. Antimicrob. Agents Chemother. Vol. 38. 1763-1772. Inhibition of human immunodeficiency virus type 1 replication by SDZ NIM 811, a nonimmunosuppressive cyclosporine analog. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Billich A., Hammerschmid F., Peichl P., Wenger R., Zenke G., Quesniaux V., Rosenwirth B. Mode of action of SDZ NIM 811, a nonimmunosuppressive cyclosporin A analog with activity against human immunodeficiency virus (HIV) type 1: interference with HIV protein-cyclophilin A interactions. J. Virol. 1995;69:2451–2461. doi: 10.1128/jvi.69.4.2451-2461.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gamble T.R., Vajdos F.F., Yoo S., Worthylake D.K., Houseweart M., Sundquist W.I., Hill C.P. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell. 1996;87:1285–1294. doi: 10.1016/s0092-8674(00)81823-1. [DOI] [PubMed] [Google Scholar]
- 9.Yoo S., Myszka D.G., Yeh C., McMurray M., Hill C.P., Sundquist W.I. Molecular recognition in the HIV-1 capsid/cyclophilin A complex. J. Mol. Biol. 1997;269:780–795. doi: 10.1006/jmbi.1997.1051. [DOI] [PubMed] [Google Scholar]
- 10.Braaten D., Ansari H., Luban J. The hydrophobic pocket of cyclophilin is the binding site for the human immunodeficiency virus type 1 Gag polyprotein. J. Virol. 1997;71:2107–2113. doi: 10.1128/jvi.71.3.2107-2113.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dorfman T., Weimann A., Borsetti A., Walsh C.T., Göttlinger H.G. Active-site residues of cyclophilin A are crucial for its incorporation into human immunodeficiency virus type 1 virions. J. Virol. 1997;71:7110–7113. doi: 10.1128/jvi.71.9.7110-7113.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bukovsky A.A., Weimann A., Accola M.A., Göttlinger H.G. Transfer of the HIV-1 cyclophilin-binding site to simian immunodeficiency virus from Macaca mulatta can confer both cyclosporin sensitivity and cyclosporin dependence. Proc. Natl. Acad. Sci. U. S. A. 1997;94:10943–10948. doi: 10.1073/pnas.94.20.10943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Braaten D., Luban J. Cyclophilin A regulates HIV-1 infectivity, as demonstrated by gene targeting in human T cells. EMBO J. 2001;20:1300–1309. doi: 10.1093/emboj/20.6.1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kootstra N.A., Munk C., Tonnu N., Landau N.R., Verma I.M. Abrogation of postentry restriction of HIV-1-based lentiviral vector transduction in simian cells. Proc. Natl. Acad. Sci. U. S. A. 2003;100:1298–1303. doi: 10.1073/pnas.0337541100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bosco D.A., Eisenmesser E.Z., Pochapsky S., Sundquist W.I., Kern D. Catalysis of cis/trans isomerization in native HIV-1 capsid by human cyclophilin A. Proc. Natl. Acad. Sci. U. S. A. 2002;99:5247–5252. doi: 10.1073/pnas.082100499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Towers G.J., Hatziioannou T., Cowan S., Goff S.P., Luban J., Bieniasz P.D. Cyclophilin A modulates the sensitivity of HIV-1 to host restriction factors. Nat. Med. 2003;9:1138–1143. doi: 10.1038/nm910. [DOI] [PubMed] [Google Scholar]
- 17.Perron M.J., Stremlau M., Song B., Ulm W., Mulligan R.C., Sodroski J. TRIM5alpha mediates the postentry block to N-tropic murine leukemia viruses in human cells. Proc. Natl. Acad. Sci. U. S. A. 2004;101:11827–11832. doi: 10.1073/pnas.0403364101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sayah D.M., Sokolskaja E., Berthoux L., Luban J. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature. 2004;430:569–573. doi: 10.1038/nature02777. [DOI] [PubMed] [Google Scholar]
