SUMMARY
eIF4E, the major cap-binding protein, has long been considered limiting for translating the mammalian genome. However, the requirement for eIF4E dose at an organismal level remains unexplored. By generating an Eif4e haploinsufficient mouse, we found that 50% reduction in eIF4E expression, while compatible with normal development and global protein synthesis, significantly impeded cellular transformation. Genome-wide translational profiling uncovered a translational program induced by oncogenic transformation and revealed a critical role for eIF4E dose specifically in translating a network of mRNAs enriched for a unique 5′UTR signature. In particular, we demonstrate that eIF4E dose is essential for translating mRNAs regulating reactive oxygen species that fuel transformation and cancer cell survival in vivo. Our findings indicate that cancer cells hijack the eIF4E level in excess for normal development to drive a translational program supporting tumorigenesis.
INTRODUCTION
For decades, the major cap binding protein eukaryotic initiation factor 4E (eIF4E) has been recognized as the quantitatively limiting initiation factor for mRNA translation (Duncan et al., 1987; Hiremath et al., 1985). eIF4E is a critical component of the eIF4F tertiary translation initiation complex, which recognizes the 5′ 7-methyl guanosine cap of mRNAs and drives their translation. In addition to eIF4E, the eIF4F complex is comprised of the DEAD-box helicase eIF4A and eIF4G, a large protein scaffold that recruits the 40S ribosome subunit to mature mRNAs (Gingras et al., 1999). As the limiting component of the eIF4F complex, eIF4E expression levels are believed to be a critical determinant for translation of eukaryotic mRNAs (De Benedetti et al., 1991). In particular, the translation of mRNAs harboring structured 5′UTRs is thought to be exquisitely sensitive to eIF4E levels due to eIF4E’s ability to both recruit the eIF4A helicase to the 5′cap and to stimulate its activity in a cap-independent manner (Feoktistova et al., 2013). Collectively, even small changes in eIF4E levels are believed to broadly impact mRNA translation and thus cellular and organismal function (Figure 1A). However, the normal physiological role and threshold for eIF4E dose in organismal development in vivo is essentially unknown. Moreover, the specific requirements for eIF4E dose in both global and specific translation of mRNAs genome-wide remain poorly defined.
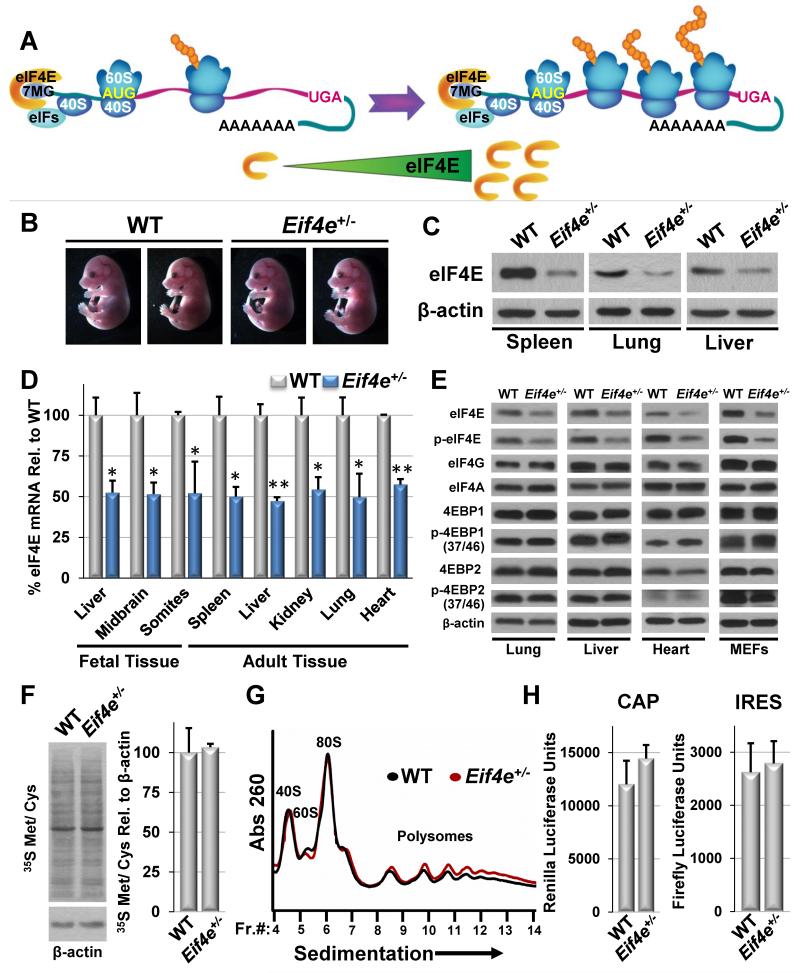
Figure 1. A 50% reduction in eIF4E dose is compatible with normal in vivo development and protein synthesis.
A) Cartoon demonstrating the current dogma of eIF4E as quantitatively limiting for protein synthesis. “7MG” denotes the 5′ 7-methylguanosine cap. “eIFs” denotes eukaryotic initiation factors that promotes the association of eIF4E with the 40S ribosomal subunit. B) Gross morphology of WT and Eif4e+/− E15.5 embryos. C) Representative western blots for eIF4E protein levels in adult tissues of mice. D) Quantitative polymerase chain reaction analysis of eIF4E mRNA levels in fetal (E11.5) and adult tissues. E) Representative western blots for eIF4F complex members and regulators in adult tissues and primary MEFs. F) Global protein synthesis measured by 35S methionine/cysteine incorporation in MEFs. G) Representative polysome profiles of WT and Eif4e+/− MEFs. “Fr.#” denotes the fraction number for sucrose gradient fractionation. Lower molecular weight (MW) complexes (40S and 60S) are on the left side of the x-axis and higher MW complexes (polysomes) are on the right. H) Cap and IRES dependent translation measured by luciferase activity in MEFs from the CMV-HCV-IREST reporter mouse. All values represent the mean +SEM. Results are representative of at least three independent experiments. See also Figures S1 and S2.
In this study, we generate a mouse model for Eif4e haploinsufficiency as a genetic tool to define the requirements for eIF4E dose in vivo. Surprisingly, we find that eIF4E exists in excess for normal development and global protein synthesis, and instead, becomes limiting under the specific condition of oncogene induced cellular transformation. Employing unbiased genome-wide profiling to identify the translational landscape of mRNAs induced by oncogenic transformation, we delineate an “oncogenic translation program” comprised of hundreds of mRNAs. Several key nodes in this program, including functional classes of mRNAs such as those involved in signaling and control of reactive oxygen species (ROS), are exquisitely sensitive to eIF4E dose.
One of the defining stress phenotypes that cancer cells encounter is the build-up of ROS, and cancer cells typically generate more ROS than normal cells (Szatrowski and Nathan, 1991). The ability of cancer cells to distinguish between ROS as a survival or apoptotic signal is controlled by the dosage and duration of ROS production, such that modest levels are required for cancer cell survival whereas excessive levels promote cell death (Trachootham et al., 2006; Weinberg et al., 2010). Here, we show that the capacity to properly translate mRNAs regulating intracellular ROS levels is critical for tumor cell survival. Indeed, we demonstrate that reductions in eIF4E cause a toxic accumulation of ROS that renders, at least in part, Eif4e haploinsufficient mice remarkably resistant to transformation. These studies thereby reveal a mechanism by which cancer cells regulate the translation of key stress response transcripts in a manner that is directly coupled to the dose of eIF4E in the cell. Collectively, these findings highlight that mammalian cells have evolved a surplus of eIF4E that exists above the required threshold for normal development and physiology and instead plays a specialized role in regulating mRNA translation under specific conditions, which has been coopted by tumor cells to weather the stress of oncogenic transformation.
RESULTS
In vivo requirements for a threshold of eIF4E
In order to address the in vivo requirements for a threshold of eIF4E, we generated an eIF4E knock-out mouse (Figure S1A-B). We reasoned that in the context of Eif4e heterozygosity (Eif4e+/−), the impact of 50% reductions in eIF4E levels could be assessed genetically at an organismal level to delineate the in vivo function of eIF4E dose. Surprisingly, Eif4e+/− mice are viable and indistinguishable from their WT littermates. For example, Eif4e+/− mice are born at normal Mendelian ratios (Table S1) and display normal body weight (Figure S1C) and survival (data not shown) within one year of age. In addition, Eif4e+/− embryos and adult mice show normal tissue architecture and development (Figure 1B and Figure S1D-E) with no obvious changes in cell size, survival, or proliferation (Figure S1F-G and Figure S2A-B). Moreover, Eif4e+/− embryos show equal sensitivity to developmental defects induced by exposure to teratogenic agents such as alcohol (Table S1).
