Summary
Specific binding proteins are crucial for the correct spatiotemporal expression of mRNA. To understand this process, a method is required to characterize RNA-protein interactions in single living cells with subcellular resolution. We combined endogenous single RNA and protein detection with two-photon fluorescence fluctuation analysis to measure the average number of proteins bound to mRNA at specific locations within live cells. We applied this to quantify the known binding of zipcode binding protein 1 (ZBP1) and ribosomes, to β-actin mRNA within subcellular compartments of primary fibroblasts and neurons. ZBP1-mRNA binding did not occur in nuclei, contrary to previous conclusions. ZBP1 interaction with β-actin mRNA was enhanced perinuclearly in neurons compared to fibroblasts. Cytoplasmic ZBP1 and ribosome binding to the mRNA were anti-correlated depending on their location in the cell. These measurements support a mechanism whereby ZBP1 inhibits translation of localizing mRNA until its release from the mRNA peripherally, allowing ribosome binding.
Keywords: beta-actin mRNA, ZBP1, ribosomes, fluorescence fluctuation spectroscopy, translational regulation
Introduction
Proteins play an essential role in the metabolism of RNA. The dynamic changes in the composition of protein-mRNA complex is critical for mRNA regulation (Buxbaum et al., 2015; Hutten et al., 2014). However, methods to investigate RNA-protein interactions using traditional biochemical or genetic approaches suffer from false positive results because many proteins nonspecifically bind mRNA and form aggregates once removed from the cellular environment. False negatives may result from some protein-mRNA complexes not surviving cell lysis. Furthermore, ensemble biochemical measurements lack the spatiotemporal information of the interaction. Colocalization of images obtained by fluorescence in situ hybridization and immunofluorescence has been used to infer the interactions between mRNA and protein. However, colocalization is a poor indicator of physical interaction due to the inherent limitations of optical resolution. Fluorescence resonance energy transfer (FRET) has been extensively used to measure protein-protein interactions, but it is limited to fluorescent molecules that are in close contact (<10nm). Single molecule techniques such as fluorescence correlation spectroscopy (FCS), fluorescence cross-correlation spectroscopy (FCCS) and the corresponding image correlation spectroscopy (ICS) overcome these limitations (Digman et al., 2005; Kim et al., 2005; Petersen et al., 1993; Sankaran et al., 2009; Schwille et al., 1997). These methodologies are widely used to detect protein mobility and protein-protein interactions, yet their potential to study intracellular protein-RNA interactions quantitatively remains unexplored. Fluorescence fluctuation spectroscopy (FFS) (Chen et al., 2003; Digman and Gratton, 2011), a technique closely related to FCS, provides an alternative measure of molecular interactions via brightness analysis. Brightness characterizes the average fluorescence intensity of a single particle and encodes the stoichiometry of a molecular complex. For instance, if two fluorescently labeled monomers form a homo-dimer, the brightness doubles. Single-color brightness analysis utilizes this effect to study protein homo-interactions directly in cellular data (Chen et al., 2003). Number and brightness (N&B) analysis have generalized the brightness to the imaging modality and promise a spatial resolution of molecular interactions (Ross et al., 2011). Two molecular species labeled with spectrally separated fluorescent probes can be resolved with dual-color FFS brightness analysis (Chen et al., 2005). Recently, we have developed a method called heterospecies partition analysis (HSP) to extract the binding curves and stoichiometry for protein hetero-interactions (Li et al., 2012; Wu et al., 2010). However, a general technique to study protein-mRNA interaction using FFS is still not available. In this work, we apply dual-color brightness and heterospecies partition analysis to study protein-mRNA interactions directly in living cells.
Localization of mRNA to specific subcellular compartments allows spatial regulation of gene expression that is required for polarized cell morphology and motility. It has been previously established that β-actin mRNA localizes to the leading edge of primary fibroblasts (Lawrence and Singer, 1986) and the growth cone of the axon (Zhang et al., 1999). A cis-acting element, termed the zipcode, in the 3′ untranslated region (3’ UTR) is necessary and sufficient for the localization phenotype (Kislauskis et al., 1994). Zipcode binding protein 1 (ZBP1/IMP1/IGF2BP1) is the transacting protein that binds to the zipcode and is necessary for β-actin mRNA localization (Ross et al., 1997). ZBP1 has a number of conserved homologs in different species, collectively known as the VICKZ RNA binding protein family (Deshler et al., 1998; Kislauskis et al., 1994; Nielsen et al., 1999; Noubissi et al., 2006; Yisraeli, 2005). The VICKZ proteins share a characteristic arrangement of three canonical RNA binding didomains with two RNA recognition motifs (RRM) followed by two hnRNP-K homology (KH) didomains (Ross et al., 1997). Biochemical analysis has shown that the KH3 and KH4 didomain of ZBP1 binds to the zipcode with the same affinity as the full length protein. Mutation of the linker region between the KH3 and KH4 disrupts the binding of KH34 to the zipcode (Chao et al., 2010). So far, the interaction between ZBP1 and β-actin mRNA has been shown in biochemical analyses with fragments of mRNA or proteins in vitro (Chao et al., 2010). The spatiotemporal distribution of this interaction in living cells has never been investigated.
To observe mRNA, we use the MS2 technology (Bertrand et al., 1998). In this method, genetically encoded MS2 binding sites (MBS) from bacteriophage are inserted into the gene of interest. Co-expression of the MS2 coat protein (MCP) fused to a fluorescent protein (FP) labels the target mRNA. Recently, we applied single color FFS to quantitatively study MCP binding on MBS. We developed a tandem dimer of MCP (tdMCP) that greatly increased the uniformity of the MS2 labeling (Wu et al., 2012). Due to significant brightness differences, the mRNA and free tdMCP could be readily resolved by FFS. This is crucial for applying HSP to study protein-mRNA interactions. With the recent advent of the β-actin-MS2 knock-in mouse ( MBS mouse) where all β-actin mRNAs have MS2 stem loops in the 3′UTR, we now have the ability to visualize and track endogenous mRNAs in real-time (Lionnet et al., 2011). Crossing the MBS mouse with a ZBP1 KO mouse (Katz et al., 2012) has enabled us to generate a cell line lacking ZBP1 expression. When mCherry labeled ZBP1 is reintroduced into the cells and expressed at endogenous levels, we are able to probe the interaction of endogenous mRNA and protein. We use the HSP analysis to study the interaction between ZBP1 and β-actin mRNA in different cellular compartments in both mouse embryonic fibroblasts (MEF) and hippocampal neurons.
