Abstract
Genetically engineered mouse models of lung adenocarcinoma have proven invaluable for understanding mechanisms of tumorigenesis, therapy response, and drug resistance. However, mechanistic studies focused on studying these processes in tumor-bearing mouse lungs are confounded by the fact that, in most cases, relevant signaling pathways are analyzed in whole-lung preparations, which are composed of a heterogeneous mixture of cells. Given our increasing knowledge about the roles played by different subpopulations of cells in the development of lung adenocarcinoma, separating the major cellular compartments of the tumor microenvironment is recommended to allow for a precise analysis of relevant pathways in each isolated cell type. In this study, we optimized magnetic- and fluorescence-based isolation protocols to segregate lung epithelial (CD326/epithelial cell adhesion molecule–positive), endothelial (CD31-positive), and immune (CD45-positive) cells, with high purity, from the lungs of transgenic mice with mutant epidermal growth factor receptor–induced lung adenocarcinomas. This approach, which can potentially be extended to additional lung adenocarcinoma models, enables delineation of the molecular features of individual cell types that can be used to gain insight into their roles in lung adenocarcinoma initiation, progression, and response to therapy.
Keywords: epithelial cell adhesion molecule, epithelial cell isolation, lung adenocarcinoma, transgenic mouse models
Clinical Relevance
This research will allow analysis of the molecular features of different cell types within the tumor microenvironment, and will ultimately provide information on how lung cancer cells interact with immune cells and endothelial cells. The information from these studies may then be used to identify new therapeutic targets and/or processes that are important for tumor progression in vivo.
The main compartments in the adult mouse lung are the trachea, bronchi, bronchioles, and alveoli. Several types of epithelial cells characterize these different compartments. Clara cells, for example, primarily found in the bronchioles, produce Clara cell secretory protein (CCSP/CC10) (1), which helps mitigate inflammation and oxidative stress in the lungs (2). The alveolar sacs, in contrast, are composed of alveolar type (AT) I cells and ATII cells. ATI cells mediate gas exchange (3); ATII cells maintain alveolar fluid balance, produce surfactant proteins, and differentiate into ATI cells (4). Surfactant protein (SP)-C in particular is a unique marker for ATII cells (5, 6). Other types of epithelial cells, such as basal, goblet, and ciliated cells, make up the bronchi and larger airways of the lung.
Lung adenocarcinoma, the most common histopathological subtype of lung cancer, is thought to arise from the atypical proliferation of cells located in the bronchioles and alveoli (7–12). In genetically engineered mouse models of lung adenocarcinoma, induced by frequently observed oncogenic drivers, such as mutant Kirsten rat sarcoma viral oncogene homolog (Kras) or epidermal growth factor receptor (EGFR), tumors are mainly composed of ATII cells (although both Clara and ATII cells have been implicated in Kras-induced models of lung adenocarcinoma) (10, 13, 14). These mouse models are extensively used to study mechanisms associated with Clara and/or ATII cell proliferation, to analyze signaling pathways important in tumorigenesis, and to identify possible molecular targets for therapies.
In addition to epithelial cells, which spur the development of lung adenocarcinoma in the Kras and EGFR mutant mouse models, hematopoietic and endothelial cells are actively involved in shaping the tumor microenvironment in the lungs. For example, it has been shown that increased activity of the signal transducers and activators of the transcription 3 (Stat3c) pathway in ATII cells promotes inflammation and immune cell infiltration in murine lung adenocarcinomas (15). It has also been shown that EGFR-mutant epithelial cells dampen the antitumor activity of immune cells through the programmed cell death-1 (PD-1) pathway, and that blocking this pathway impairs tumor growth (16). Thus, immune and lung epithelial cells are intimately linked in mice with lung adenocarcinoma. Endothelial cells also play an active role in the progression of lung cancer. There is evidence, for example, that overexpression of hypoxia-inducible factor 2 α (HIF2α) can lead to increased angiogenesis and reduced survival in mice expressing oncogenic Kras in the lungs (17). Moreover, clinical data suggest that combined inhibition of EGFR and vascular endothelial growth factor receptor is beneficial (18). In addition, immune and endothelial cells are also linked in the development of tumors. There is evidence, for example, that a subset of immune cells (Gr+CD11b+) promote angiogenesis and endothelial cell proliferation (19). Thus, being able to isolate immune, endothelial, and epithelial cells from tumor-bearing mouse lungs is important to precisely elucidate the molecular framework of the tumor microenvironment in lung adenocarcinoma.
Magnetic-activated cell sorting (MACS) is a technique in which cells can be depleted (or positively selected) by using microbeads that target specific cell surface antigens. Epithelial cells from normal mouse lungs have previously been isolated using MACS by first depleting CD45pos hematopoietic cells and then selecting for epithelial cell adhesion molecule (EpCAM/CD326)–expressing cells (20). EpCAM is expressed on Clara, ATII, and even potentially on tumor-initiating cells (20–23). Moreover, EpCAM, which can be used to isolate mouse lung epithelial cells (20–22), is overexpressed in lung adenocarcinoma (24), and is being studied as a target for cancer therapy (25). A significant fraction of EpCAM-positive cells, however, are also positive for the endothelial cell marker, CD31pos, possibly due to the very vascularized nature of the lung epithelium and the tight association of cells at the endothelial–epithelial interface (26). Thus, there is a need to effectively deplete CD31pos cells for optimal epithelial cell purity. This is even more crucial when isolating cells from tumors, which can have an increased level of angiogenesis and endothelial cell recruitment (27).
In this study, we optimized a protocol for the magnetic-based isolation of cells from the lungs of transgenic mice with lung adenocarcinomas by depleting CD45pos and CD31pos cells before the positive selection of EpCAM-expressing cells. Using this technique, we isolated high-purity fractions of immune, endothelial, and epithelial cells from the lungs of mice with lung adenocarcinoma. We compared this approach to fluorescence-activated cell sorting (FACS) of EpCAM-, CD45-, and CD31-expressing cells from lung single-cell suspensions.
Implementation of these protocols may be helpful in shedding light on the molecular signatures of the three major cellular compartments of the tumor microenvironment in lung adenocarcinoma; thus, it may contribute to delineating mechanisms of tumorigenesis, therapy response, and drug resistance.