To confirm that Eif4e+/− mice display reductions in eIF4E expression, we analyzed eIF4E mRNA and protein levels in these mice. We observe a 50% reduction in eIF4E expression in virtually all cell and tissue types examined in vivo (Figure 1C-D and Figure S2C-D). We next asked if there might be compensation for eIF4E loss of function through known translation initiation pathway regulators. However, we find no evidence of compensation in tissues or cells of Eif4e+/− mice at the level of expression or phosphorylation of any other component of the eIF4F initiation complex or of the inhibitor proteins, eIF4E Binding Protein 1 and 2 (4EBP1 and 4EBP2), which block the interaction of eIF4E with eIF4G (Figure 1E) (Bah et al., 2014; Haghighat et al., 1995). In addition, eIF4E activity can be directly regulated by phosphorylation at Serine 209 by the Mnk kinases, however, we see no signs of compensation for reduced eIF4E levels through enhanced phosphorylation (Figure 1E) (Waskiewicz et al., 1997). As expected, complete loss of function of eIF4E is not compatible with life and Eif4e−/− embryos die before E6.5 (data not shown). Together, these findings unexpectedly demonstrate that a 50% reduction of eIF4E is fully compatible with normal organismal development and physiology in vivo.
We next sought to determine the impact of a 50% reduction in eIF4E dose on protein synthesis control in vivo. Remarkably, Eif4e+/− mice display normal levels of global protein synthesis as assessed by incorporation of 35S labeled methionine in primary cells (Figure 1F and Figure S2E-F). Interestingly, reductions of eIF4E in yeast have been shown to cause a dramatic redistribution of mRNA from actively translating poly-ribosomes to monosomes with minimal impacts on global protein synthesis rates (von der Haar and McCarthy, 2002). However, we find no evidence for globally altered polysome profiles in Eif4e+/− cells under exponential growth conditions (Figure 1G and Figure S2G). Although the majority of translation is thought to be cap-dependent and thus rely on eIF4E activity, reductions in eIF4E could potentially affect internal ribosomal entry site (IRES)-mediated translation, an alternative form of translation that is independent of the 5′cap structure (Macejak and Sarnow, 1991). To address this question, we employed a transgenic mouse that stably expresses a bicistronic luciferase reporter for cap- and IRES-mediated translation in all tissues (CMV-HCV-IREST) (Bellodi et al., 2010; Hsieh et al., 2010). By crossing Eif4e+/− mice with CMV-HCV-IREST reporter mice and assaying Rluc and Fluc expression in primary MEFs, we find that Eif4e+/− mice are not impaired in either cap- or IRES-mediated translation. These findings demonstrate that reductions in eIF4E do not induce a switch from cap- to IRES-dependent translation (Figure 1H).
Eif4e haploinsufficiency during cellular transformation
We next asked if eIF4E expression levels are critical during stress conditions that impinge on protein synthesis, such as during the early steps downstream of oncogenic signaling (Barna et al., 2008). Oncogenic signaling increases expression and phosphorylation of eIF4E and leads to hyperphosphorylation and inactivation of the eIF4E inhibitor protein, 4EBP1 (Figure 2A-C) (Aoki et al., 2001; Rajasekhar et al., 2003; Rosenwald et al., 1993). Modest overexpression of an eIF4E transgene under the control of the ubiquitous β-actin promoter results in the formation of multiple cancer types (Ruggero et al., 2004) and phosphorylation of eIF4E, at Ser209, has been shown to be required for tumorigenesis (Furic et al., 2010). In addition, overexpression and hyperactivation of eIF4E is a common feature of many human cancers (De Benedetti and Graff, 2004). Yet, it remains an outstanding question how changes in the levels of eIF4E expression lead to cellular transformation and whether this is mediated through control of either global or transcript-specific mRNA translation.
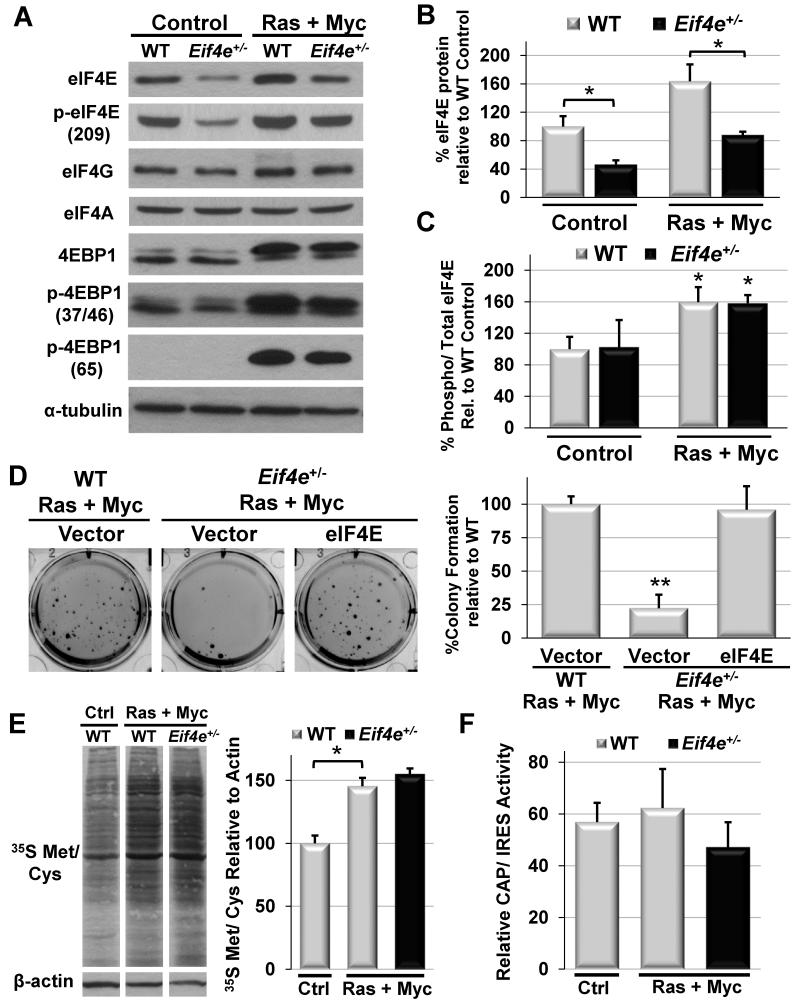
Figure 2. eIF4E is haploinsufficient for oncogenic transformation.
A) Representative western blots for eIF4F complex members and regulators in WT and Eif4e+/− MEFs before and after overexpression of HRasV12 and Myc. B) Quantification of total eIF4E protein levels relative to β-actin and normalized to primary WT MEFs. C) Quantification of eIF4E phosphorylation relative to total eIF4E levels in WT MEFs. D) Soft agar colony formation as a measure of oncogenic transformation upon overexpression of HRasV12 and Myc in WT and Eif4e+/− MEFs with and without eIF4E rescue (average WT vector colony number = 115). E) Global protein synthesis measured by 35S methionine/cysteine incorporation upon transformation in MEFs. F) Relative Cap and IRES luciferase activity in transformed MEFs derived from the CMV-HCV-IREST reporter mouse. Asterisks indicate a statistically significant change (*= p<0.05, **= p<0.01). All values represent the mean +SD. Results are representative of at least three independent experiments. See also Figure S2.
We first tested the ability of specific oncogenic insults, such as Ras and Myc as well as Ras and E1A, to transform primary Eif4e+/− mouse embryonic fibroblasts (MEFs) under conditions where eIF4E is genetically reduced by 50%. Notably, Eif4e+/− MEFs show a consistent 50% reduction in eIF4E expression levels even upon oncogenic insult (Figure 2A-B). Similar to steady state, no compensation is observed with respect to eIF4E phosphorylation or other components of the eIF4F initiation complex (Figure 2A-C). Additionally, no differences are observed in cell size or expression of the driving oncogenes in Eif4e+/− cells compared to WT (Figure S2H-I). Strikingly, Eif4e+/− MEFs are dramatically resistant to cellular transformation as measured by soft agar colony formation (Figure 2D and Figure S2J). In addition, transformation defects can be ascribed specifically to reductions in eIF4E dose as soft agar colony growth is rescued by restoring eIF4E expression levels (Figure 2D and Figure S2K). Thereby, eIF4E dose becomes critical under specific cellular stress conditions, such as during cellular transformation.
Profiling the oncogenic translation program
In order to gain insight into the underlying mechanism by which Eif4e+/− cells are resistant to oncogenic transformation, we next asked if eIF4E dose is critical for global protein synthesis during this cellular stress. Remarkably, although oncogenic transformation leads to substantial increases in global protein synthesis relative to untransformed cells, transformed Eif4e+/− MEFs show no defects in global protein synthesis rates as measured by incorporation of 35S labeled methionine or by assessing Rluc and FLuc expression in cells isolated from CMV-HCV-IREST reporter mice (Figure 2E-F). In line with this, reductions in eIF4E dose have no impact on the global redistribution of mRNAs from monosomes to actively translating polysomes during cellular transformation (Figure S2L). We therefore asked whether eIF4E dose is critical in regulating the translation of a specific subset of mRNAs integral to oncogenic transformation. We characterized the impact of oncogenic signaling on the translational landscape of gene expression by undertaking an unbiased translational profiling approach to monitor changes in poly-ribosomal associated mRNA and total transcript levels induced by cellular transformation in WT and Eif4e+/− MEFs (Figure 3A and Table S2). By comparing the relative differences in the ratio of polysomal mRNA isolated from high MW sucrose gradient fractions to total mRNA expression levels (termed translational efficiency), we identified 722 gene products whose translational efficiency (TE) was altered by oncogenic transformation with Ras and Myc in WT MEFs (Absolute fold change in TE >1.7, p <0.05; Figure 3B and Table S2). Importantly, we find the translation of a subset of 133 gene products is sensitive to eIF4E dose specifically during oncogenic transformation (Fold change in TE >1.7, p <0.05; Figure 3C, Figure S3A-B and Table S2). In order to better understand how eIF4E dose impacts expression of the translational program induced by oncogenic transformation, we performed network-based cluster analysis of interactions for the 722 genes that were either translationally induced (Figure 3D and Figure S3C) or repressed by transformation (Figure S3D). We find that gene sets translationally regulated by transformation are enriched in protein-protein interactions with several functional clusters emerging from this analysis, including those involved in cell cycle control, signaling, cell-to-cell communication, cell adhesion, and protein homeostasis. Moreover, this analysis highlights how specific subsets of these genes are translated in a manner sensitive to eIF4E dose. It is interesting to note that reductions in eIF4E dose are also associated with the increased translation of a subset of mRNAs. While the functional relevance remains to be determined, these could possibly reflect mRNAs that are translated in a cap-independent manner. Thus, employing unbiased genome-wide translational profiling, we identified a translational program comprised of 722 gene products translationally controlled during cellular transformation and demonstrated that eIF4E is critical for the translational activation of a specific subset of this ‘oncogenic translation program’.