- 19.Nisole S., Lynch C., Stoye J.P., Yap M.W. A Trim5-cyclophilin A fusion protein found in owl monkey kidney cells can restrict HIV-1. Proc. Natl. Acad. Sci. U. S. A. 2004;101:13324–13328. doi: 10.1073/pnas.0404640101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilson S.J., Webb B.L., Ylinen L.M., Verschoor E., Heeney J.L., Towers G.J. Independent evolution of an antiviral TRIMCyp in rhesus macaques. Proc. Natl. Acad. Sci. U. S. A. 2008;105:3557–3562. doi: 10.1073/pnas.0709003105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berthoux L., Sebastian S., Sokolskaja E., Luban J. Cyclophilin A is required for TRIM5{alpha}-mediated resistance to HIV-1 in Old World monkey cells. Proc. Natl. Acad. Sci. U. S. A. 2005;102:14849–14853. doi: 10.1073/pnas.0505659102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ptak R.G., Gallay P.A., Jochmans D., Halestrap A.P., Ruegg U.T., Pallansch L.A., Bobardt M.D., de Béthune M.P., Neyts J., De Clercq E., Dumont J.M., Scalfaro P., Besseghir K., Wenger R.M., Rosenwirth B. Inhibition of human immunodeficiency virus type 1 replication in human cells by Debio-025, a novel cyclophilin binding agent. Antimicrob. Agents Chemother. 2008;52:1302–1317. doi: 10.1128/AAC.01324-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chatterji U., Bobardt M.D., Stanfield R., Ptak R.G., Pallansch L.A., Ward P.A., Jones M.J., Stoddart C.A., Scalfaro P., Dumont J.M., Besseghir K., Rosenwirth B., Gallay P.A. Naturally occurring capsid substitutions render HIV-1 cyclophilin A independent in human cells and TRIM-cyclophilin-resistant in Owl monkey cells. J. Biol. Chem. 2005;280:40293–40300. doi: 10.1074/jbc.M506314200. [DOI] [PubMed] [Google Scholar]
- 24.Matreyek K.A., Engelman A. The requirement for nucleoporin NUP153 during human immunodeficiency virus type 1 infection is determined by the viral capsid. J. Virol. 2011;85:7818–7827. doi: 10.1128/JVI.00325-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schaller T., Ocwieja K.E., Rasaiyaah J., Price A.J., Brady T.L., Roth S.L., Hué S., Fletcher A.J., Lee K., KewalRamani V.N., Noursadeghi M., Jenner R.G., James L.C., Bushman F.D., Towers G.J. HIV-1 capsid-cyclophilin interactions determine nuclear import pathway, integration targeting and replication efficiency. PLoS Pathog. 2011;7:e1002439. doi: 10.1371/journal.ppat.1002439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Di Nunzio F., Danckaert A., Fricke T., Perez P., Fernandez J., Perret E., Roux P., Shorte S., Charneau P., Diaz-Griffero F., Arhel N.J. Human nucleoporins promote HIV-1 docking at the nuclear pore, nuclear import and integration. PLoS ONE. 2012;7:e46037. doi: 10.1371/journal.pone.0046037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shah V.B., Shi J., Hout D.R., Oztop I., Krishnan L., Ahn J., Shotwell M.S., Engelman A., Aiken C. The host proteins transportin SR2/TNPO3 and cyclophilin A exert opposing effects on HIV-1 uncoating. J. Virol. 2013;87:422–432. doi: 10.1128/JVI.07177-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guth C.A., Sodroski J. Contribution of PDZD8 to stabilization of the human immunodeficiency virus type 1 capsid. J. Virol. 2014;88:4612–4623. doi: 10.1128/JVI.02945-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Price A.J., Fletcher A.J., Schaller T., Elliott T., Lee K., KewalRamani V.N., Chin J.W., Towers G.J., James L.C. CPSF6 defines a conserved capsid interface that modulates HIV-1 replication. PLoS Pathog. 2012;8:e1002896. doi: 10.1371/journal.ppat.1002896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bichel K., Price A.J., Schaller T., Towers G.J., Freund S.M., James L.C. HIV-1 capsid undergoes coupled binding and isomerization by the nuclear pore protein NUP358. Retrovirology. 