Controlling mRNA localization through RNA binding protein (RBP) mediated translational repression regulates protein expression spatially. For β-actin mRNA, ZBP1 binds to the β-actin mRNA and is proposed to repress its translation until it reaches the leading edge of the fibroblast where Src phosphorylates the Tyr396 residue of ZBP1 (Huttelmaier et al., 2005) and releases the mRNA from repression. In order to verify this in fibroblasts and extend this model to neurons, we correlated the association of the β-actin mRNA with ZBP1 or ribosomes in the cellular compartments where the mRNA is predicted to be either repressed or translated. To investigate translation, current technologies have been developed to visualize newly synthesized proteins (David et al., 2012; Dieterich et al., 2010) or ribosome occupancy (Ingolia et al., 2009; Jan et al., 2014). However, a method does not exist to image translation of specific mRNA with precise subcellular resolution. The FFS technique is ideal for this purpose since mRNA-protein interactions can be determined within sub-femtoliter observation volume. We labeled ribosomes by fluorescently tagging the ribosomal protein rpL10A with mCherry. We applied HSP analysis to directly measure the association between labeled ribosomes and β-actin mRNAs as a proxy to assess their translation in different cellular compartments. This approach revealed that the association of ribosomes and ZBP1 with the mRNA were anti-correlated both near the nucleus of primary fibroblasts and neurons, and in peripheral cellular regions such as the leading edge of fibroblasts, therefore validating the hypothesis that ZBP1 spatially regulates β-actin mRNA localized translation.
Results
Heterospecies Partition Analysis for protein-mRNA interaction
The theory behind this approach is described in the Experimental Procedures. In brief, an mRNA species (M), in this case β-actin mRNA labeled by tdMCP-EGFPs binding to 24xMBS inserted in the 3′ UTR, and an RNA binding protein (P), for instance mCherry-ZBP1, are expressed in the same cell (Fig. 1A). The mixture is partitioned into three classes of pseudo-species (Fig 1B), the unbound MS2 coat protein; the heterospecies H, comprising all mRNA species associated with different numbers of RBP; and the free species F, containing RBP and its oligomers. We assume that the mRNA M is a singlet, that is, each mRNA particle contains only one mRNA molecule, which is supported by fluorescence in situ hybridization (Femino et al., 1998) and single color FFS measurements (Wu et al., 2012). Intuitively, the heterospecies can be differentiated from free unbound tdMCP-FP with single color brightness since mRNA is much brighter than single tdMCP-EGFPs in the green channel, and from free species with dual-color brightness analysis since free species does not have green color (Figs. 1B & S1A). The red channel heterospecies brightness depends on the degree of binding between M and P (Figs. S1B-E). The exact FFS parameters of the hetero-species H are derived mathematically in the Extended Experimental Procedure.
Figure 1.
The concept and validation of HSP: (A) A schematic representation of an mRNA M with binding sites for a RNA binding protein P. The mRNA has 24xMS2 inserted in the 3′ UTR and was labeled by fluorescent MS2 coat proteins. (B) The brightness of various species in a hypothetical interaction M + 2P ↔ MP2. For simplicity, we assumed that there was no spectral crosstalk. All species containing mRNA were partitioned into the heterospecies H. The free species F included all RBP and its oligomers. The free tdMCP-EGFP was the third species . (C) The plasmids CFP-12xPBS-24xMBS, NLS-tdMCP-EGFP and NLS-tdPCP-mCherry were transfected into U2OS cells. The Ti-sapphire laser was tuned to 1010nm to avoid exciting CFP. The transfected cells were measured for 3 minutes at the perinuclear region. The data were fit with the 3-species HSP model. The normalized red channel brightness represented the number of tdPCP-mCherry bound to the mRNA. To cross-validate the result, we transfected U2OS cells with CFP-12xPBS-24xMBS and NLS-tdPCP-EGFP and performed single color FFS measurements. The mRNA brightness normalized by EGFP brightness directly measured the number of NLS-tdPCP-EGFP on mRNA. The single-color and dual-color results were consistent with each other. (D) The same experiments were performed for mRNA constructs CFP-nxPBS-24xMBS (n=0,1,6,12) as in (C). Note the green channel brightness of all mRNAs was from 24xMBS and hence equal. The red channel measured the variable number of NLS-tdPCP-mCherry bound to mRNA. The more PBS in the mRNA, the more tdPCP bound to each mRNA. In both (C) and (D), each data point represents a single cell. See also Figure S1.
Validation of HSP analysis
To calibrate the FFS system for quantifying the number of RBPs bound to a single mRNA, we designed mRNAs containing two types of well characterized protein binding sites. PP7 bacteriophage coat protein (PCP) binds to its own unique stem-loop: PP7 binding site (PBS), similar to the MCP and MBS association. The interaction between PCP and PBS has been extensively described both in vitro and in vivo (Chao et al., 2008; Wu et al., 2012). Here we use PCP (tdPCP) as a model RBP and PBS as the target sites. Plasmids coding for cyan fluorescent protein (CFP) with 24xMBS inserted in the 3′ UTR to label the mRNA were generated. Between the stop codon and the MBS, variable numbers of PBS were inserted, down to a single stem-loop to mimic the binding of a single protein to a single site (Extended Experimental Procedure). The plasmid was transfected together with both tdMCP-EGFP and tdPCP-mCherry in U2OS cells. Two avalanche photodiodes (APD) detect fluorescence signals from the subfemtoliter two-photon focal volume that could be positioned accurately within the cell. The experiment was done at the two-photon laser wavelength 1010 nm so that CFP was not excited. An example of an experimental fluorescence intensity trace of 12xPBS (red)-24xMBS (green) was plotted in Fig. S1F. From the fluorescence photon counts, we calculated the bivariate fluorescence cumulants of different binning times (Wu et al., 2006; Wu and Muller, 2005). We fit the dual-color time-integrated fluorescence cumulant with the three-species HSP model. An example fit is plotted in Figs. S1 (G-J). From the fit, the brightness of the heterospecies was extracted. Normalized red channel brightness was used to directly measure the number of tdPCP-mCherry bound to mRNA (Fig. 1C). This measurement revealed that the average tdPCP bound to an mRNA was approximately half the expected full occupancy. To confirm this, we performed a single color experiment using only tdPCP-EGFP instead of both tdPCP-mCherry and tdMCP-EGFP. Single channel brightness of mRNA was compared to the brightness of the EGFP monomer, which gave an independent measurement of the number of tdPCP bound to mRNA (Wu et al., 2012). The single color and dual-color experiment provided the same number of proteins bound (Fig. 1C).
To test our ability to measure the stoichiometry of the protein-mRNA interactions, we varied the number of PBS in the 3′UTR of the reporter mRNAs. Green channel (MCP) brightness is constant as all the mRNAs measured have the same number of MBS. However, normalized brightness of the red channel directly correlated with the number of PBS stem loops in the mRNA (Fig. 1D). Importantly, only one binding site inserted into the mRNA was sufficient to distinguish it from no binding site at all. This measurement is critical because this provides an absolute value when one single RBP binds to an mRNA (see below).
