SUMMARY
Histone modification and DNA methylation are associated with varying epigenetic “landscapes”, but detailed mechanistic and functional links between the two remain unclear. Using the ATRX-DNMT3-DNMT3L (ADD) domain of the DNA methyltransferase Dnmt3a as a paradigm, we apply protein engineering to dissect the molecular interactions underlying the recruitment of this enzyme to specific regions of chromatin in mouse embryonic stem cells (ESCs). By rendering the ADD domain insensitive to histone modification, specifically H3K4 methylation or H3T3 phosphorylation, we demonstrate the consequence of dysregulated Dnmt3a binding and activity. Targeting of a Dnmt3a mutant to H3K4me3 promoters decreases gene expression in a subset of developmental genes and alters ESC differentiation, whereas aberrant binding of another mutant to H3T3ph during mitosis promotes chromosome instability. Our studies support the general view that histone modification “reading” and DNA methylation are closely coupled in mammalian cells, and suggest an avenue for the functional assessment of chromatin-associated proteins.
INTRODUCTION
Histone modification and DNA methylation comprise two distinct modes of chromatin regulation that are essential in establishing patterns of gene expression during development. Although they are often considered separately, histone modification and DNA methylation are linked in various organisms (Smith and Meissner, 2013; Suzuki and Bird, 2008; Cedar and Bergman, 2009). Two models have been proposed to explain this interrelationship. In one, DNA methylation and cis-acting DNA sequences direct the pattern of downstream histone modification; DNA methylation-reading proteins such as MeCP2 engage complexes containing histone deacetylases, which induce repressive chromatin states (Jones et al., 1998; Nan et al., 1998), and unmethylated CpG-dense DNA sequences recruit histone methyltransferases leading to ectopic trimethylated H3 at lysine 4 (H3K4me3) (Thomson et al., 2010). Alternatively, another model proposes that histone modification is required for downstream DNA methylation. Pioneering studies in fungi and plants indicate a link between histone H3 lysine 9 trimethylation (H3K9me3) and DNA methylation (Jackson et al., 2002; Tamaru and Selker, 2001). In mouse ESCs, the H3K9me3-reading protein HP1 recruits de novo DNA methyltransferases (Dnmts), and the loss of H3K9me3 decreases DNA methylation at H3K9me3-dense pericentromeric repeats (Lehnertz et al., 2003). As of yet, mechanistic insights into the molecular details underlying the relationship between DNA methylation and histone modification in support of either of these two models is lacking.
Previous biochemical studies indicate that H3K4 methylation status is closely associated with DNA methylation. Specifically, the ATRX-DNMT3-DNMT3L (ADD) domain in the de novo Dnmts preferentially binds histone peptides containing unmodified histone H3 lysine 4 (H3K4me0), but not H3K4me3 (Ooi et al., 2007; Otani et al., 2009; Zhang et al., 2010). Genomic regions enriched with H3K4me3 consistently mark CpG islands (CGI), which are located mainly at gene promoters and are mostly free of DNA methylation (Edwards et al., 2010; Meissner et al., 2008; Mikkelsen et al., 2007). Therefore, it has been suggested that H3K4me3 disrupts the binding of de novo Dnmts and maintains a hypomethylated DNA state at the majority of CGI. Besides H3K4me3, phosphorylation within the H3 N-terminus, such as at H3 threonine 3 (H3T3ph) and at serine 10 (H3S10ph), disrupt ADD binding in vitro (Zhang et al., 2010). However, the molecular basis underlying the antagonistic effect of H3 phosphorylation and ADD binding and its functional consequences in vivo remain unclear.
The relationship between histone modification and DNA methylation is of particular interest in embryonic stem cells (ESCs). These cells are uniquely capable of differentiating into all three embryonic germ layers (ectoderm, mesoderm, and endoderm) by integrating transcription-factor expression, environmental signals and chromatin modifications (Young, 2011). To direct this developmental variation, ESCs initiate de novo DNA methylation and trigger dynamic histone alterations (Smith and Meissner, 2013). ESCs express maintenance Dnmt1 and high levels of de novo Dnmts. The knockout of any single Dnmt results in embryonic or postnatal lethality in mice (Lei et al., 1996; Okano et al., 1999). However, triple knockout (TKO) ESCs lacking Dnmt1, Dnmt3a, and Dnmt3b are viable and can self-renew (Tsumura et al., 2006) suggesting that ESCs have a safeguarding mechanism against variations in DNA methylation. Once ESCs and TKO ESCs begin to differentiate, DNA methylation is essential for cell viability. Thus, TKO ESCs provide an ideal cellular model to study the potential relationships between histone modification and de novo DNA methylation and their contribution to the establishment of cellular states.
Here, by solving a co-crystal structure of the ADD of Dnmt3a (ADD3a) in complex with the H3 N-terminus, we have identified not only residues known to specify unmodified H3K4 but also an amino acid crucial for recognizing the unmodified H3T3 state. Using structure-based protein engineering, we generated separate mutations in the ADD3a that allow Dnmt3a to bind to H3K4me3 or H3T3ph. When expressed in TKO ESCs, the engineered mutant insensitive to H3K4 methylation, but not H3T3ph, binds to a subset of CGI marked with H3K4me3 and induces modest DNA methylation at targeted genomic loci. Moreover, this mutant exhibits transcriptional defects in a subset of H3K4me3-enriched developmental genes that lead to a failure to specify a lineage upon differentiation. By contrast, the ADD mutant insensitive to H3T3ph remains inappropriately bound to chromatin during mitosis, resulting in chromosome instability. Taken together, our structure-function studies with the ADD3a underscore the biological significance of the precise interplay between histone H3 modifications and DNA methylation in mammalian pluripotent cells.