Materials and Methods
Isolation of Cells from Mouse Models of EGFR-Driven Lung Adenocarcinoma
Previously described CCSP–reverse tetracycline transactivator (rtTA) (+) tetracycline-responsive element (TetO) EGFRL858R (+) bitransgenic mice that develop lung adenocarcinomas were used (see Figure E1A in the online supplement) (14). This model employs a construct in which TetO, activated by rtTA in the presence of doxycycline, is upstream of a mutant EGFR cDNA harboring a lung adenocarcinoma–associated point mutation (EGFRL858R). The CCSP-rtTA transgenic strain was used to direct expression of rtTA to the lung epithelium; in this line, rtTA is mainly expressed in ATII cells (28, 29). Expression of mutant EGFR protein in these epithelial cells, after induction with doxycycline, induces lung adenocarcinomas with bronchioloalveolar carcinoma features. Magnetic resonance imaging of bitransgenic CCSP-rtTA (+) TetOEGFRL858R (+) mice shows widespread tumorigenesis in the lungs (Figure E1C) compared with monotransgenic CCSP-rtTA (+) TetOEGFRL858R (−) mice, which do not develop tumors (Figure E1B). These transgenic mice are widely used in studies to understand the mechanistic basis of mutant EGFR–induced lung tumorigenesis, drug response, and resistance to therapies targeting EGFR (30–32).
CCSP-rtTA (+) TetOEGFRL858R (+) mice develop tumors approximately 60 days after doxycycline induction. For these studies, mice were killed by CO2 asphyxiation upon tumor development. The lungs were extracted from the chest cavity, and a lung single-cell suspension was prepared through proteolytic digestion of the lungs. Thereafter, MACS or FACS was used to isolate epithelial, endothelial, and immune cells from the lung single-cell suspension. We determined the purity of the isolated cells using Western blot, flow cytometry, and/or immunocytochemistry (ICC). The viability and yield of the isolated cells were also assessed. See the online supplement for detailed protocols of all the techniques used (including the MACS and FACS protocols). All procedures regarding the handling of the mice were performed in accordance with the guidelines established by the Institutional Animal Care and Use Committee of Yale University (New Haven, CT).
Reagents
Reagents used for MACS and/or FACS were purchased from Miltenyi Biotec Inc. (Bergisch Gladbach, Germany), and include: Mouse Lung Dissociation Kit (cat no. 130-095-927), CD31 MicroBeads (cat no. 130-097-418), CD45 MicroBeads (cat no. 130-052-301), EpCAM-phycoerythrin (PE) (130-102-265), Anti-PE MicroBeads (cat no. 130-048-801), CD31-fluorescein isothiocyanate (FITC) (cat no. 130-102-519), CD45-allophycocyanin (APC) (cat no. 130-102-544), CD45-PE (cat no. 130-102-781), CD146-APC (cat no. 130-102-846), and autoMACS Running Buffer (cat no. 130-091-221). Other reagents used include: PBS, DMEM, 1× Red Blood Cell Lysis Buffer (cat no. 00-4333; e-Biosciences, San Diego, CA), and SYTOX Red Dead Cell Stain (cat no. S34859; Life Technologies, Carlsbad, CA).
Reagents used for Western blot and/or ICC include: anti-CD45 (cat no. ab10558; Abcam, Cambridge, UK), anti-ProSurfactant protein C (cat no. ab90716; Abcam), anti–platelet endothelial cell adhesion molecule (PECAM)-1 (cat no. sc-1506; Santa Cruz Biotechnology Inc., Dallas, TX), anti-EGFRL858R (cat no. 3197S; Cell Signaling Technology, Danvers, MA), anti-Aquaporin 5 antibody (cat no. ab76013; Abcam), anti–thyroid transcription factor 1 (TTF1) (cat no. ab76013; Abcam), anti-Actin (cat no. sc-8432; Santa Cruz Biotechnology Inc.), Anti-rabbit IgG, horseradish peroxidase (HRP)-linked antibody (cat no. 7074S; Cell Signaling Technology), Alexa Fluor 594 donkey anti-rabbit IgF (cat no. A-21207; Life Technologies), and VECTASHIELD Mounting Medium with 4′,6-diamidino-2-phenylindole (cat no. H-1200; Vector Laboratories, Burlingame, CA). T1α serum was obtained from the Developmental Studies Hybridoma Bank, created by the National Institute of Child Health and Human Development of the National Institutes of Health, and maintained in the Department of Biology of the University of Iowa (Iowa City, IA).
Results
Optimization of a MACS-Based Protocol for the Isolation of Cells from Lungs of Tumor-Bearing Mice
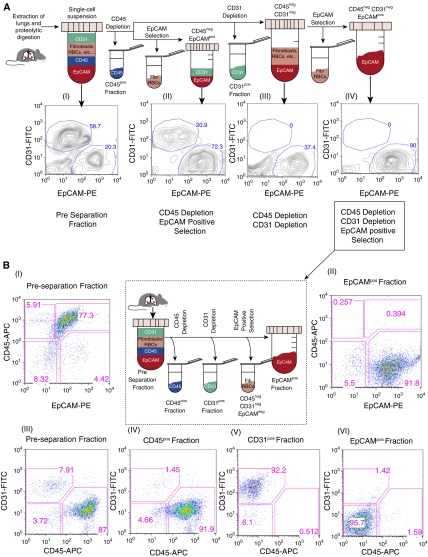
EGFRL858R mutant lung tumors grow diffusely through the lung (14); therefore, it is difficult to dissect distinct nodules of mutant epithelial cells for downstream analysis. To overcome this challenge, our initial efforts were centered on optimizing a method for the isolation of epithelial cells from whole lungs of CCSP-rtTA (+) TetOEGFRL858R (+) tumor-bearing mice. To accomplish this, we first tested the purity of epithelial cells obtained using three different MACS protocols (Figure 1A). Single-cell suspensions from tumor-bearing mouse lungs (using flow cytometry and gating on CD45neg cells) contained roughly 20% EpCAMpos epithelial cells and roughly 59% CD31pos cells (Figure 1A, I). To enrich for epithelial cells, we first tested whether depleting CD45pos cells and performing a positive selection step for EpCAM (without CD31 depletion) increased epithelial cell purity, which it did, by approximately 3.5 times (Figure 1A, II). We found that the EpCAM-positive selection step was critical to achieve high epithelial cell purity (>70%), as carrying out CD31/CD45 depletion alone (without EpCAM-positive selection) did not lead to similar levels of enrichment (Figure 1A, III). Finally, to further enrich for epithelial cells, we tested CD31/CD45 depletion (sequential or simultaneous) followed by EpCAM-positive selection. This protocol was optimal for enriching epithelial cells, such that they composed over 90% of the isolated fraction (Figure 1A, IV). If the CD31 depletion step was excluded before EpCAM-positive selection, the epithelial cell fraction contained roughly 20% endothelial cells (Figure 1A, II). Depletion of CD31pos cells ensured that almost no endothelial cells were present in the epithelial cell fraction (Figure 1A, III and IV). Therefore, EpCAM-positive selection alone does not fully distinguish between endothelial and epithelial cells, and a CD31 depletion step is necessary (before EpCAM-positive selection) for maximum epithelial cell purity. The antibodies and microbeads used for the depletion and positive selection steps were titrated for optimal cell fraction purities (see the online supplement for details).