Interaction between ZBP1 and β-actin mRNA
We applied HSP to study the interaction between ZBP1 and β-actin mRNA. To image endogenous β-actin mRNA, we used a transgenic mouse in which 24xMBS was inserted into the 3′ UTR of the β-actin gene (Lionnet et al., 2011). To image endogenous ZBP1, we used cells isolated from ZBP1−/− embryos and reintroduced a tagged variant of ZBP1. Since homozygous ZBP1−/− is perinatal lethal, it was not possible to establish a ZBP1−/− mouse line (Katz et al., 2012). Heterozygous ZBP1+/− mice (Katz et al., 2012) were crossed with the β-actin MBS mouse to generate a ZBP1+/− MBS mouse, which were mated to obtain primary or immortalized embryonic ZBP1−/− MBS cells (Extended Experimental Procedure). We introduced mCherry labeled ZBP1 into immortalized ZBP1−/− MBS MEF cells using lentivirus (Extended Experimental Procedure). In order to obtain expression of mCherry-ZBP1 equivalent to endogenous levels, we obtained stable cell lines expressing various amounts of mCherry-ZBP1 using fluorescence-activated cell sorting. We verified the levels of mCherry-ZBP1 in these cell lines by western blot and compared them to ZBP1 levels in wild type MEFs (Fig. S2). We then imaged the mRNA and ZBP1 interactions on these cells by FFS. We focused the laser in a cytoplasmic spot near the nucleus and counted photons for three minutes. The data was then analyzed with the three-species HSP model. Typically, over a three-minute observation period, we counted hundreds to thousands of mRNAs. The normalized green channel brightness of the heterospecies was 13 ± 2 (Fig. 2A), indicating that about half of the expected 24 MBS were occupied by tdMCP-EGFP. This is in agreement with the single color measurement of mRNA brightness (Wu et al., 2012).
Figure 2.
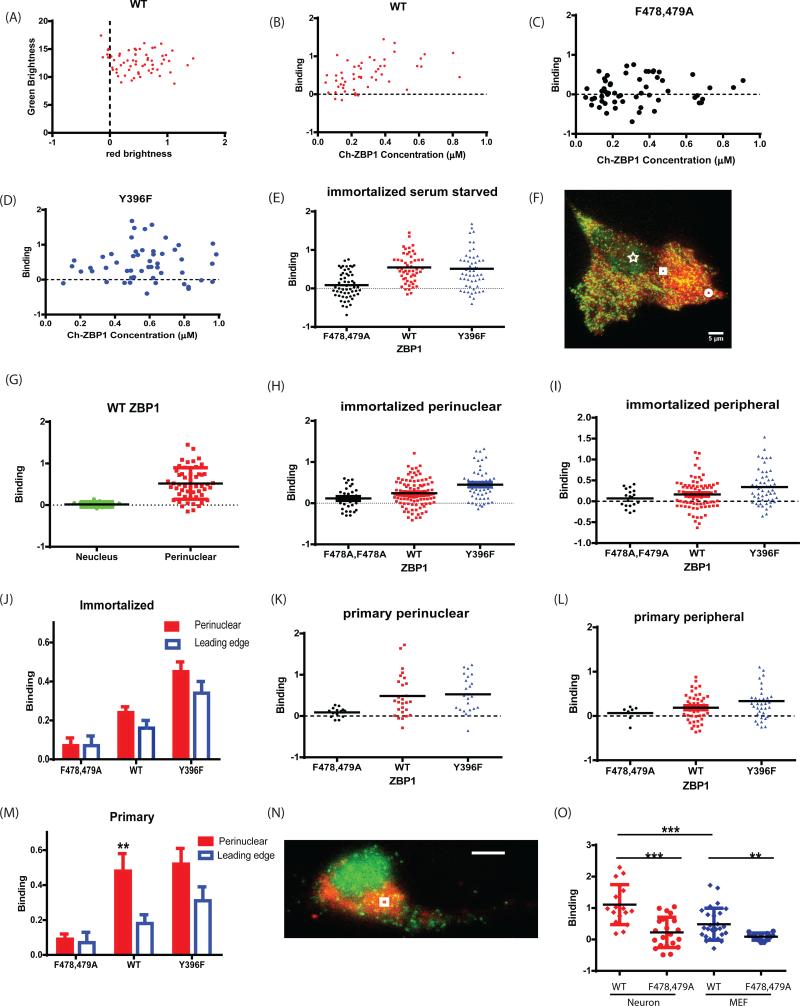
The interaction between ZBP1 and β-actin mRNA in immortalized (A-J) or in primary (J-O) cells. The ZBP1−/− MBS MEF cells were infected by lentivirus expressing NLS-tdMCP-EGFP and mCherry-Flag-ZBP1. The immortalized cells were sorted for endogenous level of ZBP1 expression (Fig. S2). (A-E) The cells were serum starved overnight and measured at the perinuclear region for 3 minutes. The data were fit by the HSP model. (A) The normalized brightness of the heterospecies. The green channel brightness scattered around 13 ± 2 MCP-EGFP per mRNA. The red channel brightness was greater than 0, indicating that ZBP1 bound to mRNA. (B) The normalized red channel brightness , which measured the average number of ZBP1 bound to an mRNA, was plotted against the concentration of mCherry-Flag-ZBP1. The same experiment was performed as in (A)-(B) except that the ZBP1 mutant mCherry-ZBP1F478A, F479A (C) or mCherry-ZBP1Y396F (D) was used to substitute for wild type ZBP1. The concentrations of ZBP1 mutant were chosen to be in the similar range (0-1μM) to that of the wild type. The binding of the ZBP1 and its mutants to the mRNA was summarized and compared (E): mCherry-ZBP1F478A, F479A did not bind to mRNA and mCherry-ZBP1Y396F bound similarly to wild type. (F-J) The interaction between ZBP1 and β-actin mRNA in different cellular compartments in immortalized MEF. (F) An example of measurement location, star: the nucleus; circle: the leading edge; square: the perinuclear region. Red: mCherry-ZBP1; Green: tdMCP-EGFP. (G) ZBP1−/− MBS MEFs were serum stimulated and FFS experiments were conducted in nucleus during the first 45 minutes post stimulation. ZBP1 does not associate with β-actin mRNA in nucleus. (H-J) In normal growth medium with 10% FBS, the average number of mCherry-ZBP1 or mutants bound to β-actin mRNA in the perinuclear region (H) and the leading edge (I) were plotted. (J) Comparison of the binding of ZBP1 to β-actin mRNA in the perinculear region to the leading edge. There was no significant difference between the perinuclear region and in the leading edge. The p-value for unpaired t-test was 0.8, 0.12 and 0.08 for WT, ZBP1Y396F and ZBP1F478A,F479A mutants respectively. (K-M) The interaction between ZBP1 and β-actin mRNA in primary MEF. ZBP1−/− MBS MEF cells were prepared and infected with NLS-tdMCP-EGFP and mCherry-Flag-ZBP1 or its mutants (Extended Experimental Procedures). The average number of ZBP1 binding to mRNA in perinuclear (J) or in the leading edge (K) were plotted for various mutant. (L) Comparison of ZBP1 interacting with β-actin mRNA in different compartments in primary MEFs. In the perinuclear region, there was a higher percentage of β-actin mRNA associated with ZBP1 than in the leading edge. (N-O) The interaction between ZBP1 and β-actin mRNA in somata of cultured primary hippocampal neurons, embryonic day 18, days in vitro (DIV) = 4-7. (N) A representative image shows the perinuclear measurement location in the soma. Green: NLS-tdMCP-GFP, Red: mCherry-ZBP1. (O) The binding of ZBP1 to β-actin mRNA in soma of neurons compared with perinuclear region in primary MEFs. (*: p<0.05, **: p<0.01, ***: p<0.001). See also Figure S2.
