Abstract
Bactericidal/permeability-increasing protein fold–containing family member A1 (BPIFA1), formerly known as SPLUNC1, is one of the most abundant proteins in respiratory secretions and has been identified with increasing frequency in studies of pulmonary disease. Its expression is largely restricted to the respiratory tract, being highly concentrated in the upper airways and proximal trachea. BPIFA1 is highly responsive to airborne pathogens, allergens, and irritants. BPIFA1 actively participates in host protection through antimicrobial, surfactant, airway surface liquid regulation, and immunomodulatory properties. Its expression is modulated in multiple lung diseases, including cystic fibrosis, chronic obstructive pulmonary disease, respiratory malignancies, and idiopathic pulmonary fibrosis. However, the role of BPIFA1 in pulmonary pathogenesis remains to be elucidated. This review highlights the versatile properties of BPIFA1 in antimicrobial protection and its roles as a sensor of environmental exposure and regulator of immune cell function. A greater understanding of the contribution of BPIFA1 to disease pathogenesis and activity may clarify if BPIFA1 is a biomarker and potential drug target in pulmonary disease.
Keywords: BPIFA1, SPLUNC1, airway epithelium, inflammation, host protection
The airway mucosa constantly adapts to our changing environment and provides a first line of defense against pathogens and irritants in contact with the respiratory tract. Airway epithelial cells form a structural first line of host protection, and secretions of epithelial cells serve to protect against pathogens, coordinate immune responses, and limit lung injury (1, 2). Respiratory secretions contain numerous proteins known for their role in mucociliary clearance, antimicrobial defense, and immune modulation, including collectins, defensins, cathelicidins, and bactericidal/permeability-increasing protein (BPI). Many of these are produced by the epithelium and have multifaceted functions, with overlapping roles in host protection (1, 3). This review focuses on BPI fold–containing family member A1 (BPIFA1), formerly known as SPLUNC1, one of the most highly expressed proteins in respiratory secretions and an increasingly identified molecule in pulmonary diseases with host-protective and immunomodulatory functions.
The BPIF and PLUNC Protein Families
The BPIF superfamily comprises a heterogeneous group of proteins that share the three-dimensional structure of the antimicrobial and immunomodulatory protein BPI (4–6). The structural arrangement of BPI includes two large, barrel-shaped domains connected by a central β-sheet. Molecules containing this arrangement are known as BPI fold–containing (BPIF) proteins (5, 7, 8). Using simple phylogenetic analysis, the members of the BPIF superfamily can be separated into two clusters: one including the lipid-transfer proteins BPI, lipopolysaccharide-binding protein (LBP), cholesterylester transfer protein, and phospholipid-transfer protein and another branch containing the Palate Lung and Nasal epithelium Clone (PLUNC) proteins, expressed almost exclusively in the respiratory tract of air-breathing vertebrates (7).
The PLUNC genes are located in chromosome 20 in humans and chromosome 2 in mice and appear to be rapidly evolving host-protective genes (7–9). The BPIF locus includes 12 genes in humans and 14 in mice, and within the human locus there are eight PLUNC genes with at least three pseudogenes (10). There is low sequence identity between members of the PLUNC and BPI families, a feature that suggests PLUNCs have host-protective properties (8).
PLUNCs are expressed largely in the respiratory tract of air-breathing vertebrates. The original nomenclature distinguishing PLUNC proteins was based on their length, and hence they were called short (S) PLUNCs (∼250 amino acids in length) or long (L) PLUNCs (∼450 amino acids). An updated nomenclature reflects the inclusion of the BPI fold in their predicted structures, with SPLUNCs being denominated with the prefix BPIFA, where “A” represents proteins containing one BPI domain, and LPLUNCs being denominated BPIFB, where “B” represents proteins containing two BPI domains. The numbering of short and long PLUNC previously assigned to these proteins was retained whenever possible (7–9). BPIFA1, or SPLUNC1 (previously known as PLUNC, LUNX, SPURT, and YY1), is one of the most widely studied PLUNC proteins. BPIFA1 was originally identified in mice in a study seeking genetic contributions to palatal malformations, and it is frequently identified in genomic and proteomic studies of healthy and diseased lung tissue and respiratory secretions (11–16).
The BPIFA1 Gene and Protein
Expression and Distribution
The BPIFA1 gene has a length of 1.2 kb in mice and 7.3 kb in humans, and it is expressed in discrete regions within the respiratory epithelium and submucosal glands of the palate, nose, trachea, and bronchi; hence, it was formerly referred to as PLUNC. The human BPIFA1 gene has over 72% sequence identity with its mouse homolog, and it contains a similar nine-exon structure in both species (17). There are multiple human BPIFA1 splice variants, but their functional significance is unknown (18, 19).
Human BPIFA1 is a 256–amino acid protein with globular shape and slightly hydrophobic properties containing a 19–amino acid secretory signal at the N-terminal domain. BPIFA1 has a calculated mass of 26.6 kD in humans and 28.6 kD in mice (11, 18, 20, 21). The crystal structure of human BPIFA1 was solved by Garland and colleagues in 2013, revealing a monomer that contains a central β-sheet flanked by six α-helices, with structural similarity to the N-terminal half of BPI (22). Bioinformatics analysis of the BPIFA1 sequence suggested N-glycosylation motifs and a series of charged amino acid residues that form an “electrostatic patch,” a region essential to the role of BPIFA1 in ion transport and airway surface liquid (ASL) regulation (11, 22).
Multiple isoforms of human BPIFA1 have been detected in respiratory secretions, likely generated through translational modifications such as glycosylation, phosphorylation, deamidation, or truncation of the larger protein (23, 24). These isoforms can be heavily glycosylated, and some researchers have proposed that changes in the concentration of glycosylated BPIFA1 isoforms may affect inflammatory responses and host defense (25). Similar to other members of the BPIF superfamily that modulate immune responses through binding to LPS, several studies have showed that BPIFA1 and its isoforms bind to LPS (24, 26–29). However, these findings have not been universally confirmed because BPIFA1 failed to compete with LBP for LPS binding in another study (23). Zhou and colleagues demonstrated that deletion of the BPI domain disrupted the colocalization of BPIFA1 with nanobacteria in airway epithelial cell cultures, suggesting that BPIFA1 binds to LPS through its BPI domain (30).
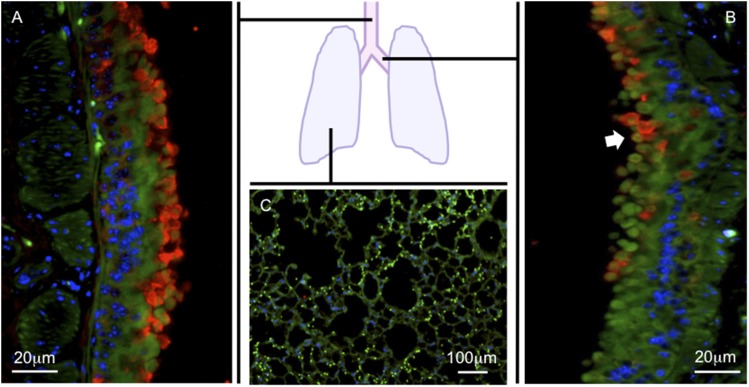
BPIFA1 gene expression is high in the proximal trachea, gradually decreases in the distal airways at the level of the bifurcation of the mainstem bronchi, and becomes undetectable in the peripheral lungs of mice and humans (Figure 1) (18, 31). BPIFA1 mRNA expression is absent in peripheral lung tissue from healthy human or mouse lungs (9, 11, 32). BPIFA1 is not expressed outside of the rodent respiratory tract, with the exception of limited mRNA expression in rat heart, mouse thymus, and olfactory mucosa (15, 33–36). In humans, limited BPIFA1 expression outside of the respiratory tract has been reported in the kidney and colon (37). BPIFA1 mRNA has not been detected in the human heart, liver, brain, stomach, small intestine, placenta, skeletal muscle, pancreas, spleen, normal lymph nodes, peripheral lymphocytes, prostate, testis, or ovary (17, 18, 31). The expression of BPIFA1 mRNA follows a similar distribution in embryonic and adult tissues (19, 36).
Figure 1.
