Summary
Insults to endoplasmic reticulum (ER) homeostasis activate the unfolded protein response (UPR), which elevates protein folding and degradation capacity and attenuates protein synthesis. While a role for ubiquitin in regulating the degradation of misfolded ER-resident proteins is well described, ubiquitin-dependent regulation of translational reprogramming during the UPR remains uncharacterized. Using global quantitative ubiquitin proteomics, we identify evolutionarily conserved, site-specific regulatory ubiquitylation of 40S ribosomal proteins. We demonstrate that these events occur on assembled cytoplasmic ribosomes and are stimulated by both UPR activation and translation inhibition. We further show that ER stress-stimulated regulatory 40S ribosomal ubiquitylation occurs on a timescale similar to eIF2α phosphorylation, is dependent upon PERK signaling, and is required for optimal cell survival during chronic UPR activation. In total, these results reveal regulatory 40S ribosomal ubiquitylation as a previously uncharacterized and important facet of eukaryotic translational control.
Introduction
Protein homeostasis is maintained through the careful balance of protein synthesis and degradation (Wolff et al., 2014). Disruption of protein homeostasis results in the coordinated regulation of protein synthesis and degradation, as exemplified by the unfolded protein response (UPR). A well-characterized and multi-tiered cellular response to proteotoxic stress in the endoplasmic reticulum (ER), the UPR elevates production of proteins whose function is to enhance protein homeostasis capacity, and attenuates protein synthesis to limit the load on protein homeostasis pathways (Back and Kaufman, 2012; Walter and Ron, 2011). Sustained activation of the UPR induces a cell death response that eliminates cells with unbalanced protein homeostasis (Sano and Reed, 2013).
Down regulation of protein synthesis upon UPR induction is mediated by phosphorylation of the translation initiation factor eIF2α catalyzed by the ER-localized kinase PERK (Harding et al., 1999). Phosphorylated eIF2α (eIF2α-P) limits the abundance of functional ternary complexes (eIF2-GTP-Met-tRNAMet) and results in translation initiation inhibition (Jackson et al., 2010). Loss of PERK activity leads to an inability to reduce translation in response to UPR activating insults and a subsequent elevation in UPR-stimulated cell death (Harding et al., 2000b). Paradoxically, the UPR allows for selective translation of specific mRNAs (Harding et al., 2000a). In the case of ATF4 mRNA, for example, UPR stimulation relieves translational repression mediated by cis-acting small upstream open reading frames (uORFs) within the 5’UTR of ATF4 (Vattem and Wek, 2004). This is thought to reflect a “leaky scanning” mechanism in which conditions that limit active ternary complex abundance allow for bypassing of inhibitory uORFs during the scanning phase of translation initiation (Jackson et al., 2010).
Global sequence analyses have revealed that ~50% of all human mRNA sequences contain at least one uORF in their 5’UTR (Calvo et al., 2009; Resch et al., 2009). Recent ribosome profiling data revealed that many 5’ uORF sequences are actively translated, suggesting that this type of cis-acting post-transcriptional regulation of protein production may be pervasive (Andreev et al., 2014; Ingolia et al., 2014). However, another study utilizing ribosome profiling to directly monitor mRNA translation during UPR activation demonstrated that many mRNAs lacking uORF elements were translationally activated upon UPR induction (Reid et al., 2014). Thus additional mechanisms likely contribute to translational reprogramming during protein homeostasis stress. Indeed, an eIF2α-independent mechanism of translational attenuation that utilizes mTOR activation to limit protein production during prolonged UPR activation was recently described (Guan et al., 2014).
Despite the importance of ubiquitin proteasome system function in facilitating the ER associated degradation (ERAD) pathway during conditions that activate the UPR, the role of ubiquitin (ub)-mediated regulation of the translational machinery during protein homeostasis stress remains poorly understood (Christianson and Ye, 2014). Ubiquitylation directs diverse functional outputs to target proteins. While lysine-48 linked poly-ubiquitylation largely targets substrates for proteasome-mediated degradation, lysine-63 linked poly-ubiquitylation and mono-ubiquitylation impart non-degradation based regulatory control (Komander and Rape, 2012). Both types of events – regulatory and degradative – can be captured by quantitative proteomic profiling of the ub-modified proteome (Carrano and Bennett, 2013).
Here, we utilize a quantitative proteomic approach to identify ub-dependent mechanisms governing the response to protein homeostasis stress. We demonstrate that canonical UPR activation via ER stressors induces alterations in the ub-modified proteome that are distinct from those observed upon direct proteasome inhibition. We identify site-specific regulatory ubiquitylation of 40S ribosomal proteins as an early event during UPR activation. We further find that regulatory 40S ribosomal ubiquitylation (RRub) is robustly induced by inhibitors of translation elongation and can be detected on assembled cytoplasmic ribosomal complexes. We show that PERK and eIF2α phosphorylation are both necessary, but not sufficient, for UPR induced RRub. Our finding that cell death upon thapsigargin-mediated UPR activation is enhanced when we prevent ubiquitylation of RPS2 on lysines 58 or 275 and lysine 8 on RPS20 demonstrates the importance of RRub in mounting a successful cellular response to agents that induce the UPR. Lastly, we demonstrate that a subset of RRub is conserved among humans, Drosophila and S. cerevisiae. Taken together, these studies identify a critical role for regulatory, non-degradative, ubiquitylation that communicates protein homeostasis dysfunction to the translational apparatus.
Results
Characterization of the ub-modified proteome upon induction of the unfolded protein response
Dynamic coordination between the protein synthesis and degradation activities of the cell maintains and rebalances protein homeostasis upon proteotoxic challenges. The well-characterized unfolded protein response (UPR) directly connects protein homeostasis dysfunction with dynamic proteome remodeling (Walter and Ron, 2011). The ubiquitin-dependent degradation of misfolded or otherwise defective proteins is an integral component of this response (Lykke-Andersen and Bennett, 2014). However, the scope of ubiquitin-dependent regulation during protein homeostasis stress is uncharacterized. To explore the ubiquitin-mediated response to protein homeostasis dysfunction, we used a quantitative proteomics approach to identify and quantify ubiquitylation events that are induced or diminished upon UPR stimulation.
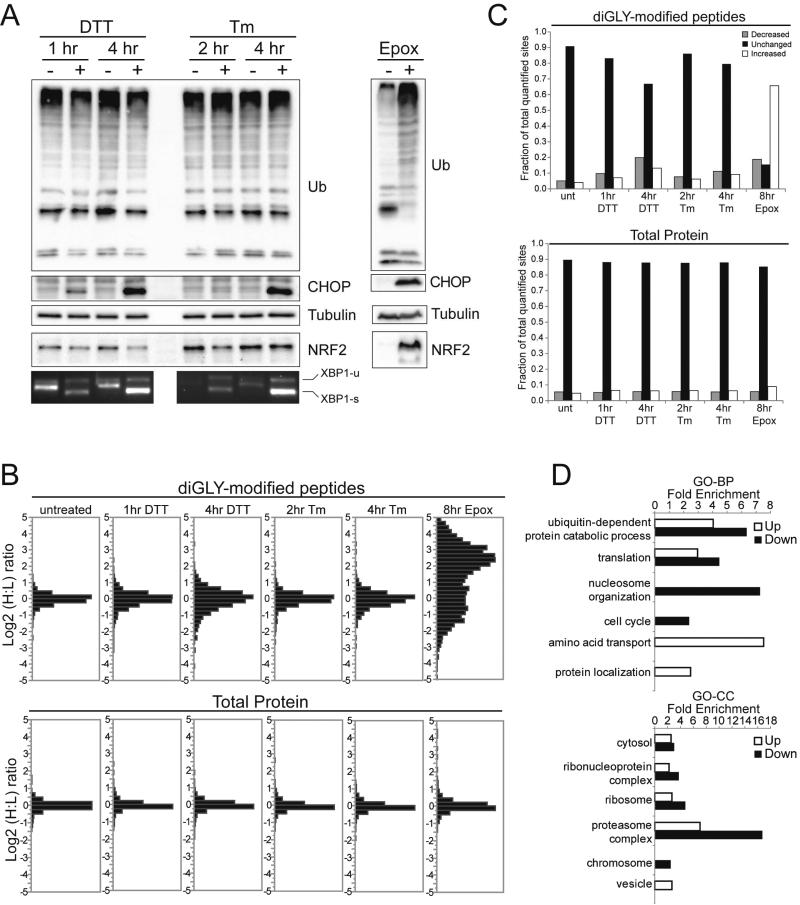
We examined the dynamics of protein ubiquitylation in response to UPR activation in HCT116 cells over an 8 hour period using two well-established UPR inducers, dithiothreitol (DTT) and tunicamycin (Tm) (Back and Kaufman, 2012). Both treatments resulted in splicing of XBP1, phosphorylation of eIF2α within 1 to 2 hours, and induction of the pro-apoptotic protein CHOP at 4 to 8 hours (Figure 1A, S1A). Examination of early and late time points for each treatment enabled us to monitor changes to the ub-modified proteome both prior to and following CHOP induction. When compared to the proteasome inhibitor epoxomicin, ER stressors did not induce global accumulation of poly-ubiquitylated proteins, nor of a known endogenous ubiquitin proteasome system substrate Nrf2 (Figure 1A) (Taguchi et al., 2011). However, both treatment with epoxomicin and the ER stressors resulted in CHOP induction (Figure 1A).
