Abstract
Antenatal corticosteroids enhance lung maturation. However, the importance of glucocorticoid genes on early lung development, asthma susceptibility, and treatment response remains unknown. We investigated whether glucocorticoid genes are important during lung development and their role in asthma susceptibility and treatment response. We identified genes that were differentially expressed by corticosteroids in two of three genomic datasets: lymphoblastoid cell lines of participants in the Childhood Asthma Management Program, a glucocorticoid chromatin immunoprecipitation/RNA sequencing experiment, or a murine model; these genes made up the glucocorticoid gene set (GCGS). Using gene expression profiles from 38 human fetal lungs and C57BL/6J murine fetal lungs, we identified developmental genes that were in the top 5% of genes contributing to the top three principal components (PCs) most highly associated with post-conceptional age. Glucocorticoid genes that were enriched in this set of developmental genes were then included in the developmental glucocorticoid gene set (DGGS). We then investigated whether glucocorticoid genes are important during lung development, and their role in asthma susceptibility and treatment response. A total of 232 genes were included in the GCGS. Analysis of gene expression demonstrated that glucocorticoid genes were enriched in lung development (P = 7.02 × 10−26). The developmental GCGS was enriched for genes that were differentially expressed between subjects with asthma and control subjects (P = 4.26 × 10−3) and were enriched after treatment of subjects with asthma with inhaled corticosteroids (P < 2.72 × 10−4). Our results show that glucocorticoid genes are overrepresented among genes implicated in fetal lung development. These genes influence asthma susceptibility and treatment response, suggesting their involvement in the early ontogeny of asthma.
Keywords: glucocorticoid genes, lung development, asthma, asthma treatment
Clinical Relevance
Using an integrative genomic approach combining publicly available genomic data, we have identified that glucocorticoid genes are overrepresented among genes implicated in fetal lung development. Developmental glucocorticoid genes also influence asthma susceptibility and treatment response, suggesting their involvement in the developmental origins of asthma.
Glucocorticoids are endogenous steroid hormones that are involved in a variety of important physiologic responses in humans, including late fetal lung development and intrauterine lung maturation (1, 2). Animal models have demonstrated that glucocorticoids affect structural lung growth and development through the up-regulation of surfactant protein production (3), the regulation of lung growth factors (4), and the ability to modulate inflammatory mediators (5). Glucocorticoids administered to pregnant women at risk of premature delivery have been shown to accelerate fetal lung maturation, to reduce the occurrence and severity of neonatal respiratory distress syndrome, and to decrease neonatal mortality (6). However, the association of antenatal glucocorticoids with the development of postnatal chronic respiratory diseases, including asthma, has also been reported (7, 8). The biologic mechanisms underlying these associations have yet to be fully elucidated.
Asthma is the most common chronic respiratory disease of childhood and is associated with airway inflammation, airway hyperresponsiveness, and reversible airflow obstruction. Inhaled glucocorticoids are essential for the treatment of asthma and have been shown to decrease asthma exacerbation rates in children (9, 10). However, epidemiologic studies suggest that in utero exposure to glucocorticoids is an independent risk factor for the development of early childhood asthma between 3 and 5 years of age (8). The biologic mechanisms underlying this association are unclear. Although it may in part be due to an underlying predisposition that results from prematurity for which the corticosteroids are most often used, it may also result from a change in the genomic signature of lung development that results from this intrauterine exposure. The application of integrative genomic analyses to lung development may allow us to investigate the role of glucocorticoid genes in lung development and their role in the developmental origins of asthma.
We hypothesized that glucocorticoid genes (i.e., genes in which changes in expression characterize the response to glucocorticoids) are important during lung development and may play a role in the developmental origins of asthma. Using an integrative genomics approach incorporating multiple genomic datasets, we tested this hypothesis by identifying a set of genes that are consistently regulated by glucocorticoids. Because the cell-specific and tissue-specific effects of glucocorticoids are well recognized, we included several publically available genomic datasets that incorporate different tissue types and experimental designs to determine a universal set of glucocorticoid response genes that are robust to tissue-type or cell-type specific changes in gene expression alone. In addition, we investigated whether the identified glucocorticoid genes are enriched during the time period of in utero airway development and examined whether glucocorticoid genes that are expressed during lung development influence asthma susceptibility and asthma treatment response.
Materials and Methods
Derivation of a Glucocorticoid Gene Set
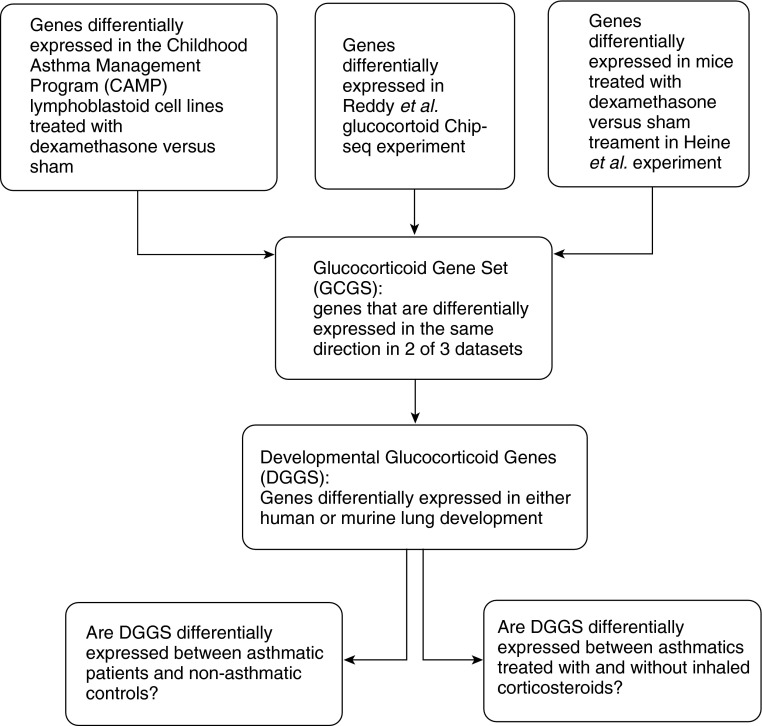
Genes were selected for the glucocorticoid gene set (GCGS) if they were consistently differentially expressed (increased or decreased) in response to dexamethasone treatment in at least two of the following three published sources (Figure 1):
-
1.
Lymphoblastoid cell lines (LCLs) from the Childhood Asthma Management Program (CAMP): we used a nonparametric Wilcoxon sign rank test to investigate for differential expression between dexamethasone and sham-treated Epstein Barr virus–transformed immortalized LCLs previously generated in a subset of subjects with mild to moderate persistent asthma participating in the CAMP study (adjusted P ≤ 1 × 10−5) (11).
-
2.
Gene expression in C57BL/J6 newborn mice treated with dexamethasone versus saline by Heine and colleagues (GSE51213) (12): we performed differential expression analysis on these data using geometric fold analysis (13).
-
3.
Chromatin immunoprecipitation and RNA-sequencing by Reddy and colleagues (14): genes significantly differentially expressed in response to dexamethasone in A549 lung epithelial carcinoma cell lines identified by Reddy and colleagues (14) were considered for inclusion
Figure 1.
Flow chart demonstrating our integrative genomic study design.
Derivation of a Developmental Glucocorticoid Gene Set
We used principal component analysis (PCA) to investigate gene expression variation from two published developmental genomic datasets: gene expression profiles from mRNA extracted from 38 early human fetal lung development tissue samples (postconceptional age, 53–154 d) (GSE14334) and genome-wide gene expression profiles from C57BL/J6 mice (GSE11539). We focused on the first three principal components (PC1–3) correlated with postconceptional age and noted the top 5% greatest contributor genes to the variation along PC1, PC2, and PC3. We also used linear models to identify developmental genes associated with postconceptional age (P < 0.05). GCGS genes were visualized in the transcriptomic developmental profile by plotting the GCGS against the two PCs most highly correlated with postconceptional age. We derived the developmental glucocorticoid gene set (DGGS) from genes in the GCGS that are represented in the top 5% of the first three PCAs (PCA1–3) of human or murine lung development or identified by linear analyses.