RESULTS
H3K4 methylation and H3T3/T6 phosphorylation disrupt ADD binding to the H3 N-terminus
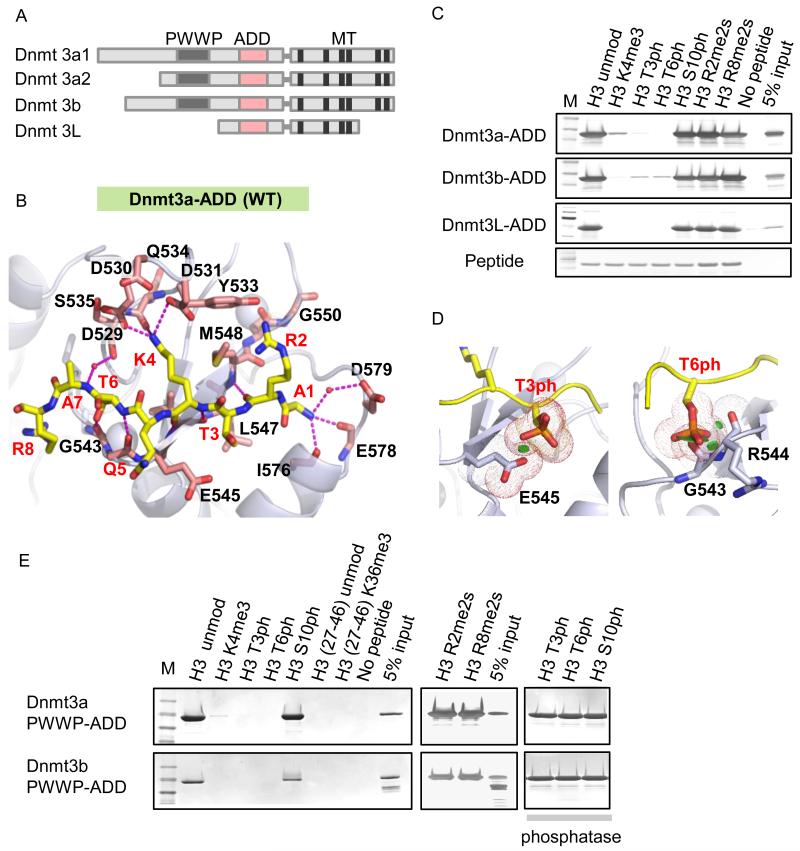
Dnmt3 family proteins contain a well-conserved ADD domain (Figure 1A), which binds to the N-terminus of unmodified H3 and is sensitive to H3 modifications, notably H3K4me3 (Ooi et al., 2007; Otani et al., 2009; Zhang et al., 2010). To assess the molecular interactions between de novo Dnmts and the H3 N-terminus, we solved the 2.4 Å crystal structure of the ADD3a in complex with unmodified H3(1-15) peptide (Figure 1B, Figure S1A-C, and Table 1). Unlike a previous study (Otani et al., 2009), in which the H3 N-terminal segment was covalently fused to the ADD3a to facilitate co-crystallization, we obtained the complex crystal by mixing a free H3 peptide with ADD3a that in principle reflects a more natural state between the two binding partners. When we traced the first eight residues, A1-R2-T3-K4-Q5-T6-A7-R8, of the bound H3 peptide (Figure 1B), the intermolecular features of ADD3a are similar overall to those of the previously reported H3 peptide-ADD3a fusion protein (Otani et al., 2009). Unmethylated K4 (H3K4me0), but not H3K4me3, forms multiple hydrogen bonding and electrostatic interactions with three surface residues (D529, D531, Q534) in the ADD3a pocket. The unmodified T6 residue forms a hydrogen bond with a nearby glycine (G543) carbonyl oxygen, enhancing ADD3a binding to H3 N-terminus. Intriguingly, the unmodified T3 residue is adjacent to negatively charged glutamate (E545), a feature that was not discussed previously.
Figure 1.
ADD binding is disrupted by H3K4me3, H3T3ph, and H3T6ph.
A, Domain architecture of the Dnmt3 family proteins. B, Recognition of an unmodified H3(1-8) peptide by the wild-type (WT) ADD3a. The histone H3 peptide is shown in yellow, side chains of the residues that comprise the binding pocket are shown in pink, small red balls are waters. C, Peptide pull-down assays with Dnmt3a-, 3b-, 3L-ADD and modified H3 peptides as indicated. D, Structural models of the WT-ADD3a in complexes with either H3T3ph or H3T6ph. Green disks indicate steric clashes. E, Peptide pull-down assays with WT-PWWP-ADD3a and modified H3 peptides with or without phosphatase treatment.
Table 1. Data Collection and Refinement Statistics.
| ADD3a(479-610) -H31-15 |
ADD3a(476-611)(G550D) -H31-10 |
ADD3a(476-611)(E545R) -H31-10T3ph |
|
|---|---|---|---|
| Data Collection | |||
|
| |||
| Wavelength (Å) | 0.9792 | 0.9793 | 0.9792 |
| Space group | P2 1 | P1 | P3121 |
|
| |||
| Cell dimensions | |||
|
| |||
| a, b, c (Å) | 41.6, 56.4, 57.3 | 36.1, 37.2, 50.1 | 57.3, 57.3, 71.9 |
| α, β, γ (°) | 90, 90.3, 90 | 78.7, 83.9, 81.6 | 90, 90, 120 |
| Resolution (Å) | 50-2.4 (2.46-2.40)a | 50 -1.9 (1.93 -1.90) | 50-1.8 (1.83-1.80) |
| No. reflections | 10,329 | 19,077 | 13,010 |
| R sym | 11.7 (41.0) | 9.6 (58.5) | 9.1 (63.3) |
| I/σI | 13.3 (2.5) | 23.3 (3.1) | 26.1 (6.4) |
| Completeness (%) | 99.9 (99.4) | 96.7 (96.0) | 99.4 (99.1) |
| Redundancy | 4.8 (4.1) | 4.4 (4.3) | 5.9 (6.0) |
|
| |||
| Refinement | |||
|
| |||
| Resolution (Å) | 50-2.4 | 50-1.9 | 50-1.8 |
| Rwork/Rfree | 17.4/23.0 | 18.1/22.5 | 16.7/20.3 |
|
| |||
| No. atoms | |||
|
| |||
| Protein | 2126 | 2160 | 1097 |
| Ligand/ion | 65/6 | 106/6 | 57/8 |
| Water | 104 | 216 | 125 |
|
| |||
| B-factors (Å2) | |||
|
| |||
| Protein | 29.8 | 21.4 | 25.9 |
| Peptide/Zinc ion | 44.2/23.6 | 26.1/16.9 | 25.1/16.6 |
| Water | 30.2 | 29.2 | 33.0 |
|
| |||
| R.m.s. deviations | |||
|
| |||
| Bond lengths (Å) | 0.009 | 0.008 | 0.011 |
| Bond angles (°) | 1.26 | 1.09 | 1.37 |
Highest resolution shell is shown in parenthesis.
To assess the impact of histone modifications on ADD binding to the H3 N-terminus, we performed a series of peptide pull-downs using recombinant ADD from Dnmt3a, 3b, and 3L. In agreement with the homology of the three domains, H3K4me3, H3T3ph and H3T6ph disrupt the binding of all three ADDs to the H3 N-terminus; however, H3R2me2, H3R8me2, and H3S10ph are compatible with binding (Figure 1C). Modeling analysis revealed both steric clashes and electrostatic repulsion between ADD3a and the H3 peptide when a bulky and negative phosphate group was introduced at either T3 or T6 (Figure 1D). Binding to a longer regulatory domain (PWWP-ADD) is still disrupted by H3K4me3, H3T3ph, and H3T6ph but not by H3S10ph, and phosphatase-treated peptides restore PWWP-ADD binding to the H3 N-terminus (Figure 1E). Thus, in addition to H3K4me3, the phosphorylation of T3 and T6, but not S10, probably acts as additional “on/off switches” (Fischle et al., 2003) to control the sites at which Dnmt3a associates with chromatin. H3T3ph appears exclusively during mitosis (Dai et al., 2005; Markaki et al., 2009) and H3T6ph occurs mainly during interphase (Garske et al., 2010; Metzger et al., 2010). Considering that both T3ph and S10ph appear during mitosis, selective disruption by T3ph may be linked to specific function.