Figure 1. Characterization of alterations in protein ubiquitylation upon UPR activation.
A) Whole cell lysates of HCT116 cells treated with dithiothreitol (DTT), tunicamycin (Tm), or epoxomicin (Epox) were immunoblotted with the indicated antibodies. XBP1 splicing was detected by PCR from cDNA produced from total mRNA.
B) SILAC-based quantitative proteomic analysis of total proteins (bottom) or diGly-enriched fractions (top) from metabolically heavy labeled cells, measured as the Log2 heavy to light (H:L) peptide or protein intensity ratio.
C) The fraction of quantified diGly-modified peptides (top) or total proteins (bottom) whose abundance increased (white bars), decreased (grey bars), or was unchanged (black bars) by the indicated treatment is depicted. The standard deviation (SD) derived from untreated controls provided the basis for determining the fraction of diGly-modified peptides or total proteins that were altered in response to the indicated treatment. Log2 ratios greater than one SD from the untreated mean are plotted separately as increased and decreased.
D) Gene ontology (GO) enrichment analysis of proteins containing diGly-modified peptides which increased (up, open bars) or decreased (down, filled bars) in abundance upon ER stress. The fold enrichment of selected biological processes (BP, top) and cellular compartments (CC, bottom) is depicted.
To interrogate site-specific alterations to individual ub-modified proteins, we utilized a SILAC-based quantitative proteomic approach coupled with immuno-affinity enrichment of diGlycine (diGly)-modified peptides (Carrano and Bennett, 2013; Kim et al., 2011). Trypsin-catalyzed cleavage of ubiquitylated proteins generates a proteotypic peptide containing the diGly remnant derived from the carboxy-terminus of ubiquitin linked to lysine residues of substrate peptides (Bustos et al., 2012). Using this approach, we could identify accumulation or depletion of specific ub-modified peptides.
In total, 5893 unique diGly-modified peptides were identified and 90% of sites were quantified in at least one experimental condition (Table S1). Of all quantified diGly-modified peptides, 33% and 20% treated for 4 hours with DTT and Tm, respectively, displayed abundance changes greater than 1.5 fold (Figure 1B,C). In contrast, epoxomicin treatment altered the abundance more than 1.5 fold for 84% of all diGly-modified peptides (Figure 1B,C). The alterations observed in response to ER stress were specific for the diGly-modified proteome as analysis of the total proteome revealed no difference in the number of proteins whose abundance was significantly altered upon treatment with the ER stressors (Figure 1B,C, Table S2,S3).
Defects in proteasome function often result in the global accumulation of poly-ubiquitin chains that target proteins for degradation (Komander and Rape, 2012). In agreement with this observation, epoxomicin treatment resulted in the accumulation of K11 and K48-linked ub peptides, indicative of degradative poly-ub chains (Figure S1B). However, treatment with ER stressors failed to induce global alterations in poly-ub chain abundance, consistent with the profiles shown in Figure 1A. Similarly, treatment with epoxomicin, but not DTT or Tm, reduced histone ubiquitylation likely arising from the redistribution of ubiquitin from mono-ubiquitylated proteins to poly-ubiquitylated proteins (Figure S1B). These results demonstrate that proteasome inhibition results in more wide-spread alterations in the abundance of ub-modified peptides compared to treatment with two distinct ER stress inducers.
Proteasome inhibitors and ER stressors induce distinct site-specific alterations to individual ub-modified proteins
ER associated degradation (ERAD) is charged with degrading ER resident proteins and secretory cargo, many of with contain membrane-spanning domains, that misfold during ER stress conditions (Christianson and Ye, 2014). To explore whether ubiquitylation of membrane-associated proteins was specifically induced by ER stress, we treated cells with DTT for 1 hour or Tm for 2 hours and observed that the abundance of individual diGly-modified peptides from all proteins containing an annotated transmembrane domain increased relative to the entire set of diGly-modified peptides quantified in each sample (Figure S1C,D). In contrast, treatment with epoxomicin resulted in a net decrease in the abundance of specific diGly-modified peptides from membrane domain-containing proteins, even though there was an overall 2-fold increase in the abundance of all diGly-modified peptides in response to proteasome inhibition (Figure S1D). Examination of a subset of known membrane-associated proteins revealed striking differences in the extent of site-specific ubiquitylation in response to the different treatments (Figure S1E, Table S4). In particular, we observed that multiple individual diGly-modified peptides from both amino acid transporters (SLC3A2, SLC7A5, SLC1A5) and cell adhesion proteins (ITGB1,ITGB4) increased in abundance in response to ER stressors, but decreased in abundance in response to epoxomicin treatment (Figure S1E). This observation was not limited to transmembrane-domain containing proteins: comparison of the SILAC abundance ratios for all diGly-modified peptides from cells treated with ER stressors and those treated with epoxomicin revealed no correlation in individual diGly-modified peptide SILAC ratios (Figure S1F). These results indicate that proteasome inhibition, with a resulting depletion of free ub levels, affects protein ubiquitylation in a manner distinct from that of other protein homeostasis stress inducers.
The ubiquitylation of specific 40S ribosomal proteins is induced by UPR activation
Separate from the degradation of damaged ER proteins, UPR activation globally down-regulates protein synthesis via phosphorylation of the translation initiation factor eIF2α (Walter and Ron, 2011). However, unlike the pathway for triggering ER protein degradation, the UPR-induced translational down-regulation of protein synthesis has not previously been linked to ubiquitylation events. Interestingly, gene-ontology analysis of proteins whose ubiquitylation was either induced or repressed upon UPR induction revealed enrichment for proteins involved in mRNA translation and localized to the cytosolic ribosome (Figure 1D). This observation prompted a closer examination of ribosome ubiquitylation upon UPR induction by either ER stressors or proteasome inhibition.
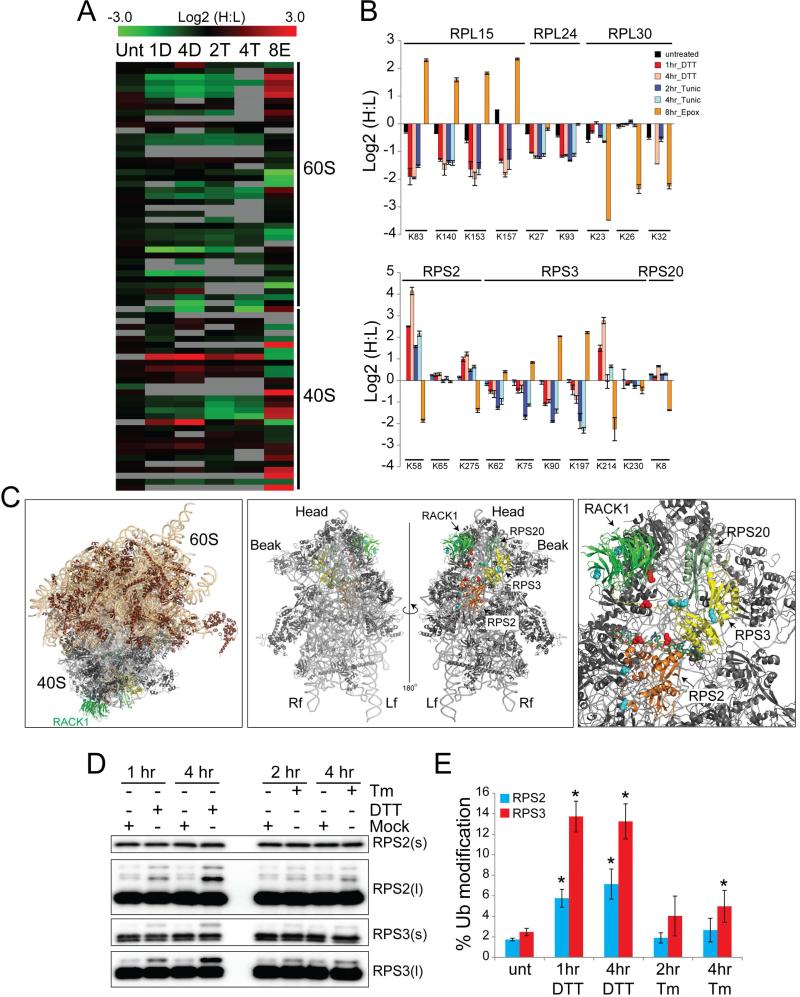
Ribosomal proteins comprising both the large (60S) and small (40S) subunits contained diGly-modified peptides that were altered in abundance upon treatment with ER stressor in a manner that was not similarly observed upon proteasome inhibition (Figure 2A, Table S5). For example, the abundance of four separate diGly-modified peptides from the 60S protein RPL15 was increased upon epoxomicin treatment but decreased upon both DTT and Tm treatment (Figure 2B). In contrast, RPL30, another 60S protein, contained individual diGly-modified peptides that were decreased by proteasome inhibition but unaltered by ER stress. Overall, these data indicate that ER stress induces specific alterations in the ubiquitylation of ribosomal proteins.
Figure 2. UPR stimulation induces site-specific ubiquitylation of 40S ribosomal proteins.
A) Heat map of the Log2 H:L ratios from all SILAC experiments for all quantified diGly-modified peptides arising from 40S or 60S ribosomal proteins. Unt=untreated; 1D and 4D =1 and 4hr DTT; 2T and 4T=2 and 4 hr Tm; 8E=8 hr Epox.