The normalized red channel brightness of heterospecies was used to measure the average number of ZBP1 proteins associated with the mRNA (Figs. 2A-B). Each symbol in the plot is a measurement of a single cell and it represents the ensemble average of interactions between ZBP1 and the mRNA in the laser focal volume (the average is 0.55 ± 0.06. A value of zero would mean no interaction and a value of one means all measured mRNAs contain one ZBP1 protein on average). The concentration of mCherry-ZBP1 was calculated using the red channel fluorescence intensity and brightness of mCherry. We compared these results to a ZBP1 mutant, ZBP1F478A, F479A, that cannot bind the mRNA. Disruption of the linker region (F478A, F479A) between KH3 and KH4 disrupts the binding of ZBP1 to β-actin mRNA (Chao et al., 2010; Ross et al., 1997). Over the same concentration range as the wild type ZBP1, mRNAs bound an average of 0.07 ± 0.04 mutant proteins (Figs. 2C,E), equivalent to a lack of interaction. We also investigated the ZBP1Y396F mutant, which cannot be phosphorylated, allowing protein release and therefore is predicted to remain bound to β-actin mRNA (Huttelmaier et al., 2005). We found that under serum starvation conditions, mRNAs bound 0.5 ZBP1, both WT or mutant ZBP1Y396F (Figs. 2D,E). However, in normal medium (10% FBS) the ZBP1Y396F mutant bound the same as in starvation conditions while the WT ZBP1 reduced its mRNA binding. This is likely due to enhanced phosphorylation of the serine 396 under normal conditions. During periods of starvation ZBP1 remains dephosphorylated leading to repression of translation.
One of the advantages of FFS is the ability to measure molecular interactions in precise cellular locations because of the femtoliter spot size. It has been previously hypothesized that β-actin binds to ZBP1 in the nucleus such that it is repressed for translation once it enters the cytoplasm (Huttelmaier et al., 2005). Therefore, we wanted to address whether ZBP1 was loaded on mRNA in the nucleus using this sensitive approach. Because nuclear β-actin mRNA concentration is typically low, to achieve sufficient mRNA counts in nucleus, we serum stimulated the cells after overnight serum starvation to induce mRNA transcription and performed FFS measurements in the nucleus (Fig. 2F star) within 45 minutes post stimulation. Unexpectedly, the data showed that ZBP1 was not bound to β-actin mRNAs in the nucleus (Fig. 2G). Therefore, our results indicate that ZBP1 must bind to mRNA either in the nuclear periphery, or after it exits the nucleus, in contrast to what has been hypothesized. Next, we focused on the interaction in the cytoplasm, more specifically, the leading edge (Fig. 2F, circle) and perinuclear regions (Fig. 2F, square). In immortalized MEFs (Figs. 2H-I), we found that ZBP1 bound β-actin mRNAs almost equally in both compartments (p>0.1) (Fig. 2J). It has been shown previously that β-actin mRNA localized to the leading edge of primary chicken fibroblasts, but this localization phenotype has been lost in immortalized cells (Park et al., 2012). Therefore we measured the interaction at the perinuclear region (Fig. 2K) and the leading edge (Fig. 2L) in primary MEFs, obtained from ZBP1−/− embryos. The mCherry-ZBP1 was introduced by lentivirus (Extended Experimental Procedure) and calibrated to be in the same concentration range as that of the immortalized cells. In primary MEFs, the average number of ZBP1 molecules bound to β-actin mRNA in the perinuclear region (0.48 ± 0.1) was double that in the immortalized cells (0.24 ± 0.03), and comparable to the binding in the serum starved condition for immortalized cells. Importantly, we found that more ZBP1 bound to β-actin mRNA in the perinuclear region than in the leading edge of the cell (0.18 ± 0.05) (Fig. 2M). Perinuclear ZBP1Y396F bound to β-actin mRNA similarly to the WT (0.52 ± 0.09), but in the leading edge, there were more ZBP1Y396F bound to β-actin mRNA (0.45 ± 0.05 ) than the WT. These results provide a quantitative corroboration that ZBP1 binds β-actin mRNAs in the nuclear periphery and that phosphorylation of Y396F on ZBP1 leads to the release of the mRNA at the leading edge. This is consistent with the hypothesis that the removal of ZBP1 asymmetrically leads to translation at the leading edge and consequent cell polarization and directed migration (Park et al., 2012).
We investigated the interaction between ZBP1 and β-actin mRNAs in cultured primary ZBP1−/− MBS hippocampal neurons (Extended Experimental Procedure). It has been shown that β-actin mRNAs localize in neuronal processes and are masked in RNA granules, possibly involving ZBP1 for enhanced translation repression compared to other cell types (Buxbaum et al., 2014). Therefore, neurons may exhibit enhanced binding of ZBP1 leading to repression of translation of β-actin mRNAs. We focused on the interaction in soma since we anticipated that ZBP1 plays an elevated role by packaging the mRNA for transport compared to non-neuronal cells (Fig. 2N). We expressed mCherry1-ZBP1 with lentivirus infection and conducted FFS experiments in primary ZBP1−/− MBS+/+neurons after 4-7 days in vitro (DIV). In contrast to fibroblasts, we measured a significantly higher interaction between WT ZBP1 and β-actin mRNAs (Fig. 2O) in neurons and on average all mRNAs were bound to a ZBP1 protein. ZBP1F478A, F479A still did not interact with the mRNA, as expected. This suggests that in the neuronal soma, ZBP1 has an enhanced participation in the formation of RNA granules, including β-actin mRNAs, and in repressing translation until the mRNA reaches its final destination.
Association of Ribosomes with β-actin mRNA
The localization and translational regulation of mRNA are coupled through the binding of ZBP1. The purpose of mRNA localization is to achieve asymmetric protein distribution. Therefore, localization is usually coupled to a mechanism that represses translation of non-localized mRNA while allowing translation of localized mRNA to occur (Huttelmaier et al., 2005). However, direct observation of this process in live cells has not been possible previously. In order to correlate localization with translation we applied HSP to measure ribosome association with β-actin mRNA. We chose the ribosomal protein rpL10A to label ribosomes (Fig. 3A). Previously, it has been shown that mice with rpL10A tagged by green fluorescent protein are viable (Heiman et al., 2008) indicating that fluorescent tagging does not alter translation efficiency. We established MBS MEF cell lines with rpL10A-mCherry stably expressed. To demonstrate that rpL10A-mCherry was successfully incorporated into ribosomes, we purified the ribosomes by ultracentrifugation followed by western blot detection of rpL10A (Fig S3A-B). Approximately 40% of ribosomes were labeled with mCherry.
Figure 3.