Engineering of ADD3a alters its binding to methylated H3K4 or phosphorylated H3T3
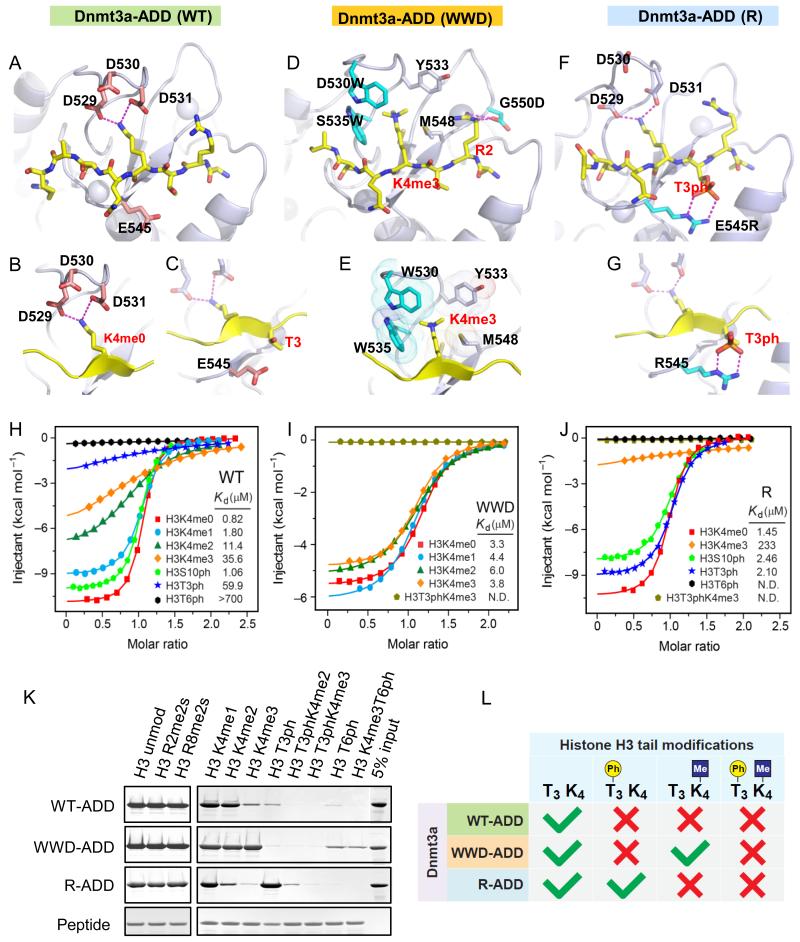
To investigate the biological significance of the relationship between H3K4me3 and unmethylated CGI, and the links between H3T3ph/H3T6ph and de novo Dnmts, we engineered the ADD3a domain to enable it to bind to H3K4me3, H3T3ph, or H3T6ph. We first sought to mirror the H3K4me3-binding site of the plant homeodomain (PHD) of the bromodomain PHD finger transcription factor (BPTF) in ADD3a (Figure S1D-G). We introduced an aromatic cage (D530W, S535W) to stabilize binding to H3K4me3, and added an acidic residue (G550D) to interact with unmodified H3R2, which has been shown to be necessary for the binding of BPTF-PHD to H3K4me3 (Li et al., 2006; Wysocka et al., 2006). This D530W, S535W, G550D triple mutant will hereafter be referred to as “WWD”. We similarly engineered ADD3a to bind to H3T3ph by replacing a negatively charged glutamate with a positively charged arginine at position 545 (E545R; hereafter called “R”). We were unable to engineer a specific ADD3a mutation that binds to H3T6ph.
Structures of the engineered ADD3a reveal two independent reading compartments
The co-crystal structure of the ADD3a-wild-type (WT) in a complex with the unmodified H3 N-terminus (1-8) shows its reading modules of H3K4me0 and unmodified H3T3 (Figure 2A-C). Energy minimization-optimized structural modeling of the ADD3a-WWD indicates that the trimethyl group of H3K4me3 is positioned within an aromatic cage formed by S535W, D530W and M548 (Figure 2D, E), contrary to ADD3a-WT. The co-crystal structure of the ADD3a (G550D) mutant in complex with unmodified H3(1-10) indicates that G550D forms charge-stabilized hydrogen bonds with H3R2 (Figure S2A), similar to the PHD finger of BPTF (Figure S1F), thereby stabilizing the binding with H3 (Figure 2D, Figure S2B). Peptide pull-downs performed with the individual mutations of WWD revealed the relative importance of each substitution, with S535W identified as the crucial site for H3K4me3 binding (data not shown). The 1.8 Å co-crystal structure of the ADD3a-R mutant in complex with H3T3ph revealed that the arginine substitution (E545R) creates a pair of charge-stabilized hydrogen bonds with the phosphate group of H3T3ph (Figure 2F, G, and Table 1), in contrast to ADD3a-WT. These structure studies strongly indicate that ADD3a-WWD and -R are “insensitive” to H3K4me3 and H3T3ph, respectively.
Figure 2.
Targeted ADD3a mutations acquire additional binding ability towards H3K4me3 or H3T3ph.
A-C, Complex structures of the WT-ADD3a domain bound to H3(1-8) peptide. In all of the panels, the peptide is shown in yellow (A). Close-up views of H3K4me0 binding pocket (B) and the residue E545 surrounding H3T3 (C). D, E, Structural model of the WWD-ADD3a domain in complex with the H3K4me3 peptide. The mutated residues (D530W, S535W, and G550D) are shown in cyan (D). Close-up view showing the positioning of H3K4me3 within an aromatic cage (E). F, G, Complex structure of the R-ADD3a domain bound to H3(1-8)T3ph peptide. The mutated residue (E545R) is shown in cyan (F). Close-up view of H3T3ph binding residue (G). H-J, Isothermal titration calorimetry (ITC) curves for WT- (H), WWD- (I), and R-ADD3a (J) with H3(1-15) peptides containing various modifications. K, Peptide pull-down assays with WT-, WWD-, R-ADD3a and modified H3 peptides. L, Schematic summary of the results obtained in the pull-down assays.
To determine their structural integrity, we performed circular dichroism (CD) on the wild-type and mutant ADD3a. All proteins showed characteristic negative ellipticities in the region of 240 nm and below, indicative of a well-folded structure (Figure S2C). Isothermal titration calorimetry (ITC) data support the structure-based observation that ADD3a-WT binding is greatly diminished by H3K4me3 (43 times lower affinity; KD: 35.6 μM for H3K4me3 vs. 0.82 μM for H3K4me0; Figure 2H). By contrast, the WWD mutant is insensitive to the H3K4 state and binds to the H3K4me3 peptide with a similar affinity as the unmodified H3 peptide (KD: 3.8 μM for H3K4me3 vs. 3.3 μM for H3K4me0; Figure 2I). Although the binding enthalpy is compromised by the WWD mutations (ΔH = −10.9 kcal/mol for WT-H3K4me0 vs. −4.9 kcal/mol for WWD-H3K4me3), the recognition of H3K4me3 by WWD is supported by entropic changes (ΔS= 8.4 cal/mol/deg for WWD-H3K4me3 vs. −8.8 cal/mol/deg for WT-H3K4me0), suggesting that favorable hydrophobic contacts are introduced by the engineered aromatic pocket (Table S1, related to Figure 2). The R mutant has a similar affinity for the H3T3ph peptide as the unmodified H3 peptide (KD: 2.10 μM for H3T3ph vs. 1.45 μM for H3T3; Figure 2J), whereas WT binds the H3T3ph peptide with 73 times lower affinity (KD: 59.9 μM for H3T3ph vs. 0.82 μM for H3T3; Figure 2H). Therefore, ITC results validate the generation of engineered ADD domains with unique molecular determinants of binding specificity.