2013;10 doi: 10.1186/1742-4690-10-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manel N., Hogstad B., Wang Y., Levy D.E., Unutmaz D., Littman D.R. A cryptic sensor for HIV-1 activates antiviral innate immunity in dendritic cells. Nature. 2010;467:214–217. doi: 10.1038/nature09337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rasaiyaah J., Tan C.P., Fletcher A.J., Price A.J., Blondeau C., Hilditch L., Jacques D.A., Selwood D.L., James L.C., Noursadeghi M., Towers G.J. HIV-1 evades innate immune recognition through specific cofactor recruitment. Nature. 2013;503:402–405. doi: 10.1038/nature12769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cox A.L., Siliciano R.F. Making sense of HIV innate sensing. Immunity. 2013;39:998–1000. doi: 10.1016/j.immuni.2013.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Z., Pan Q., Ding S., Qian J., Xu F., Zhou J., Cen S., Guo F., Liang C. The interferon-inducible MxB protein inhibits HIV-1 infection. Cell Host Microbe. 2013;14:398–410. doi: 10.1016/j.chom.2013.08.015. [DOI] [PubMed] [Google Scholar]
- 35.Gallay P.A., Ptak R.G., Bobardt M.D., Dumont J.M., Vuagniaux G., Rosenwirth B. Correlation of naturally occurring HIV-1 resistance to DEB025 with capsid amino acid polymorphisms. Viruses. 2013;5:981–997. doi: 10.3390/v5030981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Teraoka S., Mishiro S., Ebihara K., Sanaka T., Yamaguchi Y., Nakajima I., Kawai T., Yagisawa T., Honda H., Fuchinoue S. Effect of cyclosporine on proliferation of non-A, non-B hepatitis virus. Transplant. Proc. 1988;20:868–876. [PubMed] [Google Scholar]
- 37.Watashi K., Hijikata M., Hosaka M., Yamaji M., Shimotohno K. Cyclosporin A suppresses replication of hepatitis C virus genome in cultured hepatocytes. Hepatology. 2003;38:1282–1288. doi: 10.1053/jhep.2003.50449. [DOI] [PubMed] [Google Scholar]
- 38.Nakagawa M., Sakamoto N., Enomoto N., Tanabe Y., Kanazawa N., Koyama T., Kurosaki M., Maekawa S., Yamashiro T., Chen C.H., Itsui Y., Kakinuma S., Watanabe M. Specific inhibition of hepatitis C virus replication by cyclosporin A. Biochem. Biophys. Res. Commun. 2004;313:42–47. doi: 10.1016/j.bbrc.2003.11.080. [DOI] [PubMed] [Google Scholar]
- 39.Goto K., Watashi K., Murata T., Hishiki T., Hijikata M., Shimotohno K. Evaluation of the anti-hepatitis C virus effects of cyclophilin inhibitors, cyclosporin A, and NIM811. Biochem. Biophys. Res. Commun. 2006;343:879–884. doi: 10.1016/j.bbrc.2006.03.059. [DOI] [PubMed] [Google Scholar]
- 40.Ishii N., Watashi K., Hishiki T., Goto K., Inoue D., Hijikata M., Wakita T., Kato N., Shimotohno K. Diverse effects of cyclosporine on hepatitis C virus strain replication. J. Virol. 2006;80:4510–4520. doi: 10.1128/JVI.80.9.4510-4520.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakagawa M., Sakamoto N., Tanabe Y., Koyama T., Itsui Y., Takeda Y., Chen C.H., Kakinuma S., Oooka S., Maekawa S., Enomoto N., Watanabe M. Suppression of hepatitis C virus replication by cyclosporin a is mediated by blockade of cyclophilins. Gastroenterology. 2005;129:1031–1041. doi: 10.1053/j.gastro.2005.06.031. [DOI] [PubMed] [Google Scholar]
- 42.Watashi K., Ishii N., Hijikata M., Inoue D., Murata T., Miyanari Y., Shimotohno K. Cyclophilin B is a functional regulator of hepatitis C virus RNA polymerase. Mol. Cell. 2005;19:111–122. doi: 10.1016/j.molcel.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 43.Gaither L.A., Borawski J., Anderson L.J., Balabanis K.A., Devay P., Joberty G., Rau C., Schirle M., Bouwmeester T., Mickanin C., Zhao S., Vickers C., Lee L., Deng G., Baryza J., Fujimoto R.A., Lin K., Compton T., Wiedmann B. Multiple cyclophilins involved in different cellular pathways mediate HCV replication. Virology. 2010;397:43–55. doi: 10.1016/j.virol.2009.10.043. [DOI] [PubMed] [Google Scholar]