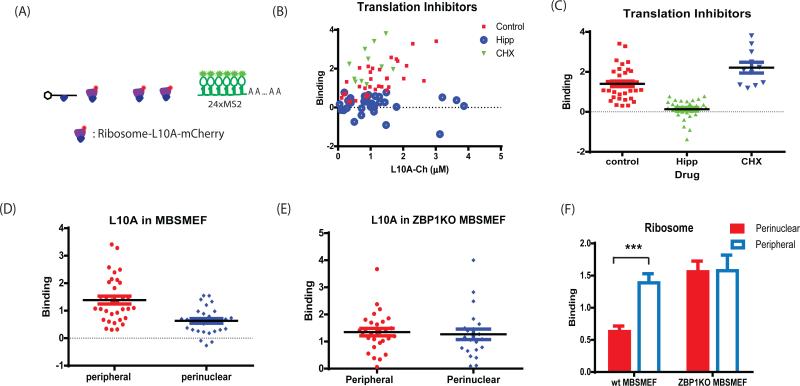
The association of ribosomes with β-actin mRNA in primary MEF. (A) The schematic shows the labeling of large ribosomal subunits by rpL10-mCherry. Primary ZBP1−/− or WT MBS MEF cells were prepared and infected with NLS-tdMCP-EGFP and rpL10A-mCherry (Methods). (B) The association of ribosomes with the mRNA in the presence of translation inhibitors in WT MBS MEF. The number of ribosomes bound per mRNA was plotted as a function of the rpL10A-mCherry concentration. Hippuristanol (Hipp: 1uM/mL) inhibited translation initiation and abolished the association of ribosomes with mRNA. In contrast, cycloheximide (CHX: 2ug/mL) increased the number of ribosomes on mRNA since it slowed down the elongation speed. (C) Summary of the average number rpL10A-mCherry associated with mRNA in the presence of translation inhibitors. Furthermore, FFS experiments were performed in different cellular compartments (D-F). The number of ribosomes binding to β-actin mRNA in the leading edge or perinuclear region was compared in primary MBS MEFs (D) and ZBP1−/− MBS MEF (E). (F) Comparison of ribosome association with mRNA in different cellular regions. There were fewer ribosomes per β-actin mRNA in perinuclear region than in the leading edge in WT MBSMEF. If ZBP1 was knocked out, however, there is no difference in ribosome loading. (*: p<0.05, **: p<0.01, ***: p<0.001). See also Figure S3.
We measured the interaction of ribosomes with β-actin mRNA in primary MBS MEF cells. The normalized red channel brightness was plotted as a function of rpL10A-mCherry concentration (Fig. 3B). As the concentration of rpL10A-mCherry increased, the brightness ranged from 0 to more than 3, with an average of 1.4 ± 0.14 fluorescent ribosomes per mRNA. Since roughly 40% of ribosomes were labeled, this corresponded to approximately 3.5 ribosomes bound to each mRNA in the leading edge. Polysome analysis showed that 80% β-actin mRNAs existed in polysomes in HeLa cells (Sanchez et al., 2007), indicating that this housekeeping gene is constitutively actively translated. We then tested the effect of various translation inhibitors on ribosome binding in MEF cells (Figs. 3B-C). Treatment with Hippuristanol (1μM), an inhibitor of the translation initiation factor eIF4A (Bordeleau et al., 2006), completely eliminated ribosome association with mRNA (Figs. 3B-C). Additionally, the diffusion coefficient of mRNA increased (Figs. S3C-D), further indicating that ribosomes did not associate with the mRNA. In contrast, when treated with a low concentration of cycloheximide (2μg/mL), an inhibitor of translation elongation, ribosome association with mRNA increased over the control (Figs 3B-C) and mRNA diffusion slowed (Figs. S3C-D).
We further compared the association of ribosomes with β-actin mRNA in different cellular compartments. We noticed that there were fewer ribosomes per mRNA (about one per mRNA correcting for the label dilution) in the perinuclear region than in the leading edge (3.5 per mRNA) in ZBP1+/+ MBS MEFs over the range of rpL10A-mCherry concentration measured (Figs. 3D, F). However, in MEFs lacking ZBP1, ribosome association in both compartments was comparable to the leading edge in ZBP1+/+ (Figs 3E, F). Therefore, the association of ribosomes with β-actin mRNA in WT cells was anti-correlated to the ZBP1 - β-actin mRNA interaction (Fig. 2M). In ZBP1−/− cells, there was increased ribosome association at the nuclear periphery, confirming that the cells were unable to establish asymmetry of translation, leading to loss of polarity and directed migration.
Discussion
Currently no method exists that can observe native protein-RNA interactions in single living cells in real time. We applied heterospecies partition analysis (HSP) of FFS, combined with single molecule mRNA detection with the MS2 system, to measure the molecular associations between a protein and an mRNA in different locations and in different cell types. This subcellular resolution, capable of determining the interaction dynamics in various cellular compartments is vital for investigating the cellular regulation of mRNA expression. Here we measure the interactions in select compartments in two cell types: fibroblasts (transformed and primary), and neurons. In principle, this method could be generalized to an imaging approach to map the interaction in each pixel of an image (Ross et al., 2011). However, the RNA concentration is in general much lower than proteins and the time for each point measurement may be prohibitive.
Because HSP measures interactions directly in single live cells, it provides information about the variance among cells in the population, and in cellular regions, resulting in the ability to correlate protein-RNA interactions with cell behavior and morphology. For instance, it has long been hypothesized that an mRNP is continuously remodeled starting from transcription to degradation, however traditional approaches lack sufficient quantitative measures and spatiotemporal precision. HSP allowed us to directly measure some of these events and build a quantitative model of β-actin mRNA metabolism. To demonstrate this, we applied HSP to study the interaction between endogenous β-actin mRNA with ZBP1 or ribosomal subunits. In contrast to our original hypothesis (Huttelmaier et al., 2005), ZBP1 did not bind to β-actin mRNA in the nucleus. Therefore, we conclude that ZBP1 is likely loaded onto the mRNA perinuclearly after export, although we cannot rule out an interaction occurring close to the nuclear pores. In neurons, we measured enhanced ZBP1-β-actin mRNA binding perinuclearly. We propose that this enhanced interaction underlies differences in the β-actin mRNA behavior in neurons, where mRNA is packaged into granules and transports long distances (Buxbaum et al., 2014). Our measurements demonstrate possible cell specific mechanisms whereby expression of the same RBPs can result in alternate mRNA metabolism. In MEFs, ZBP1 and ribosome binding to the β-actin mRNA were anti-correlated when comparing the perinuclear with the peripheral regions in fibroblasts: half of all mRNAs in the perinuclear region were associated with ZBP1 and had an average of about 1 ribosome per mRNA whereas less than 20% of mRNAs in the periphery contained ZBP1 and the average mRNA had almost 4 ribosomes associated. This is the direct quantification of ribosome loading onto a specific mRNA in living cells. Note ribosomes on RNA are not equivalent to active translation since they may be stalled. Nevertheless we are using ribosome loading as a proxy for translational activity. We thus demonstrated a spatial dependence on this translation. Furthermore, we were able to investigate how the nature of translational activity was spatially altered with the loss of ZBP1. Because of the specificity of our approach, we were also able to obtain the range of these numbers in individual cells: from 0 to 2 ribosomes perinuclearly compared to 1 to 8 ribosomes per mRNA peripherally after correction. This variance can be attributed in part to the varying amounts of ZBP1 binding or perhaps due to the stochasticity in the low number of receptors in normal cells that sense the environmental fluctuations of growth factors which induce phosphorylation of the protein. Interestingly, our measurements demonstrate that the notable difference in ZBP1−/− MEFs was enhanced ribosomal loading on perinuclear mRNA. This suggests that a loss of translational repression in the center of the cell may play a role in the loss of polarization and motility observed in ZBP1−/− MEFs (Katz et al., 2012). The temporal and spatial interactions between ZBP1 and ribosomes with β-actin mRNA are summarized in a working model (Fig. 4). Hence the method described here allows the measurement of both in single cells as well as in the population and provides a basis for modeling where and how specific mRNAs are regulated.