Figure 1.
Optimization of the magnetic-activated cell sorting (MACS) protocol for isolating lung epithelial, endothelial, and immune cells. (A) Flow cytometry analysis of single-cell suspensions from the lungs of a tumor-bearing Clara cell secretory protein (CCSP)–reverse tetracycline transactivator (rtTA) (+) tetracycline-responsive element (TetO) EGFRL858R (+) mouse (A [I]) before performing MACS-based separations. The whole single-cell suspension was then depleted of CD45pos cells, followed either directly by epithelial cell adhesion molecule (EpCAM)–positive selection (A [II]) or CD31 depletion (A [III and IV]). EpCAM-positive selection was then performed on a subset of the CD45negCD31neg cells (A [IV]). The highest epithelial cell purity was achieved using the latter protocol. All the fractions analyzed were gated on CD45neg cells. (B) Flow-based analysis of the EpCAMpos, CD31pos, and CD45pos fractions (B [II, IV–VI]) isolated using sequential CD45, CD31, and EpCAM selection (see center panel) compared with the preseparation fraction (B [I and III]). Flow analysis was performed on a FACSCalibur using CD31-fluorescein isothiocyanate (FITC), CD45-allophycocyanin (APC), and EpCAM-phycoerythrin (PE) antibodies. EGFR, epidermal growth factor receptor; Fib, fibroblasts; RBC, red blood cell.
To isolate immune, endothelial, and epithelial cells from individual mice using MACS, we had to determine the exact order in which each selection step should be performed to achieve a high purity of cells. We found that it is best to do the CD31 selection before and not after the EpCAM-positive selection. This is because, once the epithelial cells go through positive selection, they still have the microbeads adhering to their surfaces, and subsequent depletion steps attempting to negatively select CD31 cells from the epithelial cells using an external magnetic system will be unsuccessful. For the same reason, because CD31 is expressed on some immune cells, whereas endothelial cells do not express CD45, it is best to perform the CD45 selection before the CD31 selection. In summary, we obtained the highest purity of epithelial cells (along with immune and endothelial cells) from tumor-bearing mice by sequentially selecting for CD45, CD31, and EpCAM (see Figure 1B, center panel).
Assessing the Quality of the MACS-Based Protocol
To assess the purity of the sequential CD45, CD31, and EpCAM MACS-based selection strategy performed on tumor-bearing mouse lungs, we examined the different isolated fractions using flow cytometry (Figure 1B). EpCAMpos epithelial cells composed roughly 4% of the total lung cell population before separation (Figure 1B, I), but they were significantly enriched in the EpCAMpos fraction, where they represented over 90% of the cells (Figure 1B, II). On average, we observed a 24-fold enrichment of EpCAMpos epithelial cells (Table 1) using our protocol. In the preseparation fraction (Figure 1B, I), there are two subpopulations of CD45pos cells, those that express EpCAM and those that do not. On average, approximately 75% of the cells were double positive for CD45 and EpCAM in EGFRL858R tumor-bearing mice before MACS was performed. This result is consistent with studies indicating that EpCAM is expressed on a subset of hematopoietic cells of the mouse lung (33, 34). This emphasizes the need to perform the CD45 depletion before the EpCAM-positive selection to avoid positively selecting EpCAMposCD45pos immune cells along with the EpCAMpos CD45neg epithelial cells (resulting in a low purity of epithelial cells).
Table 1.
Assessment of the Purity, Viability, and Yield of Cells Isolated Using a Fluorescence-Activated Cell Sorting– or Magnetic-Activated Cell Sorting–Based Protocol from Mice with Lung Adenocarcinoma
| MACS |
FACS |
|||||
|---|---|---|---|---|---|---|
| EpCAMpos | CD45pos | CD31pos | EpCAMpos | CD45pos | CD31pos | |
| Preseparation, % of total cells | 3.8 ± 0.5 | 83.9 ± 4.0 | 5.9 ± 0.8 | 3.2 ± 0.2 | 84.0 ± 0.9 | 7.5 ± 0.8 |
| Postseparation, % of total cells | 92.9 ± 1.2 | 93.7 ± 1.7 | 90.6 ± 0.6 | 97.4 ± 0.7 | 95.8 ± 1.1 | 98.7 ± 1.0 |
| P value | 0.03* | 0.38* | 0.001* | |||
| Enrichment (post/pre) | 24.4× | 1.1× | 15× | 30× | 1.1× | 13.2× |
| Viability, % | 74.0 ± 2.4 | 96.0 ± 0.4 | 90.2 ± 2.5 | 57.2 ± 6.0 | 90.0 ± 1.2† | 65.8 ± 5.7 |
| 0.06* | 0.01* | 0.02* | ||||
| Average no. of cells per tumor-bearing lung‡ | 2.07 ± 0.49 × 106 | 6.23 ± 1.36 × 107 | 5.1 ± 0.85 × 106 | 5.6 ± 0.66 × 105 | 1.8 ± 0.37×107 | 1.5 ± 0.06 × 106 |
| P value | 0.06* | 0.05* | 0.02* | |||
Definition of abbreviations: EpCAM, epithelial cell adhesion molecule; FACS, fluorescence-activated cell sorting; MACS, magnetic-activated cell sorting.