Bactericidal/permeability-increasing protein fold–containing family member A1 (BPIFA1) distribution in the respiratory tract. BPIFA1 protein expression was detected by immunofluorescence on lung sections from naive C57BL/6 mice using anti-mouse BPIFA1 IgG (red). Antipancytokeratin IgG was used to identify epithelial cells (green). 4′,6-Diamidino-2-phenylindole (blue) was used to identify cell nuclei. The trachea (A) and mainstem bronchi (B) show abundant BPIFA1 expression. BPIFA1 expression decreases distally to mainstem bronchi (arrowhead) and becomes undetectable in the peripheral lung tissue (C).
BPIFA1 protein was detected in mucus cells of submucosal glands and their ducts, in airway luminal secretions, and in nonciliated (secretory) cells of the airway epithelium in the nasopharynx, trachea, and large airways (9, 38, 39). Whether BPIFA1 can be expressed in other secretory cell types, such as mixed mucus/serous cells, remains to be elucidated (7, 23). BPIFA1 was also detected in the parotid gland and submucosal glands of nasal polyps, tongue, and sinus tissue but not on their surface epithelium (18, 19, 36, 38). The anatomical distribution of BPIFA1 protein expression in human fetuses is similar to that observed in adults (40). In upper respiratory tract secretions, BPIFA1 can be detected in saliva, nasal lavage fluid, and middle ear effusions (9, 14, 24, 32, 34, 40, 44). In lower respiratory tract secretions, BPIFA1 has been detected in sputum, tracheal secretions, airway lining fluid, and bronchoalveolar lavage fluid (BALF) (9, 18, 23).
BPIFA1 is one of the major secretory products of tracheobronchial epithelial cells grown at the air–liquid interface (9, 18, 23). Strong BPIFA1 expression was detected in primary cultured airway and nasal epithelium, the mucoepidermoid carcinoma cell line NCI-H292, the lung adenocarcinoma cell line PC-3, the human adenosquamous carcinoma cell line NCI-H647, and the nasopharyngeal carcinoma cell line HNE1 (17, 19, 31, 45). BPIFA1 accounted for up to 10% of the total secreted protein from cell culture supernatants, and its concentration greatly increased over time once airway epithelial cells became fully differentiated (23). In addition, BPIFA1 expression is induced by all-trans retinoic acid in a dose-dependent manner, and this induction becomes readily detectable after 7 days of treatment (18, 31, 46). When cultures are allowed to dedifferentiate by removal of growth factors, the expression of BPIFA1 is lost (47). These optimal culture conditions likely stimulate epithelial differentiation toward a polarized airway cell phenotype, suggesting that BPIFA1 serves as a marker of a type of differentiated airway epithelial cell.
Outside of the respiratory tract, BPIFA1 protein was detected in neutrophil lysates from healthy volunteers, where it localized to the specific granules of neutrophils (48). In contrast, two other studies showed no BPIFA1 protein expression in circulating neutrophils, neutrophils within cystic fibrosis (CF) airway mucus plugs, or other hematopoietic cells, such as peripheral blood mononuclear cells, B cells, T cells, monocytes, and monocyte-derived macrophages (47). These discrepancies may be due to activation of BPIFA1 expression triggered by the migration of circulating cells into pulmonary tissues or antibody cross-reactivity with other members of the BPIF family, such as BPI, known to be produced by neutrophils (6, 32, 49, 50).
Biological Properties of BPIFA1
Antimicrobial
Due to its structural similarities to LPS-binding proteins BPI and LBP and its presence at critical sites of pathogen exposure, BPIFA1 was predicted to have critical host-protective functions. The majority of the studies of BPIFA1 have focused on its antimicrobial properties and identified it as a multifaceted antimicrobial protein with direct effects on bacterial growth, biofilm formation, expression of other antibacterial proteins, and mucociliary clearance (21, 51, 52).
In pulmonary infection models with Mycoplasma pneumoniae (Mp), Pseudomonas aeruginosa (Pa), Klebsiella pneumoniae (Kp), and nontypeable Haemophilus influenzae while blocking BPIFA1, using BPIFA1-deficient mice, BPIFA1 silencing, or blocking antibodies, led to increased infection, suggesting that inhibiting BPIFA1 reduces resistance to bacterial pathogens (27, 51, 53–58). Deficiency or blockade of BPIFA1 was also associated with increased inflammatory cell infiltration and cytokine concentrations when compared with control animals with normal levels of BPIFA1, suggesting that ongoing infection or BPIFA1 deficiency was responsible for excessive inflammation in these models. Increasing BPIFA1 in the airway using transgenic mice that overexpressed human BPIFA1 improved bacterial clearance, decreased lung inflammation, and improved survival in Pa infection (56).
The mechanisms by which BPIFA1 inhibits microbial growth were demonstrated in in vitro studies showing direct bacteriostatic effects of recombinant BPIFA1 on Pa and Mp as well as inhibitory effects on the replication of Epstein-Barr virus (EBV) (27, 53). BPIFA1 inhibited motility and increased cell wall permeability through the formation of small-sized pores in cell walls of Pa, reducing bacterial growth (51). These effects were reversible after removal of BPIFA1 in the cultures. BPIFA1 reduced biofilm formation of Kp and Pa, and this effect is likely mediated by its surfactant properties (51, 58, 59). In a chinchilla model of nontypeable H. influenzae otitis media, BPIFA1 silencing impaired mucus transport and ciliary clearance through the eustachian tube. This likely reflects increased surface tension and decreased ASL water content caused by loss of surfactant and ion transport properties in BPIFA1-deficient animals (51, 57). Gross and colleagues suggested that the induction of BPIFA1 expression in airway epithelial cells by β2-agonist treatment should improve Mp clearance through the mechanisms described above (60). BPIFA1 may also modulate the secretion of other antimicrobial proteins in the airway because naive BPIFA1-deficient mice exhibited decreased expression of antimicrobial peptides LL37, lactoferrin, and lyzozyme. Other genes that contribute to bacterial clearance, including MUC5AC, MUC5B, and CCSP, were reduced in BPIFA1-deficient mice (52).
BPIFA1 has effects on local immunity that may affect microbial growth and invasion. One potential mechanism is the ability of BPIFA1 to enhance bacterial uptake and destruction by phagocytic cells. Alveolar macrophages from transgenic BPIFA1 mice that overexpressed BPIFA1 exhibited increased opsonization and phagocytosis of carbon nanotubes in a sterile model of airway inflammation (61). In addition, commensal gram-negative nanobacteria present in human saliva colocalized with BPIFA1 within epithelial cells from nasopharyngeal carcinoma specimens, and the authors suggested that BPIFA1 binding to nanobacterial LPS could limit its proinflammatory effects (30).
Given the range of its antimicrobial functions and its retained structure in air-breathing vertebrates, BPIFA1 appears to have evolved to provide essential host-protective functions (62). There is significant redundancy in antimicrobial effectors in animals; yet, BPIFA1 may be essential based on its location in the proximal airways and its high level under basal conditions. Thus, it appears to serve its most potent effects to impede and clear infection before pathogen invasion. After invasion, as described below, BPIFA1 levels may drop significantly, and this fluctuation may signal to activate immunity and modulate other airway functions that are more effective at eliminating bacterial infection in the respiratory tract.
Ion Transport and ASL Regulation
BPIFA1 regulates epithelial sodium transport and thus influences the height of ASL by acting as an autocrine negative regulator of the epithelial sodium channel (ENaC) (37, 63). Adequate ENaC function is essential for the maintenance of ASL height, a prerequisite for adequate mucociliary clearance. Garcia-Caballero and colleagues showed that BPIFA1 potently decreased ENaC currents in BPIFA1-transfected Xenopus laevis oocytes. When this mechanism of sodium transport regulation was tested in airway epithelial cells, it resulted in increased ASL height, improving mucociliary clearance (37). Initial studies suggested that BPIFA1 regulated ENaC through inhibition of its cleavage by serine proteases present in the ASL. However, subsequent studies showed that BPIFA1 did not directly inhibit extracellular proteases but rather controlled ENaC function through multiple other mechanisms, including binding to ENaC and preventing its cleavage, reducing the amount of ENaC on the epithelial cell surface, and possibly by maintaining ENaC in a low-opening probability state (64, 65). BPIFA1 modulates sodium transport through ENaC in a pH-dependent manner. A more acidic environment impaired the ability of BPIFA1 to inhibit ENaC function, resulting in increased sodium transport and more water reabsorption from the airway, and this effect was reversed by raising the pH (22). S18, a peptide derived from the experimental cleavage of BPIFA1, was shown to bind to ENaC and to block its function in a pH-independent manner. The addition of the BPIFA1 S18 peptide to human CF airway epithelial cells in culture increased ASL height and prevented ASL hyperabsorption. It is possible that BPIFA1 regulates ENaC function through an interaction of the S18 sequence with this channel. The effect of BPIFA1 on ion transport regulation has been of particular interest in CF, where the inability of the CF airway epithelium to autoregulate ASL volume may contribute to the development of airway disease. The S18 regulation of ENaC was present even at high concentrations of neutrophil elastase, common in CF airways, identifying S18 as an attractive therapeutic molecule to maintain ASL height and improve airway clearance in CF (20, 22).