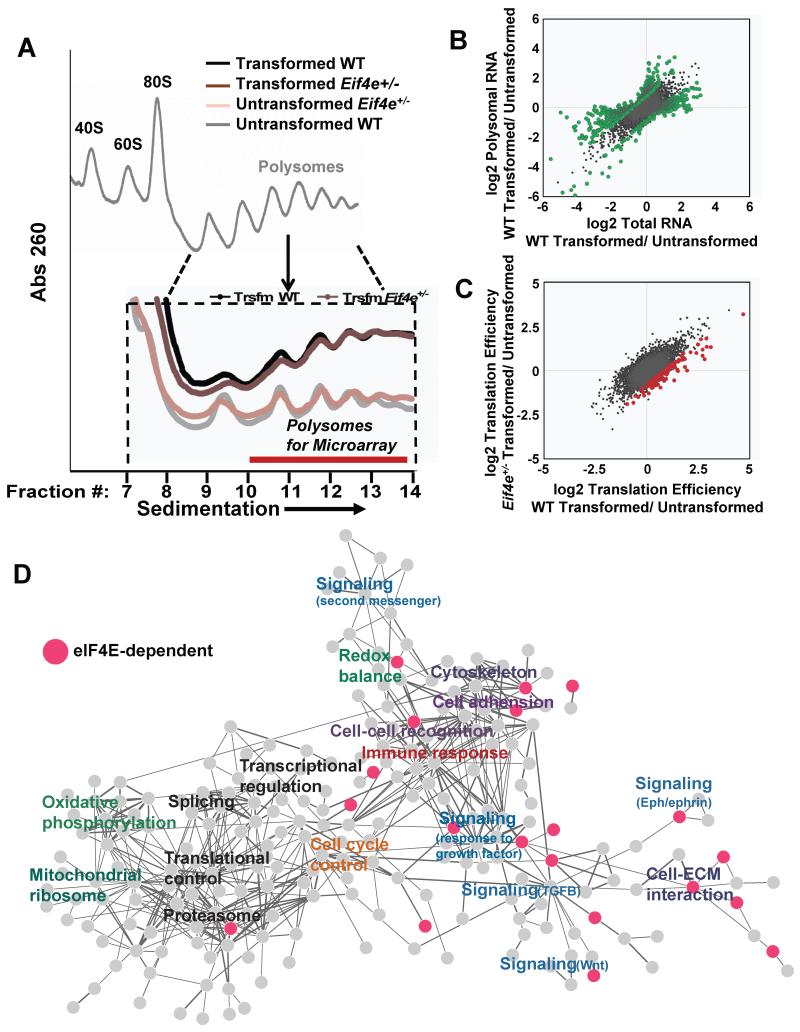
Figure 3. eIF4E is required for expression of the oncogenic translational program.
A) Representative polysome profiles of transformed and untransformed WT and Eif4e+/− MEFs separated on a sucrose gradient. Inset highlights the polysomal fractions (10-13) used for translational profiling by polysomal microarray. B) Changes in total RNA and polysome associated RNA in WT MEFs upon oncogenic transformation by HRasV12 and Myc. Green points indicate genes with a fold change in translation efficiency (polysomal/total RNA) greater than 1.7 fold, p<0.05 (See Table S3). C) Changes in translation efficiency (TE) induced by transformation in WT and Eif4e+/− MEFs. Red points indicate genes whose fold change in TE during transformation is reduced in Eif4e+/− MEFs compared to WT MEFs (>1.7 fold decrease in TE, p<0.05) (See Table S2). D) Network-based cluster analysis of genes and associated functional classes whose translation is increased upon transformation in WT MEFs. Pink nodes represent genes whose translation is impaired in transformed Eif4e+/− cells. See also Figure S3,
Since our network data analysis suggested that cellular transformation may promote the translation of distinct classes of mRNAs, we next performed gene set enrichment analysis (GSEA) to identify functional classes of genes whose translation is statistically enriched by cellular transformation and to evaluate their requirements for eIF4E dose. This analysis revealed a large and diverse group of functional ontologies whose translation was enhanced by cellular transformation, including classes of genes known to be translationally regulated downstream of oncogenic signaling, such as cell cycle, apoptosis, ribosome biogenesis, and nucleotide biosynthesis (Table S3) (Cunningham et al., 2014; Hsieh et al., 2010; Hsieh et al., 2012; Stumpf et al., 2013). In line with our network interactome data, we find that eIF4E dose is required for the expression of a subset of functional classes of genes that are translationally induced by transformation (Table S3). For example a 50% reduction of eIF4E significantly impacts the translation of genes involved in the proteasome, cell signaling, and nucleotide biosynthesis (Figure S4A and Table S4).
Strikingly, this analysis also revealed unexpected coordinate translational upregulation of genes involved in both the production of reactive oxygen species (ROS) and the regulation of ROS levels during cellular transformation. Both oncogenic signaling and deregulated mitochondrial respiration of cancer cells can contribute to ROS generation and oncogenic stress (Irani et al., 1997; Sattler et al., 2000; Vafa et al., 2002; Wallace, 2012). Importantly, the ability of cancer cells to modulate the dosage and duration of ROS production is vital for their growth and survival. Mitochondrial oxidative phosphorylation, the main source of cellular ROS (Wallace, 2012), is one of the functional classes most translationally enriched by transformation (Figure 4A, Figure S4B, and Table S5). In addition, we find that transformation enhances the translation of genes involved in detoxifying and regulating ROS levels, including glutathione metabolism genes (Figure S4B-C and Table S5). Remarkably, eIF4E dose appears to be critical during cellular transformation for select translation of mRNAs that control intracellular ROS levels but not for mRNAs associated with oxidative phosphorylation (Table S4).
Figure 4. eIF4E is critical for the translation of distinct functional classes of mRNAs induced by oncogenic transformation.
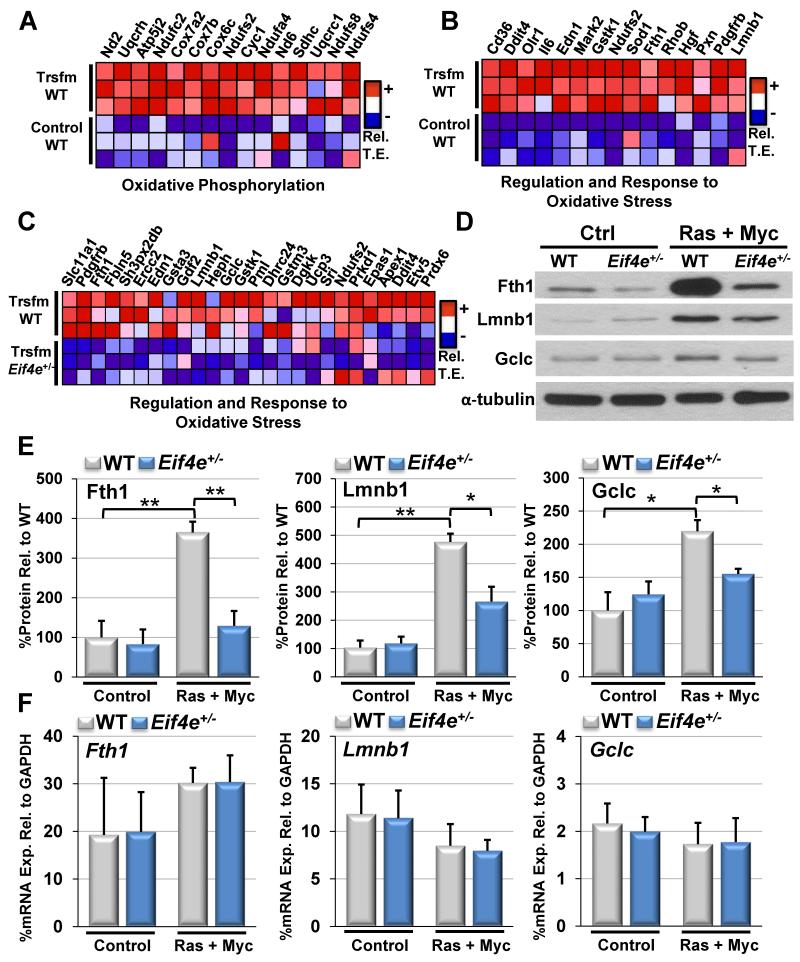
GSEA analysis demonstrating A) enrichment for genes involved in oxidative phosphorylation during cellular transformation (KEGG, FDR<0.01), B) enrichment for a broad class of genes involved in the regulation and response to oxidative stress (see Table S6), during cellular transformation (FDR<0.01), and C) Eif4e+/− MEFs are defective in a subset of translationally regulated genes involved in the regulation and response to oxidative stress during cellular transformation (see Table S6) (FDR <0.01). Each square represents the relative translational efficiency (TE) for a given gene from one individual biological replicate (n=3 per group) with red indicating relatively increased TE and blue indicating decreased TE. Top 15 genes with the highest enrichment scores are shown for A and B and all enriched genes are shown for (C). D) Western blot validation of eIF4E ROS target genes. E) Quantification of eIF4E target protein expression levels. Values represent mean +SEM. F) qPCR analysis of eIF4E target mRNA levels. Values represent mean +SD. Asterisks indicate a statistically significant change from WT samples (*= p<0.05, **= p<0.01). Results are representative of experiments from at least three sets of independently isolated primary and transformed WT and Eif4e+/− cells. See also Figures S4, S5, and S6.