Values are mean ± SEM.
P value computed using a t test comparing analogous fractions isolated using MACS and FACS.
Viability assay was performed using trypan blue staining.
The mice were on doxycycline ∼60 days.
The preseparation fraction was composed of approximately 84.0% CD45pos cells and approximately 6% CD31pos cells (Table 1 and Figure 1B, III). On the other hand, the EpCAMpos fraction had less than 2% CD45pos cells and CD31pos cells each (Figure 1B, VI). Thus, we successfully isolated the epithelial cells by depleting CD31pos and CD45pos cells and positively selecting for EpCAMpos epithelial cells (Figure 1B, II, VI). Furthermore, CD31pos and CD45pos cells were also isolated in their respective fractions with purities greater than 90% (Figure 1B, IV and V). The purities (mean ± SEM) of the different fractions are summarized in Table 1.
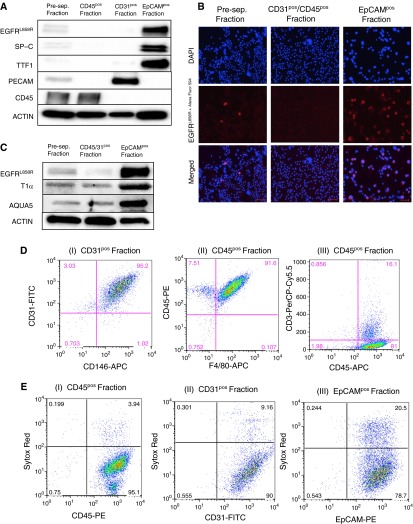
To further confirm the nature of the isolated cell fractions, we used immunoblotting with antibodies for cell type–specific markers. Western blot analysis showed that EGFRL858R and SP-C were significantly enriched in the EpCAMpos fraction when compared with the preseparation fraction (Figure 2A). Furthermore, CD45 is only localized to the CD45pos fraction, and PECAM (endothelial cell marker) is only detected in the CD31pos fraction. We also observed TTF-1 expression predominantly in the EpCAMpos fraction. TTF-1 is a transcription factor that is clinically used as a marker for lung adenocarcinoma (35). These results confirm that we successfully isolated epithelial, endothelial, and immune cells, and validate the purity analysis obtained with flow cytometry. The lack of SP-C and EGFRL858R in the CD45pos fraction supports the idea that EpCAMposCD45pos cells (Figure 1B, I) are immune cells that express EpCAM, and not epithelial cells that express CD45. We also observed that a small fraction of epithelial cells (∼5–6%) are found in the CD31pos fraction, which explains the low level of TTF1 expression in this fraction (Figure 2A). To determine whether ATI cells are also enriched with this protocol, we probed for the ATI-specific proteins, aquaporin 5 and T1α, which were both enriched in the EpCAMpos fraction compared with the preseparation fraction (Figure 2C).
Figure 2.
Molecular and cellular characterization of the MACS-isolated cell factions. (A) Western blot analysis on MACS-isolated cells from an EGFRL858R tumor-bearing mouse for surfactant protein (SP)-C, EpCAM, thyroid transcription factor-1 (TTF1), CD45, platelet endothelial cell adhesion molecule (PECAM), and EGFRL858R, as indicated. (B) Immunocytochemistry for EGFRL858R in preseparation cells, in EpCAMpos cells, and in CD31pos/CD45pos cells. Cells were incubated with an EGFRL858R primary antibody (red), followed by a secondary antibody (Alexa Fluor 594) and 4′,6-diamidino-2-phenylindole (DAPI) (blue) before being imaged (scale bars, 50 μm). (C) Lysates from cells isolated from an EGFRL858R tumor-bearing mouse probed with antibodies to aquaporin 5 (AQUA5) and T1α (alveolar type I cell markers) are shown. Of note, the center lane shows lysates of cells after CD31/CD45 simultaneous depletion (not sequential). (D) Flow cytometry analysis of isolated nonepithelial cell fractions, as indicated, to confirm their identity. (E) Sytox red staining on MACS-isolated CD45-, CD31-, and EpCAM-expressing cells from tumor-bearing (EGFRL858R) mice, as indicated. The fractions were stained with Sytox red to visualize the proportion of viable cells. Of note, the EpCAMpos fraction was isolated using CD31/CD45 simultaneous depletion (not sequential) before the EpCAM-positive selection to maximize viability.
In addition to the Western blot, ICC analysis of the different fractions further confirmed that epithelial cells expressing EGFRL858R are enriched when comparing the preseparation and the EpCAMpos fractions (Figure 2B), where approximately 77% of the epithelial cells were found to express EGFRL858R.
To further assess the identity of the isolated fractions (Figure 2D), we stained the CD31pos fraction with antibodies to CD31 and CD146 (endothelial cell marker) conjugated to fluorochromes. Using flow cytometry, we found that approximately 95% of the CD31pos fraction is double positive for the two markers (Figure 2D, I), confirming that this fraction predominantly contains endothelial cells. We also further evaluated the identity of cells in the CD45pos fraction (isolated from EGFRL858R tumor-bearing mice). We found that approximately 91% of the CD45pos cells were double positive for CD45 and F4/80 (a macrophage marker; Figure 2D, II), and approximately 16.1% of the population was double positive for CD45 and CD3 (T cell marker; Figure 2D, III). Of note, cells that express both macrophage and lymphoid markers have been described, which explains why we observed a small fraction of cells that are both F4/80pos and CD3pos (36).