Figure 4.
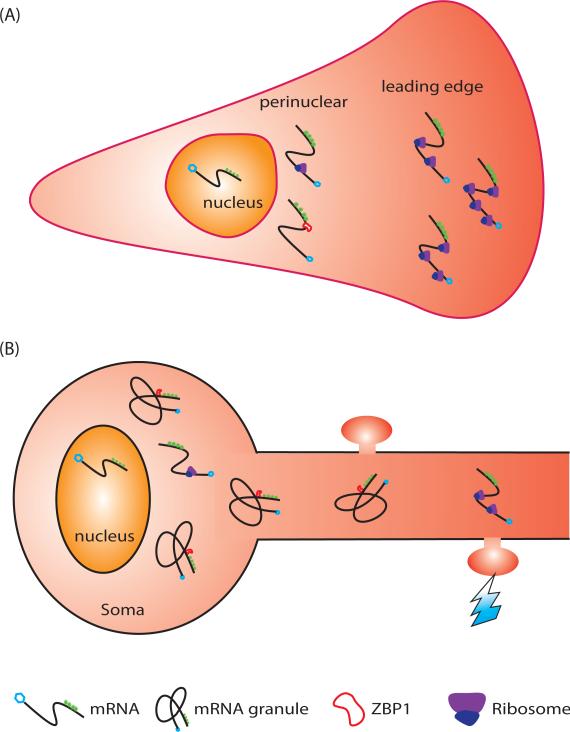
working model compatible with the localized interaction observed in Fig. 2-3. When β-actin mRNA is in the perinuclear region, ZBP1 interacts with the mRNA to repress its translation. When the mRNA reaches the leading edge of the fibroblast (A), it releases ZBP1 and translates more actively. In the soma of neurons, ZBP1 binds more effectively to β-actin mRNA in the perinuclear region (B) than in the fibroblast, likely to assemble the mRNA into granules and inhibit translation more completely until the mRNA is transported to dendrites ((Buxbaum et al., 2014)).
The single cell analysis compares the binding properties of ZBP1 in vivo with its behavior in vitro. It has been previously reported that ZBP1 KH34 binds to the β-actin zipcode with the same affinity of the full-length protein in vitro. The other domains do not bind to the zipcode. However, it is unknown whether other parts of β-actin mRNA contain unidentified binding sites for ZBP1, or other proteins may assist in its binding. The ZBP1F478A, F479A mutant disrupts the linker region between KH3 and KH4 domains and does not interact with β-actin mRNA in vitro (Chao et al., 2010). Because cellular binding was not seen as well, the in vitro and in vivo results agree and KH34 must be solely responsible for the binding of ZBP1 to β-actin mRNA in living cells.
It has been shown that β-actin mRNA localizes to the leading edge of primary fibroblasts. But in immortalized cells, this localization phenotype is diminished. The exact reason for the loss of phenotype is unclear, but perhaps is due to the high levels of phosphorylation in immortalized cells. Enhanced phosphorylation has been suggested to release ZBP1 from the mRNA (Huttelmaier et al., 2005), therefore loss of ZBP1 regulation may play a role in the difference between immortalized and primary cells. Our results show that ZBP1 interacts with β-actin mRNA less efficiently in the perinuclear region in immortalized cells than in primary MEFs. These results provide an explanation for the observation that in metastatic cells, the mRNA is unable to localize efficiently and hence maintain a polarized phenotype and proper cell adhesion, allowing the cells to disseminate (Gu et al., 2012; Katz et al., 2012; Lapidus et al., 2007).
Questions regarding RNA granule composition and heterogeneity can be addressed with the methodology described here. This approach may be generally applied to any mRNA-protein combination, and with additional fluorophores, to larger complexes of mRNAs or proteins, allowing the investigation of dependent, mutually exclusive, or symbiotic protein interactions on single mRNAs. Although we demonstrate the proof-of-principle here using a mouse knock-in for labeling the mRNA and a knock out for ZBP1, which we replaced with a fluorescent version of the protein, this would not be necessary for investigating many mRNA-protein interactions. Use of the CRISPR-Cas9 recombinase system (Mali et al., 2013) can allow the introduction of fluorescent tags and stem loops to allow imaging of endogenous molecules without generating transgenic animals. As illustrated in this work with the use of labeled ribosomal subunits, quantitative interactions can be extrapolated from the percent labeled mRNA or protein introduced exogenously compared to the endogenous levels. However, care must be taken not to increase either component substantially and thus perturb native reaction stoichiometry or kinetics. To this end, the use of shRNAs to knock down both components is a routine procedure. The HSP microscopy is not yet routine, but is achievable through various commercial two-photon microscopes capable of photon counting. The analysis tools we provide online with this publication will allow calculation of the binding constants and equilibrium values for these interactions. By this approach, important regulatory molecules and their mRNA targets discovered through high throughput approaches (Hafner et al., 2010; Ule et al., 2003) can be verified and evaluated in detail at the single cell level.
Experimental Procedures
Derivation of the Heterospecies Partition Analysis for protein-mRNA interaction
The detailed derivation and theoretical predictions were listed in the Extended Experimental Procedures. Here we define the variable and give the theoretical result essential for understanding the paper.
In dual-color FFS experiment, the brightness of species S in detection channel j is described by λj,S, where j = R for the red and j = G for the green detection channel. In a mixture of mRNA M, RNA binding protein P and their different oligomeric states, [M, P, P2, MP, MP2 ...], the hetero-species H includes M and its complexes with P (H = [M, MP, MP2, ...]) (Fig. 1A). The red channel heterospecies brightness λR,H depends on the relative concentration of each constitutive species and is used to measure the degree of binding between mRNA and protein.
where and Nj is the concentration of the j-th species. The normalized brightness measures the degree of binding y for reaction .