B) The Log2 ratio of individual diGly-modified lysine residues from representative 60S proteins (top), or 40S proteins (bottom) in each experimental condition. The positions of the diGly-modified lysines in each protein are indicated. Error bars = SEM.
C) Left: 80S ribosome structure with the positions of RPS2 (orange), RPS3 (yellow), RPS20 (pale green) and RACK1 (green) indicated. Ribosomal RNAs are shown in ribbon representation in light orange (60S) or gray (40S) (PDB: 4V6X with mRNA superimposed from PDB: 4KZZ). Middle: view of the isolated 40S from the perspective of the 60S interface (left), or the solvent exposed surface distal to the 60S interface (right) is shown. Right: magnified view of the solvent exposed surface of the 40S subunit. Side chain coloring indicates lysine residues within 40S ribosomal proteins for which a 1.5 fold increase (red) or decrease (blue) in diGly modification was detected upon proteasome inhibition.
D) Immunoblot of HCT116 whole cell lysates from cells immunoblotted with antibodies against RPS2 or RPS3 with short (s) or long (l) exposures.
E) Quantification of the fraction of RPS2 (blue bars) or RPS3 (red bars) that is ub-modified in untreated cells or cells treated as indicated. Error bars = SEM from triplicate experiments. * indicates a p-value of < 0.05 using Student's t-test.
Because proteasome inhibition leads to demodification of proteins whose ubiquitylation is regulatory in nature (Kim et al., 2011; Mimnaugh et al., 1997), we investigated whether proteasome inhibition might also trigger a decrease in ub-modification of some ribosomal proteins. For a subset of 40S proteins – RPS2, RPS3, and RPS20 – we made just such an observation: individual lysine residues were strongly demodified in response to proteasome inhibition (Figure 2B). Interestingly, ER stressors induced ubiquitylation of these same lysine residues. For example, lysine 58 (K58) on RPS2 underwent a nearly 16 fold increase in ubiquitylation upon DTT treatment, but was demodified in response to epoxomicin treatment (Figure 2B). The ubiquitylation of lysine 275 (K275) within RPS2, lysine 214 within RPS3, and K8 on RPS20 was altered in a similar manner (Figure 2B). The responses we observed were site-specific, not a general feature for the entire protein. For instance, the abundance of the diGly-modified form of K65 within RPS2 did not vary with K58, but rather was unaltered by treatment with ER stressors or epoxomicin (Figure 2B). Similarly, RPS3 contained many diGly-modified lysine residues at sites other than K214 that largely increased in abundance upon proteasome inhibition.
If site-specific ubiquitylation of 40S proteins has a regulatory role during protein biogenesis, rather than targeting ribosomal proteins for degradation, we might expect a clustering on the ribosome surface of ub-modified lysines from 40S proteins that decreased in abundance in response to proteasome inhibition. To visualize where regulatory ubiquitylation occurred on the 40S ribosome, we mapped all ub-modified lysines identified in this work, and previous studies, onto a structure of the mammalian 80S ribosome (Anger et al., 2013). We further classified the ub-modified lysines on 40S proteins of interest as being increased or decreased in abundance upon proteasome inhibition. We focused our attention on RPS2, RPS3, RPS20, and RACK1 which are clustered on the solvent-exposed surface of the 40S distal to the 60S interaction surface (Figure 2C). Lysine residues on RPS2, RPS3, and RACK1 for which ubiquitylation decreased in response to proteasome inhibition, and thus likely marked regulatory events, exhibited spatial separation from those that increased in response to proteasome inhibition (Figure 2C). These observations suggest that regulatory 40S ribosome ubiquitylation (hereafter referred to as RRub) has a functional role in controlling the response to UPR activation as well as possibly regulating overall protein biogenesis.
Regulatory ubiquitylation of RPS2 and RPS3 is an early event upon UPR induction
Previous studies have demonstrated that mono-ubiquitylated proteins become de-modified in response to proteasome inhibition (Kim et al., 2011; Mimnaugh et al., 1997). To determine whether the 40S proteins that were deubiquitylated in response to proteasome inhibition were mono-ubiquitylated upon UPR induction, we monitored RPS2 and RPS3 by immunoblotting and observed a slower-migrating band upon treatment with either DTT or Tm (Figure 2D). After 4 hours of DTT treatment, 6 and 13% of total RPS2 and RPS3, respectively, became modified (Figure 2E).
To confirm that the mobility shift observed by immunoblotting represented ub-modified RPS2 and RPS3, we used immobilized purified tandem ubiquitin binding entities (TUBEs) as an affinity capture reagent (Lopitz-Otsoa et al., 2012). Immunoblotting of TUBE eluates from DTT treated samples confirmed that the slower-migrating band was ub-modified RPS2 and RPS3 (Figure S2A). To further validate that RPS2 and RPS3 were ubiquitylated in response to UPR activation, DTT treated cells were lysed and subsequently incubated with the promiscuous deubiquitylating enzyme, Usp2cc. As predicted, Usp2cc treatment eliminated the slower-migrating, ub-modified species (Figure S2B).
To determine the timing of RRub relative to other well-characterized molecular events induced upon UPR activation, we performed a time course analysis on cells treated with DTT. RPS2 and RPS3 ubiquitylation were detected as soon as 30min after DTT treatment, a time point prior to ATF4 and CHOP induction, and coincident with eIF2α phosphorylation (Figure S2A). Further, RPS3 and RPS2 regulatory ubiquitylation could be stimulated by three distinct UPR activators at minimal concentrations required to stimulate the UPR (Figure S2C). These results suggest that RRub is an early event during UPR activation.
Diverse but specific protein homeostasis stressors stimulate RRub
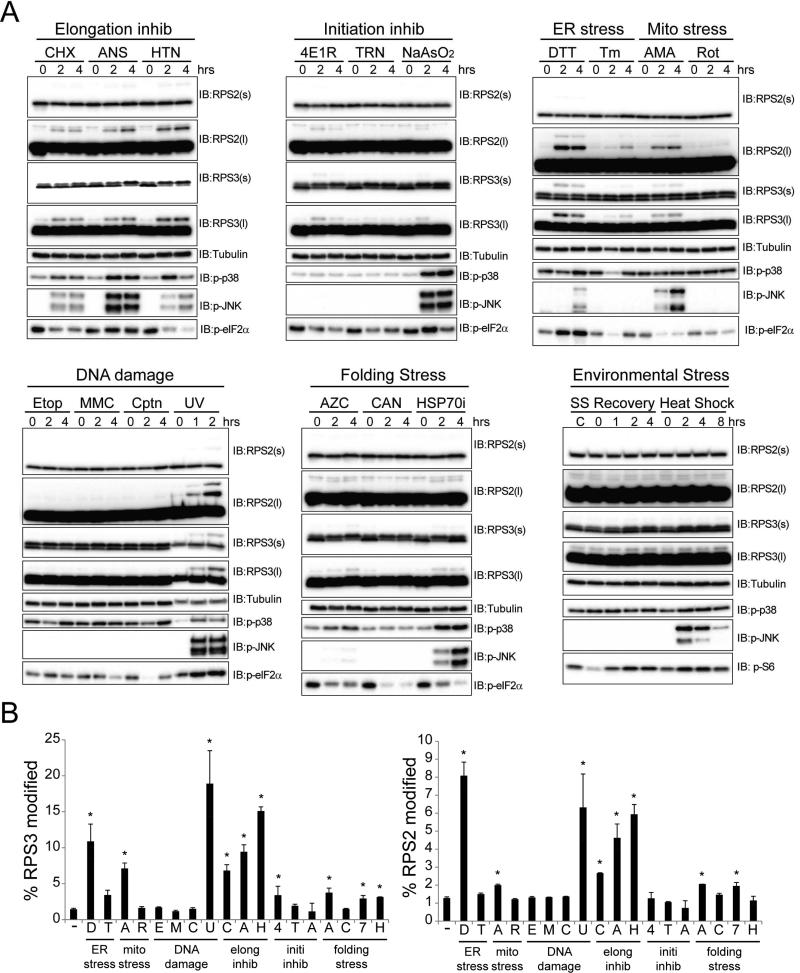
As UPR activation globally represses protein biogenesis through inhibitory eIF2α phosphorylation, we tested if other agents that attenuate translation also stimulate RRub. All three translation inhibitors tested that either inhibit translation elongation or a step immediately prior to elongation – cycloheximide (CHX), anisomycin (ANS), and harringtonine (HTN) – were potent inducers of regulatory RPS2 and RPS3 ubiquitylation (Figure 3A,B). However, inhibitors of translation initiation that act prior to 80S formation induced RPS2 or RPS3 ubiquitylation weakly or not at all (Figure 3A,B). This was surprising, given that UPR activation primarily inhibits translation initiation and was originally used to identify RRub. This result prompted us to examine RRub induction after treatment with a panel of diverse protein homeostasis stressors. The majority of protein homeostasis stressors failed to induce or weakly induced RPS2 or RPS3 ubiquitylation. These included a broad range of protein folding stressors, such as the proline analog L-Azetidine-2-carboxylic acid (AZC) and overt heat shock (Figure 3A,B). DNA damaging agents also failed to induce RRub, except for UV treatment, which activates the UPR (Wu et al., 2002). A significant portion of the cellular stressors studied induced the activation of the stress-sensitive MAPKs p38 and JNK (Figure 3A). However, some stressors that potently induced MAPK activation, such as sodium arsenite and heat shock, failed to robustly induce RPS2 or RPS3 ubiquitylation. These observations suggest that p38 or JNK activation is not sufficient to stimulate regulatory 40S ubiquitylation.