- 44.Yang F., Robotham J.M., Nelson H.B., Irsigler A., Kenworthy R., Tang H. Cyclophilin A is an essential cofactor for hepatitis C virus infection and the principal mediator of cyclosporine resistance in vitro. J. Virol. 2008;82:5269–5278. doi: 10.1128/JVI.02614-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chatterji U., Bobardt M., Selvarajah S., Yang F., Tang H., Sakamoto N., Vuagniaux G., Parkinson T., Gallay P. The isomerase active site of cyclophilin A is critical for hepatitis C virus replication. J. Biol. Chem. 2009;284:16998–17005. doi: 10.1074/jbc.M109.007625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaul A., Stauffer S., Berger C., Pertel T., Schmitt J., Kallis S., Zayas M., Lohmann V., Luban J., Bartenschlager R. Essential role of cyclophilin A for hepatitis C virus replication and virus production and possible link to polyprotein cleavage kinetics. PLoS Pathog. 2009;5:e1000546. doi: 10.1371/journal.ppat.1000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dorner M., Horwitz J.A., Donovan B.M., Labitt R.N., Budell W.C., Friling T., Vogt A., Catanese M.T., Satoh T., Kawai T., Akira S., Law M., Rice C.M., Ploss A. Completion of the entire hepatitis C virus life cycle in genetically humanized mice. Nature. 2013;501:237–241. doi: 10.1038/nature12427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ma S., Boerner J.E., TiongYip C., Weidmann B., Ryder N.S., Cooreman M.P., Lin K. NIM811, a cyclophilin inhibitor, exhibits potent in vitro activity against hepatitis C virus alone or in combination with alpha interferon. Antimicrob. Agents Chemother. 2006;50:2976–2982. doi: 10.1128/AAC.00310-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paeshuyse J., Kaul A., De Clercq E., Rosenwirth B., Dumont J.M., Scalfaro P., Bartenschlager R., Neyts J. The non-immunosuppressive cyclosporin DEBIO-025 is a potent inhibitor of hepatitis C virus replication in vitro. Hepatology. 2006;43:761–770. doi: 10.1002/hep.21102. [DOI] [PubMed] [Google Scholar]
- 50.Hopkins S., Scorneaux B., Huang Z., Murray M.G., Wring S., Smitley C., Harris R., Erdmann F., Fischer G., Ribeill Y. SCY-635, a novel nonimmunosuppressive analog of cyclosporine that exhibits potent inhibition of hepatitis C virus RNA replication in vitro. Antimicrob. Agents Chemother. 2010;54:660–672. doi: 10.1128/AAC.00660-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Flisiak R., Horban A., Gallay P., Bobardt M., Selvarajah S., Wiercinska-Drapalo A., Siwak E., Cielniak I., Higersberger J., Kierkus J., Aeschlimann C., Grosgurin P., Nicolas-Métral V., Dumont J.M., Porchet H., Crabbé R., Scalfaro P. The cyclophilin inhibitor Debio-025 shows potent anti-hepatitis C effect in patients coinfected with hepatitis C and human immunodeficiency virus. Hepatology. 2008;47:817–826. doi: 10.1002/hep.22131. [DOI] [PubMed] [Google Scholar]
- 52.Hopkins S., DiMassimo B., Rusnak P., Heuman D., Lalezari J., Sluder A., Scorneaux B., Mosier S., Kowalczyk P., Ribeill Y., Baugh J., Gallay P. The cyclophilin inhibitor SCY-635 suppresses viral replication and induces endogenous interferons in patients with chronic HCV genotype 1 infection. J. Hepatol. 2012;57:47–54. doi: 10.1016/j.jhep.2012.02.024. [DOI] [PubMed] [Google Scholar]