Immunomodulatory
BPIFA1 has pro- and anti-inflammatory effects in different mouse models of acute airway inflammation. In models of allergic airway inflammation, Nogo A/B–deficient mice, which are BPIFA1 deficient, showed increased airway inflammation with eosinophilia, mucus production, and airway hyperreactivity and higher levels of the TH2 cytokines IL-4, IL-5, and IL-13 when compared with identically treated wild-type mice. When Nogo A/B–deficient mice were cross-bred with mice that overexpressed BPIFA1 in the airway, thus complementing the deficiency in BPIFA1, the exaggerated TH2-induced inflammation was no longer present. These studies showed that BPIFA1 plays an anti-inflammatory role in allergic airway inflammation (66). These results were confirmed in the lungs of BPIFA1-deficient mice, where the eosinophil chemokine CCL24 (Eotaxin-2) was increased (67). BPIFA1 down-regulated CCL24 expression in macrophages, and this may be a mechanism by which BPIFA1 controls eosinophilia in allergic diseases (51, 67).
The proinflammatory and antifibrotic effects of BPIFA1 were recently studied in a model of acute pulmonary inflammation triggered by inhaled single-walled carbon nanotubes (SWCNTs), which increased airway leukocyte recruitment and enhanced phagocytic activity in the absence of infection, thus permitting the study of the non–infection-related, immunomodulatory properties of BPIFA1. In transgenic mice that constitutively overexpressed BPIFA1 in the airways, inhaled SWCNTs caused increased neutrophilic pulmonary inflammation and elevations in the proneutrophil cytokines TNF-α and IL-6. Thus, in contrast to studies of allergic, eosinophilic inflammation, BPIFA1 promoted neutrophilic inflammation in this model. Although more inflamed, the lungs of these mice showed significantly less fibrosis after inflammation, suggesting that BPIFA1 was protective against fibrosis (61). These results contrast with studies in idiopathic pulmonary fibrosis (IPF), in which higher levels of BPIFA1 were associated with worse disease outcomes (16).
BPIFA1 may also control airway inflammation by increasing epithelial cell apoptosis. Chen and colleagues showed that overexpression of BPIFA1 in a nasopharyngeal carcinoma cell line decreased expression of the antiapoptotic protein Bcl-2 and increased expression of proapoptotic proteins BAX, BAD, and caspases 3 and 8. Furthermore, BPIFA1 overexpression in nasopharyngeal carcinoma cells led to markedly increased apoptosis through activation of the MAPK pathway (68).
Although few studies have examined the impact of BPIFA1 in inflammation in the absence of infection, it appears that BPIFA1 has variable effects in acute inflammation and may influence different cell types. In addition to its effects on chemokine expression, one study showed that BPIFA1 had a modest chemotactic effect on macrophages and neutrophils (51). BPIFA1 limits eosinophilia in models of allergic airway inflammation, possibly through effects on an eosinophil chemokine, and may promote lung neutrophilia in an inhaled SWCNT model. BPIFA1 is concentrated in airways, and because airway macrophages and epithelial cells are major producers of chemokines, these cells may be important targets of BPIFA1.
BPIFA1 Regulation
Regulation by Respiratory Pathogens
In addition to its antimicrobial properties, BPIFA1 is tightly regulated by exposure to pathogens. BPIFA1 is present at high concentrations in airway secretions under basal conditions, and these high levels provide protection during daily, ubiquitous exposure to environmental pathogens. Pathogen exposures appear to regulate the amount of BPIFA1 in the airways because mice lacking pathogen recognition receptor toll-like receptor (TLR) 4 had 2-fold higher BPIFA1 under basal, naive conditions (31). Tonic inhibitory signals from environmental pathogen–associated molecular patterns (PAMPs) likely suppress BPIFA1 to its steady-state level. BPIFA1 is also rapidly modulated during infection. Our own studies showed that BPIFA1 protein and mRNA were significantly decreased in mouse models of acute pulmonary infection with gram-negative (Pa), gram-positive (Streptococcus pneumoniae), and viral (Influenza A virus) respiratory pathogens, whereas others showed that Mp and Kp infection led to an increase in BPIFA1 expression (31, 54, 58). To model the effect of gram-negative infection, we intranasally administered Pa-derived LPS, a TLR4 ligand, and showed that BPIFA1 increased within 3 hours of exposure, indicating release of stored protein. This was followed by a rapid 70% reduction in the airway protein by 24 hours. The decline in BPIFA1 protein in the airway was concurrent with a drop in mRNA expression. LPS-treated epithelial cells in culture also had reduced BPIFA1 protein and mRNA with a nadir observed at 48 hours (31). Mp infection was modeled with the TLR2 agonist Pam3CSK4, which increased BPIFA1 protein and mRNA expression in cultured epithelial cells, and this effect was abrogated in cultured murine TLR2-deficient epithelial cells and in normal human bronchial epithelial cells treated with TLR2-targeting short hairpin RNA (53). The TLR2-mediated induction of BPIFA1 occurred through activation of NF-κB (54, 69, 70).
Infection also leads to regulation of BPIFA1 through degradation by proteases. In vitro, neutrophil elastase applied to cultured normal human tracheobronchial epithelial cells degraded BPIFA1 in a dose-dependent fashion. In vivo, treatment with an aerosolized α-1 proteinase inhibitor increased BPIFA1 in BALF from Pa-infected mice, suggesting that increased proteinase activity during infection contributed to decreased BPIFA1 (71, 72). Collectively, these findings suggest that the regulation of BPIFA1 during infection may occur through protease activity, gene expression regulation, and specific responses to PAMPs. The limited data available to date suggest that BPIFA1 can be rapidly regulated in different directions, and this effect is dependent on the specific stimulus.
Regulation by Inflammatory Cytokines and Airway Inflammation
Inflammation, independent of infection, also regulates BPIFA1. BPIFA1 was inhibited in acute airway inflammation caused by adoptive transfer of TH1 and TH2 cells, followed by inhaled antigen (31). When we dissected the effects of individual cytokines, IFN-γ rapidly and potently inhibited BPIFA1 expression in epithelial cells, and this effect was faster and more potent than that of LPS. This raised the possibility that the rapid inhibition of BPIFA1 by LPS in vivo might be due to cytokines in addition to the effects of TLRs. In vivo, we showed that tonic inhibitory signals from IFN-γ also suppressed BPIFA1 to its baseline levels because naive IFN-γ receptor 1–deficient mice had significantly higher baseline levels of BPIFA1 compared with wild-type control mice. Other researchers have shown that cytokines induced downstream of TLR4, such as TNF-α and IL-1β, do not inhibit BPIFA1 expression in cultured human tracheal epithelial cells (17, 46, 47). However, our recent studies show that TNF-α and IL-1β inhibit BPIFA1 expression in mouse tracheal epithelial cells (unpublished observation).
BPIFA1 down-regulation is not a universal response to inflammatory cytokines. Our studies showed that IL-13–treated mouse and human tracheal epithelial cells increased BPIFA1 protein and mRNA expression, whereas others showed that IL-13 decreased BPIFA1. In contrast to the in vitro effects of a single TH2 cytokine (i.e., IL-13), TH2 inflammation in mice reduced BPIFA1, and this effect is likely due to activation of inflammatory cascades with many cytokines that dominate over the effects of IL-13 (31).
BPIFA1 was also induced by cell injury, as observed in nasal epithelial cells after olfactory bulbectomy, and in whole lung microarrays after exposure to the cytotoxic substance 1–nitronaphtalene (13, 34). Although the mechanisms of BPIFA1 induction were not investigated here, it is possible that these effects were cytokine driven or may represent other pathways by which BPIFA1 is regulated.
BPIFA1 likely serves essential functions in the respiratory tract under steady-state conditions through its antimicrobial effects and maintaining the height of ASL. In acute inflammation and acute infection, PAMPs and inflammatory signals cause fluctuations of BPIFA1. Fluctuations in BPIFA1 may regulate innate immunity through effects on granulocytes and chemokines and may alter mucociliary clearance by affecting the rheology of airway secretions through the regulation of ENaC function.