Figure 3. Regulatory RPS2 and RPS3 ubiquitylation is induced by diverse but specific cell stressors.
A) HCT116 cells were treated with a panel of cellular stressors. Whole cell lysates were analyzed by SDS-PAGE and immunoblotted with the indicated antibodies. (s) and (l) denote short and long exposures, respectively.
B) Quantification of the percent RPS3 (left) or RPS2 (right) that is ub-modified from untreated cells or cells treated with the indicated cellular stressor. Error bars represent SEM from combined time course treatments for each cellular stressor. * indicates a p-value of < 0.05 comparing untreated cells to the combined time points for each cell stressor using Student's t-test. ER stress – DTT (D), Tunicamycin (Tm, T); Mitochondrial Stress – Antimycin A (AMA, A), Rotenone (Rot, R); Translation inhibitors – Cycloheximide (CHX, C), Anisomycin (ANS, A), Harringtonine (HTN, H); Translation initiation inhibitors – 4E1RCat (4E1R, 4), Torin-1 (TRN, T), Sodium Arsenite (NaAsO2, A); DNA damage inducers – Etoposide (Etop, E), Mitomycin-C (MMC, M), Cisplatin (Cptn, C), UV (U); Protein folding stressors – Azetidine-2-carboxylic acid (AZC, A), Canavanine (CAN, C), Pifithrin-μ (HSP70i, 7), Exposure to 42°C (Heat shock, H), Serum starvation overnight followed by re-addition of full media (SS Recovery).
See also Figure S3
To test if the UV and translation inhibitor-induced ubiquitylation of RPS2 and RPS3 observed by immunoblotting corresponded to site-specific ubiquitylation of 40S ribosomal proteins detected by mass spectrometry upon DTT and Tm treatment, we profiled alterations to the ubiquitin-modified proteome upon UV, HTN, and ANS treatment. This analysis revealed that UV treatment resulted in a time dependent increase not only in known UV-stimulated ubiquitylation events on XPC, FANCD2, and PCNA, but also in previously unseen ubiquitylation events on K58 and K275 on RPS2, K214 and K230 on RPS3, as well as K8 on RPS20 (Figure S3B,C). Immunoblotting confirmed that RPS2 and RPS3 ubiquitylation was rapidly induced upon UV treatment (Figure S3A). Similarly, treatment with HTN or ANS induced the site-specific ubiquitylation of RPS2, RPS3, and RPS20 (Figure S3D,E). These results demonstrate that diverse activators of the UPR, as well as direct inhibitors of translation elongation, result in site-specific RRub.
RPS2 and RPS3 ubiquitylation occurs on monosomes and polysomes
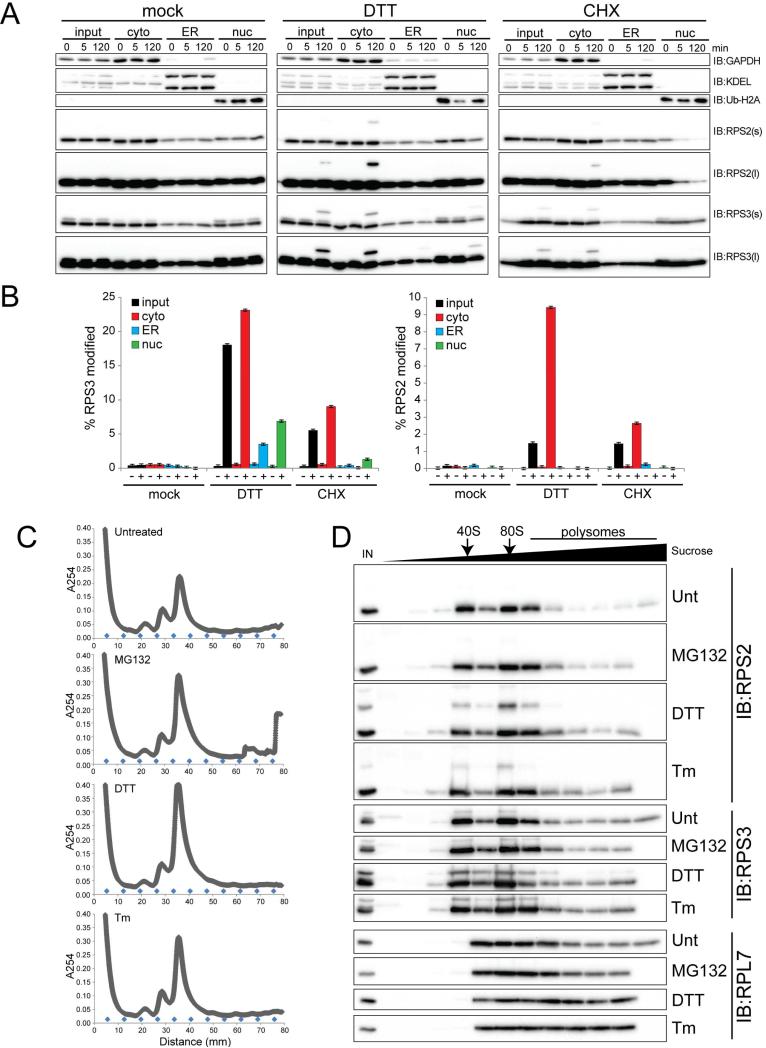
Ribosomes are localized to diverse cellular compartments. Given that ubiquitylation of ribosomal proteins contributes to ribosome biogenesis, the observed ubiquitylation of 40S proteins might be part of the ribosome assembly cascade (Shcherbik and Pestov, 2010). Given our observation that diverse ER stressors induce RRub, it is also possible that RRub occurs on ER-docked ribosomes. Furthermore, RRub could take place on unassembled ribosomal proteins rather than on assembled ribosomes. To evaluate these possibilities, we used subcellular fractionation to directly observe where in the cell RRub occurs. Using an established differential detergent fractionation technique, we separated cells into cytosolic, membrane-enriched (ER included), and nuclear fractions (Jagannathan et al., 2011). Compared to mock treated cells, DTT treatment resulted in robust ubiquitylation of RPS2 and RPS3 predominantly within the cytoplasmic fraction (Figure 4A). Longer exposures of the immunoblots revealed that a smaller portion of RPS3, but not RPS2, was ubiquitylated within the ER and nuclear-enriched fractions (Figure 4A,B). This result was specific for DTT treatment as CHX treatment resulted in RPS2 and RPS3 ubiquitylation that was detectable almost exclusively in the cytoplasmic fraction (Figure 4A,B). The observation that a minor portion of RPS3 ubiquitylation was detectable in non-cytoplasmic fractions could be due to technical limitations of the fractionation procedure or that regulatory ubiquitylation of RPS3 has separate functions within isolated sub-cellular compartments. However, these results clearly indicate that RRub occurs primarily on cytosolic ribosomes.
Figure 4. Cytosolic elongating ribosomes and free 40S subunits contain ubiquitylated RPS2 and RPS3.
A) 293T cells treated as indicated were separated into cytoplasmic (cyto), endoplasmic reticulum (ER), and nuclear (nuc) enriched fractions using differential detergent-based fractionation. Whole cell lysates and separated fractions were blotted with the indicated antibodies.
B) The percent RPS3 (left) or RPS2 (right) that is ubiquitin-modified in each fraction was quantified at the 120 minute time point from the immunoblots depicted in panel A.
C) HCT116 cells were untreated or treated with DTT, Tm, or MG132. Whole cell lysates were separated on a 10-50% linear sucrose gradient. The rRNA absorbance (A254) reading during subsequent fraction collection is depicted. The blue diamonds indicate the position of each fraction.
D) Sucrose gradient fractions from cells treated as indicated were analyzed by SDS-PAGE and immunoblotted with the indicated antibodies. IN represents the whole cell lysate input. Arrows depict the position of the 40S and 80S particles.
See also Figure S4
Cytosolic ribosomes are present in distinct 40S, 60S, 80S, and polysomal ribosome fractions. To examine if RPS2 and RPS3 ubiquitylation was present on elongating ribosomes or solely on free 40S ribosomal subunits, we separated native cell lysates on sucrose density gradients prior to immunoblotting. Cells were either untreated or treated with DTT, Tm, or the proteasome inhibitor MG132 prior to fractionation using sucrose density gradients. As expected, all treatments resulted in an accumulation of the 80S monosome population, as indicated by rRNA absorbance readings across the gradient (Figure 4B). Examining 40S protein ubiquitylation, we found that treatment with DTT, and to a lesser extent Tm, stimulated RPS2 and RPS3 ubiquitylation in fractions enriched for isolated 40S and 80S particles (Figure 4C). RPS2 and RPS3 ubiquitylation within polysomal fractions was observed in longer exposures of the immunoblots, as well as when cycloheximide was added to the lysis buffer to stabilize polysomes (Figure S4). Together, these findings reveal that UPR-induced ubiquitylation of distinct lysine residues on 40S proteins occurs on both assembled and elongating ribosomes.