- 53.Hopkins S., Gallay P. Cyclophilin inhibitors: an emerging class of therapeutics for the treatment of chronic hepatitis C infection. Viruses. 2012;4:2558–2577. doi: 10.3390/v4112558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gallay P.A., Lin K. Profile of alisporivir and its potential in the treatment of hepatitis C. Drug Des. Dev. Ther. 2013;7:105–115. doi: 10.2147/DDDT.S30946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Robida J.M., Nelson H.B., Liu Z., Tang H. Characterization of hepatitis C virus subgenomic replicon resistance to cyclosporine in vitro. J. Virol. 2007;81:5829–5840. doi: 10.1128/JVI.02524-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fernandes F., Poole D.S., Hoover S., Middleton R., Andrei A.C., Gerstner J., Striker R. Sensitivity of hepatitis C virus to cyclosporine A depends on nonstructural proteins NS5A and NS5B. Hepatology. 2007;46:1026–1033. doi: 10.1002/hep.21809. [DOI] [PubMed] [Google Scholar]
- 57.Goto K., Watashi K., Inoue D., Hijikata M., Shimotohno K. Identification of cellular and viral factors related to anti-hepatitis C virus activity of cyclophilin inhibitor. Cancer Sci. 2009;100:1943–1950. doi: 10.1111/j.1349-7006.2009.01263.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Puyang X., Poulin D.L., Mathy J.E., Anderson L.J., Ma S., Fang Z., Zhu S., Lin K., Fujimoto R., Compton T., Wiedmann B. Mechanism of resistance of hepatitis C virus replicons to structurally distinct cyclophilin inhibitors. Antimicrob. Agents Chemother. 2010;54:1981–1987. doi: 10.1128/AAC.01236-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu Z., Yang F., Robotham J.M., Tang H. Critical role of cyclophilin A and its prolyl-peptidyl isomerase activity in the structure and function of the hepatitis C virus replication complex. J. Virol. 2009;83:6554–6565. doi: 10.1128/JVI.02550-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Foster T.L., Gallay P., Stonehouse N.J., Harris M. Cyclophilin A interacts with domain II of hepatitis C virus NS5A and stimulates RNA binding in an isomerase-dependent manner. J. Virol. 2011;85:7460–7464. doi: 10.1128/JVI.00393-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nag A., Robotham J.M., Tang H. Suppression of viral RNA binding and the assembly of infectious hepatitis C virus particles in vitro by cyclophilin inhibitors. J. Virol. 2012;86:12616–12624. doi: 10.1128/JVI.01351-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Heck J.A., Meng X., Frick D.N. Cyclophilin B stimulates RNA synthesis by the HCV RNA dependent RNA polymerase. Biochem. Pharmacol. 2009;77:1173–1180. doi: 10.1016/j.bcp.2008.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hanoulle X., Badillo A., Wieruszeski J.M., Verdegem D., Landrieu I., Bartenschlager R., Penin F., Lippens G. Hepatitis C virus NS5A protein is a substrate for the peptidyl-prolyl cis/trans isomerase activity of cyclophilins A and B. J. Biol. Chem. 2009;284:13589–13601. doi: 10.1074/jbc.M809244200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang F., Robotham J.M., Grise H., Frausto S., Madan V., Zayas M., Bartenschlager R., Robinson M., Greenstein A.E., Nag A., Logan T.M., Bienkiewicz E., Tang H. A major determinant of cyclophilin dependence and cyclosporine susceptibility of hepatitis C virus identified by a genetic approach. PLoS Pathog. 2010;6:e1001118. doi: 10.1371/journal.ppat.1001118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Grisé H., Frausto S., Logan T., Tang H. A conserved tandem cyclophilin-binding site in hepatitis C virus nonstructural protein 5A regulates Alisporivir susceptibility. J. Virol. 