In order to better understand the requirements for eIF4E in translational control of this distinct class of genes, we next expanded our functional analysis to more broadly examine genes involved in the regulation and response to ROS and we identified 99 genes translationally induced upon oncogenic transformation (Figure 4B, Figure S4D, and Table S6). Strikingly, we observe that a large fraction, 26, of these genes are sensitive to eIF4E dose (Figure 4C, Figure S4E, and Table S6), including the ferritin heavy chain (Fth1), the catalytic subunit of glutamate-cysteine ligase (Gclc), and lamin B1 (Lmnb1) (Figure 4D-F) (Malhas et al., 2009; Pham et al., 2004; Shi et al., 2000). Importantly, we find that the translation of genes involved in the regulation and response to ROS is significantly reduced in Eif4e+/− cells only upon cellular transformation (Figure S4F-G). Moreover, qPCR analysis of polysomal mRNA distribution confirmed that this gene class is indeed translationally enhanced by transformation in an eIF4E dependent manner, as visualized by a diminished shift of mRNAs into actively translating high MW polysomes in Eif4e+/− cells upon transformation (Figure S5 and Figure S6). Thus, this analysis identifies groups of functional gene classes whose translation is induced by transformation in an eIF4E-dependent manner, including a class of mRNAs involved in the response to oxidative stress.
A 5′UTR Signature of eIF4E-dependent mRNAs
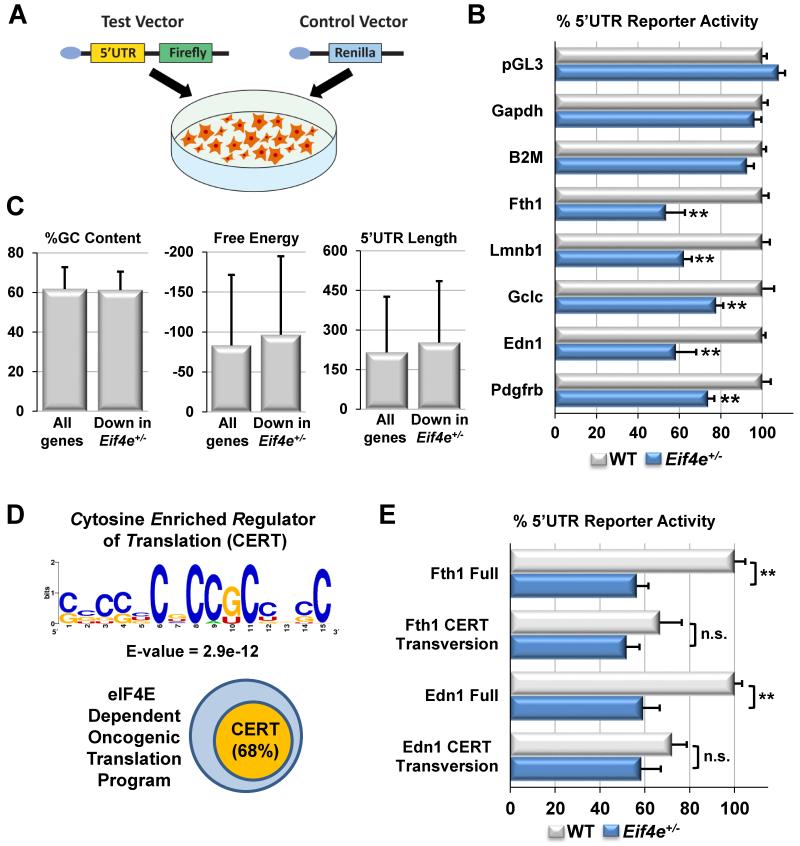
It has been previously proposed that long 5′UTRs and complex secondary structures embedded within them, revealed by high GC content and low free energies, render certain transcripts sensitive to eIF4E activity (Pickering and Willis, 2005). Importantly, 5′UTR luciferase reporter assays demonstrate that the 5′UTR of eIF4E target mRNAs, such as Fth1, Lmnb1, and Edn1, is sufficient to confer translational sensitivity to eIF4E expression levels (Figure 5A-B). Surprisingly, however, we see no major differences in 5′UTR length, GC content, or free energy in eIF4E translational targets compared to the whole genome (Figure 5C). This suggests that additional features of specific 5′UTRs may sensitize them to changes in eIF4E expression levels. In order to identify potential cis-acting regulatory elements that may confer sensitivity to eIF4E dose during oncogenic transformation, we performed an unbiased Multilpe Em for Motif Elicitation (MEME) search for sequence-specific motifs present in the 5′UTRs of eIF4E sensitive mRNAs. This analysis identified a cytosine-rich 15-nucleotide motif that we have termed the Cytosine Enriched Regulator of Translation (CERT) domain (Figure 5D and Table S7).
Figure 5. The 5′UTR confers translational sensitivity to eIF4E target mRNAs.
A) Diagram of the 5′UTR luciferase reporter assay. B) Requirements for eIF4E in 5′UTR mediated translation of target mRNAs (Fth1, Lmnb1, Gclc, Edn1, and Pdgfrb) and control mRNAs (Gapdh and B2M) by 5′UTR luciferase reporter assay in transformed MEFs. “pGL3” denotes the test vector lacking a 5′UTR. Results are normalized to 5′UTR reporter activity in transformed WT cells. C) Comparison of canonical 5′UTR features between all mouse genes and the subset whose translation efficiency is reduced in Eif4e+/− during oncogenic transformation. D) Consensus sequence and enrichment value (E-value) of the Cytosine Enriched Regulator of Translation (CERT) motif identified by MEME analysis along with a diagram illustrating the frequency of eIF4E target mRNAs containing a CERT. E) Effect of C to G transversion mutations of the CERT domains in Fth1 (+153 nt from 5′cap and −1nt from the ATG) and in Edn1 (+422 nt from the 5′cap and −188nt from the ATG) on 5′UTR luciferase reporter activity in transformed WT and Eif4e+/− MEFs. Results are normalized to non-mutated 5′UTR reporter activity in transformed WT cells. Asterisks indicate a statistically significant change (*= p<0.05, **= p<0.01). “n.s.” = not significant. All values represent the mean +SEM except for (C) which represent mean +SD.
The CERT domain is present in nearly 70% of eIF4E target mRNAs and is preferentially enriched (81%, p<0.05) in the subset of eIF4E targets increased by oncogenic transformation, including oxidative stress response genes like Fth1 (Figure 5D and Table S7). Notably, this motif is located on average 143 nucleotides downstream of the 5′cap and 146 nucleotides upstream of the AUG start codon and can be present at multiple copies within the 5′UTR of target genes. We next directly ascertained the possible functional significance of the CERT domain within the context of eIF4E-sensitized mRNAs identified in this study. Mutations of individual motifs in the 5′UTR of the eIF4E targets Fth1 and Edn1 are sufficient to restore translational differences conferred by eIF4E levels, and thus the CERT domain has the potential to explain, at least in part, target mRNAs that are sensitized to eIF4E expression levels (Figure 5E). Notably, not all eIF4E targets contain a CERT motif, thus, it is likely that other context dependent features of the 5′UTR can confer functionality towards eIF4E selective translation and potentially work in concert with the CERT. Collectively, these data demonstrate remarkable translational reprogramming of genes involved in control of ROS upon oncogenic transformation that are sensitized to eIF4E dose and marked by the presence of the 5′UTR cis-acting motif.