PERK and eIF2α phosphorylation are necessary but not sufficient for ER stress induced RRub
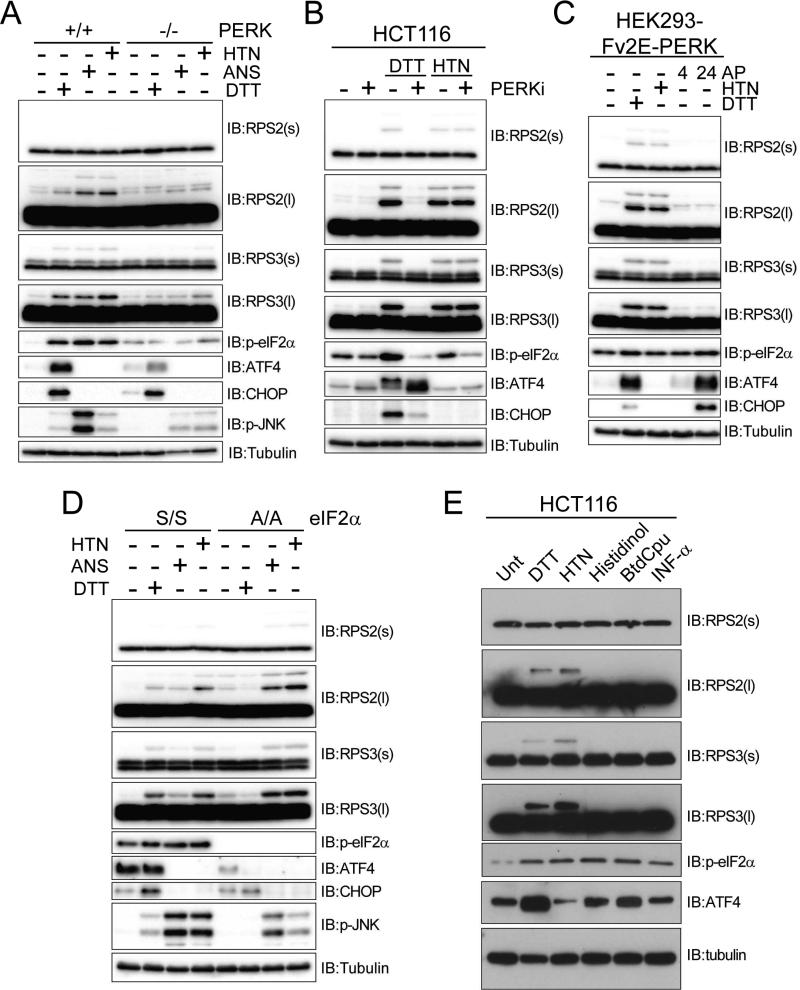
The UPR consists of three main branches with distinct mechanisms of activation and downstream effectors (Back and Kaufman, 2012). One branch is defined by the activation of the resident ER kinase PERK (EIF1AK3). To test if RRub was dependent on the PERK branch of the UPR, we examined RPS2 and RPS3 ubiquitylation in mouse embryonic fibroblasts (MEFs) derived from PERK knockout (KO) mice (Harding et al., 2000b). RPS2 and RPS3 ubiquitylation induced by DTT, or by HTN or ANS, was significantly impaired in MEFs lacking PERK compared to control MEFs (Figure 5A). This result was specific to the PERK branch of the UPR, as DTT-induced RRub was not dependent upon the IRE1 or ATF6 branch of the UPR pathway (Figure S5A,B) (Lee et al., 2002; Wu et al., 2007). To control for possible differences between PERK KO and control MEFs arising from chronic loss of PERK function, we treated HCT116 cells with a small molecule inhibitor of PERK (Harding et al., 2012). Pharmacological PERK inhibition completely blocked DTT, but not HTN-induced RPS2 and RPS3 ubiquitylation (Figure 5B). Surprisingly, PERK activation using engineered cell lines expressing ligand-dimerizable PERK in the absence of overt UPR stimulation was insufficient to induce RRub despite clear PERK activation (Figure 5C) (Lin et al., 2009; Lu et al., 2004). Stimulation of downstream UPR signaling without overt UPR activation was also insufficient to induce RRub, suggesting that RRub is a proximal event within the UPR that requires full UPR activation (Figure S5C) (Shoulders et al., 2013). Together, these results demonstrate that ER-stress induced RRub is dependent upon PERK, but PERK activation alone is not sufficient for RRub. These results also suggest that RRub can be activated in a PERK independent manner through direct inhibition of translation.
Figure 5. PERK mediated eIF2α phosphorylation is necessary but not sufficient for UPR-induced regulatory 40S ribosomal ubiquitylation.
A) Whole cell lysates from immortalized mouse embryonic fibroblasts derived from wildtype or PERK knockout mice treated with DTT, harringtonine (HTN), or anisomycin (ANS) were analyzed by SDS-PAGE and immunoblotted with the indicated antibodies.
B) Whole cell lysates from HCT116 cells treated with DTT or HTN alone or in combination with a PERK inhibitor were analyzed by SDS-PAGE and immunoblotted with the indicated antibodies.
C) HEK293 cells expressing Fv2E-PERK were treated with DTT or HTN for 4 hrs, or the dimerizing ligand AP20187 (AP) for 4 or 24 hrs. Whole cell lysates were analyzed by SDS-PAGE and immunoblotted with the indicated antibodies.
D) Whole cell lysates from immortalized mouse embryonic fibroblasts derived from mice expressing wildtype eIF2α (S/S) or S51A mutant eIF2α (A/A) were treated as indicated and analyzed by SDS-PAGE and immunoblotted with the indicated antibodies.
E) HCT116 cells were treated with a panel of individual eIF2α kinase inducers. Whole cell lysates were analyzed by SDS-PAGE and immunoblotted with the indicated antibodies. See also Figure S5
Similar to what we observed with pharmacological PERK inhibition, MEFs containing a mutant eIF2α unable to be phosphorylated upon UPR induction (A/A) were deficient in DTT-induced RRub (Figure 5D) (Scheuner et al., 2001). HTN and ANS were able to induce RRub in both wildtype and mutant eIF2α MEFs, indicating that eIF2α phosphorylation, like PERK, is only necessary for ER stress-induced RRub (Figure 5D). Our demonstration that sodium arsenite treatment, a potent activator of eIF2α phosphorylation, did not robustly induce RPS2 or RPS3 ubiquitylation suggested that eIF2α phosphorylation alone was not sufficient to induce RRub (Figure 3A). Since eIF2α can also be phosphorylated by kinases other than PERK as part of the integrated stress response, we tested whether agents that activate GCN2/EIF2AK4 (histidinol), HRI/EIF2AK1 (BTdCPU), or PKR/EIF2AK2 (α-interferon) were capable of inducing RRub (Harding et al., 2003). Although treatment with each resulted in eIF2α phosphorylation and ATF4 induction, they failed to induce or weakly induced RPS2 or RPS3 ubiquitylation (Figure 5E, S5D).
Defective RRub results in an elevated sensitivity to UPR-induced cell death
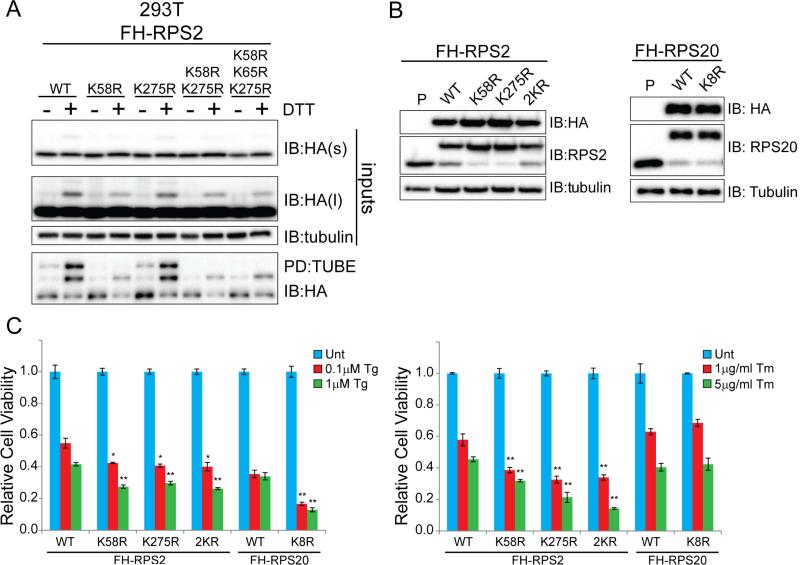
An inability to reduce protein synthesis upon UPR activation results in enhanced cell death upon exposure to UPR inducers (Harding et al., 2000b; Scheuner et al., 2001). To test if an inability to catalyze RRub similarly alters the cell death response upon UPR induction, we mutated individual lysine residues on 40S proteins demonstrated to be ubiquitylated in response to UPR activation. We mutated lysine residues 58 and 275 to arginine in RPS2, singly and in combination. We also mutated lysine 214 in RPS3 and lysine 8 in RPS20 and stably expressed the mutated forms, as well as wild type RPS2, RPS3, and RPS20 in mammalian cells (Figure 6A,B, S6A-E). Mutation of lysine 58, but not 275, substantially decreased the abundance of DTT-induced RPS2 mono-ubiquitylation in both whole cell lysates and TUBE enriched fractions (Figure 6A, S6A,B). A closer examination of RPS2 ubiquitylation revealed that the K58R mutation resulted in a near complete loss of the doubly ubiquitin (bi-ubiquitin) modified form of RPS2 (Figure S6A,B). Mutation of the neighboring ubiquitylated lysine residue, K65, did not further diminish the residual RPS2 mono-ubiquitylation observed upon K58R mutation. Taken together, these results suggest that lysine 58 largely contributes to the regulatory RPS2 ubiquitylation observed by immunoblotting. Mutation of lysine 214 in RPS3 completely blocked DTT-induced RPS3 ubiquitylation (Figure S6D,E). Sucrose density gradient analysis confirmed incorporation of exogenous wild-type and mutant RPS2, RPS3, and RPS20 into assembled ribosomal complexes (data not shown).