DGGS Genes and Their Association with Asthma and Treatment Response
Using a sign rank test at P < 0.05, we tested for differential expression and overrepresentation of the DGGS in three sets of publicly available gene expression data: (1) the MRC UK National Family Collection study including LCLs in 95 subjects with asthma and their 95 unaffected sibling control subjects (GSE8052) (15), (2) bronchial brushing specimens before and after treatment with inhaled fluticasone (GSE4302) in 19 subjects with asthma, and (3) bronchial biopsy specimens from 12 pairs of subjects with asthma before and after treatment with inhaled fluticasone (GSE23611).
Replication of Differential Expression of DGGS Genes between Subjects with Asthma and Nonasthmatic Control Subjects in Asthma BRIDGE
DGGS gene expression, indicating initial association with asthma (CEBPD, DDIT4, and FKBP5), was compared in an independent set of subjects with asthma (n = 865) and nonasthmatic control subjects (n = 116) using gene expression profiles generated from whole blood as part of the Asthma BioRepository for Integrative Genomic Exploration (Asthma BRIDGE), a multicenter effort to develop a well-characterized translational genomic dataset in asthma (16). Differential expression was determined using linear models adjusted for age, gender, and race. Additional details of the genomic datasets and analytic methodologies are available in the online supplement.
Results
Differential Expression by Dexamethasone Treatment in LCLs from Subjects with Asthma Participating in CAMP
Using LCLs from subjects with asthma participating in CAMP, we identified 4,271 genes that were differentially expressed between dexamethasone and sham treatment. Gene ontology (GO) pathway enrichment for this set of differentially expressed genes demonstrated significant associations with multiple biological processes, including programmed cell death, apoptosis, and response to DNA damage stimulus (false discovery rate [FDR]-adjusted P < 2 × 10−8). These 4,271 genes were considered for inclusion in the GCGS.
Glucocorticoid Genes Identified through Chromatin Immunoprecipitation and RNA Sequencing
Reddy and colleagues reported that approximately 30% of regulated genes had glucocorticoid receptor binding sites within 10 kb of the promoter. In addition, RNA sequencing revealed that 234 genes demonstrated significant changes in expression in response to dexamethasone (FDR-adjusted P < 0.05) (14). Of these, only the 209 genes that mapped to Entrez Gene identifiers were considered for inclusion in the GCGS.
Gene Expression in C57BL/J6 Newborn Mice Treated with Dexamethasone
Differential gene expression analysis between dexamethasone-treated and control C57BL/J6 mouse lung demonstrated evidence of differential expression for 1,290 genes. Of these genes, 704 demonstrated decreased and 586 demonstrated increased gene expression after dexamethasone treatment. These were considered for inclusion in the GCGS.
GCGS
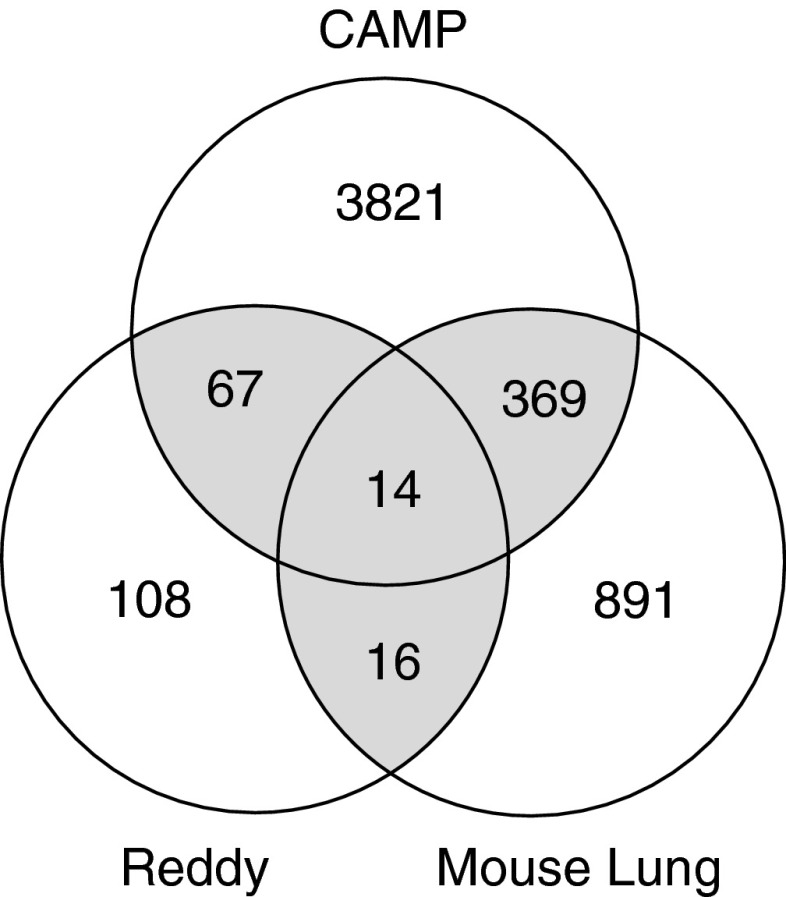
In total, 232 genes were significantly differentially expressed in the same direction in at least two of the three genomic datasets and were thus included in the GCGS (Figure 2 and see Table E1 in the online supplement). Of these genes, 126 consistently demonstrated increased expression, and 106 showed down-regulation of gene expression after treatment with glucocorticoids in both datasets. In total, eight genes (BIRC3, CEBPD, DDIT4, FKBP5, KLF9, PER1, ENC1, and SOX4) demonstrated consistent differential expression after dexamethasone treatment in all three datasets.
Figure 2.
Diagram demonstrating the genes identified by the independent genomic methodologies used for the identification of the GCGS. Genes shown demonstrate genes that were differentially expressed independent of direction. Only those genes that demonstrated consistent expression (either increased or decreased expression) after corticosteroid treatment in both datasets were included in the GCGS.
Results of the GO pathway enrichment analysis of the GCGS genes relative to the human transcriptome performed using DAVID are presented in Table 1. GO analysis demonstrated enrichment of genes involved in several biologic processes, including those involved in the regulation of phosphorylation, phosphorus metabolic processes, and response to oxygen species (FDR-adjusted P < 0.05). The 126 genes that demonstrated increased gene expression after treatment with corticosteroids in the GCGS were enriched for genes involved in the response to reactive oxygen species, hydrogen peroxide, and inorganic substances (FDR-adjusted P < 0.05). The 106 genes that consistently demonstrated decreased gene expression after treatment with dexamethasone were enriched for genes involved in the mitotic cell cycle, lymphocyte differentiation, mitosis, and nuclear division (FDR-adjusted P < 0.05).
Table 1.
Gene Ontology Pathway Enrichment of Genes in the Developmental Glucocorticoid Gene Set
| GO Pathway | GO Term | Fold Enrichment | FDR-Adjusted P value | Representative Genes |
|---|---|---|---|---|
| Regulation of phosphorylation | GO:0042325 | 3.07 | 0.01 | ITGA1, BCL2, SPRY1, SPRY2 |
| Regulation of phosphorus metabolic process | GO:0051174 | 2.96 | 0.02 | ITGA1, BCL2, ERN1, SPRY1 |
| Response to oxygen species | GO:0000302 | 7.69 | 0.03 | PRDX6, DUSP1, OLR1, TXNIP |
| Response to hydrogen peroxide | GO:0042542 | 9.16 | 0.04 | PRDX6, DUSP1, OLR1, TXNIP, BCL2, SOD2, SDC1 |
Definition of abbreviations: FDR, false discovery rate; GO, gene ontology.