To determine the viability of MACS-isolated cells, we labeled cells from the CD31pos, CD45pos, and EpCAMpos fractions with the SYTOX Red Dead Cell stain (Figure 2E). Cells that were positive for SYTOX Red were not viable. We found that 96.0 (±0.4)% of the CD45pos cells (Table 1; Figure 2E, I) and 90.2 (±2.5)% of the CD31pos cells (Table 1; Figure 2E, II) were viable. We observed that the shorter the total time of isolation, the higher the viability. The best viability for EpCAMpos cells was obtained when CD45pos and CD31pos cells were depleted simultaneously, which significantly decreases the total isolation time (by ∼40 min) and avoids extra stress on the cells, due to two sequential depletion steps before the EpCAM-positive selection. We observed that this protocol noticeably increased the viability for EpCAMpos cells without significantly affecting the purity or yield. Using this protocol, the viability of EpCAMpos cells was 74.0 (±2.4)% (Table 1; Figure 2E, III), the disadvantage being that the CD31pos and CD45pos cells cannot be isolated separately using this approach.
At the end of the isolation process, there is a fourth fraction, characterized as CD45neg CD31neg EpCAMneg. We observed that this fraction is mainly composed of lingering red blood cells, fibroblasts, and possibly other stromal cells. Flow, Western, and ICC analysis indicated that these cells do not express candidate proteins characteristic of epithelial, endothelial, or immune cells (data not shown).
Comparison of MACS- versus FACS-Based Cell Isolation
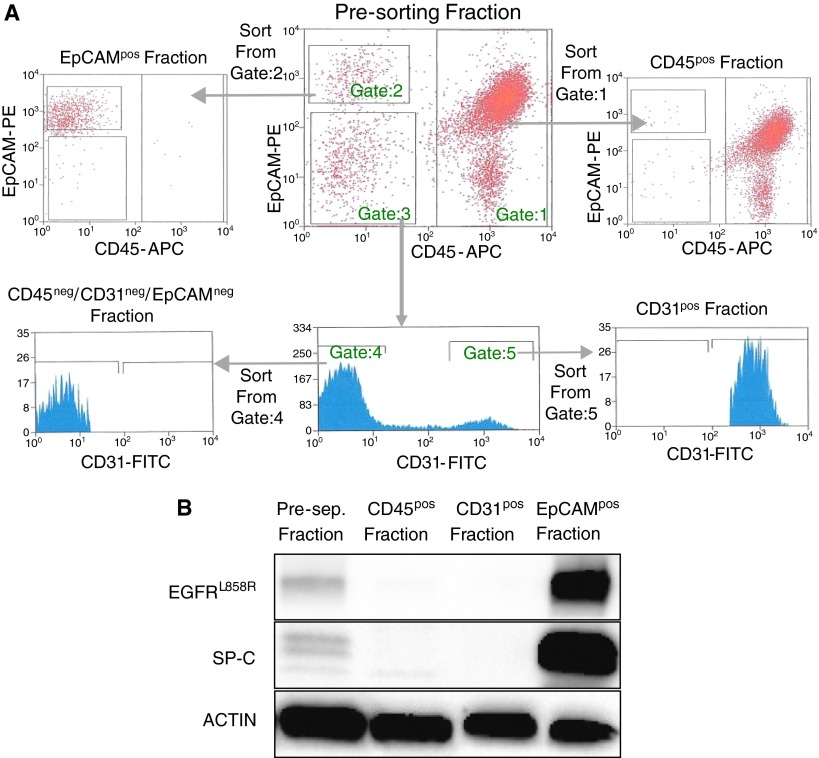
An alternative approach to separate cell populations is that of using FACS. To compare and contrast MACS versus FACS, we used CD31-FITC, CD45-APC, and EpCAM-PE to sort cells from the lungs of EGFRL858R tumor-bearing mice based on a gating scheme as illustrated in Figure 3A. Western blot analysis of the sorted fractions revealed that the EpCAMpos fraction is enriched for SP-C and EGFRL858R (Figure 3B). We also observed that the CD31 fraction is enriched for PECAM, and the CD45 fraction is enriched for the CD45 protein (data not shown). Because all the markers are localized to their respective fractions with high specificity, we conclude that the FACS-based protocol is also successful in isolating endothelial, epithelial, and immune cells from mouse lungs.
Figure 3.
Fluorescence-activated cell sorting (FACS)–based isolation of EpCAMpos, CD45pos, and CD31pos cells from tumor-bearing (EGFRL858R) mouse lungs. (A) Cells from a single-cell suspension of lung tissue were labeled with EpCAM-PE, CD31-FITC, and CD45-APC before being sorted using the BD FACSAria. Cells were gated as follows: CD45-APC–expressing cells (Gate:1), EpCAM-PE expressing cells (Gate:2), and EpCAM-PE/CD45APC double-negative cells (Gate:3). To isolate the CD31-FITC–expressing cells, we visualized the EpCAM-PE/CD45-APC double-negative cells on a CD31-FITC histogram and identified cells positive for CD31-FITC (Gate:5) separately from the cells that were CD31-FITC, EpCAM-PE, and CD45-APC triple negative (Gate:4). Therefore, to isolate CD45pos, EpCAMpos, CD31pos, and triple-negative cells, we sorted from gates 1, 2, 5, and 4, respectively. (B) Western blot analysis on FACS-isolated cells from an EGFRL858R tumor-bearing mouse for SP-C and EGFRL858R.
To define the similarities and differences between the two methods, we compared the purities, viabilities, and yields from the sorted fractions, as summarized in Table 1. The MACS protocol takes approximately 5–6 hours to complete per mouse, but it is possible to perform isolations from more than one mouse concurrently, provided that sufficient magnets are available. On the other hand, the FACS protocol takes approximately 3 hours per mouse, but samples from only one mouse at a time can be isolated. The FACS-isolated EpCAMpos and CD31pos fractions (Table 1) had slightly higher purities and less debris compared with those isolated by MACS. On the other hand, we observed higher viabilities and yield using the MACS protocol compared with those of the FACS protocol for all the isolated fractions. Although other reasons inherent to the FACS protocol might account for the yield discrepancy, one explanation is that we used doublet discrimination gating to avoid sorting cells that adhere to one another (which otherwise might result in fractions with low purity). Quantitative and qualitative comparisons of the two protocols are summarized in Table 1 and Table 2, respectively.
Table 2.