The engineered ADD3a mutants act independently and acquire a T3phos/K4methyl switch
To provide additional insight into ADD-mediated interactions with chromatin, we performed peptide pull-down assays and showed that wild-type and mutants bind only to the extreme N-terminus of histone H3 and not to any other histone regions (Figure S2D). ADD3a-WT and R exhibited gradually reduced binding to the H3 N-terminus in the order mono-, di- and trimethylation of K4. By contrast, the ADD3a-WWD bound all of these methylation states similarly, suggesting that methylation at H3K4 does not affect WWD binding (Figure 2K). Focusing on H3T3ph, the binding of ADD3a-WT and WWD is markedly decreased, but R can bind H3T3 regardless of its phosphorylation status. For the H3T6ph, ADD3a-WT and R do not bind, but the ADD3a-WWD shows a weak signal similar to wild-type binding to T3ph or K4me3 (Figure 2K). Importantly, the access of the WWD mutant to H3K4me3 is disrupted by H3T3ph, and R mutant access to H3T3ph is disrupted by H3K4me3; thus, each mutant acts independently (Figure 2K, L). These results indicate that both ADD mutants, unlike endogenous ADD, have distinct binding properties (Figure 2L). Although phos/methyl “switching” is well documented for HP1 binding at H3K9me3/S10ph (Fischle et al., 2005), to our knowledge, no previous study has systematically generated mutants that perturb these relationships. The insensitivity of WWD and R to modified H3K4 and H3T3, respectively, is unaffected by other potential H3 N-terminal methylation states such as H3R2me2 and H3R8me2 (Figure 2K). Moreover, this distinct mutant binding pattern is maintained in the context of a longer regulatory domain (PWWP-ADD) and a full-length Dnmt3a protein (Figure S2E-G). Assessment of the wild-type and mutant ADD binding to native mono-nucleosomes showed the same results as peptides (Figure S2H). Taken together, our targeted mutational approach therefore transformed the ADD3a domain into a “reading” module that is insensitive to H3K4 methylation (WWD) or H3T3 phosphorylation (R) (see summary in Figure 2L).
Generation of epigenome maps in ESCs expressing wild-type and mutant Dnmt3a2
We next sought to develop a cellular system to examine the genomic distribution and the biological function of wild-type and engineered Dnmt3a mutants. ESCs initiate de novo DNA methylation and trigger dynamic epigenetic alterations that are required for developmental variation. ESCs express high levels of de novo Dnmts, particularly a short N-terminal isoform of Dnmt3a, Dnmt3a2, which is associated with active euchromatin (Chen et al., 2002). To explore its impact on de novo DNA methylation and gene transcription, we stably introduced FLAG-tagged, full-length WT or mutant Dnmt3a2 into ESCs lacking all Dnmts (TKO ESCs) (Tsumura et al., 2006) using the PiggyBac transposon system (Wang et al., 2008). Clonally derived ESC lines for each WT-, WWD-, and R-Dnmt3a2 have the same growth rate and show comparable levels of Dnmt3a2 mRNA and protein expression without global changes in H3K4me3 (Figure S3A, B). Notably, the expression of the WWD mutant was somewhat lower (~2-fold by RT-qPCR) due to fewer instances of transposition. Sequence analysis of the integration sites revealed that the ectopic genes are located in intra- or intergenic regions, verifying the legitimacy of the clones.
To quantitatively measure genome-wide Dnmt3a2 binding and activity, ESC lines were subjected to chromatin immunoprecipitation followed by high-throughput DNA sequencing (ChIP-seq). Genomic maps of DNA methylation at single-base, CpG (cytosine) resolution with enriched CGI were obtained using an enhanced form of reduced representation bisulfite sequencing (RRBS). In parallel, genome-wide transcription profiles were determined using RNA-seq. After verifying the correlation between biological replicates (Pearson’s r > 0.95, see Extended Experimental Procedures, Table S2 related to Figure 3, and Figure S3C-E), we performed a data analysis.
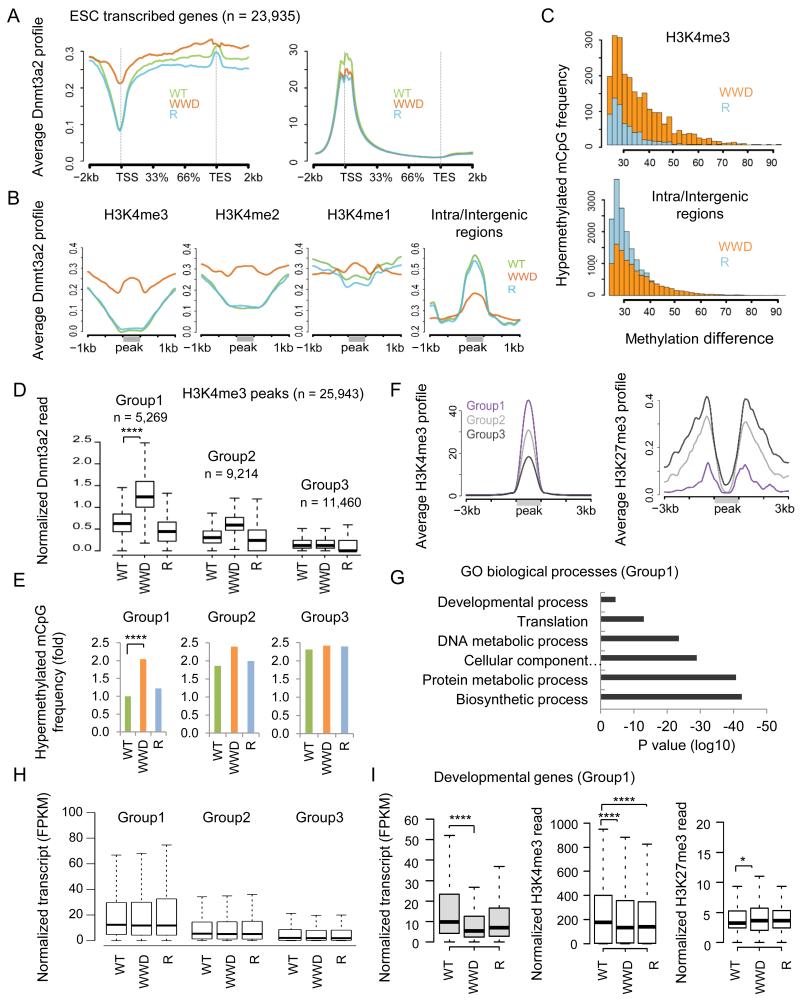
Figure 3.
WWD-Dnmt3a2 exhibits a relative loss of the signal at inter/intragenic regions but significant gain in H3K4me2/3
A, ChIP-seq profiles of WT and mutant Dnmt3a2 (left) and matched H3K4me3 (right) at transcribed genes in ESCs. B, Average ChIP-seq profiles of WT and mutant Dnmt3a2 binding at genomic regions enriched for each H3K4 methylation or inter/intragenic regions. Gray lines indicate the H3K4me3/2/1 peaks and inter/intragenic regions. C, Distributions of hypermethylated mCpG sites in WWD and R compared to WT (mCpG; ≥10× reads coverage across samples; ≥ 25% relative to WT). D, Levels of ChIP-seq enrichment of WT and mutant Dnmt3a2 in groups divided with the density of WWD-Dnmt3a binding. H3K4me3 peaks were assigned to Groups 1, 2 and 3 based on high, moderate and low levels of WWD-Dnmt3a2 binding, respectively using K-means clustering. E, Hypermethylated mCpG frequency in WT-, WWD-, or R-Dnmt3a2, compared to TKO at the matched groups. F, Average H3K4me3 and H3K27me3 ChIP-seq profiles of Groups 1, 2 and 3, obtained from WWD-Dnmt3a2-expressing ESCs. G, Gene ontology (GO) classification of the Group 1 genes (n = 3,583). H, RNA-seq analysis of the genes associated with the matched groups. I, RNA-seq analysis (left), H3K4me3 ChIP-seq (middle), and H3K27me3 ChIP-seq (right) of the embryonic developmental genes (n = 377) belong to Group1. Fragments Per Kilobase of exon per Million fragments mapped (FPKM). ChIP-seq read counts on the y-axis are normalized to 10 million reads. Box plots show the central 50% of the data (filled box), the median (central line) and 1.5× the interquartile range (whiskers). ****p ≤ 2.2 × 10−16, *p < 0.05.