eIF4E dependent control of ROS during oncogenic transformation and in vivo tumorigenesis
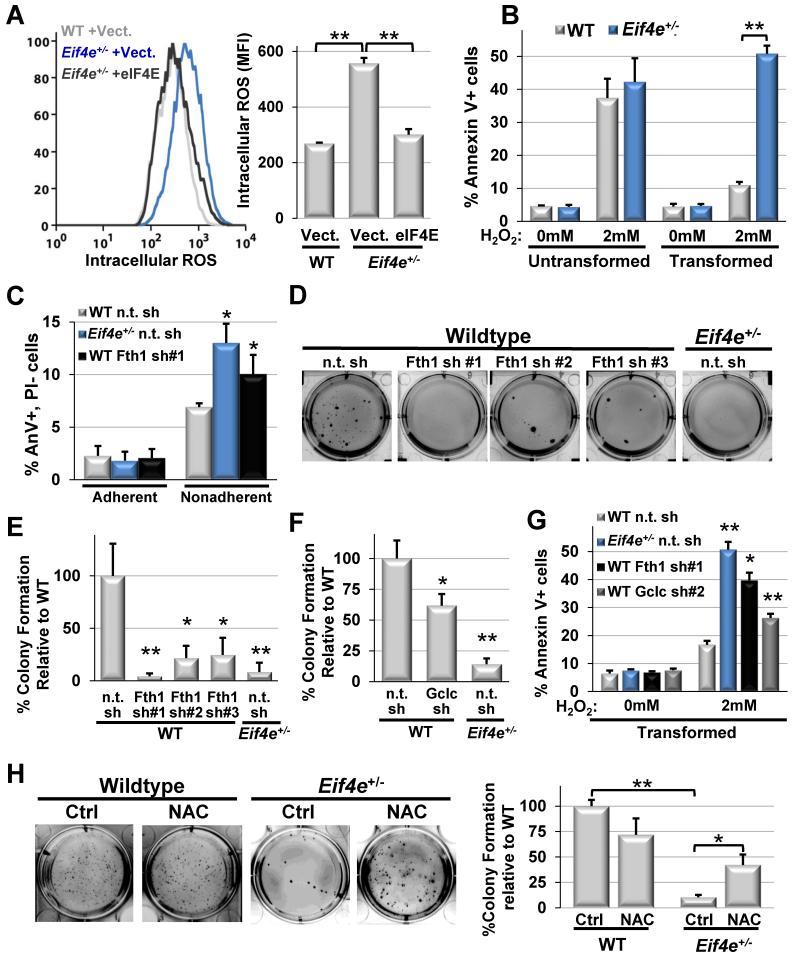
Given the unexpected functional role for eIF4E in the expression of genes involved in the control of reactive oxygen species during oncogenic transformation, we next examined the ability of Eif4e+/− cells to regulate and respond to ROS. The ability to recognize and detoxify ROS is critical for the process of oncogenic transformation (DeNicola et al., 2011). In particular, the accumulation of ROS is associated with the high proliferative capacity and altered mitochondrial respiration of cancer cells and needs to be detoxified in order to maintain tumor growth (Raj et al., 2011; Trachootham et al., 2006). In line with decreased translation of ROS target genes, fluorescent staining with CM-H2DCFDA reveals that Eif4e+/− cells display elevated levels of intracellular ROS downstream of oncogenic transformation by Ras and Myc (Figure 6A). Importantly, we find that transformed Eif4e+/− cells are significantly more sensitive to both the addition of exogenous ROS and treatment with a redox-cycling agent, DMNQ, which induces superoxide formation (Figure 6B and Figure S7A). Indeed, we find that transformed Eif4e+/− cells undergo programmed cell death under anchorage independent growth conditions known to induce the accumulation of ROS (Schafer et al., 2009), suggesting that eIF4E is required to maintain accurate levels of ROS compatible with cancer cell survival (Figure 6C).
Figure 6. eIF4E-dependent control of reactive oxygen species is critical for cellular transformation.
A) CM-H2DCFDA mean fluorescence intensity (MFI) quantification by FACS as a relative measure of intracellular ROS levels in transformed WT and Eif4e+/− MEFs with and without eIF4E rescue. B) Annexin V staining as a measure of apoptosis in response to exogenous ROS (2mM H2O2) in untransformed and transformed MEFs. C) Apoptosis under adherent and non-adherent growth conditions in transformed WT and Eif4e+/− MEFs expressing either a non-targeting (n.t) shRNA or a Fth1 targeting shRNA. D) Soft agar colony formation upon shRNA-mediated knock down (k.d.) of three independent Fth1 shRNAs (average WT n.t. shRNA colony number = 115). E) Quantification of soft agar colony formation in (D). F) Quantification of soft agar colony formation upon shRNA-mediated k.d. of Gclc (average WT n.t. shRNA colony number = 97). G) Annexin V staining in response to the addition of exogenous ROS (2mM H2O2) in transformed WT and Eif4e+/− n.t. shRNA MEFs compared to Fth1 or Gclc shRNA mediated k.d. H) Restoration of soft agar colony forming potential in Eif4e+/− MEFs upon addition of 1mM of the antioxidant N-acetyl cysteine (NAC) (average WT control colony number = 121). Asterisks indicate a statistically significant change from WT samples (*= p<0.05, **= p<0.01). All values represent the mean +SD. Results are representative of at least three independent experiments. See also Figure S7.
To extend these findings to specific genes that control ROS levels, we examined the roles of Fth1 and Gclc during the process of oncogenic transformation. Fth1 is an iron storage protein that possesses an intrinsic ferroxidase activity allowing it to neutralize toxic reactive oxygen species (Pham et al., 2004). Gclc is the catalytic subunit of glutamate-cysteine ligase, the rate-limiting enzyme for the synthesis of glutathione used by glutathione peroxidase to neutralize ROS (Shi et al., 2000). Importantly, translation of Fth1 and Gclc is significantly enhanced by oncogenic signaling and highly sensitized to eIF4E expression levels (Figure 4D and 4E). Strikingly, we find that knockdown of either Fth1 or Gclc is sufficient to inhibit oncogenic transformation, mirroring the resistance to cellular transformation and sensitivity to ROS observed in Eif4e+/− cells (Figure 6D-G and Figure S7B-H). Therefore, we asked if the inability to regulate ROS underlies the transformation defect observed in Eif4e+/− cells. Remarkably, we demonstrate that addition of the antioxidants N-acetyl Cysteine (NAC) or Trolox, a water soluble Vitamin E analog, is able to significantly restore the tumorigenic potential of Eif4e+/− cells (Figure 6H and Figure S7I-J).
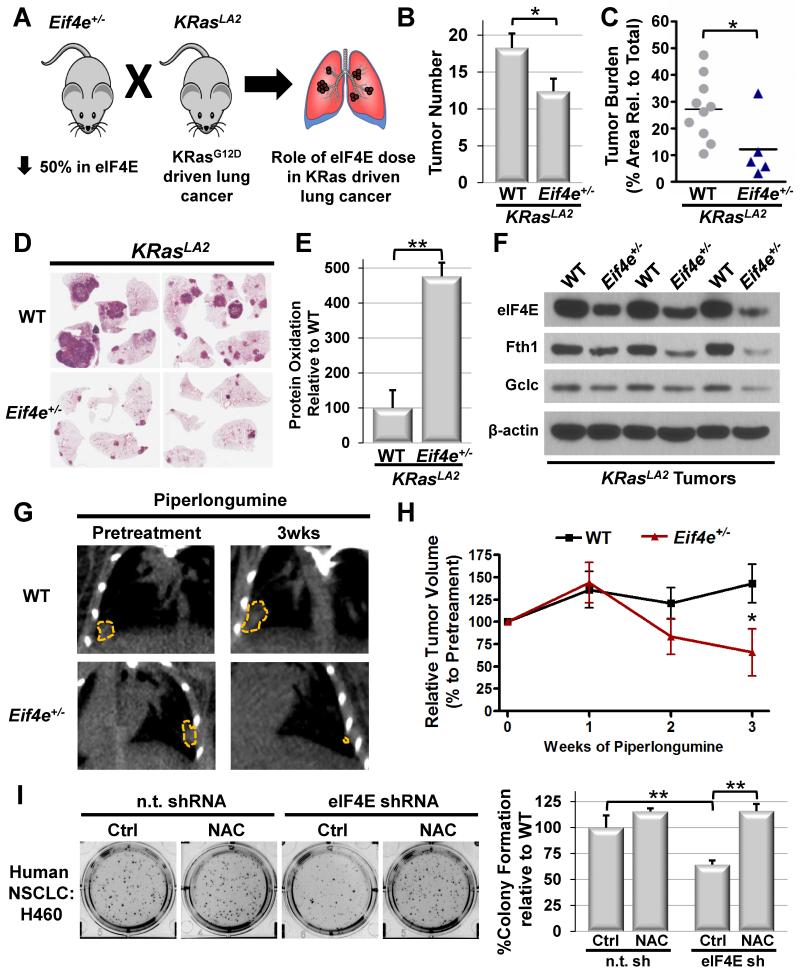
We next tested the requirements for eIF4E dose in the setting of in vivo tumorigenesis driven by endogenous expression of oncogenic KRas. Importantly, the ability to properly respond to and control ROS is critical for lung cancer driven by endogenous expression of oncogenic KRas (DeNicola et al., 2011; Sayin et al., 2014). Thus, we asked what effect genetically reducing eIF4E dose by 50% would have in the KRasLA2 model of lung cancer (Figure 7A) (Johnson et al., 2001). We find that limiting eIF4E levels has a dramatic effect on KRas-driven tumor initiation as revealed by a significant reduction in tumor number by 12 weeks of age in Eif4e+/− mice (Figure 7B). Strikingly, this manifests in a more than two-fold reduction in tumor burden in Eif4e+/− mice at one year of age (Figure 7C and 7D). Importantly, we find that reductions in tumor burden and number are associated with significant increases in oxidative stress, as measured by the protein oxidation marker dityrosine (Figure 7E). Moreover, we demonstrate that expression of eIF4E-dependent ROS target genes, such as Fth1 and Gclc, is significantly reduced in tumors of Eif4e+/− mice (Figure 7F and Figure S7K), further revealing that eIF4E is critically required for the control of ROS downstream of cellular transformation in vivo. We next asked if reductions in eIF4E dose could confer KRas driven lung cancers with enhanced sensitivity to pharmacological induction of ROS in vivo. We treated KRasLA2 mice with the small molecule piperlongumine (Raj et al., 2011), which selectively increases levels of ROS, and monitored the response of WT and Eif4e+/− tumors by micro x-ray computed tomography. We find that Eif4e haploinsufficiency renders KRasLA2 tumors remarkably susceptible to ROS induction compared to WT tumors, leading to significant reductions in tumor volumes upon treatment that are independent from tumor size (Figure 7G-H and Figure S7L).