2012;86:4811–4822. doi: 10.1128/JVI.06641-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Verdegem D., Badillo A., Wieruszeski J.M., Landrieu I., Leroy A., Bartenschlager R., Penin F., Lippens G., Hanoulle X. Domain 3 of NS5A protein from the hepatitis C virus has intrinsic alpha-helical propensity and is a substrate of cyclophilin A. J. Biol. Chem. 2011;286:20441–20454. doi: 10.1074/jbc.M110.182436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ansari I.U., Striker R. Subtype specific differences in NS5A domain II reveals involvement of proline at position 310 in cyclosporine susceptibility of hepatitis C virus. Viruses. 2012;4:3303–3315. doi: 10.3390/v4123303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chatterji U., Lim P., Bobardt M.D., Wieland S., Cordek D.G., Vuagniaux G., Chisari F., Cameron C.E., Targett-Adams P., Parkinson T., Gallay P.A. HCV resistance to cyclosporin A does not correlate with a resistance of the NS5A-cyclophilin A interaction to cyclophilin inhibitors. J. Hepatol. 2010;53:50–56. doi: 10.1016/j.jhep.2010.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gregory M.A., Bobardt M., Obeid S., Chatterji U., Coates N.J., Foster T., Gallay P., Leyssen P., Moss S.J., Neyts J., Nur-e-Alam M., Paeshuyse J., Piraee M., Suthar D., Warneck T., Zhang M.Q., Wilkinson B. Preclinical characterization of naturally occurring polyketide cyclophilin inhibitors from the sanglifehrin family. Antimicrob. Agents Chemother. 2011;55:1975–1981. doi: 10.1128/AAC.01627-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Coelmont L., Hanoulle X., Chatterji U., Berger C., Snoeck J., Bobardt M., Lim P., Vliegen I., Paeshuyse J., Vuagniaux G., Vandamme A.M., Bartenschlager R., Gallay P., Lippens G., Neyts J. DEB025 (Alisporivir) inhibits hepatitis C virus replication by preventing a cyclophilin A induced cis-trans isomerisation in domain II of NS5A. PLoS ONE. 2010;5:e13687. doi: 10.1371/journal.pone.0013687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Garcia-Rivera J.A., Bobardt M., Chatterji U., Hopkins S., Gregory M.A., Wilkinson B., Lin K., Gallay P.A. Multiple mutations in hepatitis C virus NS5A domain II are required to confer a significant level of resistance to alisporivir. Antimicrob. Agents Chemother. 2012;56:5113–5121. doi: 10.1128/AAC.00919-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Anderson L.J., Lin K., Compton T., Wiedmann B. Inhibition of cyclophilins alters lipid trafficking and blocks hepatitis C virus secretion. Virol. J. 2011;8:329. doi: 10.1186/1743-422X-8-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Romero-Brey I., Bartenschlager R. Membranous replication factories induced by plus-strand RNA viruses. Viruses. 2014;6:2826–2857. doi: 10.3390/v6072826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Madan V., Paul D., Lohmann V., Bartenschlager R. Inhibition of HCV replication by cyclophilin antagonists is linked to replication fitness and occurs by inhibition of membranous web formation. Gastroenterology. 2014;146:1361–1372. doi: 10.1053/j.gastro.2014.01.055. (e1-9) [DOI] [PubMed] [Google Scholar]
- 75.Chatterji U., Bobardt M.D., Wood M., Gallay P.A. 2014. Alisporivir specifically eliminates HCV-created double membrane vesicles in infected hepatocytes by disrupting CypA-NS5A interactions. (21th International Symposium on Hepatitis C Virus And Related Viruses, Banff Canada). (Abstract # P.3.54) [Google Scholar]