Altered binding and activity of WWD-Dnmt3a2 in the ESC genome extends to H3K4me2/3 loci
Globally, H3K4me3 is often coincident with CGI and marks ~75% of transcription start sites (TSS) in mammalian cells (Mikkelsen et al., 2007; Guenther et al., 2007). We therefore asked whether the binding of the WWD mutant is increased at TSS as a result of its acquired ability to recognize H3K4me3. ChIP-seq reads over all ESC transcribed genes (n = 23,935) revealed that WWD-Dnmt3a2 shows a higher signal at TSS and up to transcription end site (TES) compared with WT- and R-Dnmt3a2 (Figure 3A, left). Matched ChIP-seq reads of H3K4me3 show a prominent enrichment at TSS although the signals in both WWD and R are mildly decreased (Figure 3A, right), suggesting a common change that is responsible for maintaining H3K4me3 levels. Next, we segmented the genome enriched in each H3K4 methylation state (H3K4me1/2/3) using ENCODE/LICR data (Shen et al., 2012) and profiled ChIP-seq signals of each Dnmt3a2. Wild-type and R-Dnmt3a2 show a graded decrease in genomic regions containing H3K4me1, me2, and me3; however, WWD-Dnmt3a2 demonstrates similar levels in all the regions, directly reflecting the insensitive binding properties of WWD (Figure 3B; WT versus WWD; p ≤ 2.2 × 10−16; Wilcoxon rank-sum test). WT- and R-Dnmt3a2 are enriched at inter/intragenic regions, consistent with their binding to the unmodified H3 N-terminus. WWD-Dnmt3a2 binding is relatively decreased in inter/intragenic regions (Figure 3B), consistent with its lower expression and lower binding affinity to the unmodified H3 N-terminus (Figure S3A and Figure 2H, I,). Given that these regions occupy a large portion of the genome, the lower binding of WWD to the unmodified H3 and intra/intergenic regions could be coupled to a positive aspect to binding that directs the WWD mutant to TSSs and other K4 methylation-enriched regions (see Discussion). ChIP-qPCR validation at individual loci confirmed an increase in WWD-Dnmt3a2 binding at H3K4me3 peaks/CGI, but not at adjacent sites upstream/downstream (± 2kb) of the tested peaks, nor at gene deserts (Figure S3F, G).
Global levels of CpG methylation (mCpG) correlate with Dnmt3a2 expression. In WT and R, ~41% of CpG are methylated, whereas it drops to ~26% and 0.6% in WWD and TKO, respectively (≥ 10× reads coverage, minimal CpG methylation level of 5%). When we compare mCpG levels between WT and the mutants, all differences were locus-specific and relatively modest, and the distribution of mCpG overall mirrored the protein binding of Dnmt3a2. Specifically, at H3K4me3 peaks, hypermethylated mCpG in WWD were more abundant than those in R, whereas at inter/intragenic regions, hypermethylated mCpG in R outnumbered those in WWD (≥10× reads coverage across samples; mCpG ≥ 25% relative to WT; WWD versus R; p ≤ 2.2 × 10−16; Figure 3C). Assembled data at selected loci illustrate that WWD-, but not WT- or R-Dnmt3a2, colocalizes with H3K4me3 peaks and increases DNA methylation at CGI with only a minor perturbation, if any, of gene transcription (Figure S3H). Taken together, these genome-wide results support the biochemical properties of the engineered ADD3a domain.
Developmental genes, but not housekeeping genes, are sensitive to DNA hypermethylation at TSS
To dissect the WWD bound loci, we further segmented the H3K4me3 peaks (n = 25,943) according to WWD-Dnmt3a2 read densities (K-means clustering), and classed them into Groups 1, 2 and 3 representing high, moderate, and low enrichment of WWD-Dnmt3a2, respectively (Figure 3D). Group 1 loci, defined by a high signal of WWD-Dnmt3a2, includes ~20% of H3K4me3 peaks (n = 5,269) and exhibits significantly increased levels of hypermethylation at mCpG in WWD compared with WT and R (≥10× reads coverage across samples; mCpG relative to TKO; p ≤ 2.2 × 10−16; Friedman rank-sum test; Figure 3E). Group1 also shows a higher H3K4me3 and lower H3K27me3 enrichment in all ESC lines (shown by ChIP-seq in WWD; Figure 3F), which is compatible with an antagonism between H3K27me3 and DNA methylation (Lister et al., 2009; Meissner et al., 2008). Group 1-linked genes (TSS located within ± 1kb peaks; n = 3,583) are associated with metabolic processes, translation and embryo development (Figure 3G), suggesting that mainly active housekeeping genes and a subset of developmental genes are accessible to WWD-Dnmt3a2. No specific DNA sequence elements were found at the WWD-Dnmt3a2-bound loci (data not shown). Group 2 showed intermediate enrichment of H3K4me3 and H3K27me3, and included more developmental genes, and Group 3 – containing the most bivalent genes – exhibited a lower H3K4me3 and higher H3K27me3 enrichment (Figure 3F). Thus, the engineered WWD-Dnmt3a2 acquires the ability to bind to H3K4me3, apparent in Group 1, although the lack of WWD enrichment at Group 3 peaks suggests that other aspects of the chromatin environment (e.g., H3K27me3, other chromatin-associated factors, etc.) have an effect in vivo.
To correlate gene expression with the matched group, we selected the genes associated with H3K4me3 peaks (TSS located within ± 1kb peaks; Group 1, n = 3,583; Group 2, n = 5,152; Group 3, n = 3,940) and measured average transcript levels in the wild-type and mutant ESCs. WWD-Dnmt3a2 does not lower average gene expression levels in Group 1 (Figure 3H) consistent with the safeguarding property of the ESC state against epigenetic transformation (Gifford et al., 2013; Xie et al., 2013). Alternatively, the relocation of WWD-Dnmt3a2 and the subsequent DNA hypermethylation might be insufficient to trigger transcriptional changes in actively expressed housekeeping genes. Interestingly, when we examined developmental genes (n = 377) belonging to Group 1, a significant decrease in gene expression was observed (WT versus WWD; p ≤ 2.2 × 10−16; Figure 3I), suggesting that they are more susceptible to epigenetic changes. H3K4me3 levels are decreased in these genes in both WWD and R mutants, correlate with their general reduction. The H3K27me3 level is modestly increased in ESCs expressing the WWD mutant (WT versus WWD; p = 0.012; Figure 3I), but not R. Thus, WWD-Dnmt3a2 binding to CGI increases DNA methylation, which may in turn increase H3K27me3. These two epigenetic signals could contribute to a decrease in gene expression in the WWD mutant.