Figure 6. Loss of site-specific regulatory ubiquitylation of RPS2 and RPS20 enhances ER-stress induced cell death.
A) 293T cells stably expressing Flag-HA (FH) tagged wild type or mutant RPS2 were untreated or treated with DTT. Whole cell lysates (inputs) or TUBE-enriched fractions were analyzed by SDS-PAGE and immunoblotted with the indicated antibodies.
B) Whole cell lysates from 293T cells stably expressing Flag-HA (FH) tagged wild type RPS2 (WT), RPS20 (WT), RPS2K58R, RPS2K275R, RPS2K58RK275R (2KR), or RPS20K8R were analyzed by SDS-PAGE and immunoblotted with the indicated antibodies. P=parental 293T cell line. C) 293T cells with stable expression of the indicated wild type or mutant ribosomal proteins were either untreated (Unt) or treated with thapsigargin (Tg, left) or tunicamycin (Tm,right) at the indicated concentrations for 48hrs. Cell viability was measured using Celltiter-Glo. Error bars represent SEM from triplicate measurements for each condition. *, and ** indicates a p-value of < 0.05 and 0.01, respectively, compared to the respective wild type control cell line using Student's t-test.
See also Figure S6
Interestingly, expression of epitope-tagged versions of RPS2 and RPS20, but not RPS3, resulted in a substantial reduction in the overall protein abundance of the endogenous versions of the 40S proteins (Figure 6B, S6C). This finding allowed us to test the phenotypic consequences associated with the loss of site-specific regulatory ubiquitylation events on RPS2 and RPS20. Examination of UPR activation upon Tg treatment in RPS2K58R and RPS2K275R expressing cell lines did not reveal any obvious defects in eIF2α phosphorylation or ATF4 and CHOP induction compared to cell lines expressing wild type RPS2 (Figure S6F). Overall UPR activation upon Tg treatment was similarly unaffected in RPS20K8R expressing cells compared to wild type controls (Figure S6F). Despite having no delay in ATF4 or CHOP activation, RPS2K58R, RPS2K275R, and RPS20K8R expressing cell lines were sensitive to thapsigargin-induced cell death (Figure 6C). Interestingly, RPS2K58R and RPS2K275R, but not RPS20K8R expressing cell lines were also sensitive to tunicamycin-induced cell death (Figure 6C). We observed no enhancement in cell death upon DTT treatment in any of the mutant ribosome expressing cell lines, likely due to the inherent profound cytotoxicity observed upon DTT treatment (Figure S6G). Together, these results demonstrate that failure to ubiquitylate RPS2 at either K58 or K275 enhances sensitivity to ER-stress induced cell death using two distinct ER stressors.
RRub is conserved in fruit flies and yeast
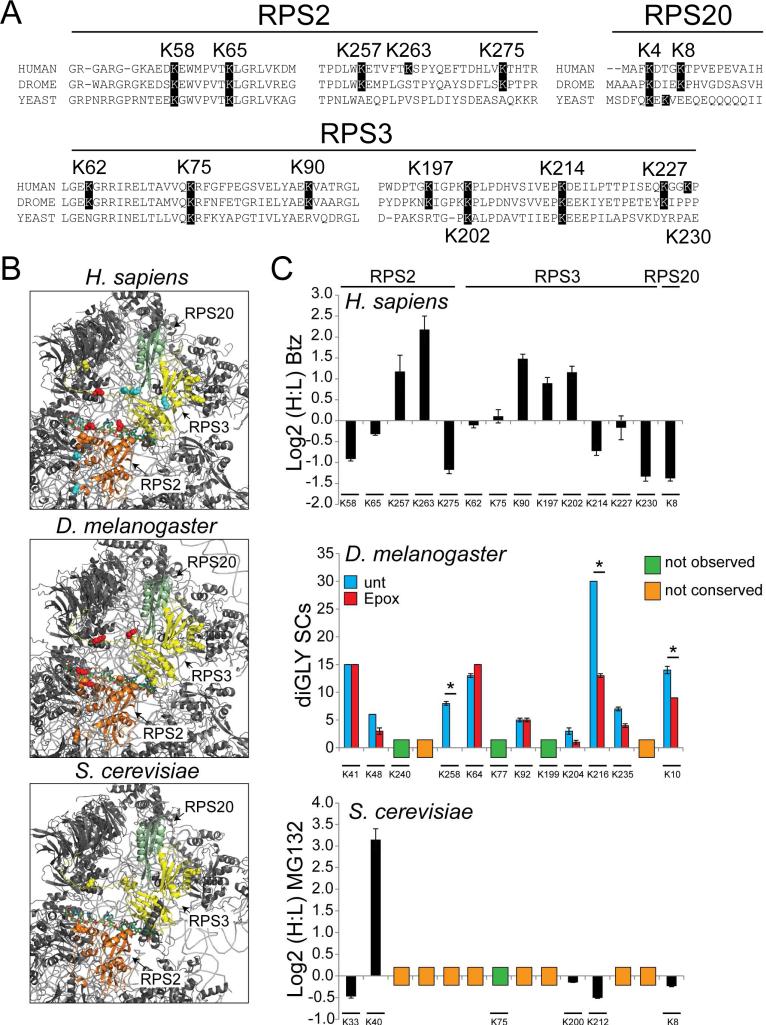
Previous studies have suggested that lysine residues found to be targets for ubiquitylation were not more likely to be conserved than non-ubiquitin modified lysines residues (Beltrao et al., 2012). However, it remains possible that ubiquitylation events that are regulatory in nature are more conserved than ubiquitylation events that target the protein for degradation. Due to the site-specific nature of regulatory 40S ubiquitylation, we examined if the observed human 40S ubiquitylation was conserved in other species. Ribosomal proteins are generally well-conserved with more than 60% sequence identity observed between human and S. cerevisiae RPS2, RPS3, and RPS20 proteins (Figure S7A). A subset of observed diGly-modified lysine residues on the 40S proteins RPS2, RPS3, and RPS20, whose abundance was previously quantified in response to proteasome inhibition (Kim et al., 2011), were variably conserved at the sequence level (Figure 7A). The positions of regulatory ub-modified lysine residues on RPS2 and RPS3 within the 40S structure is highly similar in human, fly, and yeast ribosomal structures (Anger et al., 2013; Ben-Shem et al., 2011) (Figure 7B).
Figure 7. 40S regulatory ubiquitylation is conserved across eukaryotes.
A) Multiple sequence alignments of segments of RPS2, RPS3, and RPS20 from H. sapiens, D. melanogaster, and S. cerevisiae that contain ub-modified lysine residues. The highlighted lysine positions indicate lysine residues observed to be ub-modified in human cells (position is human numbering).
B) Structural representation of the solvent exposed surface of the 40S subunit from H. sapiens (PDB:4V6X), D. melanogaster (PDB:1V6W), and S. cerevisiae (PDB:4V7R). Representations and coloring are as in Fig 2C.
C) Top: SILAC (H:L) Log2 ratios for selected RPS2, RPS3, and RPS20 ub-modified peptides from HCT116 cells treated with bortezomib for 8 hours. Primary data is extracted from published datasets (Kim et al., 2011). Error bars represent SEM from multiple peptide MS quantifications. Middle: Spectral counts (SCs) for ub-modified peptides from untreated (blue bars) and epoxomicin treated (red bars) Drosophila S2 cells. Error bars represent SEM from triplicate measurements. * indicates a p-value of < 0.05 using Student's t-test. Bottom: SILAC (H:L) Log2 ratios for selected diGly-modified peptides from heavy labeled yeast cultures treated with MG132 prior to mixing with untreated, unlabeled control cells. Error bars represent SEM from multiple peptide MS quantifications. The position of the ub-modified lysine is aligned to the analogous human position. The lysine position within the D. melanogaster or S. cerevisiae 40S protein sequences is shown below. Lysine residues not found to be ub-modified or conserved are indicated.
To determine whether for lysines that were conserved, the ub-modification event was also conserved, we utilized our diGly-modified peptide enrichment approach to directly interrogate individual diGly-modified lysines in Drosophila S2 cells and S. cerevisiae. To assay how proteasome inhibition altered the abundance of individual diGly-modified peptides, cells from both species were either untreated or treated with a proteasome inhibitor prior to harvesting.
Drosophila cells, like human cells, responded to epoxomicin treatment with a robust increase in polyubiquitylated material detected by immunoblotting, as well as an increase in the ubiquitylation of the small heat shock protein Hsp23 detected by mass spectrometry (Figure S7B,C). Using the Drosophila lysates, we directly evaluated the same set of diGly-modified lysine residues on the 40S proteins RPS2, RPS3, and RPS20 for which we had previously quantified the alteration in abundance in human cells (Kim et al., 2011) (Figure 7C, Table S6). In general, ubiquitylation of RPS2, RPS3, and RPS20 was well-conserved between fly and humans, as 75% of the sequence conserved lysine residues were diGly-modified in Drosophila (Figure 7C). Interestingly, ubiquitylation of Drosophila RPS2 K258, RPS3 K216, and RPS20 K10 decreased in response to proteasome inhibition, similar to what we observed for the equivalent residues on human RPS2 K275, RPS3 K214, and RPS20 K8, respectively (Figure 7C).