Figure 7. eIF4E is required for lung tumorigenesis and regulation of the oxidative stress response in vivo.
A) Cartoon outlining experimental mouse crosses used to test the requirements for eIF4E dose during in vivo lung tumorigenesis. B) Counts of superficial KRasLA2 lung tumors in WT KRasLA2 mice (n=11) and Eif4e+/−; KRasLA2 mice (n=10) at 12wks of age. C) Representative cross-section of lungs from 1yr old KRasLA2 mice. D) Quantification of tumor burden in 1yr old KRasLA2 mice (WT n=10 mice; Eif4e+/− n=5 mice). E) Quantification of protein expression for the oxidative stress marker dityrosine in 12wk old lung tumors (WT n=3 tumors, Eif4e+/− n=3 tumors). F) Representative western blots of eIF4E ROS target gene expression in three independent WT KRasLA2 tumors and three independent Eif4e+/−; KRasLA2 tumors. G) Representative micro x-ray computed tomography (μCT) cross sections of established size matched WT and Eif4e+/− tumors from 8mo old KRasLA2 mice prior to and after 3wks of treatment with the ROS inducer piperlongumine (tumors are outlined in yellow). H) Relative tumor volumes of established KRasLA2 tumors followed longitudinally by μCT imaging upon treatment with piperlongumine (WT n=8 tumors, Eif4e+/− n=6 tumors). (I) Soft agar colony formation potential in the H460 human NSCLC line is decreased upon shRNA-mediated k.d. of eIF4E and can be rescued by addition of 0.5mM of the antioxidant NAC (average WT control colony number = 139). Asterisks indicate a statistically significant change (*= p<0.05, **= p<0.01). All values represent the mean +SEM except for (I) which represent mean +SD.
To extend our observations to human disease, we asked if reducing eIF4E dose could inhibit the oncogenic potential of human KRas-driven lung cancer cells through ROS control. We find that shRNA mediated knock down of eIF4E in H460 and A549 non-small cell lung cancer (NSCLC) cells causes a potent block in their oncogenic potential as measured by soft agar colony formation (Figure 7I and Figure S7M-N). Furthermore, we demonstrate that this block in tumorigenicity is not only associated with a marked decrease in the expression of eIF4E-dependent ROS target genes, but can also be rescued, at least in part, by the addition of the antioxidant NAC (Figure 7I and Figure S7M-N). Together, these data reveal that Eif4e dose becomes critical during tumorigenesis and delineate an eIF4E-dependent translational program underlying regulation and response to ROS during cellular transformation.
DISCUSSION
Collectively, our data unexpectedly uncover that eIF4E expression levels can be maintained at half of the normal genetic dosage without apparent consequences to normal organismal development and global mRNA translation. This finding is consistent with previous cell based observations showing that depletion of eIF4E by 80-90% in rabbit reticulocyte extracts has only a moderate effect on protein synthesis (Rau et al., 1996). Interestingly, shRNA mediated knock down of eIF4E by greater than 90% in the HeLa cancer cell line was also shown to have minimal impacts on global protein synthesis rates, however, this study demonstrated that sustained protein synthesis rates could be explained by compensation for reductions in eIF4E through selective degradation of phospho-4EBP1 (Yanagiya et al., 2012). Although it is unclear if this compensation may be a feature specific to cancer cell lines addicted to high global protein synthesis rates. Importantly, we find no evidence for similar alterations in levels or phosphorylation of 4EBPs in primary Eif4e+/− cells and mice, however, we cannot formally exclude the potential for unknown compensatory mechanisms arising from reductions in eIF4E levels. In this regard, it is interesting to note that eIF4E hyperactivation, both through direct overexpression or through deletion of 4EBPs, is also compatible with normal organismal development and does not induce any apparent compensatory feedback mechanisms (Blackshear et al., 1997; Ruggero et al., 2004). Surprisingly, our data suggest that mammalian systems have evolved surplus levels of eIF4E beyond that required for normal cellular function. Therefore, it remains an outstanding question what benefit an excess of eIF4E might convey at the organismal level under normal cellular conditions. While it is possible that additional translation-independent eIF4E functions, such as mRNA transport (von der Haar et al., 2004), could require higher levels of eIF4E, our data suggest that eIF4E dose may predominately be required to buffer against specific stress conditions, a function which is in turn coopted by cancer cells during the oncogene induced stress response that underlies transformation.
Deregulated translation control is a hallmark of human cancers and is critical for tumorigenesis downstream of multiple oncogenic signaling pathways (Barna et al., 2008; Hsieh et al., 2010). However, the full repertoire of mRNAs translationally altered by oncogenic signaling and the underlying molecular mechanisms that direct their translational control remain poorly understood. Employing unbiased genome-wide profiling we have uncovered an oncogenic translational program comprised of hundreds of mRNAs and find that a specific subset of transcripts within this group is exquisitely sensitized to eIF4E dose. Importantly, we find that translational changes induced by oncogenic signaling affect many functional gene classes, such as those involved in cell signaling, apoptosis, ribosome biogenesis, the proteasome, nucleotide biosynthesis, oxidative phosphorylation, and the oxidative stress response, which may in concert act to promote tumorigenesis. This data further suggests that translational control of mRNAs comprising multiple cellular functions are likely to underlie the remarkable resistance to transformation in Eif4e+/− cells and mice. In this regard, we have functionally delineated the impact of translation control of at least one of these functional classes, a group of mRNAs involved in the regulation and response to ROS, towards cellular transformation. In particular, we show that key eIF4E-dependent ROS targets, such as Fth1 and Gclc, are critically required for cellular transformation and, moreover, demonstrate that antioxidant suppression of ROS in Eif4e+/− cells rescues to a large extent the cellular transformation capacity of these cells. Future studies will be needed to further validate the role of other eIF4E-dependent functional gene classes during tumorigenesis and to elucidate specific conditions where eIF4E-dependent translation of these mRNAs becomes important.
Oxidative stress is one of the defining stress phenotypes encountered during tumorigenesis and cancer cells have been shown to generate more ROS than normal cells (Szatrowski and Nathan, 1991). In this regard, it has been historically thought that ROS plays a distinctly pro-tumorigenic role. Recent studies have also shown that physiological expression of oncogenes may drive down intracellular ROS levels, suggesting that complex regulation of ROS levels may occur during different stages of in vivo tumorigenesis as well as downstream of progressive genomic aberrations, such as oncogene amplification. Thereby, our understanding of the key molecular mechanisms responsible for intricately maintaining ROS levels compatible with tumor cell survival is incomplete. Here, we find that genes regulating ROS are under exquisite translational regulation during oncogenic transformation and are sensitized to eIF4E dose, revealing an unexpected post-transcriptional circuitry regulating ROS levels. For example, Fth1, an iron-storage protein that protects against the formation of hydroxyl radicals, and Gclc, the rate-limiting enzyme for the synthesis of glutathione used to neutralize ROS, are translationally induced by cellular transformation in an eIF4E-dependent manner. Importantly, we show that reductions in eIF4E dose lead to significant increases in intracellular ROS downstream of ectopic oncogene expression, sensitizing tumor cells to ROS induced apoptosis and blocking cellular transformation. In addition, we demonstrate that eIF4E is required for proper expression of the ROS response and control of intratumoral oxidative stress underlying in vivo lung tumorigenesis driven by endogenous expression of KRas. Thereby, our data suggest that the regulation of ROS levels through eIF4E-mediated translational control may provide a molecular rheostat that fine-tunes the levels of oxidative species that are selectively required to maintain cancer cell survival.