Gene expression decrease and DNA hypermethylation at promoter CGI intersect at a subset of developmental genes
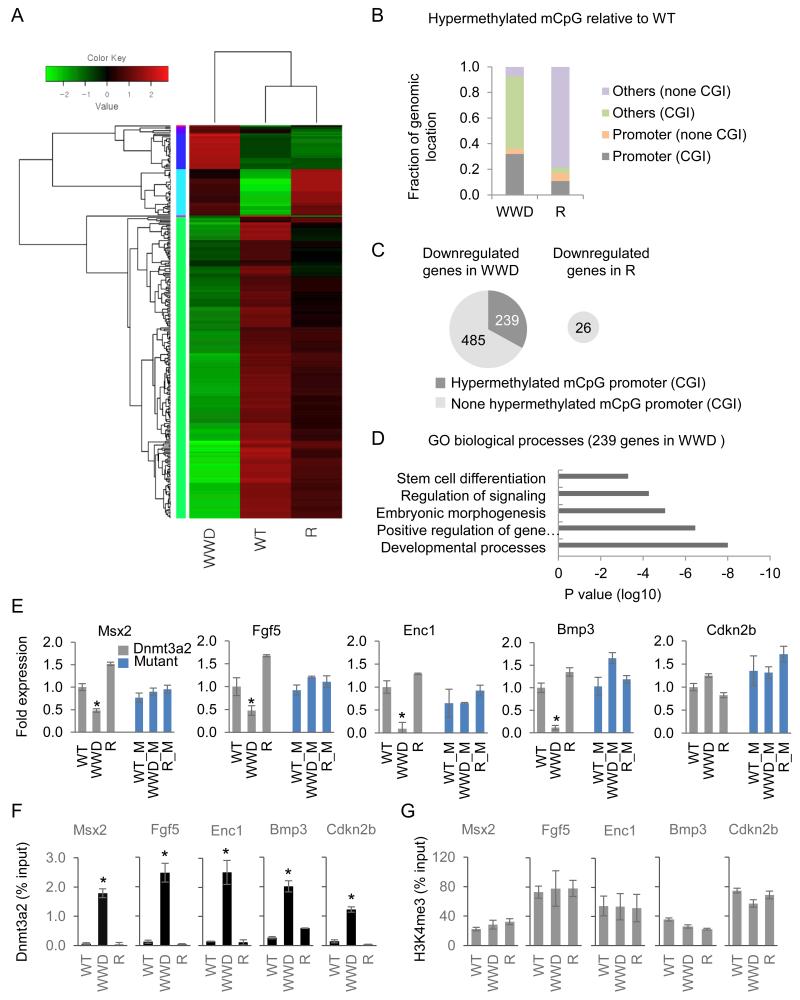
To analyze the gene expression effects more thoroughly, we performed unbiased RNA-seq comparisons with WT, WWD, and R. As shown in a heat map, we revealed a downregulation of 724 and an upregulation of 375 genes in the WWD mutant and a downregulation of 26 and an upregulation of 24 genes in the R mutant (1.5 fold change; q < 0.05; FDR-adjusted p value; Figure 4A). Further validation of multiple hits by RT-qPCR (Figure S4A, 4E) indicates a strong positive correlation between the RNA-seq analysis and the RT-qPCR (18 out of 20 genes). In parallel, we unbiasely choose significantly hypermethylated mCpG in the WWD and R mutants relative to WT and annotated their locations in the genome. 32% and 56% of hypermethylated mCpG overlap with promoter CGI and non-promoter CGI respectively in WWD, whereas only 14% of hypermethylated mCpG in R overlap with any CGI (Figure 4B). Bisulfate mass array of the target CGI validated modest but significant locus-specific hypermethylation (e.g., Socs3, Figure S4B; other loci are averaged, Figure S4C). Levels of mCpG in R-Dnmt3a2 are similar, but not identical to WT-Dnmt3a2, and interestingly, 26% of hypermethylated mCpG in R overlap with repeat elements. Thus, we conclude that relative to WT, WWD- and R-Dnmt3a2 are recruited to distinct genomic regions of H3K4me3peak/CGI and repeat/mCpG, respectively, and induce a modest increase of de novo hypermethylation therein.
Figure 4.
A subset of developmental genes downregulated in WWD-Dnmt3a2 coincides with mCpG hypermethylation in promoter CGI.
A, Heat map representation of the differentially expressed genes (1.5 fold change, FDR < 0.05) in wild-type and mutant Dnmt3a2 ESCs. Scale bar shows the color-coded differential expression from the mean with red and green indicating higher and lower expression, respectively. B, Genomic distribution of hypermethylated mCpG sites in ESCs expressing WWD or R compared to WT (mCpG; ≥10× reads coverage across samples; p < 0.05 relative to WT). C, Fraction of downregulated genes overlapping with hypermethylated CGI (dark grey) at the promoter. D, Gene ontology (GO) classification of genes that had hypermethylated CGI at the promoter and were downregulated in WWD compared to WT and R. E, RT-qPCR analysis of the developmental genes (Msx2, Fgf5, Enc1, Bmp3) in wild-type and mutant Dnmt3a2 (grey bars), and additional catalytic mutation-expressing ESCs (blue bars, denoted as M). Relative expression levels are normalized to Gapdh and the level of gene expression in the wild-type Dnmt3a2-expressing ESCs is set as 1. Cdkn2b serves as a non-developmental gene showing the WWD mutant enrichment at promoter CGI without transcript changes. Data are mean ± s.d., n = 3. F, Dnmt3a2 ChIP-qPCR analysis at promoter CGI of the indicative genes. G, H3K4me3 ChIP-qPCR analysis at promoter CGI of the indicative genes. Data are represented as a percentage of input DNA. Data are mean ± s.d., n = 3. *p < 0.05.
In WWD-Dnnmt3a2 ESCs, identifying the genes with both decreased expression and hypermethylated CGI at their promoters produced a subset of “DNA methylation-sensitive” genes (n = 239 out of 724 downregulated genes in WWD; Figure 4C), which include a number of genes involved in cell-lineage specification (e.g. Msx2, Fgf5, Enc1, and Bmp3). A gene ontology (GO) analysis indicates that 239 genes are highly associated with developmental processes (Figure 4D), involved in ectoderm and mesoderm development. In contrast, we failed to identify a gene in R-Dnnmt3a2 ESCs that exhibited decreased expression and hypermethylated CGI at the promoter, consistent with the infrequent binding of R-Dnmt3a2 to promoter CGI. Our finding that 33% of downregulated genes in WWD-Dnmt3a2 coincide with the occurrence of CpG hypermethylation at the promoter CGI suggests that WWD-Dnmt3a2-mediated epigenetic changes contribute to transcriptional output in ESCs.
Additional catalytic mutation abolishes the downregulation of developmental genes in WWD
Thus far, our data demonstrate that engineered Dnmt3a2, particularly WWD-Dnmt3a2, redistributes within the genome, induces a modest “ectopic” CGI methylation, and perturbs a subset of developmental genes. We next wanted to determine whether the changes in gene expression are caused by WWD-Dnmt3a2 activity. To this end, we generated additional mutants that combine a catalytic mutation in the full-length WT-, WWD-, and R-Dnmt3a2 and examined the expression of developmental genes targeted by WWD-Dnmt3a2 (Figure 4E-G, gray bars) in a parallel set of catalytic mutants (Figure 4E, blue bars). A previous study of a DNA methyltransferase reported that a single replacement of cysteine at position 706 by a serine (C706S) abolished its catalytic activity (Hsieh, 1999; Wyszynski et al., 1992). We found that additional catalytic mutations indeed revert the gene-expression decrease in the WWD mutant (Figure 4E, blue bars, shown with C706S). Together, these results support the conclusion that the gene-expression defects are indeed caused by DNA methylation.