The Developmental Profile of GCGS Genes
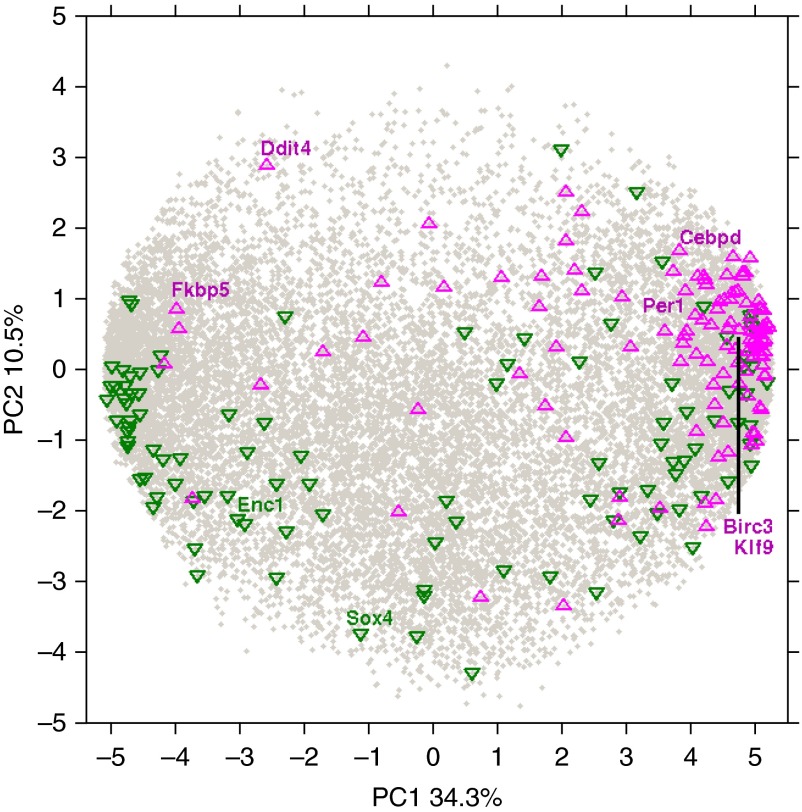
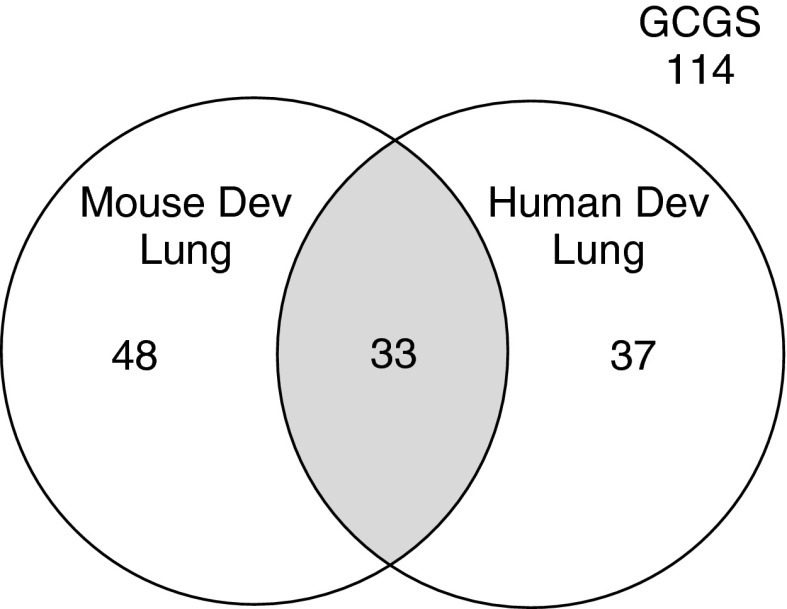
To investigate the developmental signature of the GCGS genes in early and late lung development, we tested for enrichment of this gene set in the genes involved in development using gene expression profiles of murine and human fetal lung. Because access to late-stage human fetal tissues is limited, human fetal lung tissues from the early pseudoglandular and canalicular stages of development were used to investigate the enrichment of GCGS genes in early human lung development. Gene expression patterns of the entire murine lung development trajectory were also analyzed to identify the role of GCGS during early and late lung development. PCA was performed to investigate the variance in gene expression across the murine and human fetal lung samples. The first three PCs (PC1–3), which explain greater than 50% of the variance in gene expression in murine and human fetal lung, were significantly correlated with postconceptional age. To visualize the potential effects of GCGS genes in the developing lung, we plotted the GCGS genes against the two PCs most highly correlated with postconceptional age to examine the expression of these genes during murine lung development (Figure 3). Genes shown in green are down-regulated by dexamethasone treatment across the developmental timeline. Although these genes are expressed during early gestation and in the early postnatal period, a larger number of genes that demonstrate decreased gene expression after corticosteroid exposure are associated with early lung development. GCGS genes that are up-regulated by dexamethasone treatment are shown in magenta. There is a notable increase in the expression of genes up-regulated by corticosteroids during the late gestational periods, including the saccular stage of lung development, suggesting that these genes may be implicated in lung maturation and provide a genomic basis for the use of prenatal corticosteroids. Using the top 5% greatest loading magnitude genes of the first three PCs that were found to be correlated with postconceptional age (PC1–3) to represent the developing lung time series, we found that GCGS genes were significantly enriched in murine lung development (odds ratio [OR], 4.51; P = 1.40 × 10−22) and in early lung development in the human fetal lung (OR, 3.15; P = 3.22 × 10−13). We also identified an enrichment for GCGS genes in the set of genes identified to be differentially expressed using linear models in human lung development (OR, 1.91; 95% confidence interval [CI], 1.36–2.69). Combined, GCGS genes were substantially enriched in murine and human lung development (OR; 4.23; 95% CI, 3.23–5.52) (Table 2). The 118 GCGS genes that were enriched in either murine (n = 81) or human lung development (n = 70) were included in the DGGS, including 33 genes that were in both developmental datasets (Figure 4; Table E1). Furthermore, of the previously identified eight genes that showed consistent differential expression after dexamethasone treatment that were common to all three initial genomic datasets used to identify the initial GCGS, six were included in the DGGS (CEBPD, DDIT4, FKBP5, KLF9, ENC1, and SOX4). GO enrichment analysis of the DGGS genes demonstrates enrichment of genes in cell cycle (GO:0007049, fold enrichment: 3.21; FDR P = 0.01), nuclear cell division (GO:0000280, fold enrichment: 4.56; FDR P = 0.026), and organelle fission (GO:0048285, fold enrichment: 5.32; FDR P = 0.03). The DGGS were then tested for association with asthma disease susceptibility and asthma treatment response. We maintained inclusion of the murine dataset for our disease association studies given its more comprehensive coverage of the developmental timeline.
Figure 3.
Graph demonstrating the first two principal components (PCs) of gene expression of murine lung development. PC1, which explains 34.3% of mouse gene expression, is correlated with gestational age. The gray dots represent all genes in the murine genome that are expressed in the prenatal and postnatal periods. Genes delineated in green demonstrate decreased expression after corticosteroid treatment; genes delineated in magenta demonstrate increased gene expression in our DGGS. Early murine lung development is enriched for genes that are down-regulated by corticosteroids, whereas genes that demonstrate increased expression after corticosteroid treatment are enriched in later murine development.
Table 2.
Enrichment of Glucocorticoid Genes in Human or Murine Lung Development*
| Relevance of Gene to Lung Development | In DGGS | Not in DGGS | OR | 95% CI |
|---|---|---|---|---|
| Involved in lung development | 118 | 3,960 | 4.23 | 3.23–5.52 |
| Not involved in lung development | 114 | 16,169 |
Definition of abbreviations: CI, confidence interval; DGGS, developmental glucocorticoid gene set; OR, odds ratio.
For gene lists, see Table E1 in the online supplement.
Figure 4.
Diagram demonstrating the selection of the GCGS genes that are also important in lung development. The genes identified in these analyses were represented in the top 5% of genes in the first three PCs of lung development.
Association of DGGS Genes with Asthma Susceptibility and Treatment Response
Differential expression of DGGS genes in asthma
Using publicly available genome-wide gene expression data from LCLs obtained from 95 unaffected and asthma-affected sib-pairs recruited through a proband with asthma (GSE8052), we identified 2,732 genes that were differentially expressed between subjects with asthma and nonasthmatic control subjects (P < 0.05). This set of differentially expressed genes was enriched for genes in our DGGS (OR, 1.92; 95% CI, 1.22–2.99) (Table 3). Furthermore, the enrichment of DGGS genes appears to be stronger than that of GCGS genes alone (OR, 1.61; P = 6.38 × 10−3), suggesting a possible role for this subset of genes in the pathobiology of asthma disease susceptibility and providing evidence for the fetal origins of postnatal disease. GO pathway enrichment analysis of the 27 DGGS genes (Table E2) that are associated with asthma susceptibility includes the cell cycle pathway (GO:0007049) and the regulation of apoptosis (GO:0042981) (FDR-adjusted P < 0.05).