Precisely how distinct subsets of mRNAs become sensitized to eIF4E dose during oncogenic transformation remains an important area of investigation. Classically, long, structured 5′UTRs with high GC content and low free energy have been thought to be the only features that confer sensitivity to eIF4E dose. Interestingly, these 5′UTR features were not altered in the subset of eIF4E-dependent genes translationally activated by oncogenic transformation. Although we cannot exclude a possible role for distinct subregions of secondary structures within the 5′UTRs of eIF4E sensitive genes, our data suggests that alternative modes of translational control may also be important. Recently, there has been an emerging appreciation for the role of shorter, sequence-specific elements embedded within the 5′UTR in directing mRNA translation (Hsieh et al., 2012; Thoreen et al., 2012; Wolfe et al., 2014; Xue et al., 2015). Along these lines, we find that the majority of eIF4E-dependent mRNAs induced by transformation are marked by the presence of a sequence-specific CERT motif within their 5′UTRs, suggesting that this motif may have broader impacts towards eIF4E-dependent translation. Indeed, within the context of target mRNAs, such as Fth1 and Edn1, the CERT motif is required to confer eIF4E sensitivity. However, it remains an outstanding question how the CERT domain acts mechanistically to couple eIF4E dose to translational control of specific mRNAs. It is possible, for example, that this motif serves as a docking site for RNA binding proteins that interact either directly or indirectly with eIF4E, potentially to stabilize eIF4E cap-binding or to promote eIF4F activity. Future studies will be required to address the biochemical mechanisms by which eIF4E interfaces with the CERT motif and how other context dependent features of the 5′UTR influence CERT activity.
Collectively, our data surprisingly show that the long held dogma that eIF4E dose is critically required for translating the mammalian genome needs to be revisited. Maintaining eIF4E dose at 50% normal levels does not appear to be detrimental for normal mammalian development and even more strikingly global protein synthesis control. Our findings further point to the possibility that cancer cells have specifically usurped excess eIF4E levels to promote their growth and survival. The fact that reducing eIF4E expression is not detrimental for normal mammalian physiology reveals a potent therapeutic window for targeting the eIF4E-dependent oncogenic translational program, which may be greatly enhanced in combination with drugs that exploit the sensitivity of cancer cells to ROS.
EXPERIMENTAL PROCEDURES
Protein, RNA, cellular, metabolic, in vivo tumorigenesis, and statistical analysis can be found in Extended Experimental Procedures along with additional experimental details.
Mice
Eif4e genetic loss-of-function mice were generated using embryonic stem cells from BayGenomics (line RRO036) and validated for gene-trap insertion by PCR and Southern blot analysis. Eif4e+/− and CMV-HCV-IREST mice were maintained on a C57BL/6J background. F1 littermates from crosses of Eif4e+/− mice and KRasLA2 mice on an FVB/n background were used for in vivo tumorigenesis experiments. Mice were maintained under specific pathogen-free conditions and all experiments were performed in compliance with guidelines approved by the Institutional Animal Care and Use Committee of UCSF.
Polysome Fractionation and Isolation of Polysome Associated RNA for Microarray
Primary MEFs, transformed MEFs, and LPS-stimulated B-cells were treated with 100μg/ml cyclohexamide (Sigma) on ice in PBS for 10min. Cells were pelleted and lysed in 10mM Tris-HCl pH8, 140mM NaCl, 1.5mM MgCl2, 0.25% NP-40, 0.1% Triton-X 100, 50mM DTT, 150μg/ml cyclohexamide, 640U/ml Rnasin for 30min. Lysates were cleared, separated on a 10-50% sucrose gradient by ultracentrifugation, and fractionated using an ISCO gradient fractionation system. For polysome microarrays, RNA was isolated from high MW polysome fractions (fractions 10-13) of three sets of independently isolated primary and matched transformed WT and Eif4e+/− MEFs.
Analysis of Microarray Data Set
Affymetrix Mouse Gene 1.0 ST array data were preprocessed by RMA using the aroma.affymetrix R package. Polysomal and Total RNA datasets were quantile normalized separately. The difference in log2 intensity between matched polysomal RNA and total RNA was taken to quantify translational efficiency (TE).
Gene Set Enrichment Analysis
Gene set enrichment analysis (GSEA) was performed using version 2.0.12 of the GSEA desktop application. Log2 values of translational efficiency (TE) were input into GSEA and analyzed for enrichment in Kyoto Encyclopedia of Genes and Genomes and Gene Ontology Biological Process gene set collections.
Motif analysis
Multiple Em for Motif Elicitation (MEME) was performed using version 4.9.1 of the MEME browser application. The training set used to identify the CERT (Table S7) was compiled from the longest known 5′UTR for RefSeq annotated mRNAs called from the UCSC Genome Browser. Gene sets were evaluated for the presence of the consensus CERT motif using Find Individual Motif Occurrences (FIMO).
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of the Ruggero lab for critical discussion and Adam Olshen for reading the manuscript. This work was supported by National Institutes of Health (NIH) Director’s New Innovator Award 7DP2OD00850902 (M.B.), NIH R01CA140456 (D.R.), NIH R01CA154916 (D.R.), NIH R01CA184624 (D.R.), NIH 1F32CA189696 (C.S.C.), NIH P30CA82103 (T.T.), Alfred P. Sloan Research Fellowship (M.B), Mallinckrodt Foundation Award (M.B.), Pew Scholars Award (M.B.), American Cancer Society PF-14-212-01-RMC (C.S.C.), and the Life Science Research Foundation Postdoctoral Fellowship (Z.S). D.R. is a Leukemia & Lymphoma Society Scholar. M.B. is a Pew Scholar and Alfred P. Sloan Research Fellow. D.R. is a shareholder of eFFECTOR Therapeutics, Inc. and a member of its scientific advisory board. This work was supported in part by the Onyx Oncology Innovation Alliance Award (D.R.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS:
M.L.T., M.B., and D.R designed the research. M.L.T., C.S.C., Z.S., X.P., A.M.C., Y.S., M.B., and D.R. conducted the experiments and analyzed the data. M.L.T., C.S.C., and X.P. designed experimental tools/ new reagents. M.L.T., T.T., and Z.S. performed array and bioinformatic analysis. M.L.T., C.S.C, Z.S., M.B., and D.R. prepared the figures. M.L.T., M.B., and D.R. wrote the manuscript. C.S.C., Z.S., and A.M.C edited the manuscript.
ACCESSION NUMBERS:
REFERENCES
- Aoki M, Blazek E, Vogt PK. A role of the kinase mTOR in cellular transformation induced by the oncoproteins P3k and Akt. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:136–141. doi: 10.1073/pnas.011528498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bah A, Vernon RM, Siddiqui Z, Krzeminski M, Muhandiram R, Zhao C, Sonenberg N, Kay LE, Forman-Kay JD. Folding of an intrinsically disordered protein by phosphorylation as a regulatory switch. Nature. 2014 doi: 10.1038/nature13999. [DOI] [PubMed] [Google Scholar]
- Barna M, Pusic A, Zollo O, Costa M, Kondrashov N, Rego E, Rao PH, Ruggero D. Suppression of Myc oncogenic activity by ribosomal protein haploinsufficiency. Nature. 2008;456:971–975. doi: 10.1038/nature07449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellodi C, Kopmar N, Ruggero D. Deregulation of oncogene-induced senescence and p53 translational control in X-linked dyskeratosis congenita. The EMBO journal. 2010;29:1865–1876. doi: 10.1038/emboj.2010.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackshear PJ, Stumpo DJ, Carballo E, Lawrence JC., Jr. Disruption of the gene encoding the mitogen-regulated translational modulator PHAS-I in mice. The Journal of biological chemistry. 1997;272:31510–31514. doi: 10.1074/jbc.272.50.31510. [DOI] [PubMed] [Google Scholar]
- Cunningham JT, Moreno MV, Lodi A, Ronen SM, Ruggero D. Protein and nucleotide biosynthesis are coupled by a single rate-limiting enzyme, PRPS2, to drive cancer. Cell. 2014;157:1088–1103. doi: 10.1016/j.cell.2014.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Benedetti A, Graff JR. eIF-4E expression and its role in malignancies and metastases. Oncogene. 2004;23:3189–3199. doi: 10.1038/sj.onc.1207545. [DOI] [PubMed] [Google Scholar]
- De Benedetti A, Joshi-Barve S, Rinker-Schaeffer C, Rhoads RE. Expression of antisense RNA against initiation factor eIF-4E mRNA in HeLa cells results in lengthened cell division times, diminished translation rates, and reduced levels of both eIF-4E and the p220 component of eIF-4F. Molecular and cellular biology. 1991;11:5435–5445. doi: 10.1128/mcb.11.11.5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeNicola GM, Karreth FA, Humpton TJ, Gopinathan A, Wei C, Frese K, Mangal D, Yu KH, Yeo CJ, Calhoun ES, et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 2011;475:106–109. doi: 10.1038/nature10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan R, Milburn SC, Hershey JW. Regulated phosphorylation and low abundance of HeLa cell initiation factor eIF-4F suggest a role in translational control. Heat shock effects on eIF-4F. The Journal of biological chemistry. 1987;262:380–388. [PubMed] [Google Scholar]