Fluorescence fluctuation spectroscopy and data analysis
The FFS microscope, built around an Olympus IX-71 stand, has been discussed previously (Wu et al., 2012). Briefly, a 60x Plan-Apo oil immersion objective (NA=1.4, Olympus) was used to focus the mode locked Ti:Sapphire laser (Chameleon Ultra, Coherent) and collect fluorescence. The back scattered laser was eliminated by two short-pass filters (ET680sp-2p8, Chroma). The fluorescence was separated into two detection channels with a dichroic mirror (565DCXR, Chroma). Each channel was equipped with a band pass filter (FF01-525/50-25 for green and FF01-630/92-25, Semrock). The spectra of EGFP and mCherry were plotted in Fig. S1A with the transmission curves of dichroic mirror and filters. With this choice of filter sets, the fluorescence of mCherry in the green channel could be completely eliminated. But there was a small residual signal for EGFP in the red channel. Two avalanche photodiodes (APD) (SPCM-AQR-14, Perkin-Elmer) detected photons in each channel. The output of the APD, which produced TTL pulses, was directly connected to a two-channel data acquisition card (FLEX02, Correlator.com). The recorded photon counts were stored and later analyzed with programs written in IDL (ITT Exelis).
The brightness in each channel of EGFP (λR,EGFP, λG,EGFP) and mCherry (λR,mCherry, 0) were determined by independent measurements of cells expressing each protein alone. Since the measured brightness depended on the thickness of the cell (Macdonald et al., 2010), we calibrated the brightness separately in the perinuclear and in the peripheral regions of the cell. The brightness of EGFP and mCherry in each compartment then served as a standard and were used to predict the brightness of the hetero complex MPn according to the two-brightness-state model of mCherry (Wu et al., 2009). Occasionally, there were unknown bright red autofluorescent particles passing through the laser focus, which produced large red fluorescent spikes. These spikes were removed in the post-acquisition analysis, as demonstrated in Fig. S1F. The FFS data were analyzed by dual-color time-integrated fluorescence cumulant analysis (TIFCA) (Wu et al., 2006) using the three-species HSP model. The first species was tdMCP-EGFP, which was a monomer and had the same brightness with EGFP. For simplicity and robustness of fitting, the brightness and diffusion time of this species was fixed to the value from independent measurement, but the number of molecules was left as a free fitting parameter. The second species was the free RBP-mCherry. By definition, its green brightness was fixed to zero. The third species was the heterospecies, whose FFS parameters were set as free fitting parameters. The brightness of heterospecies was the major focus for our analysis. For better interpretation, we presented the normalized brightness, defined as and , which described the number of EGFP and mCherry in the mRNP respectively (Wu et al., 2009; Wu et al., 2010).
Generation of ZBP1−/− MBS mouse cells
The mouse with 24xMBS knocked into the 3′ UTR of the Actb gene (MBS mouse) was described previously (Lionnet et al., 2011). Crossing the MBS mouse with a ZBP1 heterozygous mouse enables us to generate the ZBP1+/− MBS mouse. These ZBP1 heterozygous mice were mated to generate ZBP1−/− MBS embryos as shown in the extended experimental procedure.
Supplementary Material
Acknowledgments
We would like to thank Jeffrey A Chao for the construct of ZBP1F478A, F479A mutant and Xiuhua Meng for cloning the plasmids used in the manuscript. The translation initiation inhibitor hippuristanol is a generous gift from Dr. Jerry Pelletier. This work was supported by NIH grant R01 NS083085 to RHS.
References
- Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM. Localization of ASH1 mRNA particles in living yeast. Molecular cell. 1998;2:437–445. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- Bordeleau ME, Mori A, Oberer M, Lindqvist L, Chard LS, Higa T, Belsham GJ, Wagner G, Tanaka J, Pelletier J. Functional characterization of IRESes by an inhibitor of the RNA helicase eIF4A. Nat Chem Biol. 2006;2:213–220. doi: 10.1038/nchembio776. [DOI] [PubMed] [Google Scholar]
- Buxbaum AR, Haimovich G, Singer RH. In the right place at the right time: Visualizing and understanding mRNA localization. Nature Reviews Molecular and Cellular Biology. 2015 doi: 10.1038/nrm3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxbaum AR, Wu B, Singer RH. Single beta-actin mRNA detection in neurons reveals a mechanism for regulating its translatability. Science. 2014;343:419–422. doi: 10.1126/science.1242939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao JA, Patskovsky Y, Almo SC, Singer RH. Structural basis for the coevolution of a viral RNA-protein complex. Nat Struct Mol Biol. 2008;15:103–105. doi: 10.1038/nsmb1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao JA, Patskovsky Y, Patel V, Levy M, Almo SC, Singer RH. ZBP1 recognition of beta-actin zipcode induces RNA looping. Genes Dev. 2010;24:148–158. doi: 10.1101/gad.1862910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Tekmen M, Hillesheim L, Skinner J, Wu B, Muller JD. Dual-color photon-counting histogram. Biophys J. 2005;88:2177–2192. doi: 10.1529/biophysj.104.048413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Wei LN, Muller JD. Probing protein oligomerization in living cells with fluorescence fluctuation spectroscopy. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:15492–15497. doi: 10.1073/pnas.2533045100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David A, Dolan BP, Hickman HD, Knowlton JJ, Clavarino G, Pierre P, Bennink JR, Yewdell JW. Nuclear translation visualized by ribosome-bound nascent chain puromycylation. J Cell Biol. 2012;197:45–57. doi: 10.1083/jcb.201112145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshler JO, Highett MI, Abramson T, Schnapp BJ. A highly conserved RNA-binding protein for cytoplasmic mRNA localization in vertebrates. Curr Biol. 1998;8:489–496. doi: 10.1016/s0960-9822(98)70200-3. [DOI] [PubMed] [Google Scholar]
- Dieterich DC, Hodas JJ, Gouzer G, Shadrin IY, Ngo JT, Triller A, Tirrell DA, Schuman EM. In situ visualization and dynamics of newly synthesized proteins in rat hippocampal neurons. Nature neuroscience. 2010;13:897–905. doi: 10.1038/nn.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digman MA, Gratton E. Lessons in fluctuation correlation spectroscopy. Annu Rev Phys Chem. 2011;62:645–668. doi: 10.1146/annurev-physchem-032210-103424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digman MA, Sengupta P, Wiseman PW, Brown CM, Horwitz AR, Gratton E. Fluctuation correlation spectroscopy with a laser-scanning microscope: exploiting the hidden time structure. Biophys J. 2005;88:L33–36. doi: 10.1529/biophysj.105.061788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Femino AM, Fay FS, Fogarty K, Singer RH. Visualization of single RNA transcripts in situ. Science. 1998;280:585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- Gu W, Katz Z, Wu B, Park HY, Li D, Lin S, Wells AL, Singer RH. Regulation of local expression of cell adhesion and motility-related mRNAs in breast cancer cells by IMP1/ZBP1. J Cell Sci. 2012;125:81–91. doi: 10.1242/jcs.086132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr., Jungkamp AC, Munschauer M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiman M, Schaefer A, Gong S, Peterson JD, Day M, Ramsey KE, Suarez-Farinas M, Schwarz C, Stephan DA, Surmeier DJ, et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135:738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttelmaier S, Zenklusen D, Lederer M, Dictenberg J, Lorenz M, Meng X, Bassell GJ, Condeelis J, Singer RH. Spatial regulation of beta-actin translation by Src-dependent phosphorylation of ZBP1. Nature. 2005;438:512–515. doi: 10.1038/nature04115. [DOI] [PubMed] [Google Scholar]