Qualitative Comparison of Fluorescence-Activated Cell Sorting– and Magnetic-Activated Cell Sorting–Based Isolations from Lungs of Mice with Adenocarcinoma
| MACS | FACS | |
|---|---|---|
| Total time of Isolation/mouse | 5–6 h (possible to do simultaneous isolations from several mice) | 3 h (allows for isolation from only one mouse at a time) |
| Yield | Comparatively higher yield | Lower yield |
| Viability | Higher viability* | Lower viability |
| Purity | Lower overall purity | Higher purity for EpCAMpos and CD31pos fractions |
For definition of abbreviations see Table 1.
Epithelial cell viability higher when doing CD45/CD31 simultaneous depletion.
Discussion
As our understanding of the complexity of the tumor microenvironment grows, interest in the molecular features of individual cell types that make up tumors is also increasing. Such studies require the isolation of different cell types for precise and detailed molecular and cellular investigations to elucidate mechanisms that underlie inflammation, tumor initiation, angiogenesis, and drug resistance.
In this study, we optimized a MACS-based protocol to isolate the major cellular compartments of the lung from tumor-bearing transgenic mice with lung adenocarcinomas, the major histopathological subtype of lung cancer. As a proof of principle for our study, we used transgenic mice that develop lung adenocarcinomas upon expression of an EGFR mutant (EGFRL858R) found in human tumors (14). We sequentially performed CD45 selection, CD31 selection, and EpCAM-positive selection on tumor-bearing lungs to respectively isolate immune, endothelial, and epithelial cells from the same tumor-bearing lungs: a protocol that may help studies aimed at understanding the relationship between these cell types in tumor initiation and progression (19, 37). Furthermore, we found that we could achieve high numbers of viable cells in all the isolated cell fractions using the MACS protocol, which is promising for any downstream studies that require the use of viable cells, such as cell culture (37). Our approach is generalizable, and can potentially be expanded to other mouse models of lung cancer.
In addition to optimizing the MACS protocol, we used antibodies conjugated to fluorochromes that recognize EpCAM, CD45, and CD31 to perform FACS-based isolations analogous to those of the MACS protocol. We found that the MACS protocol was generally superior in generating higher cell viabilities and yields compared with those of the FACS protocol (Tables 1 and 2). On the other hand, the overall purities obtained with FACS for EpCAMpos and CD31pos cells were better than when magnetic beads were used.
There are drawbacks inherent in using FACS/MACS. The length of time that is required to process tissue and isolate cells means that gene expression and signaling pathway activation may change during the isolation process. When comparing experiments, therefore, at a minimum, it is important to take steps to make sure that the amount of time required for isolation is kept constant for each mouse studied. Moreover, analysis of signaling events without cell isolation (using flow cytometry) can be performed and compared with results obtained from isolated cell populations as a control.
We also isolated immune, endothelial, and epithelial cells from lungs of wild-type EGFRL858R (−) mice, and found that the yields for all the cell types were considerably decreased compared with cells isolated from tumor-bearing lungs (Figure E2), potentially due to increased epithelial cell proliferation, inflammation, and angiogenesis in tumor-bearing lungs. However, Western blot analysis of the cells isolated from wild-type mice showed a similar level of enrichment compared with cells isolated from tumor-bearing lungs (data not shown).
It is worth noting that there are discrepant findings regarding the proportion of epithelial cells, in particular ATII cells, in relation to the total lung cellularity in normal mice. Roper and colleagues (38) used the SP-C promoter to drive the expression of enhanced green fluorescent protein, and found that ATII cells make up less than 1% of total lung cells. This finding is congruent with our study, in which we identified approximately 1.1% EpCAM-positive cells in normal lungs (data not shown) and approximately 4% in EGFRL858R tumor-bearing lungs (Figure 1B, I). A similar study that used the SP-C promoter to drive histone 2B–green fluorescent protein (H2B-GFP) expression indicated that ATII cells make up 6–8% of normal lung cells (39). On the other hand, studies using pap staining to label ATII cells found that they make up 19.5% (40) and 40% (41) of the total lung cell population in normal mice. The lungs of mice with EGFR-induced tumors exhibit a very high proportion of inflammatory cells, which might also contribute to the low percentage of epithelial cells identified in these lungs before cell separation. More studies are needed to accurately determine the exact proportion of epithelial cells in the lungs of normal and tumor-bearing mice.
EpCAM, in addition to being expressed on Clara and ATII cells, is also expressed on terminally differentiated lung epithelial cells, such as ATI and ciliated bronchiolar cells. By using this marker to isolate epithelial cells, we do not necessarily distinguish between tumor cells and normal epithelial cells (and indeed enrich for these cells as well; e.g., Figure 2C), although ICC for EGFRL858R showed that most of these cells are positive for the oncogene. Another caveat of using EpCAM is that cancer cells that undergo epithelial to mesenchymal transition may lose EpCAM expression, and thus, EpCAM-based enrichment methods lack the ability to capture these cells. Future experiments to refine strategies to capture these cells will be important.
The ability to separate the major populations of cells from tumor-bearing mouse lungs is likely to contribute to studies aimed at determining mechanisms of carcinogenesis and drug resistance. Because, in the tumor microenvironment, many cell subpopulations have overlapping signaling mechanisms relevant to lung cancer, deconstructing the lung into its basic cellular components and analyzing characteristics of these cellular fractions separately is more advantageous than studying the tumor environment in its totality (which introduces confounding factors due to cellular heterogeneity). This approach can be applied to many lung cancer models, and will allow for more accurate signaling, gene expression, and sequencing studies.
Acknowledgments
Acknowledgments
The authors thank Mary Ann Melnick, Deborah Ayeni, and Zimei Zhang for their assistance. They also extend their appreciation to the Dhodapkar laboratory for allowing them to use their FACSCalibur, Gouzel Tokmoulina for her technical expertise at the Yale Cell Sorter Core Facility, and Dr. Rakesh Verma for his insightful input.
Footnotes
This work was supported by National Institutes of Health/National Cancer Institute grant R01 120247, the Canary Foundation and the Labrecque Foundation (K.P.), and the Yale Cancer Center and Uniting Against Lung Cancer (X.S.).