For S. cerevisiae, overall sequence conservation with humans of ub-modified lysines is lower. However, five out of six sequence conserved lysines in the subset of interrogated 40S ribosomal protein lysines were ubiquitylated in S. cerevisiae (Figure 7C). We confirmed that MG132 treatment impaired proteasome function in S. cerevisiae by evaluating the abundance of an exogenously expressed truncated version of Gnd1 (tGnd1) that is unstable and a ubiquitin proteasome system target. MG132 treatment resulted in both the accumulation of tGnd1 and an increase in the abundance of diGly-modified ub peptides which confirmed proteasome inhibition (Figure S7D,E). Proteasome inhibition also resulted in a decrease in the ubiquitylation of S. cerevisiae Rps2 K33, RPS3 K200, and RPS20 K8, indicating possible regulatory ubiquitylation events similar to what was observed in human cells (Figure 7C, Table S6). In sum, these results suggest that regulatory ubiquitylation of the solvent exposed 40S ribosome surface distal to the 60S interface is conserved in a site-specific manner from yeast to man and represents an important facet of eukaryotic translational control.
Discussion
ER stress activates the UPR, which both increases the protein folding and degradation capacity of the ER, and globally decreases protein synthesis (Walter and Ron, 2011). This reduction of protein synthesis occurs through phosphorylation of eIF2α and the subsequent inhibition of translation initiation. Here, we have identified a second mechanism that signals ER stress to the translational apparatus through site-specific regulatory ubiquitylation of 40S ribosomal proteins (RRub). Our discovery of RRub demonstrates the power of quantitative proteomics combined with ub-modified peptide enrichment strategies to evaluate site-specific alterations in protein ubiquitylation upon protein homeostasis stress.
The canonical, and most well-studied, output of protein ubiquitylation is proteasome-dependent degradation. However, it is clear that a fraction of protein ubiquitylation does not target substrates for degradation, but rather imparts regulatory control similar to protein phosphorylation (Komander and Rape, 2012). In fact, the abundance of more than 30% of all quantified diGly-modified peptides is reduced or unchanged upon treatment with a proteasome inhibitor (Kim et al., 2011). This observation suggests that regulatory ubiquitylation is pervasive and plays a substantial role in the post-translational control of protein activity. Our results suggest that a subset of 40S ubiquitylation events is regulatory in nature. Several observations support this claim. First, the abundance of site-specific ubiquitylation events within 40S proteins is decreased upon proteasome inhibition. This proteasome inhibitor-induced deubiquitylation has been observed for nearly every protein known to be ub-modified in a regulatory fashion (Kim et al., 2011). Second, RRub occurs on assembled and elongating ribosomes, and not on free ribosomal proteins. Lastly, mutation of K58 or K275 in RPS2, K8 in RPS20, or K214 in RPS3 does not alter ribosomal protein stability. These observations argue that other ubiquitylation events on ribosomal proteins that similarly decrease in abundance upon proteasome inhibition may serve an uncharacterized regulatory function.
While four separate activators of the UPR (DTT, Tm, Tg, and UV) stimulate RRub, they do so with varying robustness (Figure 3, S2). Each of the canonical ER stressors activate the UPR to similar degrees as judged by ATF4 and CHOP induction, however DTT routinely stimulates RRub to a greater extent. These observations indicate that other upstream factors, distinct from the well-studied UPR components, may contribute to the signaling events leading to RRub. This hypothesis is substantiated by our demonstration that PERK and eIF2α phosphorylation are both necessary but not sufficient events for RRub. Further, direct inhibition of translation potently induces RRub in a manner that is not dependent upon PERK or eIF2α phosphorylation (Figure 5). It is possible either that UPR activation and translation inhibition represents two distinct RRub activation pathways, or that each perturbation converges upon an unknown regulatory step during the translation cycle. A simple hypothesis that any block in translation can activate RRub is insufficient to describe why agents that inhibit translation initiation at the level of eIF4 complex association, or overt stimulation of eIF2α phosphorylation do not induce RRub (Figure 3). A more detailed examination of the cellular signaling events that distinguish between activators and non-activators of RRub is required to precisely determine the upstream components that mediate and execute RRub.
Our demonstration that failure to ubiquitylate specific lysine residues on individual 40S ribosomal protein results in enhanced sensitivity to UPR-induced cell death suggests that RRub is a functional component within the UPR (Figure 6). This observation begs the question: what is the purpose of RRub during the UPR? We envision three possible roles for RRub during the UPR. First, RRub may simply represent a previously uncharacterized mechanism to reversibly inhibit translation during UPR activation. It is also possible that RRub plays pleiotropic roles during the translation cycle. Recent structural characterization of the ribosomal pre-initiation complex (PIC) indicates that ubiquitylated lysine residues within the 40S proteins described within this study may be occluded within the PIC (Erzberger et al., 2014; Hashem et al., 2013; Hussain et al., 2014). As such, 40S ubiquitylation might help prevent PIC assembly and translation initiation during protein homeostasis stress.
A second possibility is that RRub assists in translational reprogramming during the UPR by catalyzing translation of specific mRNAs. Recent ribosome profiling studies indicate that hundreds of mRNAs, many of which do not contain uORFs, display increased ribosome density upon UPR activation (Reid et al., 2014). It is possible that RRub plays a role in targeting these mRNAs for translation during the UPR.
Third, RRub may nucleate a signaling cascade that assists in restoring protein homeostasis upon UPR activation. Ubiquitylated ribosomes may recruit factors that assist in mRNA or protein destruction on stalled ribosomal complexes. Our observations that RRub can occur on elongating 80S complexes, and that translation elongation inhibitors stimulate regulatory 40S ubiquitylation suggests that translational stalling might be a trigger for RRub. A substantial fraction of translation reactions terminate with ub-dependent removal of defective nascent chains (Duttler et al., 2013; Schubert et al., 2000; Wang et al., 2013). RRub may play a quality control role by nucleating complexes required for either defective mRNA or nascent chain degradation.
The apparent conservation of regulatory 40S ubiquitylation across all eukaryotes suggests that these ubiquitylation events play a critical role in mediating control of protein biogenesis during protein homeostasis dysfunction. This reversible ubiquitin-dependent mechanism to regulate UPR function represents a new path toward modulating the response to protein homeostasis stress. We establish RRub as direct connection between the ubiquitin proteasome system and translational control.
Experimental Procedures
SILAC-based quantitative diGly proteomics
Preparation of cell extracts for diGly-peptide immunoaffinity enrichment and total proteome analysis was performed essentially as described (Kim et al., 2011).
Sucrose density gradient separation
Cells were lysed in polysome lysis buffer both with and without 100μg/ml cycloheximide in the lysis buffer. Clarified lysates were then separated on a linear 10-50% sucrose gradient with manual fraction collection.
Cell death assay
Cells expressing either Flag-HA-tagged wild type or mutant RPS2 or RPS20 were treated with thapsigargin at the indicated concentration for 48 hours. Cell viability was measured using Celltiter-Glo (Promega) with a GloMax microplate luminometer (Promega).
Supplementary Material
Highlights.
ER stressors induce distinct alterations to the ubiquitin modified proteome.
UPR activation induces site specific regulatory ubiquitylation of ribosomal proteins.
Regulatory ubiquitylation of individual 40S proteins is conserved from yeast to man
Failure to ubiquitylate individual 40S proteins enhances UPR stimulated cell death
Acknowledgements
We thank S. Beausoleil (Cell Signaling Technologies), J. W. Harper and A. Ordureau (Harvard Medical School), R. Kaufman (Sanford-Burnham Medical Research Institute), D. Ron (Univ. of Cambridge), M. Niwa-Rosen and A. Tam (UCSD), L. Wiseman (Scripps Research Institute) for reagents and G. Yeo, K. Kapeli, J. Lykke-Andersen and M. Arribas-Layton (UCSD) for assistance with sucrose gradient separation. This work was supported by New Scholar awards from the Sidney Kimmel Foundation for Cancer Research and the Ellison Medical Foundation and a Hellman Fellowship (E.J.B.), a seed grant from the San Diego Center for Systems Biology (R.H.), NIH 5R01GM092748-04 (R.Y.H), and NIH RO1 GM05054516 (S.A.W.). N.Z. is supported by the UCSD Cell and Molecular Genetics Training Program (T32 GM007240).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
J.M.G and E.J.B conceived the study. J.M.G. performed the SILAC-based proteomic experiments with assistance from K.W. J.M.G, R.H., L.R., and R.M. performed all immunoblotting, immunoprecipitation, and TUBE enrichment experiments. R.M. performed the cell fractionation experiments. P.R., L.R., and R.H. generated point mutations and stable cell expression lines. N.Z. and R.H. performed the sucrose gradient analysis. S.E.K. assisted in ribosome structural analysis. E.F. and J.C.C. performed the eIF2α kinase activator experiments. X.T., R.Y.H, S.A.L, and S.A.W provided yeast and fly samples and assisted B.Y. in subsequent proteomic analysis. J.M.G, R.H., and E.J.B. performed data analysis and E.J.B. wrote the paper with input from J.M.G., R.H., J.C.C., and S.A.W.
See supplemental experimental procedures for detailed methods.