- Hutten S, Sharangdhar T, Kiebler M. Unmasking the messenger. RNA biology. 2014;11 doi: 10.4161/rna.32091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan CH, Williams CC, Weissman JS. Principles of ER cotranslational translocation revealed by proximity-specific ribosome profiling. Science. 2014;346:1257521. doi: 10.1126/science.1257521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz ZB, Wells AL, Park HY, Wu B, Shenoy SM, Singer RH. beta-Actin mRNA compartmentalization enhances focal adhesion stability and directs cell migration. Genes Dev. 2012;26:1885–1890. doi: 10.1101/gad.190413.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SA, Heinze KG, Bacia K, Waxham MN, Schwille P. Two-photon cross-correlation analysis of intracellular reactions with variable stoichiometry. Biophys J. 2005;88:4319–4336. doi: 10.1529/biophysj.104.055319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kislauskis EH, Zhu X, Singer RH. Sequences responsible for intracellular localization of beta-actin messenger RNA also affect cell phenotype. J Cell Biol. 1994;127:441–451. doi: 10.1083/jcb.127.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapidus K, Wyckoff J, Mouneimne G, Lorenz M, Soon L, Condeelis JS, Singer RH. ZBP1 enhances cell polarity and reduces chemotaxis. J Cell Sci. 2007;120:3173–3178. doi: 10.1242/jcs.000638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence JB, Singer RH. Intracellular localization of messenger RNAs for cytoskeletal proteins. Cell. 1986;45:407–415. doi: 10.1016/0092-8674(86)90326-0. [DOI] [PubMed] [Google Scholar]
- Li J, Barylko B, Johnson J, Mueller JD, Albanesi JP, Chen Y. Molecular brightness analysis reveals phosphatidylinositol 4-Kinase IIbeta association with clathrin-coated vesicles in living cells. Biophys J. 2012;103:1657–1665. doi: 10.1016/j.bpj.2012.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionnet T, Czaplinski K, Darzacq X, Shav-Tal Y, Wells AL, Chao JA, Park HY, de Turris V, Lopez-Jones M, Singer RH. A transgenic mouse for in vivo detection of endogenous labeled mRNA. Nat Methods. 2011;8:165–170. doi: 10.1038/nmeth.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald PJ, Chen Y, Wang X, Mueller JD. Brightness analysis by Z-scan fluorescence fluctuation spectroscopy for the study of protein interactions within living cells. Biophys J. 2010;99:979–988. doi: 10.1016/j.bpj.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Esvelt KM, Church GM. Cas9 as a versatile tool for engineering biology. Nat Methods. 2013;10:957–963. doi: 10.1038/nmeth.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen J, Christiansen J, Lykke-Andersen J, Johnsen AH, Wewer UM, Nielsen FC. A family of insulin-like growth factor II mRNA-binding proteins represses translation in late development. Mol Cell Biol. 1999;19:1262–1270. doi: 10.1128/mcb.19.2.1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noubissi FK, Elcheva I, Bhatia N, Shakoori A, Ougolkov A, Liu J, Minamoto T, Ross J, Fuchs SY, Spiegelman VS. CRD-BP mediates stabilization of betaTrCP1 and c-myc mRNA in response to beta-catenin signalling. Nature. 2006;441:898–901. doi: 10.1038/nature04839. [DOI] [PubMed] [Google Scholar]
- Park HY, Trcek T, Wells AL, Chao JA, Singer RH. An unbiased analysis method to quantify mRNA localization reveals its correlation with cell motility. Cell Rep. 2012;1:179–184. doi: 10.1016/j.celrep.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen NO, Hoddelius PL, Wiseman PW, Seger O, Magnusson KE. Quantitation of membrane receptor distributions by image correlation spectroscopy: concept and application. Biophys J. 1993;65:1135–1146. doi: 10.1016/S0006-3495(93)81173-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross AF, Oleynikov Y, Kislauskis EH, Taneja KL, Singer RH. Characterization of a beta-actin mRNA zipcode-binding protein. Mol Cell Biol. 1997;17:2158–2165. doi: 10.1128/mcb.17.4.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross JA, Digman MA, Wang L, Gratton E, Albanesi JP, Jameson DM. Oligomerization state of dynamin 2 in cell membranes using TIRF and number and brightness analysis. BiophysJ. 2011;100:L15–17. doi: 10.1016/j.bpj.2010.12.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez M, Galy B, Muckenthaler MU, Hentze MW. Iron-regulatory proteins limit hypoxia-inducible factor-2alpha expression in iron deficiency. Nat Struct Mol Biol. 2007;14:420–426. doi: 10.1038/nsmb1222. [DOI] [PubMed] [Google Scholar]
- Sankaran J, Manna M, Guo L, Kraut R, Wohland T. Diffusion, transport, and cell membrane organization investigated by imaging fluorescence cross-correlation spectroscopy. Biophys J. 2009;97:2630–2639. doi: 10.1016/j.bpj.2009.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwille P, Meyer-Almes FJ, Rigler R. Dual-color fluorescence cross-correlation spectroscopy for multicomponent diffusional analysis in solution. Biophys J. 1997;72:1878–1886. doi: 10.1016/S0006-3495(97)78833-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, Darnell RB. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302:1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- Wu B, Chao JA, Singer RH. Fluorescence fluctuation spectroscopy enables quantitative imaging of single mRNAs in living cells. Biophys J. 2012;102:2936–2944. doi: 10.1016/j.bpj.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Chen Y, Muller JD. Dual-color time-integrated fluorescence cumulant analysis. Biophys J. 2006;91:2687–2698. doi: 10.1529/biophysj.106.086181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Chen Y, Muller JD. Fluorescence fluctuation spectroscopy of mCherry in living cells. Biophys J. 2009;96:2391–2404. doi: 10.1016/j.bpj.2008.12.3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Chen Y, Müller JD. Heterospecies partition analysis reveals binding curve and stoichiometry of protein interactions in living cells. Proceedings of the National Academy of Sciences. 2010;107:4117–4122. doi: 10.1073/pnas.0905670107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Muller JD. Time-integrated fluorescence cumulant analysis in fluorescence fluctuation spectroscopy. Biophys J. 2005;89:2721–2735. doi: 10.1529/biophysj.105.063685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yisraeli JK. VICKZ proteins: a multi-talented family of regulatory RNA-binding proteins. Biol Cell. 2005;97:87–96. doi: 10.1042/BC20040151. [DOI] [PubMed] [Google Scholar]
- Zhang HL, Singer RH, Bassell GJ. Neurotrophin regulation of beta-actin mRNA and protein localization within growth cones. J Cell Biol. 1999;147:59–70. doi: 10.1083/jcb.147.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.