Author Contributions: conception and design—A.B., X.S., and K.P.; performed experiments—A.B.; analysis and interpretation—A.B., X.S., and K.P.; manuscript writing and editing—A.B., X.S., and K.P.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2014-0312MA on October 27, 2014
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Reynolds SD, Malkinson AM. Clara cell: progenitor for the bronchiolar epithelium. Int J Biochem Cell Biol. 2010;42:1–4. doi: 10.1016/j.biocel.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Broeckaert F, Bernard A. Clara cell secretory protein (CC16): characteristics and perspectives as lung peripheral biomarker. Clin Exp Allergy. 2000;30:469–475. doi: 10.1046/j.1365-2222.2000.00760.x. [DOI] [PubMed] [Google Scholar]
- 3.Ward HE, Nicholas TE. Alveolar type I and type II cells. Aust N Z J Med. 1984;14:731–734. [PubMed] [Google Scholar]
- 4.Fehrenbach H. Alveolar epithelial type II cell: defender of the alveolus revisited. Respir Res. 2001;2:33–46. doi: 10.1186/rr36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Phelps DS, Floros J. Localization of pulmonary surfactant proteins using immunohistochemistry and tissue in situ hybridization. Exp Lung Res. 1991;17:985–995. doi: 10.3109/01902149109064330. [DOI] [PubMed] [Google Scholar]
- 6.Kalina M, Mason RJ, Shannon JM. Surfactant protein C is expressed in alveolar type II cells but not in Clara cells of rat lung. Am J Respir Cell Mol Biol. 1992;6:594–600. doi: 10.1165/ajrcmb/6.6.594. [DOI] [PubMed] [Google Scholar]
- 7.Sutherland KD, Berns A. Cell of origin of lung cancer. Mol Oncol. 2010;4:397–403. doi: 10.1016/j.molonc.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sullivan JP, Minna JD, Shay JW. Evidence for self-renewing lung cancer stem cells and their implications in tumor initiation, progression, and targeted therapy. Cancer Metastasis Rev. 2010;29:61–72. doi: 10.1007/s10555-010-9216-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giangreco A, Groot KR, Janes SM. Lung cancer and lung stem cells: strange bedfellows? Am J Respir Crit Care Med. 2007;175:547–553. doi: 10.1164/rccm.200607-984PP. [DOI] [PubMed] [Google Scholar]
- 10.Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, Jacks T, Tuveson DA. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thaete LG, Malkinson AM. Cells of origin of primary pulmonary neoplasms in mice: morphologic and histochemical studies. Exp Lung Res. 1991;17:219–228. doi: 10.3109/01902149109064413. [DOI] [PubMed] [Google Scholar]
- 12.Gunning WT, Stoner GD, Goldblatt PJ. Glyceraldehyde-3-phosphate dehydrogenase and other enzymatic activity in normal mouse lung and in lung tumors. Exp Lung Res. 1991;17:255–261. doi: 10.3109/01902149109064416. [DOI] [PubMed] [Google Scholar]
- 13.Sutherland KD, Song JY, Kwon MC, Proost N, Zevenhoven J, Berns A. Multiple cells-of-origin of mutant K-Ras–induced mouse lung adenocarcinoma. Proc Natl Acad Sci USA. 2014;111:4952–4957. doi: 10.1073/pnas.1319963111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Politi K, Zakowski MF, Fan PD, Schonfeld EA, Pao W, Varmus HE. Lung adenocarcinomas induced in mice by mutant EGF receptors found in human lung cancers respond to a tyrosine kinase inhibitor or to down-regulation of the receptors. Genes Dev. 2006;20:1496–1510. doi: 10.1101/gad.1417406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, Du H, Qin Y, Roberts J, Cummings OW, Yan C. Activation of the signal transducers and activators of the transcription 3 pathway in alveolar epithelial cells induces inflammation and adenocarcinomas in mouse lung. Cancer Res. 2007;67:8494–8503. doi: 10.1158/0008-5472.CAN-07-0647. [DOI] [PubMed] [Google Scholar]
- 16.Akbay EA, Koyama S, Carretero J, Altabef A, Tchaicha JH, Christensen CL, Mikse OR, Cherniack AD, Beauchamp EM, Pugh TJ, et al. Activation of the PD-1 pathway contributes to immune escape in EGFR-driven lung tumors. Cancer Discov. 2013;3:1355–1363. doi: 10.1158/2159-8290.CD-13-0310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim WY, Perera S, Zhou B, Carretero J, Yeh JJ, Heathcote SA, Jackson AL, Nikolinakos P, Ospina B, Naumov G, et al. HIF2alpha cooperates with RAS to promote lung tumorigenesis in mice. J Clin Invest. 2009;119:2160–2170. doi: 10.1172/JCI38443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kato T, Seto T, Goto K, Atagi S, Hosomi Y, Yamamoto N, Hida T, Maemondo M, Nakagawa K, Nagase S, et al. Eroltinib plus bevacizumab (EB) versus erlotinib alone (E) as first-line treatment for advanced EGFR mutation–positive nonsquamous non-small cell lung cancer (NSCLC): an open-label randomized trial. ASCO Annual Meeting. Chicago, IL, 2014
- 19.Yang L, DeBusk LM, Fukuda K, Fingleton B, Green-Jarvis B, Shyr Y, Matrisian LM, Carbone DP, Lin PC. Expansion of myeloid immune suppressor Gr+CD11b+ cells in tumor-bearing host directly promotes tumor angiogenesis. Cancer Cell. 2004;6:409–421. doi: 10.1016/j.ccr.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 20.Messier EM, Mason RJ, Kosmider B. Efficient and rapid isolation and purification of mouse alveolar type II epithelial cells. Exp Lung Res. 2012;38:363–373. doi: 10.3109/01902148.2012.713077. [DOI] [PubMed] [Google Scholar]
- 21.Cho HC, Lai CY, Shao LE, Yu J. Identification of tumorigenic cells in Kras(G12D)-induced lung adenocarcinoma. Cancer Res. 2011;71:7250–7258. doi: 10.1158/0008-5472.CAN-11-0903. [DOI] [PubMed] [Google Scholar]