- Feoktistova K, Tuvshintogs E, Do A, Fraser CS. Human eIF4E promotes mRNA restructuring by stimulating eIF4A helicase activity. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:13339–13344. doi: 10.1073/pnas.1303781110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furic L, Rong L, Larsson O, Koumakpayi IH, Yoshida K, Brueschke A, Petroulakis E, Robichaud N, Pollak M, Gaboury LA, et al. eIF4E phosphorylation promotes tumorigenesis and is associated with prostate cancer progression. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:14134–14139. doi: 10.1073/pnas.1005320107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annual review of biochemistry. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- Haghighat A, Mader S, Pause A, Sonenberg N. Repression of cap-dependent translation by 4E-binding protein 1: competition with p220 for binding to eukaryotic initiation factor-4E. The EMBO journal. 1995;14:5701–5709. doi: 10.1002/j.1460-2075.1995.tb00257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiremath LS, Webb NR, Rhoads RE. Immunological detection of the messenger RNA cap-binding protein. The Journal of biological chemistry. 1985;260:7843–7849. [PubMed] [Google Scholar]
- Hsieh AC, Costa M, Zollo O, Davis C, Feldman ME, Testa JR, Meyuhas O, Shokat KM, Ruggero D. Genetic dissection of the oncogenic mTOR pathway reveals druggable addiction to translational control via 4EBP-eIF4E. Cancer cell. 2010;17:249–261. doi: 10.1016/j.ccr.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh AC, Liu Y, Edlind MP, Ingolia NT, Janes MR, Sher A, Shi EY, Stumpf CR, Christensen C, Bonham MJ, et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature. 2012;485:55–61. doi: 10.1038/nature10912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irani K, Xia Y, Zweier JL, Sollott SJ, Der CJ, Fearon ER, Sundaresan M, Finkel T, Goldschmidt-Clermont PJ. Mitogenic signaling mediated by oxidants in Ras-transformed fibroblasts. Science. 1997;275:1649–1652. doi: 10.1126/science.275.5306.1649. [DOI] [PubMed] [Google Scholar]
- Johnson L, Mercer K, Greenbaum D, Bronson RT, Crowley D, Tuveson DA, Jacks T. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature. 2001;410:1111–1116. doi: 10.1038/35074129. [DOI] [PubMed] [Google Scholar]
- Macejak DG, Sarnow P. Internal initiation of translation mediated by the 5′ leader of a cellular mRNA. Nature. 1991;353:90–94. doi: 10.1038/353090a0. [DOI] [PubMed] [Google Scholar]
- Malhas AN, Lee CF, Vaux DJ. Lamin B1 controls oxidative stress responses via Oct-1. The Journal of cell biology. 2009;184:45–55. doi: 10.1083/jcb.200804155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham CG, Bubici C, Zazzeroni F, Papa S, Jones J, Alvarez K, Jayawardena S, De Smaele E, Cong R, Beaumont C, et al. Ferritin heavy chain upregulation by NF-kappaB inhibits TNFalpha-induced apoptosis by suppressing reactive oxygen species. Cell. 2004;119:529–542. doi: 10.1016/j.cell.2004.10.017. [DOI] [PubMed] [Google Scholar]
- Pickering BM, Willis AE. The implications of structured 5′ untranslated regions on translation and disease. Seminars in cell & developmental biology. 2005;16:39–47. doi: 10.1016/j.semcdb.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Raj L, Ide T, Gurkar AU, Foley M, Schenone M, Li X, Tolliday NJ, Golub TR, Carr SA, Shamji AF, et al. Selective killing of cancer cells by a small molecule targeting the stress response to ROS. Nature. 2011;475:231–234. doi: 10.1038/nature10167. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Rajasekhar VK, Viale A, Socci ND, Wiedmann M, Hu X, Holland EC. Oncogenic Ras and Akt signaling contribute to glioblastoma formation by differential recruitment of existing mRNAs to polysomes. Molecular cell. 2003;12:889–901. doi: 10.1016/s1097-2765(03)00395-2. [DOI] [PubMed] [Google Scholar]
- Rau M, Ohlmann T, Morley SJ, Pain VM. A reevaluation of the cap-binding protein, eIF4E, as a rate-limiting factor for initiation of translation in reticulocyte lysate. The Journal of biological chemistry. 1996;271:8983–8990. doi: 10.1074/jbc.271.15.8983. [DOI] [PubMed] [Google Scholar]
- Rosenwald IB, Rhoads DB, Callanan LD, Isselbacher KJ, Schmidt EV. Increased expression of eukaryotic translation initiation factors eIF-4E and eIF-2 alpha in response to growth induction by c-myc. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:6175–6178. doi: 10.1073/pnas.90.13.6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggero D, Montanaro L, Ma L, Xu W, Londei P, Cordon-Cardo C, Pandolfi PP. The translation factor eIF-4E promotes tumor formation and cooperates with c-Myc in lymphomagenesis. Nature medicine. 2004;10:484–486. doi: 10.1038/nm1042. [DOI] [PubMed] [Google Scholar]
- Sattler M, Verma S, Shrikhande G, Byrne CH, Pride YB, Winkler T, Greenfield EA, Salgia R, Griffin JD. The BCR/ABL tyrosine kinase induces production of reactive oxygen species in hematopoietic cells. The Journal of biological chemistry. 2000;275:24273–24278. doi: 10.1074/jbc.M002094200. [DOI] [PubMed] [Google Scholar]
- Sayin VI, Ibrahim MX, Larsson E, Nilsson JA, Lindahl P, Bergo MO. Antioxidants accelerate lung cancer progression in mice. Science translational medicine. 2014;6:221ra215. doi: 10.1126/scitranslmed.3007653. [DOI] [PubMed] [Google Scholar]
- Schafer ZT, Grassian AR, Song L, Jiang Z, Gerhart-Hines Z, Irie HY, Gao S, Puigserver P, Brugge JS. Antioxidant and oncogene rescue of metabolic defects caused by loss of matrix attachment. Nature. 2009;461:109–113. doi: 10.1038/nature08268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi ZZ, Osei-Frimpong J, Kala G, Kala SV, Barrios RJ, Habib GM, Lukin DJ, Danney CM, Matzuk MM, Lieberman MW. Glutathione synthesis is essential for mouse development but not for cell growth in culture. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:5101–5106. doi: 10.1073/pnas.97.10.5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpf CR, Moreno MV, Olshen AB, Taylor BS, Ruggero D. The translational landscape of the mammalian cell cycle. Molecular cell. 2013;52:574–582. doi: 10.1016/j.molcel.2013.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szatrowski TP, Nathan CF. Production of large amounts of hydrogen peroxide by human tumor cells. Cancer research. 1991;51:794–798. [PubMed] [Google Scholar]
- Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485:109–113. doi: 10.1038/nature11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trachootham D, Zhou Y, Zhang H, Demizu Y, Chen Z, Pelicano H, Chiao PJ, Achanta G, Arlinghaus RB, Liu J, et al. Selective killing of oncogenically transformed cells through a ROS-mediated mechanism by beta-phenylethyl isothiocyanate. Cancer cell. 2006;10:241–252. doi: 10.1016/j.ccr.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Vafa O, Wade M, Kern S, Beeche M, Pandita TK, Hampton GM, Wahl GM. c-Myc can induce DNA damage, increase reactive oxygen species, and mitigate p53 function: a mechanism for oncogene-induced genetic instability. Molecular cell. 2002;9:1031–1044. doi: 10.1016/s1097-2765(02)00520-8. [DOI] [PubMed] [Google Scholar]
- von der Haar T, Gross JD, Wagner G, McCarthy JE. The mRNA cap-binding protein eIF4E in post-transcriptional gene expression. Nature structural & molecular biology. 2004;11:503–511. doi: 10.1038/nsmb779. [DOI] [PubMed] [Google Scholar]
- von der Haar T, McCarthy JE. Intracellular translation initiation factor levels in Saccharomyces cerevisiae and their role in cap-complex function. Molecular microbiology. 2002;46:531–544. doi: 10.1046/j.1365-2958.2002.03172.x. [DOI] [PubMed] [Google Scholar]
- Wallace DC. Mitochondria and cancer. Nat Rev Cancer. 2012;12:685–698. doi: 10.1038/nrc3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waskiewicz AJ, Flynn A, Proud CG, Cooper JA. Mitogen-activated protein kinases activate the serine/threonine kinases Mnk1 and Mnk2. The EMBO journal. 1997;16:1909–1920. doi: 10.1093/emboj/16.8.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg F, Hamanaka R, Wheaton WW, Weinberg S, Joseph J, Lopez M, Kalyanaraman B, Mutlu GM, Budinger GR, Chandel NS. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:8788–8793. doi: 10.1073/pnas.1003428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe AL, Singh K, Zhong Y, Drewe P, Rajasekhar VK, Sanghvi VR, Mavrakis KJ, Jiang M, Roderick JE, Van der Meulen J, et al. RNA G-quadruplexes cause eIF4A-dependent oncogene translation in cancer. Nature. 2014;513:65–70. doi: 10.1038/nature13485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue S, Tian S, Fujii K, Kladwang W, Das R, Barna M. RNA regulons in Hox 5′ UTRs confer ribosome specificity to gene regulation. Nature. 2015;517:33–38. doi: 10.1038/nature14010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagiya A, Suyama E, Adachi H, Svitkin YV, Aza-Blanc P, Imataka H, Mikami S, Martineau Y, Ronai ZA, Sonenberg N. Translational homeostasis via the mRNA cap-binding protein, eIF4E. Molecular cell. 2012;46:847–858. doi: 10.1016/j.molcel.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.