Table 3.
Enrichment of the Developmental Glucocorticoid Gene Set in Subjects with Asthma Compared with Nonasthmatic Control Subjects (GSE8052)*
| Asthma Status | In DGGS | Not in DGGS | OR | 95% CI |
|---|---|---|---|---|
| Subjects with asthma | 27 | 2,705 | 1.92 | 1.22–2.99 |
| Nonasthmatic control subjects | 91 | 17,538 |
For gene list, see Table E2 in the online supplement.
Association of DGGS Genes in Asthma Treatment Response
Given the integral role of inhaled glucocorticoids in the treatment of asthma, we next tested the hypothesis that DGGS genes also influence asthma treatment response. Using two publically available genomic datasets, we assessed for differential expression of DGGS genes between bronchial brushings (GEO, GSE4302) and bronchial biopsy specimens (GEO, GSE23611) from subjects with asthma treated with and without inhaled fluticasone. Differential expression analysis from 19 subjects with asthma from whom bronchial brushing specimens were obtained before and after treatment with inhaled fluticasone (GSE4302) demonstrated that DGGS genes were more likely to be differentially expressed by inhaled corticosteroid treatment in bronchial brushings of subjects with asthma (OR, 2.18; 95% CI, 1.45–3.29) (Table 4). The 36 DGGS genes that are associated with asthma treatment response in bronchial brushing specimens are shown in Table E3. DGGS genes were also significantly overrepresented in the set of genes that were differentially expressed in 12 pairs of bronchial biopsy specimens from subjects with asthma (GSE23611) treated with and without inhaled corticosteroids (OR, 5.58; 95% CI, 3.82–8.15) (Table 5). The 55 DGGS genes that are associated with asthma treatment response in bronchial biopsy specimens are shown in Table E4. These results support a role for DGGS genes in early asthma treatment response. GO pathway enrichment analysis of the DGGS genes that are associated with asthma treatment response in bronchial brushing and/or bronchial biopsy specimens includes mitotic cell cycle (GO:0000278) (FDR, P < 0.05).
Table 4.
Enrichment of the Developmental Glucocorticoid Gene Set in Pretreated versus Inhaled Corticosteroid–Treated Bronchial Brushing Specimens of Subjects with Asthma (GSE4302)
| Asthma Treatment | In DGGS | Not in DGGS | OR | 95% CI |
|---|---|---|---|---|
| Inhaled corticosteroid treatment | 36 | 3,387 | 2.18 | 1.45–3.29 |
| No inhaled corticosteroid treatment | 82 | 16,856 |
For gene list, see Table E3 in the online supplement.
Table 5.
Enrichment of the Developmental Glucocorticoid Gene Set in Pretreated versus Inhaled Corticosteroid–Treated Bronchial Biopsy Specimens of Subjects with Asthma (GSE23611)
| Asthma Treatment | In DGGS | Not in DGGS | OR | 95% CI |
|---|---|---|---|---|
| Inhaled corticosteroid treatment | 55 | 2,739 | 5.58 | 3.82–8.15 |
| No inhaled corticosteroid treatment | 63 | 17,504 |
For gene lists, see Table E4 in the online supplement.
Of the eight genes that showed consistent differential expression after dexamethasone treatment in all three initial genomic datasets used to identify the GCGS, six genes were also enriched in lung development and were thus included in DGGS (CEBPD, DDIT4, FKBP5, KLF9, ENC1, and SOX4). The two genes that were consistently differentially expressed by dexamethasone treatment but were not enriched in lung development (BIRC3 and PER1) have been shown to be associated with asthma pathobiology (17) and circadian rhythms (18), suggesting their potential importance in asthma biology. Each of these developmental glucocorticoid genes demonstrated additional associations with asthma susceptibility and/or inhaled corticosteroid treatment response in subjects with asthma (Table 6). Furthermore, three developmental glucocorticoid genes (CEBPD, DDIT4, and FKBP5) were associated with asthma susceptibility and inhaled corticosteroid treatment in bronchial brushing and bronchial biopsy specimens of subjects with asthma. These three genes likely represent an interesting and potentially important subset of developmentally relevant glucocorticoid genes that may be involved in the early ontogeny of asthma.
Table 6.
Association of Eight Common Glucocorticoid Gene Set Genes with Lung Development, Asthma, and Asthma Treatment Response
| Gene* | Developmental Gene | Asthma Association | Association with Asthma Treatment in Bronchial Brushings | Association with Asthma Treatment in Bronchial Biopsies |
|---|---|---|---|---|
| BIRC3 | No | No | No | No |
| CEBPD | Yes | Yes | Yes | Yes |
| DDIT4 | Yes | Yes | Yes | Yes |
| ENC1 | Yes | No | No | Yes |
| FKBP5 | Yes | Yes | Yes | Yes |
| KLF9 | Yes | No | Yes | Yes |
| PER1 | No | No | No | No |
| SOX4 | Yes | No | No | Yes |
The eight common glucocorticoid gene set (GCGS) genes demonstrated the same direction of gene expression in each of the three GCGS genomic datasets: lymphoblastoid cell lines of participants in the Childhood Asthma Management Program, a glucocorticoid chromatin immunoprecipitation/RNA sequencing experiment, and a murine model of lung development.
Validation: Differential Expression of CEBPD and DDIT4 between Subjects with Asthma and Nonasthmatic Control Subjects in Asthma BRIDGE
Having demonstrated that CEBPD, DDIT4, and FKBP5 are developmental glucocorticoid genes that are associated with asthma in our sib-pair analysis, we assessed whether the expression of these genes was similarly correlated with asthma status in an independent population of subjects with asthma and nonasthmatic control subjects. We assessed the expression of these genes (CEBPD, DDIT4, and FKBP5) in 865 subjects with asthma and in 116 nonasthmatic control subjects participating in Asthma BRIDGE with available expression data in whole blood (see Table E5 for baseline characteristics of the cohort used for this analysis). Of the three genes evaluated, CEBPD mRNA expression was significantly increased in subjects with asthma compared with nonasthmatic control subjects (P = 0.0003). DDIT4 expression was also modestly increased in subjects with asthma compared with control subjects (P = 0.02). There was no evidence of asthma-related differential expression of FKBP5 (P = 0.69).
Discussion
Corticosteroids are potent antiinflammatory agents that enhance late fetal lung maturation in pregnant women at risk for premature labor. Corticosteroids decrease the incidence of respiratory distress syndrome of the newborn. However, their use has been associated with asthma susceptibility in the postnatal period. The biologic mechanisms underlying this association are unknown. In addition, inhaled corticosteroids are commonly prescribed treatments for asthma and are associated with improved asthma outcomes (19). Based on these observations, we hypothesized that glucocorticoid genes are important in lung development and that developmentally relevant glucocorticoid genes are associated with asthma susceptibility and treatment response. We used an integrative genomic approach using multiple genomic datasets to investigate the role of glucocorticoid response genes in early lung development, including the time period in which in utero airway development occurs and whether these genes influence asthma susceptibility and response to asthma therapy. Our study design focused on the gene expression differences between untreated/sham treatment and dexamethasone treatment in three data sets used to develop the GCGS. Even in the initial murine data set (GSE51213), we focus on the strongest differences between corticosteroid-treated and untreated cells irrespective of their role in development. Our methodology stipulates that genes identified demonstrate a consistent difference in expression after glucocorticoid treatment in at least two of these datasets and thus minimizes the inclusion of genes demonstrating only a cell- or tissue-specific response to glucocorticoids or under a single experimental condition. In addition, we ultimately focus on the genes that were identified in all three genomic datasets, including genes that are differentially expressed by dexamethasone in LCLs, which is a dataset without any known developmental implications. We then demonstrate the enrichment of glucocorticoid genes in lung development throughout the developmental timeframe (DGGS). Although glucocorticoid receptor signaling has been clearly shown to be active in the developing lung, it has been associated with the later stages of development in previously published studies. Our study demonstrates the relevance of these genes in early-stage lung development. Furthermore, using several publicly available genomic datasets, we have demonstrated that the identified developmentally relevant DGGS genes are associated with differences in expression between patients with asthma and nonasthmatic control subjects and are correlated with the response to inhaled corticosteroids in subjects with asthma later in life. In addition, we have demonstrated consistent differential expression of CEBPD and DDIT4 between subjects with asthma and nonasthmatic control subjects in two independent data sets. These results support a possible role of glucocorticoid genes in the developmental origins of asthma susceptibility and asthma treatment response and provide motivation for future investigation of these genes.