A Dnmt3a2 mutant insensitive to H3K4 methylation state leads to a major defect in ESC differentiation
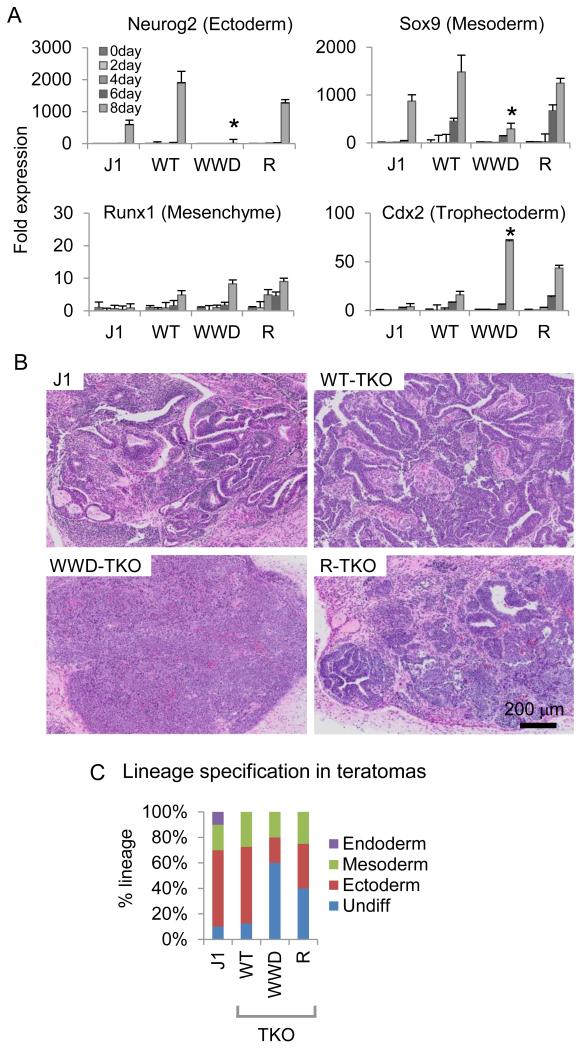
Given the disruption of developmental gene expression observed in WWD-Dnmt3a2, we next assessed the ability of WT vs. mutant Dnmt3a2 ESCs to differentiate in culture and form teratomas. We first induced ESCs to differentiate into neuronal precursors cells (NPCs) (Bibel et al., 2007) and compared the resulting lineages by profiling the expression of lineage-specifying genes. After 8 days of differentiation, the expression of self-renewal genes such as Oct4 and Nanog was markedly decreased similarly across all of the NPCs (Figure S5A). However, the expression of genes required for ectoderm and mesoderm differentiation, such as Neurog2 and Sox9, were significantly impaired in WWD-, but not in R- nor WT-Dnmt3a2-expressing NPCs. Concurrently, Cdx2 (a marker for extraembryonic tissues) expression was increased in the WWD mutant without changes in other lineage-specific markers (e.g., Runx1; hematopoietic lineage, Olig1; glial lineage) (Figure 5A and S5A).
Figure 5.
Changes in Dnmt3a2 location and function affect lineage specification in ESCs.
A, Relative expression levels of lineage-specific genes in TKO, WT-, WWD- and R-Dnmts3a ESCs. Error bars are s.e.m. of three experiments, *p < 0.05. B, Images of hematoxylin and eosin (H&E)-stained cross sections of teratomas. C, Quantification of differentiated lineage portions of the sections of teratomas (average of 4 teratomas from J1, WT-, WWD-, R-Dnmt3a2). Teratomas were evaluated by a pathologist for the differentiation of the injected ESCs into derivatives that correspond to the three germ layers.
We next conducted in vivo teratoma formation with WT and mutant ESCs. Control teratomas derived from wild-type J1 parental ESCs show representative populations of the three germ layers, ectoderm, mesoderm, and endoderm (Figure 5B, C), whereas TKO ESCs were unable to form visible teratomas (Figure S5B). TKO ESCs expressing Dnmt3a2 gave rise to teratomas despite their slow formation compared to the J1 parental line (3 weeks versus 5 weeks; Figure S5B). Interestingly, WT-Dnmt3a2 produced ectoderm and mesoderm, but not endoderm, whereas the ability of WWD- and R-Dnmt3a2 to specify lineage was reduced, resulting in 60% and 40% undifferentiated cells respectively (compared to ~10% of undifferentiated cells in WT; Figure 5B, C). These results indicate that mutant Dnmt3a2 alters the epigenetic landscape in ways that cause defects in lineage commitment during ESC differentiation and in a teratoma assay.
Because WWD-Dnmt3a2 expression is lower than WT and R-Dnmt3a2, we next set out to determine if the cell-lineage defects in the teratoma are merely due to the hypomethylation of mCpG in inter/intragenic regions. We prepared matched lines (WT, WWD, R) in a normal C57Bl/6 (C6) ESC background containing all Dnmts and repeated the teratoma assay. Similar to the results obtained in the TKO background, teratomas derived from WWD-Dnmt3a2-expressing C6 ESCs demonstrated an increase in undifferentiated cells (20% compared to 5% in WT; Figure S5C) and the presence of extra-embryonic tissue. Notably, in those C6 ESC-derived teratomas, the ectodermal defect was modest compared to the TKO ESC-derived teratoma, suggesting that non-promoter DNA methylation by Dnmt3a is necessary for neuro-ectodermal differentiation. A previous report (Wu et al., 2010) also supports this point. Taken together, these results indicate that inter/intragenic hypermethylation is required for ectodermal differentiation of ESCs and that promoter CGI hypermethylation mediated by WWD at key developmental genes probably leads to defects in differentiation potential.