BPIFA1 in Human Disease
BPIFA1 expression profiles have been studied in multiple lung diseases. In healthy humans, BPIFA1 is predominantly produced by epithelial cells of the upper and proximal lower respiratory tract. Measurements of BPIFA1 in human disease have included detection of protein by Western blot, immunohistochemistry, and ELISA. BPIFA1 gene expression patterns have been studied in lung biopsies, primary human airway epithelial cells, and human airway epithelial cell lines. There has been a high degree of variability in the choice of location to sample BPIFA1 expression, ranging from nasal lavage or brushings to BALF collection and lung tissue sampling from lung explants. Given its very restricted expression pattern, the biological material used to measure BPIFA1 is an important consideration in assessing these studies. For example, sputum and airway lavage fluid would be the best samples in which to measure BPIFA1 in the lower respiratory tract in most diseases. RNA from lung biopsies that contain samples of tracheal epithelial cells will have a dramatically increased BPIFA1 compared with biopsies of peripheral lung tissue in most diseases. We report here compilations of data published to date that begin to characterize BPIFA1 in disease and further our understanding of its functions, modulation, and contribution to pathology. Some of these studies are limited in subject number and serve only to pique our interest in additional investigations.
Allergy and Airway Irritants
BPIFA1 was decreased in nasal lavage fluid after acute and chronic exposures to allergens and irritants, including cigarette smoke (23, 25, 73). Subjects with allergic rhinitis who were tested during high-pollen season had reduced BPIFA1 when compared with samples from the same subjects out of season and from healthy control subjects (25). BPIFA1 was also reduced in nasal lavage fluid in subjects with perennial allergic rhinitis compared with those of patients with intermittent allergic rhinitis out of allergy season or with those of healthy control subjects. The lower level of BPIFA1 in the perennial allergy group suggests that active airway inflammation reduces nasal BPIFA1 (74). Tsou and colleagues showed reduced BPIFA1 mRNA and protein in surgical biopsies from subjects with chronic rhinosinusitis colonized with Pa compared with noncolonized subjects, indicating that pathogens further inhibit BPIFA1 in the setting of chronic inflammation (75).
Multiple studies showed that cigarette smoking decreased BPIFA1 in respiratory secretions (24, 72, 74, 76). Chronic exposure to airway irritants in the workplace, including exposure to metal-working fluid and epoxy, were associated with reduced BPIFA1 in the respiratory tract (24, 43, 73). Ghafouri and colleagues also showed changes in the proportions of different BPIFA1 isoforms in nasal lavage fluid in smokers compared with nonsmokers, suggesting that irritants and/or inflammation can modify BPIFA1 structure; however, the function of these isoforms remains unknown (14, 24). These data suggest that acute and chronic irritant–induced airway inflammation is associated with reduced BPIFA1 in the respiratory tract, paralleling observations of acute exposures in mice and in vitro studies of epithelial cells.
COPD
BPIFA1 expression was variable in subjects with COPD. In one study, lung biopsies from four subjects with uncharacterized COPD showed higher BPIFA1 mRNA expression in the airway epithelium by in situ hybridization, and Western blots showed increased BPIFA1 staining compared with healthy control subjects (18). Subsequent studies failed to show differences in the distribution of BPIFA1 protein in tissue sections from 10 patients with emphysema undergoing surgery for lung cancer. However, these studies did not quantify BPIFA1 protein (47). In smokers with and without COPD, BPIFA1 protein in sputum was comparably reduced (72). The differences in BPIFA1 expression in these studies may be explained by small sample size, inconsistent sampling locations, varying degrees of COPD severity, smoking status, and/or inflammation. Additional studies that better characterize these factors are necessary to assess BPIFA1 and its role in COPD.
CF
BPIFA1 mRNA and protein expression were increased in the respiratory tract of patients with CF, and BPIFA1 was more frequently detected in microarrays of human airway epithelium from patients with CF than from non-CF patients (12, 77). BPIFA1 protein staining was increased in the small airways of lung explants from patients with end-stage CF undergoing lung transplantation, and this pattern of expression was different from that of normal lungs, where BPIFA1 is not detectable distal to the mainstem bronchi. BPIFA1 was also detected in mucus plugs obtained from those subjects. The staining did not colocalize with inflammatory cells within the plugs, supporting other investigators’ reports that BPIFA1 is not readily detectable in neutrophils (32, 47). Expression of BPIFA1 in the distal, small airways in CF has only been investigated in highly diseased lungs; thus, it is unclear if this is a generalized finding in CF or if it is an adaptation to chronic airway inflammation and/or infection.
Elevation of BPIFA1 may be of great importance in CF because BPIFA1 increases ASL height, rehydrating mucus secretions and improving clearance of bacteria. In addition to its direct antibacterial effects, the surfactant properties of BIFA1 may improve the rheology of mucus and limit bacterial biofilm formation by mechanisms described above. However, the beneficial effects of BPIFA1 may be hampered by decreased pH and increased neutrophil elastase in the CF airways, which could disrupt its ion transport/ASL regulation properties (52, 57–59, 63). Several questions remain about the dysregulation of BPIFA1 in CF, including whether BPIFA1 is a marker of advanced disease or of a dysfunctional epithelium and whether elevation of BPIFA1 itself contributes to the development of CF lung disease.
Malignancies
A number of studies have investigated expression of BPIFA1 in respiratory tract malignancies. In lung cancers, only large-cell carcinomas and adenocarcinomas expressed BPIFA1, whereas squamous cell, small cell carcinoma, and mesothelioma samples did not (38, 45). It is conceivable that malignancy-associated changes in expression of BPIFA1 reflect the state of differentiation or the malignant cell type. Although BPIFA1 is normally undetectable outside of the respiratory tract, it was detected in lymph nodes, circulating metastatic cells, and pleural effusions of patients with advanced respiratory malignancies, including some non–small cell carcinomas and nasopharyngeal carcinoma (38, 45, 78–82). In this context, BPIFA1 detection likely represents respiratory epithelial-derived metastatic cells. In non–small cell carcinoma, BPIFA1-positive lymph nodes were associated with decreased survival, highlighting a potential role for BPIFA1 as a clinical biomarker that affects tumor staging and survival (83, 84). It is unclear if expression of BPIFA1 in some epithelial-derived malignancies drives metastases or leads to more aggressive/resistant disease.
In upper respiratory tract malignancies, BPIFA1 down-regulation may play a role in the development of nasopharyngeal carcinoma, which has been linked strongly to EBV infection. In one in vitro study, the addition of recombinant BPIFA1 resulted in increased cell death of EBV-infected lymphocytes, limiting infection. In the same study, BPIFA1-treated lymphocytes had decreased expression of the viral oncogene LMP1; thus, the authors concluded that BPIFA1 could protect against EBV-induced nasopharyngeal carcinoma through negative regulation of LMP1 (27). Some researchers have proposed that BPIFA1 may down-regulate the expression of mir-141, an oncogene linked to nasopharyngeal carcinoma, and through mir-141 would protect against tumorigenesis (85). Additional support for the idea that BPIFA1 may affect nasopharyngeal carcinoma pathogenesis comes from studies showing down-regulation of BPIFA1 expression in nasopharyngeal carcinoma and small-nucleotide polymorphisms in the BPIFA1 gene associated with increased susceptibility to nasopharyngeal carcinoma (19, 32, 86). In addition to this malignancy, BPIFA1 was decreased in rare squamous cell hypopharyngeal tumor tissues (87).
In contrast with nasopharyngeal carcinoma, rare tumors (e.g., mucoepidermoid carcinomas and papillary cystadenocarcinomas of the salivary glands) exhibited increased BPIFA1 expression (39, 88). In one study, BPIFA1 expression decreased with higher-grade tumors, again suggesting that BPIFA1 may be a useful marker of tumor stage or grade of differentiation (88). These differences in BPIFA1 expression patterns in different malignancies suggest that respiratory malignancies can transform airway epithelial cell populations, in some cases increasing and in others decreasing the expression of BPIFA1.
IPF
One study to date has investigated BPIFA1 in IPF. BPIFA1 mRNA expression was up-regulated 9-fold in subjects with rapidly progressive IPF compared with subjects with stable disease. Immunoreactive BPIFA1 was localized to columnar airway epithelial cells not only in the proximal airways in but also in the distal, peripheral airways, where BPIFA1 is not normally expressed (16). As noted in CF, this indicates a change in the expression pattern of airway epithelial cells with up-regulation of BPIFA1 expression in a chronic disease state. The triggering events that cause this to occur are unknown but may include infections and a response to injury. Although there are no data to support a mechanism linking BPIFA1 to IPF pathogenesis, the up-regulation of BPIFA1 and its distal expression may not only indicate that it is a marker of active disease but suggest a potential role for BPIFA1 in the development of IPF.