The importance of glucocorticoids in normal fetal lung maturation has been established by the glucocorticoid receptor knockout mouse, which dies at birth due to abnormal lung development (20). Although animal models suggest that antenatal corticosteroids enhance late fetal lung maturation by stimulating the production of pulmonary surfactants (21), the underlying biologic mechanisms to explain their associations with the development of postnatal chronic respiratory diseases like asthma have yet to be fully elucidated. Recent evidence suggests that the ethnicity of the pregnant woman influences the infant’s risk of developing neonatal respiratory distress after antenatal corticosteroid use, suggesting a role for genetics in addition to in utero exposures in the development of respiratory disease (22). Using genomic approaches to investigate glucocorticoid response, we have identified a set of genes that demonstrate consistent differential expression after dexamethasone treatment in at least two genomic datasets, have shown that these genes are expressed during normal murine and early human lung development, and have shown an association of these developmentally relevant glucocorticoid genes with asthma susceptibility and inhaled corticosteroid treatment response in subjects with asthma. These results suggest that intrauterine steroid deficiency may result in a genomic signature that reflects a more immature lung, which may be a possible mechanism for the role of developmental glucocorticoid genes in the predisposition to postnatal disease. Furthermore, these developmental glucocorticoid genes may provide further insight into novel therapeutic targets for inflammatory diseases like asthma.
Murine models of lung development have demonstrated that antenatal corticosteroids are associated with gene expression changes that result in abnormal alveolarization at birth (23). Furthermore, in preterm animal models of lung development, antenatal corticosteroid treatment has been shown to increase surfactant protein expression, to decrease vascular permeability, and to enhance clearance of extracellular fluid (24). Our genomic analysis in murine lung development confirms the up-regulation of these genes in the later stages of lung development. In addition, using genome-wide gene expression profiles of early human lung development, we have shown that glucocorticoid genes are enriched during early development, including the histologic stages during which airway branching morphogenesis occurs. Although our results corroborate the findings of late murine development, we extend previous knowledge about these genes by demonstrating their importance during the early stages of human lung development.
Given the importance of glucocorticoids in the treatment of airway inflammation associated with asthma, we also sought to determine whether developmental glucocorticoid genes influence postnatal respiratory disease susceptibility. By demonstrating enrichment for the DGGS genes in the set of genes that demonstrate differential expression between subjects with asthma and nonasthmatic control subjects, we provide some evidence for the developmental origin of asthma.
Animal models demonstrate changes in lung structure due to antenatal corticosteroid use, which results in improved lung mechanics and decreased airways resistance at 28 days (25). Human data corroborate these findings: antenatal corticosteroid therapy has been shown to increase functional residual capacity in preterm neonates (26) and to result in overall improved respiratory system compliance (27). Although these findings suggest that antenatal corticosteroids may decrease the risk of the development of asthma, epidemiologic data demonstrate an increased risk of early childhood asthma between 3 and 5 years of age in individuals with in utero exposure to corticosteroids (8, 28). This may partially be explained by animal studies that show that antenatal corticosteroids result in early suppression of inflammation but a subsequent augmentation of inflammation with increased alveolar neutrophils and expression of proinflammatory cytokines 5 days after endotoxin exposure, suggesting that glucocorticoids may impair the ability of the preterm lung to down-regulate inflammation even after fetal clearance of the glucocorticoids (29). However, treatment with betamethasone 7 days after LPS exposure resulted in increased surfactant protein expression in a similar lamb model of development (30), suggesting that, in the setting of antenatal corticosteroid use, the timing of LPS exposure and other potential in utero exposures may have implications in terms of overall lung development and the possibility of subsequent respiratory disease.
Our work identified three developmental glucocorticoid genes (CEBPD, DDIT4, and FKBP5) that are of particular interest because of their association with asthma susceptibility and inhaled corticosteroid treatment response in bronchial brushing and bronchial biopsy specimens of subjects with asthma treated with inhaled corticosteroids.
Of the three candidate genes identified, the most biologic data supporting a role in lung development and for establishing a potential biologic link to asthma are available for CCAAT/enhancer-binding protein δ (C/EBPD). C/EBPD expression has been shown to be the highest in the lung tissue and increases in response to dexamethasone treatment (31). C/EBPD is involved in surfactant protein synthesis and is expressed at a low level in fetal type II cells of the alveolar epithelium (32). Pulmonary surfactant, a phospholipid-rich lipoprotein that is synthesized by alveolar type II cells, reduces surface tension at the alveolar air–liquid interface. In a rabbit model of lung development, C/EBPD expression has been demonstrated as early as Day 19 of gestation and has been shown to increase approximately 3.7-fold to reach its peak expression level by Day 28 of gestation (33). The transcriptomic profile of C/EBPD mirrors the developmental time course of surfactant protein production. Furthermore, C/EBPD expression has been shown to increase after 12 hours of organ culture and demonstrates an additional increase after treatment with dexamethasone and cAMP (33). These results suggest that C/EBPD may mediate the hormone regulation of surfactant protein synthesis and type II pneumocyte differentiation (34, 35). Furthermore, recent work has demonstrated a greater induction of C/EBPD expression in fetal versus adult alveolar type II cells upon treatment with dexamethasone and cAMP (36). Treated fetal cells have up-regulated expression of surfactant proteins, an increased rate of phosphatidylcholine synthesis, and altered phospholipid concentrations, suggesting that the expression of these genes in the context of fetal development are hormone induced (37).
Although there is no direct evidence implicating C/EBPD in the pathogenesis of asthma, there is increasing support for a role for its family of transcription factors in the abnormal response of the asthmatic airway epithelium to infection. Several studies have demonstrated an exaggerated response of human airway smooth muscle (HASM) cells of patients with asthma to rhinovirus infection (38, 39). These differences appear to be related in part to rhinovirus-induced IL-6 secretion in the HASM cells in subjects with asthma. The transcriptional regulation of IL-6, a proinflammatory cytokine, involves the binding of transcription factors to the C/EBP binding site, which has been shown to increase transcription of IL-6 and other inflammatory cytokines (39). Furthermore, C/EBPα, a protein that is essential for the suppression of inflammation and proliferative responses, has been found to be deficient in HASM cells of subjects with asthma, suggesting that the disruption of the balance between excitatory and inhibitory CEBPs may play a role in the differential response of the HASM cells to rhinovirus infection compared with nonasthmatic control subjects (39). Based on our results and the additional supporting evidence of its importance, further investigation of the role of C/EBPD in asthma susceptibility and asthma treatment response is warranted.
We have also shown that DNA damage-inducible transcript 4 (DDIT4) is a DGGS gene that is associated with asthma susceptibility and treatment response. DDIT4, which encodes development and DNA damage responses 1 (REDD1), is a stress-response gene that decreases protein synthesis by repressing the mTOR pathway (40). We have shown that DDIT4 expression is consistently increased after treatment with dexamethasone in our three initial genomic datasets and that it is involved in lung development (Table E1). These results are similar to those reported in rat fetal lung fibroblast cultures in which there is a significant increase in DDIT4 expression after 48 hours of treatment with 50 nM of dexamethasone (41).
We have also shown that DDIT4 expression is increased in subjects with asthma compared with nonasthmatic control with asthma in two populations and in bronchial brush and bronchial biopsy specimens of subjects with asthma after treatment with inhaled corticosteroids. Although DDIT4 has yet to be implicated in the pathobiology of asthma, Yoshida and colleagues have shown that the protein coded by DDIT4 (REDD1) is increased in individuals with advanced emphysema (42). Furthermore, murine models investigating oxidative stress have shown that this protein mediates the effects of cigarette smoke–induced pulmonary pathology by demonstrating complete protection against cigarette smoke–induced acute inflammation in its knockout mouse (42). Their work demonstrates that this protein plays an important role in amplifying inflammation and cell death by negatively regulating mTOR signaling and activating NF-κB (42). These same mechanisms may provide a potential biologic basis for the association of DDIT4 with asthma and asthma treatment response.