References
- Andreev DE, O'Connor PB, Fahey C, Kenny EM, Terenin IM, Dmitriev SE, Cormican P, Morris DW, Shatsky IN, Baranov PV. Translation of 5′ leaders is pervasive in genes resistant to eIF2 repression. eLife. 2014;4:e03971. doi: 10.7554/eLife.03971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anger AM, Armache JP, Berninghausen O, Habeck M, Subklewe M, Wilson DN, Beckmann R. Structures of the human and Drosophila 80S ribosome. Nature. 2013;497:80–85. doi: 10.1038/nature12104. [DOI] [PubMed] [Google Scholar]
- Back SH, Kaufman RJ. Endoplasmic reticulum stress and type 2 diabetes. Annu Rev Biochem. 2012;81:767–793. doi: 10.1146/annurev-biochem-072909-095555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrao P, Albanese V, Kenner LR, Swaney DL, Burlingame A, Villen J, Lim WA, Fraser JS, Frydman J, Krogan NJ. Systematic functional prioritization of protein posttranslational modifications. Cell. 2012;150:413–425. doi: 10.1016/j.cell.2012.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M. The structure of the eukaryotic ribosome at 3.0 A resolution. Science. 2011;334:1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- Bustos D, Bakalarski CE, Yang Y, Peng J, Kirkpatrick DS. Characterizing ubiquitination sites by peptide-based immunoaffinity enrichment. Molecular & cellular proteomics : MCP. 2012;11:1529–1540. doi: 10.1074/mcp.R112.019117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo SE, Pagliarini DJ, Mootha VK. Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:7507–7512. doi: 10.1073/pnas.0810916106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrano AC, Bennett EJ. Using the ubiquitin-modified proteome to monitor protein homeostasis function. Molecular & cellular proteomics : MCP. 2013;12:3521–3531. doi: 10.1074/mcp.R113.029744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson JC, Ye Y. Cleaning up in the endoplasmic reticulum: ubiquitin in charge. Nature structural & molecular biology. 2014;21:325–335. doi: 10.1038/nsmb.2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duttler S, Pechmann S, Frydman J. Principles of cotranslational ubiquitination and quality control at the ribosome. Molecular cell. 2013;50:379–393. doi: 10.1016/j.molcel.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erzberger JP, Stengel F, Pellarin R, Zhang S, Schaefer T, Aylett CH, Cimermancic P, Boehringer D, Sali A, Aebersold R, et al. Molecular architecture of the 40SeIF1eIF3 translation initiation complex. Cell. 2014;158:1123–1135. doi: 10.1016/j.cell.2014.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan BJ, Krokowski D, Majumder M, Schmotzer CL, Kimball SR, Merrick WC, Koromilas AE, Hatzoglou M. Translational control during endoplasmic reticulum stress beyond phosphorylation of the translation initiation factor eIF2alpha. J Biol Chem. 2014;289:12593–12611. doi: 10.1074/jbc.M113.543215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Molecular cell. 2000a;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Bertolotti A, Zeng H, Ron D. Perk is essential for translational regulation and cell survival during the unfolded protein response. Molecular cell. 2000b;5:897–904. doi: 10.1016/s1097-2765(00)80330-5. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Molecular cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zyryanova AF, Ron D. Uncoupling proteostasis and development in vitro with a small molecule inhibitor of the pancreatic endoplasmic reticulum kinase, PERK. J Biol Chem. 2012;287:44338–44344. doi: 10.1074/jbc.M112.428987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashem Y, des Georges A, Dhote V, Langlois R, Liao HY, Grassucci RA, Hellen CU, Pestova TV, Frank J. Structure of the mammalian ribosomal 43S preinitiation complex bound to the scanning factor DHX29. Cell. 2013;153:1108–1119. doi: 10.1016/j.cell.2013.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain T, Llacer JL, Fernandez IS, Munoz A, Martin-Marcos P, Savva CG, Lorsch JR, Hinnebusch AG, Ramakrishnan V. Structural changes enable start codon recognition by the eukaryotic translation initiation complex. Cell. 2014;159:597–607. doi: 10.1016/j.cell.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Brar GA, Stern-Ginossar N, Harris MS, Talhouarne GJ, Jackson SE, Wills MR, Weissman JS. Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell reports. 2014;8:1365–1379. doi: 10.1016/j.celrep.2014.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagannathan S, Nwosu C, Nicchitta CV. Analyzing mRNA localization to the endoplasmic reticulum via cell fractionation. Methods Mol Biol. 2011;714:301–321. doi: 10.1007/978-1-61779-005-8_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ, et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Molecular cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- Lee K, Tirasophon W, Shen X, Michalak M, Prywes R, Okada T, Yoshida H, Mori K, Kaufman RJ. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes & development. 2002;16:452–466. doi: 10.1101/gad.964702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JH, Li H, Zhang Y, Ron D, Walter P. Divergent effects of PERK and IRE1 signaling on cell viability. PloS one. 2009;4:e4170. doi: 10.1371/journal.pone.0004170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopitz-Otsoa F, Rodriguez-Suarez E, Aillet F, Casado-Vela J, Lang V, Matthiesen R, Elortza F, Rodriguez MS. Integrative analysis of the ubiquitin proteome isolated using Tandem Ubiquitin Binding Entities (TUBEs). Journal of proteomics. 2012;75:2998–3014. doi: 10.1016/j.jprot.2011.12.001. [DOI] [PubMed] [Google Scholar]
- Lu PD, Jousse C, Marciniak SJ, Zhang Y, Novoa I, Scheuner D, Kaufman RJ, Ron D, Harding HP. Cytoprotection by pre-emptive conditional phosphorylation of translation initiation factor 2. The EMBO journal. 2004;23:169–179. doi: 10.1038/sj.emboj.7600030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen J, Bennett EJ. Protecting the proteome: Eukaryotic cotranslational quality control pathways. The Journal of cell biology. 2014;204:467–476. doi: 10.1083/jcb.201311103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimnaugh EG, Chen HY, Davie JR, Celis JE, Neckers L. Rapid deubiquitination of nucleosomal histones in human tumor cells caused by proteasome inhibitors and stress response inducers: effects on replication, transcription, translation, and the cellular stress response. Biochemistry. 1997;36:14418–14429. doi: 10.1021/bi970998j. [DOI] [PubMed] [Google Scholar]
- Reid DW, Chen Q, Tay AS, Shenolikar S, Nicchitta CV. The unfolded protein response triggers selective mRNA release from the endoplasmic reticulum. Cell. 2014;158:1362–1374. doi: 10.1016/j.cell.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resch AM, Ogurtsov AY, Rogozin IB, Shabalina SA, Koonin EV. Evolution of alternative and constitutive regions of mammalian 5′UTRs. BMC genomics. 2009;10:162. doi: 10.1186/1471-2164-10-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano R, Reed JC. ER stress-induced cell death mechanisms. Biochim Biophys Acta. 2013;1833:3460–3470. doi: 10.1016/j.bbamcr.2013.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuner D, Song B, McEwen E, Liu C, Laybutt R, Gillespie P, Saunders T, Bonner-Weir S, Kaufman RJ. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Molecular cell. 2001;7:1165–1176. doi: 10.1016/s1097-2765(01)00265-9. [DOI] [PubMed] [Google Scholar]
- Schubert U, Anton LC, Gibbs J, Norbury CC, Yewdell JW, Bennink JR. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature. 2000;404:770–774. doi: 10.1038/35008096. [DOI] [PubMed] [Google Scholar]
- Shcherbik N, Pestov DG. Ubiquitin and ubiquitin-like proteins in the nucleolus: multitasking tools for a ribosome factory. Genes & cancer. 2010;1:681–689. doi: 10.1177/1947601910381382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoulders MD, Ryno LM, Genereux JC, Moresco JJ, Tu PG, Wu C, Yates JR, 3rd, Su AI, Kelly JW, Wiseman RL. Stress-independent activation of XBP1s and/or ATF6 reveals three functionally diverse ER proteostasis environments. Cell reports. 2013;3:1279–1292. doi: 10.1016/j.celrep.2013.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi K, Motohashi H, Yamamoto M. Molecular mechanisms of the Keap1-Nrf2 pathway in stress response and cancer evolution. Genes Cells. 2011;16:123–140. doi: 10.1111/j.1365-2443.2010.01473.x. [DOI] [PubMed] [Google Scholar]
- Vattem KM, Wek RC. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:11269–11274. doi: 10.1073/pnas.0400541101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- Wang F, Durfee LA, Huibregtse JM. A cotranslational ubiquitination pathway for quality control of misfolded proteins. Molecular cell. 2013;50:368–378. doi: 10.1016/j.molcel.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff S, Weissman JS, Dillin A. Differential scales of protein quality control. Cell. 2014;157:52–64. doi: 10.1016/j.cell.2014.03.007. [DOI] [PubMed] [Google Scholar]
- Wu J, Rutkowski DT, Dubois M, Swathirajan J, Saunders T, Wang J, Song B, Yau GD, Kaufman RJ. ATF6alpha optimizes long-term endoplasmic reticulum function to protect cells from chronic stress. Developmental cell. 2007;13:351–364. doi: 10.1016/j.devcel.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Wu S, Hu Y, Wang JL, Chatterjee M, Shi Y, Kaufman RJ. Ultraviolet light inhibits translation through activation of the unfolded protein response kinase PERK in the lumen of the endoplasmic reticulum. J Biol Chem. 2002;277:18077–18083. doi: 10.1074/jbc.M110164200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.