Conclusions
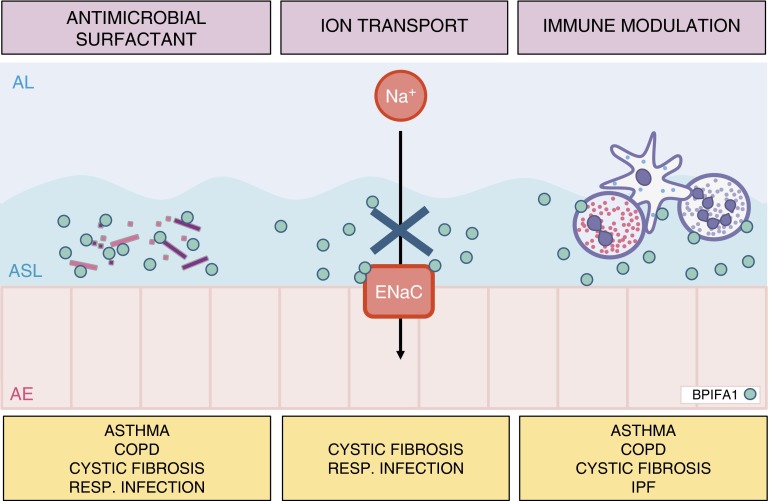
BPIFA1 is a multifunctional protein secreted by epithelial cells in the upper and proximal lower respiratory tract. Its localization at sites of pathogen entry and its conservation in air-breathing vertebrates suggest it serves crucial host-protective functions. BPIFA1 has antimicrobial effects against bacteria and viruses, and, similar to other antimicrobial proteins produced in the respiratory tract, it has multiple capabilities. BPIFA1 regulates immunity through effects on granulocytes and macrophages, partly through cytokines, and modulates the height of the ASL by inhibiting the function of the major sodium channel in the airway epithelium, affecting mucociliary clearance (Figure 2). Numerous elegant studies have contributed to our understanding of the diverse functions of BPIFA1.
Figure 2.
Pulmonary diseases associated with biological properties of BPIFA1. The antimicrobial and surfactant properties of BPIFA1 limit bacterial growth, impair bacterial biofilm formation, and may increase bacterial opsonization for phagocytosis. BPIFA1 negatively regulates epithelial sodium channel (ENaC) function, affecting the height of the airway surface liquid, which is essential for adequate mucociliary clearance. To modulate immunity, BPIFA1 serves as a neutrophil chemoattractant, regulates expression of chemokines, and modulates immune cell function. These effects appear to be different in eosinophilic and neutrophilic models of acute airway inflammation. AE, airway epithelium; AL, airway lumen; ASL, airway surface liquid; COPD, chronic obstructive pulmonary disease; IPF, idiopathic pulmonary fibrosis.
Most interesting to our laboratory has been the high level of BPIFA1 present in the airways under basal conditions and its rapid fluctuation in response to environmental exposures and airway inflammation. BPIFA1 levels in an individual rise and fall, and this may help to adjust the local milieu to optimize protective functions in the airways. The antimicrobial functions of BPIFA1 may be most critical for pathogen removal and limitation of pathogen expansion under basal conditions. The dramatic drop in BPIFA1 upon infection with respiratory pathogens instructs us that BPIFA1 is not necessary as an antimicrobial agent once a host becomes infected and high levels of PAMPs make contact with airway epithelial cells. However, this response is pathogen and receptor specific: TLR4 activation decreases BPIFA1 and TLR2 activation increases its expression. The fluctuation of BPIFA1 in response to environmental stimuli may work as a signal that helps coordinate an appropriate airway inflammatory response. Thus, BPIFA1 is regulated to optimally perform its many functions at environmental–mucosal interfaces in the respiratory tract.
BPIFA1 appears to be modulated in acute and chronic pulmonary diseases. Studies to date do not provide a clear understanding of its role in disease pathogenesis, but they do suggest an emerging role as a marker of pulmonary disease. It is too early to know how alterations of BPIFA1 reflect disease activity and if changes in BPIFA1 contribute to or protect from disease. The high levels and easy access to respiratory tract samples make standardization and routine measurement of BPIFA1 possible and will be important in future clinical applications. Our evolving understanding of BPIFA1 may position this abundant, versatile airway protein as a biomarker and potential drug target in pulmonary disease.
Footnotes
This work was supported by National Institutes of Health grants R01-HL081160 and R21-AI083475 (L.C.) and T32-HL007778 (C.B.) and by a Cystic Fibrosis Foundation’s Fifth Year Clinical Fellowship (C.B.).
Originally Published in Press as DOI: 10.1165/rcmb.2014-0297RT on September 29, 2014
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Steinstraesser L, Kraneburg U, Jacobsen F, Al-Benna S. Host defense peptides and their antimicrobial-immunomodulatory duality. Immunobiology. 2011;216:322–333. doi: 10.1016/j.imbio.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 2.Waterer GW. Airway defense mechanisms. Clin Chest Med. 2012;33:199–209. doi: 10.1016/j.ccm.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 3.Guani-Guerra E, Santos-Mendoza T, Lugo-Reyes SO, Teran LM. Antimicrobial peptides: general overview and clinical implications in human health and disease. Clin Immunol. 2010;135:1–11. doi: 10.1016/j.clim.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 4.Aichele D, Schnare M, Saake M, Rollinghoff M, Gessner A. Expression and antimicrobial function of bactericidal permeability-increasing protein in cystic fibrosis patients. Infect Immun. 2006;74:4708–4714. doi: 10.1128/IAI.02066-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beamer LJ, Carroll SF, Eisenberg D. Crystal structure of human bpi and two bound phospholipids at 2.4 angstrom resolution. Science. 1997;276:1861–1864. doi: 10.1126/science.276.5320.1861. [DOI] [PubMed] [Google Scholar]
- 6.Canny G, Levy O, Furuta GT, Narravula-Alipati S, Sisson RB, Serhan CN, Colgan SP. Lipid mediator-induced expression of bactericidal/permeability-increasing protein (BPI) in human mucosal epithelia. Proc Natl Acad Sci USA. 2002;99:3902–3907. doi: 10.1073/pnas.052533799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bingle CD, Bingle L, Craven CJ. Distant cousins: genomic and sequence diversity within the BPI fold-containing (BPIF)/PLUNC protein family. Biochem Soc Trans. 2011;39:961–965. doi: 10.1042/BST0390961. [DOI] [PubMed] [Google Scholar]
- 8.Bingle CD, Seal RL, Craven CJ. Systematic nomenclature for the plunc/psp/bsp30/smgb proteins as a subfamily of the BPI fold-containing superfamily. Biochem Soc Trans. 2011;39:977–983. doi: 10.1042/BST0390977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barnes FA, Bingle L, Bingle CD. Pulmonary genomics, proteomics, and pluncs. Am J Respir Cell Mol Biol. 2008;38:377–379. doi: 10.1165/rcmb.2007-0388TR. [DOI] [PubMed] [Google Scholar]
- 10.HUGO Gene Nomenclature Committee at the European Bioinformatics Institute. Symbol Report: BPIFA1. Cambridge, UK; 2014 [accessed 2014 Mar 14]. Available from: http://www.genenames.org/cgi-bin/gene_symbol_report?hgnc_id=HGNC:15749 [Google Scholar]
- 11.Weston WM, LeClair EE, Trzyna W, McHugh KM, Nugent P, Lafferty CM, Ma L, Tuan RS, Greene RM. Differential display identification of plunc, a novel gene expressed in embryonic palate, nasal epithelium, and adult lung. J Biol Chem. 1999;274:13698–13703. doi: 10.1074/jbc.274.19.13698. [DOI] [PubMed] [Google Scholar]
- 12.Scheetz TE, Zabner J, Welsh MJ, Coco J, Eyestone Mde F, Bonaldo M, Kucaba T, Casavant TL, Soares MB, McCray PB., Jr Large-scale gene discovery in human airway epithelia reveals novel transcripts. Physiol Genomics. 2004;17:69–77. doi: 10.1152/physiolgenomics.00188.2003. [DOI] [PubMed] [Google Scholar]
- 13.Shultz MA, Zhang L, Gu YZ, Baker GL, Fannuchi MV, Padua AM, Gurske WA, Morin D, Penn SG, Jovanovich SB, et al. Gene expression analysis in response to lung toxicants: I. Sequencing and microarray development. Am J Respir Cell Mol Biol. 2004;30:296–310. doi: 10.1165/rcmb.2003-0214OC. [DOI] [PubMed] [Google Scholar]