We also identified FK506-binding protein 5 (FKBP5) as a developmentally relevant glucocorticoid gene, whose expression was induced in bronchial brushings and bronchial biopsy specimens of subjects with asthma treated with inhaled corticosteroids, suggesting a possible role for it in the developmental origin of asthma and inhaled corticosteroid treatment response. This gene has been previously associated with the subcellular localization of the glucocorticoid receptor (43). The glucocorticoid receptor is a hormone-activated transcription factor that requires hormone-initiated transport to its site of action in the nucleus. Davies and colleagues identified that a substitution of FKBP5 for another immunophilin is one of the earliest events in steroid receptor signaling and controls subcellular localization and transport of the steroid receptor (43). Previous studies have also shown that FKBP5 is one of the most highly induced genes in a several pulmonary epithelial cell lines treated with dexamethasone (44, 45). Based on its role in glucocorticoid signaling, FKBP5 expression has been previously studied in bronchial biopsy specimens in subjects with asthma. Similar to our findings in bronchial brushing and biopsy specimens, inhaled budesonide treatment has previously been shown to increase FKBP5 expression in bronchial biopsy specimens of subjects with asthma (46). However, in a previous genetic association study, there was no association of genetic polymorphisms in FKBP5 with improved lung function in subjects with asthma treated with inhaled corticosteroids (47). Given its role in glucocorticoid signaling and its reproducible demonstration of induction of expression after inhaled corticosteroid treatment in subjects with asthma, FKBP5 warrants further investigation for its involvement in asthma pathogenesis and treatment response.
Although we undertook a comprehensive analysis of the developmental signature of glucocorticoid genes, several limitations of our analysis exist. Our gene expression analysis of human lung development was limited to fetal lung tissue samples from the early stages of development. Therefore, we only investigated the transcriptomic profile of corticosteroid genes during early human lung development. Although our investigation of gene expression during the later stages of human lung development may allow us to gain further insights into the pathogenesis of respiratory disease, the availability of this data is limited. Therefore, the use of publically available murine developmental data allows us to investigate the entire developmental timeline by evaluating the transcriptomic changes that occur during later stages of gestation and into the postnatal period. Furthermore, we recognize that demonstration of differential expression of corticosteroid genes between subjects with asthma and nonasthmatic control subjects does not confirm that these genes are causally related to the development of asthma. However, these results do suggest that further investigation of these genes in asthma susceptibility and asthma treatment response is warranted.
In this study we identified a set of glucocorticoid genes that are involved in early and late lung development and demonstrated that these genes are associated with asthma and treatment response in patients with asthma. Our results provide preliminary evidence for a role for developmental glucocorticoid genes in the fetal origins of asthma and treatment response. Although our work focused on a subset of developmental glucocorticoid genes, additional insights regarding the complex biology underlying asthma susceptibility may be gleaned from further investigation of additional genes identified herein. Furthermore, this work should motivate future investigations of the role of these genes in the pathogenesis of asthma and in other respiratory diseases in which corticosteroids have been shown to have therapeutic implications.
Acknowledgments
Acknowledgments
The authors thank all subjects for their participation in the Asthma BRIDGE. The authors acknowledge the Asthma BRIDGE investigators and research team, supported by NHLBI, for collection of study data.
The Asthma BRIDGE Consortium
Asthma BRIDGE Data Coordinating Center
Childhood Asthma Management Program (CAMP) Genetics Ancillary Study
Brigham and Women’s Hospital, Harvard Medical School, Boston MA
Benjamin A. Raby, M.D., M.P.H., principal investigator (PI); Scott T. Weiss, M.D., M.Sc. (PI); Vincent Carey, Ph.D.; WeiliangQiu, Ph.D.; Roxanne Kelly, B.Sc.; Jody Sylvia Senter, M.S.; John Ziniti, B.Sc.; Diana Tubbs; Brooke Schumann; and Damien C. Croteau-Chonka, Ph.D.
Childhood Asthma Research and Education (CARE) Network
Arizona Respiratory Center, University of Arizona (Coordinating Center)
Fernando Martinez, M.D. (PI); Wayne Morgan, M.D.; James Goodwin, Ph.D.; Anthony Bosco, Ph.D. (also at Telethon Institute for Child Health Research, Centre for Child Health Research, University of Western Australia); Monica Vasquez, M.P.H.; Rosemary Weese, R.N.; Silvia Lopez, R.N.; Jesus Wences, B.S.; Monica Varela, L.P.N.; Janette Priefert; Katherine Chee; Samira Ehteshami, B.S.; and Xiaobing Liu, B.S.
National Jewish Health, Denver, CO
Andy Liu, M.D. (PI); Allison Schlitz, B.A.; Julie Henley, C.C.R.C.; D.A. Sundström, B.A.; Melanie Phillips, B.S.; Sakari Graves, B.A.; Phillip Lopez, B.S.; Liliana Soto, C.R.C.
University of Wisconsin–Madison, Madison, WI
Robert F. Lemanske, Jr., M.D. (PI); Theresa W. Guilbert, M.D.; Sarah Sund, B.S., M.T. (ASCP); Tiffany Huard, B.S., C.C.R.C.; and Elizabeth A. Schwantes, B.S.
Washington University School of Medicine, St. Louis MO
Robert C. Strunk, MD (PI); Tina Norris (was Oliver) CCRP, CRT; Wanda Caldwell, RRT; Cynthia Moseid;
Chicago Asthma Genetics (CAG) Study
University of Chicago, Chicago, IL
Carole Ober, Ph.D. (PI); Dan Nicolae, Ph.D.; Julian Solway, M.D.; Jerry Krishnan, M.D., Ph.D.; White Steve, M.D.; Kyle Hogarth, M.D.; John McConville, M.D.; Rebecca Anderson, M.S.; Jyotsna Sudi, M.S.; Lourdes Norwick, B.S.N., R.N.; and Myers Rachel, Ph.D.
Genomic Research on Asthma in the African Diaspora (GRAAD)
Johns Hopkins University, Baltimore, MD
Kathleen C. Barnes, Ph.D. (PI); Nadia Hansel, M.D., M.P.H.; John T. Schroeder, Ph.D.; Chris Cheadle, Ph.D.; Rasika A. Mathias, Sc.D.; Alan E. Berger, Ph.D.; Jinshui Fan, M.D., Ph.D.; CandelariaVergara, M.D., M.Sc.; Cassandra Foster; Meher Boorgula; Ria Barkataki; Li Gao, M.D., Ph.D.; Joseph Potee, M.S.; Terri H. Beaty, Ph.D.; Ingo Ruczinski, Ph.D.; and Jeff Leek, Ph.D.
Mexico City Childhood Asthma Study (MCCAS)
National Institute of Environmental Health Sciences, National Institute of Public Health of Mexico, Hospital Infantil de Mexico Federico Gomez
Stephanie J. London, M.D., Dr.P.H.; Albino BarrazaVillarreal, M.Sc., Dr.P.H.; Leticia Hernandez Cadena, M.Sc., Dr.P.H.; Efrain Navarro Olivos, M.D., M.Sc.; Isabelle Romieu, M.D., M.P.H., Dr.S.; Juan Jose SienraMonge, M.D.; Blanca Estela del Río Navarro, M.D.; Isabelle García; and Cynthia Hernandez
Children’s Health Study (CHS)
University of Southern California, Los Angeles, CA
Frank D. Gilliland, M.D., Ph.D. (PI); Talat Islam, M.B.B.S., Ph.D.; Carrie V. Breton, Sc.D.; Muhammad T. Salam, M.B.B.S., Ph.D.; Kimberly D. Siegmund, Ph.D.; Xinhui Wang, M.S.; and Xia Li, M.S.