- 22.Fujino N, Kubo H, Ota C, Suzuki T, Suzuki S, Yamada M, Takahashi T, He M, Suzuki T, Kondo T, et al. A novel method for isolating individual cellular components from the adult human distal lung. Am J Respir Cell Mol Biol. 2012;46:422–430. doi: 10.1165/rcmb.2011-0172OC. [DOI] [PubMed] [Google Scholar]
- 23.Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8:755–768. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- 24.Kim Y, Kim HS, Cui ZY, Lee HS, Ahn JS, Park CK, Park K, Ahn MJ. Clinicopathological implications of EpCAM expression in adenocarcinoma of the lung. Anticancer Res. 2009;29:1817–1822. [PubMed] [Google Scholar]
- 25.Di Paolo C, Willuda J, Kubetzko S, Lauffer I, Tschudi D, Waibel R, Pluckthun A, Stahel RA, Zangemeister-Wittke U. A recombinant immunotoxin derived from a humanized epithelial cell adhesion molecule–specific single-chain antibody fragment has potent and selective antitumor activity. Clin Cancer Res. 2003;9:2837–2848. [PubMed] [Google Scholar]
- 26.Yamamoto H, Yun EJ, Gerber HP, Ferrara N, Whitsett JA, Vu TH. Epithelial–vascular cross talk mediated by VEGF-A and HGF signaling directs primary septae formation during distal lung morphogenesis. Dev Biol. 2007;308:44–53. doi: 10.1016/j.ydbio.2007.04.042. [DOI] [PubMed] [Google Scholar]
- 27.Moon WS, Park HS, Yu KH, Park MY, Kim KR, Jang KY, Kim JS, Cho BH. Expression of betacellulin and epidermal growth factor receptor in hepatocellular carcinoma: implications for angiogenesis. Hum Pathol. 2006;37:1324–1332. doi: 10.1016/j.humpath.2006.04.022. [DOI] [PubMed] [Google Scholar]
- 28.Tichelaar JW, Lu W, Whitsett JA. Conditional expression of fibroblast growth factor-7 in the developing and mature lung. J Biol Chem. 2000;275:11858–11864. doi: 10.1074/jbc.275.16.11858. [DOI] [PubMed] [Google Scholar]
- 29.Fisher GH, Wellen SL, Klimstra D, Lenczowski JM, Tichelaar JW, Lizak MJ, Whitsett JA, Koretsky A, Varmus HE. Induction and apoptotic regression of lung adenocarcinomas by regulation of a K-Ras transgene in the presence and absence of tumor suppressor genes. Genes Dev. 2001;15:3249–3262. doi: 10.1101/gad.947701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Politi K, Fan PD, Shen R, Zakowski M, Varmus H. Erlotinib resistance in mouse models of epidermal growth factor receptor–induced lung adenocarcinoma. Dis Model Mech. 2010;3:111–119. doi: 10.1242/dmm.003681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Regales L, Gong Y, Shen R, de Stanchina E, Vivanco I, Goel A, Koutcher JA, Spassova M, Ouerfelli O, Mellinghoff IK, et al. Dual targeting of EGFR can overcome a major drug resistance mutation in mouse models of EGFR mutant lung cancer. J Clin Invest. 2009;119:3000–3010. doi: 10.1172/JCI38746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Bruin EC, Cowell C, Warne PH, Jiang M, Saunders RE, Melnick MA, Gettinger S, Walther Z, Wurtz A, Heynen GJ, et al. Reduced NF1 expression confers resistance to EGFR inhibition in lung cancer. Cancer Discov. 2014;4:606–619. doi: 10.1158/2159-8290.CD-13-0741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nelson AJ, Dunn RJ, Peach R, Aruffo A, Farr AG. The murine homolog of human Ep-CAM, a homotypic adhesion molecule, is expressed by thymocytes and thymic epithelial cells. Eur J Immunol. 1996;26:401–408. doi: 10.1002/eji.1830260220. [DOI] [PubMed] [Google Scholar]
- 34.Gautier EL, Shay T, Miller J, Greter M, Jakubzick C, Ivanov S, Helft J, Chow A, Elpek KG, Gordonov S, et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat Immunol. 2012;13:1118–1128. doi: 10.1038/ni.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stenhouse G, Fyfe N, King G, Chapman A, Kerr KM. Thyroid transcription factor 1 in pulmonary adenocarcinoma. J Clin Pathol. 2004;57:383–387. doi: 10.1136/jcp.2003.007138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baba T, Ishizu A, Iwasaki S, Suzuki A, Tomaru U, Ikeda H, Yoshiki T, Kasahara M. CD4+/CD8+ macrophages infiltrating at inflammatory sites: a population of monocytes/macrophages with a cytotoxic phenotype. Blood. 2006;107:2004–2012. doi: 10.1182/blood-2005-06-2345. [DOI] [PubMed] [Google Scholar]
- 37.Zhao T, Ding X, Du H, Yan C. Myeloid-derived suppressor cells are involved in lysosomal acid lipase deficiency–induced endothelial cell dysfunctions. J Immunol. 2014;193:1942–1953. doi: 10.4049/jimmunol.1301941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roper JM, Staversky RJ, Finkelstein JN, Keng PC, O'Reilly MA. Identification and isolation of mouse type II cells on the basis of intrinsic expression of enhanced green fluorescent protein. Am J Physiol Lung Cell Mol Physiol. 2003;285:L691–L700. doi: 10.1152/ajplung.00034.2003. [DOI] [PubMed] [Google Scholar]
- 39.Lee JH, Kim J, Gludish D, Roach RR, Saunders AH, Barrios J, Woo AJ, Chen H, Conner DA, Fujiwara Y, et al. Surfactant protein-C chromatin–bound green fluorescence protein reporter mice reveal heterogeneity of surfactant protein C–expressing lung cells. Am J Respir Cell Mol Biol. 2013;48:288–298. doi: 10.1165/rcmb.2011-0403OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harrison JH, Jr, Porretta CP, Leming K. Purification of murine pulmonary type II cells for flow cytometric cell cycle analysis. Exp Lung Res. 1995;21:407–421. doi: 10.3109/01902149509023716. [DOI] [PubMed] [Google Scholar]
- 41.Corti M, Brody AR, Harrison JH. Isolation and primary culture of murine alveolar type II cells. Am J Respir Cell Mol Biol. 1996;14:309–315. doi: 10.1165/ajrcmb.14.4.8600933. [DOI] [PubMed] [Google Scholar]