- 14.Ghafouri B, Stahlbom B, Tagesson C, Lindahl M. Newly identified proteins in human nasal lavage fluid from non-smokers and smokers using two-dimensional gel electrophoresis and peptide mass fingerprinting. Proteomics. 2002;2:112–120. [PubMed] [Google Scholar]
- 15.Genter MB, Van Veldhoven PP, Jegga AG, Sakthivel B, Kong S, Stanley K, Witte DP, Ebert CL, Aronow BJ. Microarray-based discovery of highly expressed olfactory mucosal genes: potential roles in the various functions of the olfactory system. Physiol Genomics. 2003;16:67–81. doi: 10.1152/physiolgenomics.00117.2003. [DOI] [PubMed] [Google Scholar]
- 16.Boon K, Bailey NW, Yang J, Steel MP, Groshong S, Kervitsky D, Brown KK, Schwarz MI, Schwartz DA. Molecular phenotypes distinguish patients with relatively stable from progressive idiopathic pulmonary fibrosis (IPF) PLoS One. 2009;4:e5134. doi: 10.1371/journal.pone.0005134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bingle CD, Bingle L. Characterisation of the human plunc gene, a gene product with an upper airways and nasopharyngeal restricted expression pattern. Biochim Biophys Acta. 2000;1493:363–367. doi: 10.1016/s0167-4781(00)00196-2. [DOI] [PubMed] [Google Scholar]
- 18.Di YP, Harper R, Zhao Y, Pahlavan N, Finkbeiner W, Wu R. Molecular cloning and characterization of spurt, a human novel gene that is retinoic acid-inducible and encodes a secretory protein specific in upper respiratory tracts. J Biol Chem. 2003;278:1165–1173. doi: 10.1074/jbc.M210523200. [DOI] [PubMed] [Google Scholar]
- 19.Zhang B, Nie X, Xiao B, Xiang J, Shen S, Gong J, Zhou M, Zhu S, Zhou J, Qian J, et al. Identification of tissue-specific genes in nasopharyngeal epithelial tissue and differentially expressed genes in nasopharyngeal carcinoma by suppression subtractive hybridization and cDNA microarray. Genes Chromosomes Cancer. 2003;38:80–90. doi: 10.1002/gcc.10247. [DOI] [PubMed] [Google Scholar]
- 20.Hobbs CA, Blanchard MG, Alijevic O, Tan CD, Kellenberger S, Bencharit S, Cao R, Kesimer M, Walton WG, Henderson AG, et al. Identification of the SPLUNC1 ENaC-inhibitory domain yields novel strategies to treat sodium hyperabsorption in cystic fibrosis airway epithelial cultures. Am J Physiol Lung Cell Mol Physiol. 2013;305:L990–L1001. doi: 10.1152/ajplung.00103.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Di YP. Functional roles of SPLUNC1 in the innate immune response against gram-negative bacteria. Biochem Soc Trans. 2011;39:1051–1055. doi: 10.1042/BST0391051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garland AL, Walton WG, Coakley RD, Tan CD, Gilmore RC, Hobbs CA, Tripathy A, Clunes LA, Bencharit S, Stutts MJ, et al. Molecular basis for pH-dependent mucosal dehydration in cystic fibrosis airways. Proc Natl Acad Sci USA. 2013;110:15973–15978. doi: 10.1073/pnas.1311999110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Campos MA, Abreu AR, Nlend MC, Cobas MA, Conner GE, Whitney PL. Purification and characterization of PLUNC from human tracheobronchial secretions. Am J Respir Cell Mol Biol. 2004;30:184–192. doi: 10.1165/rcmb.2003-0142OC. [DOI] [PubMed] [Google Scholar]
- 24.Ghafouri B, Kihlstrom E, Stahlbom B, Tagesson C, Lindahl M. Plunc (palate, lung and nasal epithelial clone) proteins in human nasal lavage fluid. Biochem Soc Trans. 2003;31:810–814. doi: 10.1042/bst0310810. [DOI] [PubMed] [Google Scholar]
- 25.Ghafouri B, Irander K, Lindbom J, Tagesson C, Lindahl M. Comparative proteomics of nasal fluid in seasonal allergic rhinitis. J Proteome Res. 2006;5:330–338. doi: 10.1021/pr050341h. [DOI] [PubMed] [Google Scholar]
- 26.Ghafouri B, Kihlstrom E, Tagesson C, Lindahl M. Plunc in human nasal lavage fluid: multiple isoforms that bind to lipopolysaccharide. Biochim Biophys Acta. 2004;1699:57–63. doi: 10.1016/j.bbapap.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 27.Zhou HD, Li XL, Li GY, Zhou M, Liu HY, Yang YX, Deng T, Ma J, Sheng SR. Effect of SPLUNC1 protein on the pseudomonas aeruginosa and epstein-barr virus. Mol Cell Biochem. 2008;309:191–197. doi: 10.1007/s11010-007-9659-3. [DOI] [PubMed] [Google Scholar]
- 28.Zhou HD, Wu MH, Shi L, Zhou M, Yang YX, Zhao J, Deng T, Li XL, Sheng SR, Li GY. Effect of growth inhibition of the secretory protein SPLUNC1 on Pseudomonas aeruginosa [in Chinese] Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2006;31:464–469. [PubMed] [Google Scholar]
- 29.Tsou YA, Huang HJ, Lin WW, Chen CY. Investigation of anti-infection mechanism of lactoferricin and splunc-1. Evid Based Complement Alternat Med. 2014;2014:907028. doi: 10.1155/2014/907028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou HD, Li GY, Yang YX, Li XL, Sheng SR, Zhang WL, Zhao J. Intracellular co-localization of SPLUNC1 protein with nanobacteria in nasopharyngeal carcinoma epithelia HNE1 cells depended on the bactericidal permeability increasing protein domain. Mol Immunol. 2006;43:1864–1871. doi: 10.1016/j.molimm.2005.10.021. [DOI] [PubMed] [Google Scholar]
- 31.Britto CJ, Liu Q, Curran DR, Patham B, Dela Cruz CS, Cohn L. Short palate, lung, and nasal epithelial clone-1 is a tightly regulated airway sensor in innate and adaptive immunity. Am J Respir Cell Mol Biol. 2013;48:717–724. doi: 10.1165/rcmb.2012-0072OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bingle L, Bingle CD. Distribution of human PLUNC/BPI fold-containing (BPIF) proteins. Biochem Soc Trans. 2011;39:1023–1027. doi: 10.1042/BST0391023. [DOI] [PubMed] [Google Scholar]
- 33.Andrault JB, Gaillard I, Giorgi D, Rouquier S. Expansion of the BPI family by duplication on human chromosome 20: characterization of the RY gene cluster in 20q11.21 encoding olfactory transporters/antimicrobial-like peptides. Genomics. 2003;82:172–184. doi: 10.1016/s0888-7543(03)00102-2. [DOI] [PubMed] [Google Scholar]
- 34.Sung YK, Moon C, Yoo JY, Moon C, Pearse D, Pevsner J, Ronnett GV. Plunc, a member of the secretory gland protein family, is up-regulated in nasal respiratory epithelium after olfactory bulbectomy. J Biol Chem. 2002;277:12762–12769. doi: 10.1074/jbc.M106208200. [DOI] [PubMed] [Google Scholar]
- 35.LeClair EE, Nguyen L, Bingle L, MacGowan A, Singleton V, Ward SJ, Bingle CD. Genomic organization of the mouse plunc gene and expression in the developing airways and thymus. Biochem Biophys Res Commun. 2001;284:792–797. doi: 10.1006/bbrc.2001.5024. [DOI] [PubMed] [Google Scholar]
- 36.Leclair EE. Four BPI (bactericidal/permeability-increasing protein)-like genes expressed in the mouse nasal, oral, airway and digestive epithelia. Biochem Soc Trans. 2003;31:801–805. doi: 10.1042/bst0310801. [DOI] [PubMed] [Google Scholar]
- 37.Garcia-Caballero A, Rasmussen JE, Gaillard E, Watson MJ, Olsen JC, Donaldson SH, Stutts MJ, Tarran R. SPLUNC1 regulates airway surface liquid volume by protecting ENaC from proteolytic cleavage. Proc Natl Acad Sci USA. 2009;106:11412–11417. doi: 10.1073/pnas.0903609106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bingle L, Cross SS, High AS, Wallace WA, Devine DA, Havard S, Campos MA, Bingle CD. SPLUNC1 (PLUNC) is expressed in glandular tissues of the respiratory tract and in lung tumours with a glandular phenotype. J Pathol. 2005;205:491–497. doi: 10.1002/path.1726. [DOI] [PubMed] [Google Scholar]