Footnotes
This work is supported by grant R01 HL097144, R01 HL092197, U01 HL065899 from the National Heart, Lung, and Blood Institute, National Institutes of Health (NIH/NHLBI). The Asthma BRIDGE work was supported by R01 HL086601 and RC2 HL101543 from the National Heart, Lung, and Blood Institute, National Institutes of Health (NIH/NHLBI). S.S. receives additional support from K08 HL096833 from NIH/NHLBI. A.T.K. receives support from support from K25 HL091124. S.J.L. is supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (ZIA ES049019).
Author contributions: R.G., C.A.V., J.S.L, A.B.-V., S.J.L., F.G., B.A.R., S.T.W., and K.G.T aided with data collection. S.S., A.K.T., D.C., and W.Q. performed data analysis. All authors contributed to data interpretation and manuscript preparation.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2014-0109OC on September 5, 2014
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Bolt RJ, van Weissenbruch MM, Lafeber HN, Delemarre-van de Waal HA. Glucocorticoids and lung development in the fetus and preterm infant. Pediatr Pulmonol. 2001;32:76–91. doi: 10.1002/ppul.1092. [DOI] [PubMed] [Google Scholar]
- 2.Fowden AL, Forhead AJ. Hormones as epigenetic signals in developmental programming. Exp Physiol. 2009;94:607–625. doi: 10.1113/expphysiol.2008.046359. [DOI] [PubMed] [Google Scholar]
- 3.Wang JY, Yeh TF, Lin YC, Miyamura K, Holmskov U, Reid KB. Measurement of pulmonary status and surfactant protein levels during dexamethasone treatment of neonatal respiratory distress syndrome. Thorax. 1996;51:907–913. doi: 10.1136/thx.51.9.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jaskoll T, Choy HA, Melnick M. The glucocorticoid-glucocorticoid receptor signal transduction pathway, transforming growth factor-beta, and embryonic mouse lung development in vivo. Pediatr Res. 1996;39:749–759. doi: 10.1203/00006450-199605000-00002. [DOI] [PubMed] [Google Scholar]
- 5.Grigsby PL, Novy MJ, Sadowsky DW, Morgan TK, Long M, Acosta E, Duffy LB, Waites KB. Maternal azithromycin therapy for Ureaplasma intraamniotic infection delays preterm delivery and reduces fetal lung injury in a primate model. Am J Obstet Gynecol. 2012;207:475, e471–475, e414. doi: 10.1016/j.ajog.2012.10.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crowley P, Chalmers I, Keirse MJ. The effects of corticosteroid administration before preterm delivery: an overview of the evidence from controlled trials. Br J Obstet Gynaecol. 1990;97:11–25. doi: 10.1111/j.1471-0528.1990.tb01711.x. [DOI] [PubMed] [Google Scholar]
- 7.Seliem WA, Falk MC, Shadbolt B, Kent AL. Antenatal and postnatal risk factors for neonatal hypertension and infant follow-up. Pediatr Nephrol. 2007;22:2081–2087. doi: 10.1007/s00467-007-0603-2. [DOI] [PubMed] [Google Scholar]
- 8.Pole JD, Mustard CA, To T, Beyene J, Allen AC. Antenatal steroid therapy for fetal lung maturation and the subsequent risk of childhood asthma: a longitudinal analysis. J Pregnancy. 2010;2010:789748. doi: 10.1155/2010/789748. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Ververeli K, Chipps B. Oral corticosteroid-sparing effects of inhaled corticosteroids in the treatment of persistent and acute asthma. Ann Allergy Asthma Immunol. 2004;92:512–522. doi: 10.1016/S1081-1206(10)61758-9. [DOI] [PubMed] [Google Scholar]
- 10.Pauwels RA, Pedersen S, Busse WW, Tan WC, Chen YZ, Ohlsson SV, Ullman A, Lamm CJ, O’Byrne PM START Investigators Group. Early intervention with budesonide in mild persistent asthma: a randomised, double-blind trial. Lancet. 2003;361:1071–1076. doi: 10.1016/S0140-6736(03)12891-7. [DOI] [PubMed] [Google Scholar]
- 11.Tantisira KG, Lasky-Su J, Harada M, Murphy A, Litonjua AA, Himes BE, Lange C, Lazarus R, Sylvia J, Klanderman B, et al. Genomewide association between GLCCI1 and response to glucocorticoid therapy in asthma. N Engl J Med. 2011;365:1173–1183. doi: 10.1056/NEJMoa0911353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heine VM, Rowitch DH. Hedgehog signaling has a protective effect in glucocorticoid-induced mouse neonatal brain injury through an 11betaHSD2-dependent mechanism. J Clin Invest. 2009;119:267–277. doi: 10.1172/JCI36376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kho AT, Zhao Q, Cai Z, Butte AJ, Kim JY, Pomeroy SL, Rowitch DH, Kohane IS. Conserved mechanisms across development and tumorigenesis revealed by a mouse development perspective of human cancers. Genes Dev. 2004;18:629–640. doi: 10.1101/gad.1182504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reddy TE, Pauli F, Sprouse RO, Neff NF, Newberry KM, Garabedian MJ, Myers RM. Genomic determination of the glucocorticoid response reveals unexpected mechanisms of gene regulation. Genome Res. 2009;19:2163–2171. doi: 10.1101/gr.097022.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, Heath S, Depner M, von Berg A, Bufe A, Rietschel E, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–473. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 16.Qiu W, Koppelman GH, Carey VJ, Ziniti J, London S, Barnes KC, Gilliland FD, Lemanske R, Guilbert TW, Liu A, et al. American Thoracic Society International Conference; Philadelphia, PA: Expression quantitative trait locus (eqtl) mapping in diverse populations and cell types identifies numerous asthma-associated regulatory variants. May 18–22, 2012. p. A2521. [Google Scholar]
- 17.Roscioli E, Hamon R, Ruffin RE, Lester S, Zalewski P. Cellular inhibitor of apoptosis-2 is a critical regulator of apoptosis in airway epithelial cells treated with asthma-related inflammatory cytokines. Physiol Rep. 2013;1:e00123. doi: 10.1002/phy2.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burioka N, Fukuoka Y, Takata M, Endo M, Miyata M, Chikumi H, Tomita K, Kodani M, Touge H, Takeda K, et al. Circadian rhythms in the CNS and peripheral clock disorders: function of clock genes: influence of medication for bronchial asthma on circadian gene. J Pharmacol Sci. 2007;103:144–149. doi: 10.1254/jphs.fmj06003x4. [DOI] [PubMed] [Google Scholar]
- 19.Bateman ED, Bousquet J, Busse WW, Clark TJ, Gul N, Gibbs M, Pedersen S GOAL Steering Committee and Investigators. Stability of asthma control with regular treatment: an analysis of the Gaining Optimal Asthma controL (GOAL) study. Allergy. 2008;63:932–938. doi: 10.1111/j.1398-9995.2008.01724.x. [DOI] [PubMed] [Google Scholar]
- 20.Cole TJ, Blendy JA, Monaghan AP, Krieglstein K, Schmid W, Aguzzi A, Fantuzzi G, Hummler E, Unsicker K, Schütz G. Targeted disruption of the glucocorticoid receptor gene blocks adrenergic chromaffin cell development and severely retards lung maturation. Genes Dev. 1995;9:1608–1621. doi: 10.1101/gad.9.13.1608. [DOI] [PubMed] [Google Scholar]
- 21.Kotas RV, Fletcher BD, Torday J, Avery ME. Evidence for independent regulators of organ maturation in fetal rabbits. Pediatrics. 1971;47:57–64. [PubMed] [Google Scholar]
- 22.Haas DM, Sischy AC, McCullough W, Simsiman AJ. Maternal ethnicity influences on neonatal respiratory outcomes after antenatal corticosteroid use for anticipated preterm delivery. J Matern Fetal Neonatal Med. 2011;24:516–520. doi: 10.3109/14767058.2010.506228. [DOI] [PubMed] [Google Scholar]