- 76.Tian X., Zhao C., Zhu H., She W., Zhang J., Liu J., Li L., Zheng S., Wen Y.M., Xie Y. Hepatitis B virus (HBV) surface antigen interacts with and promotes cyclophilin a secretion: possible link to pathogenesis of HBV infection. J. Virol. 2010;84:3373–3381. doi: 10.1128/JVI.02555-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hansson M.J., Moss S.J., Bobardt M., Chatterji U., Coates N., Garcia-Rivera J.A., Elmér E., Kendrew S., Leyssen P., Nur-E-Alam M., Warneck T., Wilkinson B., Gallay P.A., Gregory M.A. Bioengineering and semisynthesis combined to generate an optimized natural product cyclophilin inhibitor for treatment of chronic HBV, HCV and HIV infection. Chem. Biol. 2014 doi: 10.1016/j.chembiol.2014.10.023. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chokshi S., Phillips S., Riva A., Naoumov N. Alisporivir inhibition of cellular cyclophilins disrupts hepatitis B virus (HBV) replication and this effect is further enhanced in combination with direct antiviral targeting HBV-DNA polymerase in vitro. Gut. 2012;61:A11. doi: 10.1136/gutjnl-2012-302514a.25. [DOI] [Google Scholar]
- 79.Carbajo-Lozoya J., Ma-Lauer Y., Malešević M., Theuerkorn M., Kahlert V., Prell E., von Brunn B., Muth D., Baumert T.F., Drosten C., Fischer G., von Brunn A. Human coronavirus NL63 replication is cyclophilin A-dependent and inhibited by non-immunosuppressive cyclosporine A-derivatives including Alisporivir. Virus Res. 2014;184:44–53. doi: 10.1016/j.virusres.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.de Wilde A.H., evenhoven-Dobbe J.C., van der Meer Y., Thiel V., Narayanan K., Makino S., Snijder E.J., van Hemert M.J. Cyclosporin A inhibits the replication of diverse coronaviruses. J. Gen. Virol. 2011;92:2542–2548. doi: 10.1099/vir.0.034983-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kawasaki H., Mocarski E.S., Kosugi I., Tsutsui Y. Cyclosporine inhibits mouse cytomegalovirus infection via a cyclophilin-dependent pathway specifically in neural stem/progenitor cells. J. Virol. 2007;81:9013–9023. doi: 10.1128/JVI.00261-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Keyes L.R., Bego M.G., Soland M., St Jeor S. Cyclophilin A is required for efficient human cytomegalovirus DNA replication and reactivation. J. Gen. Virol. 2012;93:722–732. doi: 10.1099/vir.0.037309-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kambara H., Tani H., Mori Y., Abe T., Katoh H., Fukuhara T., Taguwa S., Moriishi K., Matsuura Y. Involvement of cyclophilin B in the replication of Japanese encephalitis virus. Virology. 2011;412:211–219. doi: 10.1016/j.virol.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 84.Bienkowska-Haba M., Patel H.D., Sapp M. Target cell cyclophilins facilitate human papillomavirus type 16 infection. PLoS Pathog. 2009;5:e1000524. doi: 10.1371/journal.ppat.1000524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bose S., Mathur M., Bates P., Joshi N., Banerjee A.K. Requirement for cyclophilin A for the replication of vesicular stomatitis virus New Jersey serotype. J. Gen. Virol. 2003;84:1687–1699. doi: 10.1099/vir.0.19074-0. [DOI] [PubMed] [Google Scholar]
- 86.Liu X., Zhao Z., Xu C., Sun L., Chen J., Zhang L., Liu W. Cyclophilin A restricts influenza A virus replication through degradation of the M1 protein. PLoS ONE. 2012;7:e31063. doi: 10.1371/journal.pone.0031063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu X., Zhao Z., Li Z., Xu C., Sun L., Chen J., Liu W. Cyclosporin A inhibits the influenza virus replication through cyclophilin A-dependent and -independent pathways. PLoS ONE. 2012;7:e37277. doi: 10.1371/journal.pone.0037277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Qing M., Yang F., Zhang B., Zou G., Robida J.M., Yuan Z., Tang H., Shi P.Y. Cyclosporine inhibits flavivirus replication through blocking the interaction between host cyclophilins and viral NS5 protein. Antimicrob. Agents Chemother. 2009;53:3226–3235. doi: 10.1128/AAC.00189-09. [DOI] [PMC free article] [PubMed] [Google Scholar]