- 39.Vargas PA, Speight PM, Bingle CD, Barrett AW, Bingle L. Expression of PLUNC family members in benign and malignant salivary gland tumours. Oral Dis. 2008;14:613–619. doi: 10.1111/j.1601-0825.2007.01429.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou HD, Fan SQ, Zhao J, Huang DH, Zhou M, Liu HY, Zeng ZY, Yang YX, Huang H, Li XL, et al. Tissue distribution of the secretory protein, SPLUNC1, in the human fetus. Histochem Cell Biol. 2006;125:315–324. doi: 10.1007/s00418-005-0070-4. [DOI] [PubMed] [Google Scholar]
- 41.Vitorino R, Lobo MJ, Ferrer-Correira AJ, Dubin JR, Tomer KB, Domingues PM, Amado FM. Identification of human whole saliva protein components using proteomics. Proteomics. 2004;4:1109–1115. doi: 10.1002/pmic.200300638. [DOI] [PubMed] [Google Scholar]
- 42.Cole AM, Liao HI, Stuchlik O, Tilan J, Pohl J, Ganz T. Cationic polypeptides are required for antibacterial activity of human airway fluid. J Immunol. 2002;169:6985–6991. doi: 10.4049/jimmunol.169.12.6985. [DOI] [PubMed] [Google Scholar]
- 43.Lindahl M, Stahlbom B, Tagesson C. Identification of a new potential airway irritation marker, palate lung nasal epithelial clone protein, in human nasal lavage fluid with two-dimensional electrophoresis and matrix-assisted laser desorption/ionization-time of flight. Electrophoresis. 2001;22:1795–1800. doi: 10.1002/1522-2683(200105)22:9<1795::AID-ELPS1795>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 44.Casado B, Pannell LK, Iadarola P, Baraniuk JN. Identification of human nasal mucous proteins using proteomics. Proteomics. 2005;5:2949–2959. doi: 10.1002/pmic.200401172. [DOI] [PubMed] [Google Scholar]
- 45.Iwao K, Watanabe T, Fujiwara Y, Takami K, Kodama K, Higashiyama M, Yokouchi H, Ozaki K, Monden M, Tanigami A. Isolation of a novel human lung-specific gene, LUNX, a potential molecular marker for detection of micrometastasis in non-small-cell lung cancer. Int J Cancer. 2001;91:433–437. doi: 10.1002/1097-0215(200002)9999:9999<::aid-ijc1059>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 46.Bingle CD, Craven CJ. Comparative analysis of the PLUNC (palate, lung and nasal epithelium clone) protein families. Biochem Soc Trans. 2003;31:806–809. doi: 10.1042/bst0310806. [DOI] [PubMed] [Google Scholar]
- 47.Bingle L, Barnes FA, Cross SS, Rassl D, Wallace WA, Campos MA, Bingle CD. Differential epithelial expression of the putative innate immune molecule splunc1 in cystic fibrosis. Respir Res. 2007;8:79. doi: 10.1186/1465-9921-8-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bartlett JA, Hicks BJ, Schlomann JM, Ramachandran S, Nauseef WM, McCray PB., Jr PLUNC is a secreted product of neutrophil granules. J Leukoc Biol. 2008;83:1201–1206. doi: 10.1189/jlb.0507302. [DOI] [PubMed] [Google Scholar]
- 49.Krasity BC, Troll JV, Weiss JP, McFall-Ngai MJ. LBP/BPI proteins and their relatives: conservation over evolution and roles in mutualism. Biochem Soc Trans. 2011;39:1039–1044. doi: 10.1042/BST0391039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weiss J. Bactericidal/permeability-increasing protein (BPI) and lipopolysaccharide-binding protein (LBP): structure, function and regulation in host defence against gram-negative bacteria. Biochem Soc Trans. 2003;31:785–790. doi: 10.1042/bst0310785. [DOI] [PubMed] [Google Scholar]
- 51.Sayeed S, Nistico L, St Croix C, Di YP. Multifunctional role of human SPLUNC1 in Pseudomonas aeruginosa infection. Infect Immun. 2013;81:285–291. doi: 10.1128/IAI.00500-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu Y, Di ME, Chu HW, Liu X, Wang L, Wenzel S, Di YP. Increased susceptibility to pulmonary Pseudomonas infection in Splunc1 knockout mice. J Immunol. 2013;191:4259–4268. doi: 10.4049/jimmunol.1202340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chu HW, Thaikoottathil J, Rino JG, Zhang G, Wu Q, Moss T, Refaeli Y, Bowler R, Wenzel SE, Chen Z, et al. Function and regulation of SPLUNC1 protein in Mycoplasma infection and allergic inflammation. J Immunol. 2007;179:3995–4002. doi: 10.4049/jimmunol.179.6.3995. [DOI] [PubMed] [Google Scholar]
- 54.Chu HW, Gally F, Thaikoottathil J, Janssen-Heininger YM, Wu Q, Zhang G, Reisdorph N, Case S, Minor M, Smith S, et al. SPLUNC1 regulation in airway epithelial cells: role of toll-like receptor 2 signaling. Respir Res. 2010;11:155. doi: 10.1186/1465-9921-11-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gally F, Di YP, Smith SK, Minor MN, Liu Y, Bratton DL, Frasch SC, Michels NM, Case SR, Chu HW. SPLUNC1 promotes lung innate defense against Mycoplasma pneumoniae infection in mice. Am J Pathol. 2011;178:2159–2167. doi: 10.1016/j.ajpath.2011.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lukinskiene L, Liu Y, Reynolds SD, Steele C, Stripp BR, Leikauf GD, Kolls JK, Di YP. Antimicrobial activity of PLUNC protects against Pseudomonas aeruginosa infection. J Immunol. 2011;187:382–390. doi: 10.4049/jimmunol.1001769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McGillivary G, Bakaletz LO. The multifunctional host defense peptide SPLUNC1 is critical for homeostasis of the mammalian upper airway. PLoS One. 2010;5:e13224. doi: 10.1371/journal.pone.0013224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu Y, Bartlett JA, Di ME, Bomberger JM, Chan YR, Gakhar L, Mallampalli RK, McCray PB, Jr, Di YP. SPLUNC1/BPIFA1 contributes to pulmonary host defense against klebsiella pneumoniae respiratory infection. Am J Pathol. 2013;182:1519–1531. doi: 10.1016/j.ajpath.2013.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gakhar L, Bartlett JA, Penterman J, Mizrachi D, Singh PK, Mallampalli RK, Ramaswamy S, McCray PB., Jr PLUNC is a novel airway surfactant protein with anti-biofilm activity. PLoS One. 2010;5:e9098. doi: 10.1371/journal.pone.0009098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gross CA, Bowler RP, Green RM, Weinberger AR, Schnell C, Chu HW. Beta2-agonists promote host defense against bacterial infection in primary human bronchial epithelial cells. BMC Pulm Med. 2010;10:30. doi: 10.1186/1471-2466-10-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Di YP, Tkach AV, Yanamala N, Stanley S, Gao S, Shurin MR, Kisin ER, Kagan VE, Shvedova A. Dual acute proinflammatory and antifibrotic pulmonary effects of short palate, lung, and nasal epithelium clone-1 after exposure to carbon nanotubes. Am J Respir Cell Mol Biol. 2013;49:759–767. doi: 10.1165/rcmb.2012-0435OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bingle CD, LeClair EE, Havard S, Bingle L, Gillingham P, Craven CJ. Phylogenetic and evolutionary analysis of the plunc gene family. Protein Sci. 2004;13:422–430. doi: 10.1110/ps.03332704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tarran R, Redinbo MR. Mammalian short palate lung and nasal epithelial clone 1 (SPLUNC1) in pH-dependent airway hydration. Int J Biochem Cell Biol. 2014;52:130–135. doi: 10.1016/j.biocel.