- 23.San Feliciano L, Remesal A, Isidoro-García M, Ludeña D. Dexamethasone and betamethasone for prenatal lung maturation: differences in vascular endothelial growth factor expression and alveolarization in rats. Neonatology. 2011;100:105–110. doi: 10.1159/000323490. [DOI] [PubMed] [Google Scholar]
- 24.Beers MF, Shuman H, Liley HG, Floros J, Gonzales LW, Yue N, Ballard PL. Surfactant protein B in human fetal lung: developmental and glucocorticoid regulation. Pediatr Res. 1995;38:668–675. doi: 10.1203/00006450-199511000-00007. [DOI] [PubMed] [Google Scholar]
- 25.Jobe AH, Polk D, Ikegami M, Newnham J, Sly P, Kohen R, Kelly R. Lung responses to ultrasound-guided fetal treatments with corticosteroids in preterm lambs. J Appl Physiol (1985) 1993;75:2099–2105. doi: 10.1152/jappl.1993.75.5.2099. [DOI] [PubMed] [Google Scholar]
- 26.McEvoy C, Bowling S, Williamson K, Stewart M, Durand M. Functional residual capacity and passive compliance measurements after antenatal steroid therapy in preterm infants. Pediatr Pulmonol. 2001;31:425–430. doi: 10.1002/ppul.1070. [DOI] [PubMed] [Google Scholar]
- 27.McEvoy C, Schilling D, Peters D, Tillotson C, Spitale P, Wallen L, Segel S, Bowling S, Gravett M, Durand M. Respiratory compliance in preterm infants after a single rescue course of antenatal steroids: a randomized controlled trial. Am J Obstet Gynecol. 2010;202:544, e541–e549. doi: 10.1016/j.ajog.2010.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pole JD, Mustard CA, To T, Beyene J, Allen AC. Antenatal steroid therapy for fetal lung maturation: is there an association with childhood asthma? J Asthma. 2009;46:47–52. doi: 10.1080/02770900802262795. [DOI] [PubMed] [Google Scholar]
- 29.Kallapur SG, Kramer BW, Moss TJ, Newnham JP, Jobe AH, Ikegami M, Bachurski CJ. Maternal glucocorticoids increase endotoxin-induced lung inflammation in preterm lambs. Am J Physiol Lung Cell Mol Physiol. 2003;284:L633–L642. doi: 10.1152/ajplung.00344.2002. [DOI] [PubMed] [Google Scholar]
- 30.Kuypers E, Collins JJ, Kramer BW, Ofman G, Nitsos I, Pillow JJ, Polglase GR, Kemp MW, Newnham JP, Gavilanes AW, et al. Intra-amniotic LPS and antenatal betamethasone: inflammation and maturation in preterm lamb lungs. Am J Physiol Lung Cell Mol Physiol. 2012;302:L380–L389. doi: 10.1152/ajplung.00338.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cao Z, Umek RM, McKnight SL. Regulated expression of three C/EBP isoforms during adipose conversion of 3T3-L1 cells. Genes Dev. 1991;5:1538–1552. doi: 10.1101/gad.5.9.1538. [DOI] [PubMed] [Google Scholar]
- 32.Gonzales LW, Guttentag SH, Wade KC, Postle AD, Ballard PL. Differentiation of human pulmonary type II cells in vitro by glucocorticoid plus cAMP. Am J Physiol Lung Cell Mol Physiol. 2002;283:L940–L951. doi: 10.1152/ajplung.00127.2002. [DOI] [PubMed] [Google Scholar]
- 33.Breed DR, Margraf LR, Alcorn JL, Mendelson CR. Transcription factor C/EBPdelta in fetal lung: developmental regulation and effects of cyclic adenosine 3′,5′-monophosphate and glucocorticoids. Endocrinology. 1997;138:5527–5534. doi: 10.1210/endo.138.12.5637. [DOI] [PubMed] [Google Scholar]
- 34.Mendelson CR, Johnston JM, MacDonald PC, Snyder JM. Multihormonal regulation of surfactant synthesis by human fetal lung in vitro. J Clin Endocrinol Metab. 1981;53:307–317. doi: 10.1210/jcem-53-2-307. [DOI] [PubMed] [Google Scholar]
- 35.Odom MJ, Snyder JM, Boggaram V, Mendelson CR. Glucocorticoid regulation of the major surfactant associated protein (SP-A) and its messenger ribonucleic acid and of morphological development of human fetal lung in vitro. Endocrinology. 1988;123:1712–1720. doi: 10.1210/endo-123-4-1712. [DOI] [PubMed] [Google Scholar]
- 36.Ballard PL, Lee JW, Fang X, Chapin C, Allen L, Segal MR, Fischer H, Illek B, Gonzales LW, Kolla V, et al. Regulated gene expression in cultured type II cells of adult human lung. Am J Physiol Lung Cell Mol Physiol. 2010;299:L36–L50. doi: 10.1152/ajplung.00427.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wade KC, Guttentag SH, Gonzales LW, Maschhoff KL, Gonzales J, Kolla V, Singhal S, Ballard PL. Gene induction during differentiation of human pulmonary type II cells in vitro. Am J Respir Cell Mol Biol. 2006;34:727–737. doi: 10.1165/rcmb.2004-0389OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hakonarson H, Maskeri N, Carter C, Hodinka RL, Campbell D, Grunstein MM. Mechanism of rhinovirus-induced changes in airway smooth muscle responsiveness. J Clin Invest. 1998;102:1732–1741. doi: 10.1172/JCI4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oliver BG, Johnston SL, Baraket M, Burgess JK, King NJ, Roth M, Lim S, Black JL. Increased proinflammatory responses from asthmatic human airway smooth muscle cells in response to rhinovirus infection. Respir Res. 2006;7:71. doi: 10.1186/1465-9921-7-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brugarolas J, Lei K, Hurley RL, Manning BD, Reiling JH, Hafen E, Witters LA, Ellisen LW, Kaelin WG., Jr Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev. 2004;18:2893–2904. doi: 10.1101/gad.1256804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Otulakowski G, Duan W, Sarangapani A, Gandhi S, O’Brodovich H. Glucocorticoid-mediated repression of REDD1 mRNA expression in rat fetal distal lung epithelial cells. Pediatr Res. 2009;65:514–519. doi: 10.1203/PDR.0b013e3181998db6. [DOI] [PubMed] [Google Scholar]
- 42.Yoshida T, Mett I, Bhunia AK, Bowman J, Perez M, Zhang L, Gandjeva A, Zhen L, Chukwueke U, Mao T, et al. Rtp801, a suppressor of mTOR signaling, is an essential mediator of cigarette smoke-induced pulmonary injury and emphysema. Nat Med. 2010;16:767–773. doi: 10.1038/nm.2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davies TH, Ning YM, Sánchez ER. A new first step in activation of steroid receptors: hormone-induced switching of FKBP51 and FKBP52 immunophilins. J Biol Chem. 2002;277:4597–4600. doi: 10.1074/jbc.C100531200. [DOI] [PubMed] [Google Scholar]
- 44.Chivers JE, Gong W, King EM, Seybold J, Mak JC, Donnelly LE, Holden NS, Newton R. Analysis of the dissociated steroid RU24858 does not exclude a role for inducible genes in the anti-inflammatory actions of glucocorticoids. Mol Pharmacol. 2006;70:2084–2095. doi: 10.1124/mol.106.025841. [DOI] [PubMed] [Google Scholar]
- 45.Kaur M, Chivers JE, Giembycz MA, Newton R. Long-acting beta2-adrenoceptor agonists synergistically enhance glucocorticoid-dependent transcription in human airway epithelial and smooth muscle cells. Mol Pharmacol. 2008;73:203–214. doi: 10.1124/mol.107.040121. [DOI] [PubMed] [Google Scholar]
- 46.Kelly MM, King EM, Rider CF, Gwozd C, Holden NS, Eddleston J, Zuraw B, Leigh R, O’Byrne PM, Newton R. Corticosteroid-induced gene expression in allergen-challenged asthmatic subjects taking inhaled budesonide. Br J Pharmacol. 2012;165:1737–1747. doi: 10.1111/j.1476-5381.2011.01620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hawkins GA, Lazarus R, Smith RS, Tantisira KG, Meyers DA, Peters SP, Weiss ST, Bleecker ER. The glucocorticoid receptor heterocomplex gene STIP1 is associated with improved lung function in asthmatic subjects treated with inhaled corticosteroids. J Allergy Clin Immunol. 2009;123:1376–1383, e1377. doi: 10.1016/j.jaci.2009.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]