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rollins BM, Garcia-Caballero A, Stutts MJ, Tarran R. SPLUNC1 expression reduces surface levels of the epithelial sodium channel (ENaC) in Xenopus laevis oocytes. Channels (Austin) 2010;4:255–259. doi: 10.4161/chan.4.4.12255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gaillard EA, Kota P, Gentzsch M, Dokholyan NV, Stutts MJ, Tarran R. Regulation of the epithelial Na+ channel and airway surface liquid volume by serine proteases. Pflugers Arch. 2010;460:1–17. doi: 10.1007/s00424-010-0827-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wright PL, Yu J, Di YP, Homer RJ, Chupp G, Elias JA, Cohn L, Sessa WC. Epithelial reticulon 4b (Nogo-b) is an endogenous regulator of Th2-driven lung inflammation. J Exp Med. 2010;207:2595–2607. doi: 10.1084/jem.20100786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thaikoottathil JV, Martin RJ, Di PY, Minor M, Case S, Zhang B, Zhang G, Huang H, Chu HW. SPLUNC1 deficiency enhances airway eosinophilic inflammation in mice. Am J Respir Cell Mol Biol. 2012;47:253–260. doi: 10.1165/rcmb.2012-0064OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen P, Guo X, Zhou H, Zhang W, Zeng Z, Liao Q, Li X, Xiang B, Yang J, Ma J, et al. SPLUNC1 regulates cell progression and apoptosis through the miR-141-PTEN/p27 pathway, but is hindered by LMP1. PLoS One. 2013;8:e56929. doi: 10.1371/journal.pone.0056929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thaikoottathil J, Chu HW. MAPK/AP-1 activation mediates TLR2 agonist-induced SPLUNC1 expression in human lung epithelial cells. Mol Immunol. 2011;49:415–422. doi: 10.1016/j.molimm.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jiang D, Nelson ML, Gally F, Smith S, Wu Q, Minor M, Case S, Thaikoottathil J, Chu HW. Airway epithelial NF-κB activation promotes Mycoplasma pneumoniae clearance in mice. PLoS ONE. 2012;7:e52969. doi: 10.1371/journal.pone.0052969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jiang D, Persinger R, Wu Q, Gross A, Chu HW. α1-Antitrypsin promotes SPLUNC1-mediated lung defense against Pseudomonas aeruginosa infection in mice. Respir Res. 2013;14:122. doi: 10.1186/1465-9921-14-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jiang D, Wenzel SE, Wu Q, Bowler RP, Schnell C, Chu HW. Human neutrophil elastase degrades SPLUNC1 and impairs airway epithelial defense against bacteria. PLoS One. 2013;8:e64689. doi: 10.1371/journal.pone.0064689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fornander L, Graff P, Wahlen K, Ydreborg K, Flodin U, Leanderson P, Lindahl M, Ghafouri B. Airway symptoms and biological markers in nasal lavage fluid in subjects exposed to metalworking fluids. PLoS One. 2013;8:e83089. doi: 10.1371/journal.pone.0083089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Irander K, Borres MP, Ghafouri B. The effects of physical exercise and smoking habits on the expression of SPLUNC1 in nasal lavage fluids from allergic rhinitis subjects. Int J Pediatr Otorhinolaryngol. 2014;78:618–622. doi: 10.1016/j.ijporl.2014.01.014. [DOI] [PubMed] [Google Scholar]
- 75.Tsou YA, Chen CM, Lin TC, Hu FW, Tai CJ, Chen HC, Yeh TH, Harn HJ, Tsai MH, Jan CI. Decreased SPLUNC1 expression is associated with Pseudomonas infection in surgically treated chronic rhinosinusitis patients who may require repeated sinus surgery. Laryngoscope. 2013;123:845–851. doi: 10.1002/lary.23871. [DOI] [PubMed] [Google Scholar]
- 76.Steiling K, Kadar AY, Bergerat A, Flanigon J, Sridhar S, Shah V, Ahmad QR, Brody JS, Lenburg ME, Steffen M, et al. Comparison of proteomic and transcriptomic profiles in the bronchial airway epithelium of current and never smokers. PLoS ONE. 2009;4:e5043. doi: 10.1371/journal.pone.0005043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Roxo-Rosa M, da Costa G, Luider TM, Scholte BJ, Coelho AV, Amaral MD, Penque D. Proteomic analysis of nasal cells from cystic fibrosis patients and non-cystic fibrosis control individuals: search for novel biomarkers of cystic fibrosis lung disease. Proteomics. 2006;6:2314–2325. doi: 10.1002/pmic.200500273. [DOI] [PubMed] [Google Scholar]
- 78.Mitas M, Cole DJ, Hoover L, Fraig MM, Mikhitarian K, Block MI, Hoffman BJ, Hawes RH, Gillanders WE, Wallace MB. Real-time reverse transcription-PCR detects KS1/4 mRNA in mediastinal lymph nodes from patients with non-small cell lung cancer. Clin Chem. 2003;49:312–315. doi: 10.1373/49.2.312. [DOI] [PubMed] [Google Scholar]
- 79.Cheng M, Chen Y, Yu X, Tian Z, Wei H. Diagnostic utility of LunX mRNA in peripheral blood and pleural fluid in patients with primary non-small cell lung cancer. BMC Cancer. 2008;8:156. doi: 10.1186/1471-2407-8-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mitas M, Hoover L, Silvestri G, Reed C, Green M, Turrisi AT, Sherman C, Mikhitarian K, Cole DJ, Block MI, et al. Lunx is a superior molecular marker for detection of non-small cell lung cancer in peripheral blood [corrected] J Mol Diagn JMD. 2003;5:237–242. doi: 10.1016/s1525-1578(10)60480-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Katseli A, Maragos H, Nezos A, Syrigos K, Koutsilieris M. Multiplex PCR-based detection of circulating tumor cells in lung cancer patients using CK19, PTHrP, and LUNX specific primers. Clin Lung Cancer. 2013;14:513–520. doi: 10.1016/j.cllc.2013.04.007. [DOI] [PubMed] [Google Scholar]
- 82.Tang Y, Xu L. Superiority and clinical significance of Lunx mRNA in the diagnosis of malignant pleural effusion caused by pulmonary carcinoma. J Exp Clin Cancer Res. 2013;32:37. doi: 10.1186/1756-9966-32-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Benlloch S, Galbis-Caravajal JM, Alenda C, Peiro FM, Sanchez-Ronco M, Rodriguez-Paniagua JM, Baschwitz B, Rojas E, Massuti B. Expression of molecular markers in mediastinal nodes from resected stage I non-small-cell lung cancer (NSCLC): prognostic impact and potential role as markers of occult micrometastases. Ann Oncol. 2009;20:91–97. doi: 10.1093/annonc/mdn538. [DOI] [PubMed] [Google Scholar]
- 84.Li J, Shi SB, Shi WL, Wang Y, Yu LC, Zhu LR, Ge LP. Lunx mRNA-positive cells at different time points predict prognosis in patients with surgically resected nonsmall cell lung cancer. Transl Res. 2014;163:27–35. doi: 10.1016/j.trsl.2013.09.010. [DOI] [PubMed] [Google Scholar]
- 85.Zhang L, Deng T, Li X, Liu H, Zhou H, Ma J, Wu M, Zhou M, Shen S, Li X, et al. Microrna-141 is involved in a nasopharyngeal carcinoma-related genes network. Carcinogenesis. 2010;31:559–566. doi: 10.1093/carcin/bgp335. [DOI] [PubMed] [Google Scholar]
- 86.He Y, Zhou G, Zhai Y, Dong X, Lv L, He F, Yao K. Association of PLUNC gene polymorphisms with susceptibility to nasopharyngeal carcinoma in a Chinese population. J Med Genet. 2005;42:172–176. doi: 10.1136/jmg.2004.022616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lemaire F, Millon R, Young J, Cromer A, Wasylyk C, Schultz I, Muller D, Marchal P, Zhao C, Melle D, et al. Differential expression profiling of head and neck squamous cell carcinoma (HNSCC) Br J Cancer. 2003;89:1940–1949. doi: 10.1038/sj.bjc.6601373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gonzalez-Arriagada WA, Santos-Silva AR, Ito FA, Vargas PA, Speight PM, Bingle L, Lopes MA. Expression pattern of PLUNC proteins as an auxiliary tool for the diagnosis of high-grade mucoepidermoid carcinoma of the salivary gland. J Oral Pathol Med. 2012;41:589–597. doi: 10.1111/j.1600-0714.2012.01145.x. [DOI] [PubMed] [Google Scholar]