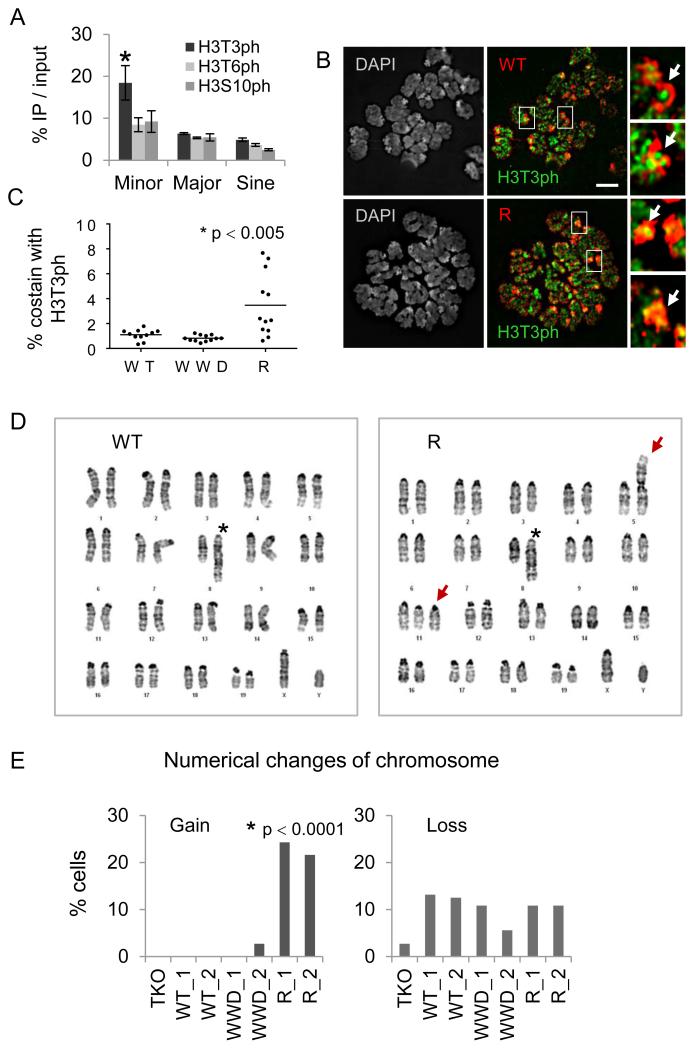
A Dnmt3a2 mutant insensitive to H3T3 phosphorylation increases chromosomal instability
Previous studies indicate that H3T3ph is found at mitotic centromeres and attracts the chromosome passenger complex (Kelly et al., 2010; Wang et al., 2010; Yamagishi et al., 2010). In agreement with these data, ChIP-qPCR analysis of the ESCs showed that H3T3ph, but not H3T6ph nor H3S10ph, is significantly enriched in minor satellite repeats indicative of centromeres (Figure 6A). As H3T3ph occurs exclusively during mitosis, we hypothesized that R-ADD3a might bind to mitotic H3T3ph and be retained at mitotic chromosomes including centromeres. To test this idea, we performed co-immunofluorescence of metaphase chromosomes with antibodies against H3T3ph and tagged Dnmt3a2. High levels of H3T3ph were located at DAPI-dense centromeric regions consistent with ChIP-qPCR. Both R- and WT-Dnmt3a2 were densely localized at pericentromeric regions where reduced levels of H3T3ph appear (Figure 6B). Interestingly, R-Dnmt3a2, but not WT-Dnmt3a2, occasionally colocalized with H3T3ph at centromeric regions (see arrows in Figure 6B, quantification in Figure 6C). To gain insight into the functional relationship between H3T3ph and Dnmt3a2, we focused on genes that are mis-regulated in the R mutant and discovered that few genes linked to chromosome segregation and fertility (e.g., Smc1b, Rhox2, Usp26) are selectively downregulated in R-, but not WWD-Dnmt3a2.
Figure 6.
The R-Dnmt3a2 mutant insensitive to mitotic H3T3ph causes chromosomal abnormalities.
A, ChIP-qPCR of H3T3ph, H3T6ph, and H3S10ph signals in repeat regions. Error bars represent s.e.m. of three experiments, *p < 0.05. B, Immunofluorescence of metaphase chromosomes reacted with antibodies against H3T3ph (green) and FLAG-tagged WT- or R-Dnmt3a2 (red). Arrows indicate pericentromeric and centromeric regions. Scale bar, 5 μm. C, Quantification of FLAG signal colocalized with H3T3ph at DAPI-dense regions (counts are based on 11~12 metaphase cells in individual clones). D, Representative karyotype images of WT- or R-Dnmt3a2-expressing ESCs. Asterisks in left “WT” and in right “R” mark a partial duplication on chromosome 8. Arrows in right “R” indicate Robertsonian fusion on chromosome 5 and trisomies on chromosome 11. E, Changes in chromosome numbers in ESCs expressing WT-, WWD-, or R-Dnmt3a2 (counts are based on 36~40 metaphase cells in individual clones). *p < 0.0001, Pearson’s Chi-squared test.
Given that R-Dnmt3a2 results in a defect in chromosome segregation genes and mis-localizes to mitotic centromeres, we next evaluated whether the R mutant exhibited signs of genomic instability. To examine the number and appearance of chromosomes, we conducted karyotype analyses with passage-matched ESCs (≤ 8 passages after establishing ESC lines). All ESCs possessed a partial duplication on passage 8 (asterisk, Figure 6D) resulting from an interstitial inverted duplication in the background line (TKO), and some loss of the Y chromosome (Figure 6E). Whereas the karyotypes of TKO, WT- and WWD-Dnmt3a2 seem relatively homogenous and stable (Figure 6D), we observed a number of chromosomal abnormalities in R-Dnmt3a2 (Table S3, related to Figure 6). Specifically, mitotic chromosome spreads from ESCs expressing R-Dnmt3a2 were heterogeneous and exhibited trisomies and Robertsonian fusions (see arrows in Figure 6D) indicative of centromeric dysfunction. These results support that in ESCs expressing R-Dnmt3a2, chromosome pairs fail to separate properly during mitosis.
DISCUSSION
Despite considerable progress, much is unknown concerning how the interplay between histone and DNA modifications affects the stable epigenetic states that underlie normal cellular differentiation, and whether aberrant links between these two distinct modes of epigenomic variation contribute to abnormal cellular and developmental outcomes. Using molecular engineering and genome-wide approaches, we show that the ADD domain of Dnmt3a is capable of sensing, and therefore integrating, the status of multiple H3 modifications (e.g. H3K4me3, H3T3ph, and H3T6ph) that, in turn, dictates the in vivo localization of full-length Dnmt3a2. To our knowledge, this study is one of the first to systematically engineer a “reader pocket” such that it redirects a downstream “writer” to new genomic loci with important biological downstream consequences.
In addition to the sensitivity of ADD3a to H3K4me3 and H3T3ph, its binding affinity for the unmodified H3 N-terminus is crucial for the in vivo distribution and function of Dnmt3a2. Our results suggest a double-guard model in which the ADD3a domain maintains the global distribution of Dnmt3a2: first, the higher affinity of ADD3a for the unmodified H3 N-terminus anchors Dnmt3a2 to large domains of inter/intragenic chromatin regions. Second, when it is somewhat mobile, wild-type ADD3a avoids H3K4me3/CGI maintaining relative enrichment in inter/intragenic regions. We envision that our engineered WWD mutant alters this balance such that its binding to the unmodified H3 N-terminus (0.82 μM in WT vs. 3.3 μM in WWD) is equal to its binding to H3K4me3 (35.6 μM in WT vs. 3.8 μM in WWD) (see Figure 2H, I), leading to a change in the overall localization of Dnmt3a2. Given that the size of inter/intragenic regions far exceeds that of H3K4me3/CGI, we suggest that wild-type and R mutant bind to the unmodified H3 N-terminus with higher affinity and so have less opportunity to relocate. Alterations in this balance, leading to even modest changes in de novo DNA methylation, may contribute to human pathologies, as mutations in ADD3a are found in numerous diseases such as acute myeloid leukemia (Yan et al., 2011) and overgrowth syndrome associated with intellectual disability (Tatton-Brown et al., 2014). To our knowledge, the exact mutations engineered in our study have not yet been associated with disease.
Previous studies have shown that the frequency of promoter CGI and its length correlate with the timing of developmental gene expression in ESCs. Specifically, genes preferentially expressed in early embryonic lineages tend to contain more CGI at their promoters than genes expressed at relatively late stages of development (Xie et al., 2013). Furthermore, somatic-tissue-specific promoters are marked by CpG-poor sequences (Schug et al., 2005). In line with this classification, our results show that WWD-Dnmt3a2 preferentially binds to a subset of promoter CGI with higher H3K4me3 and lower H3K27me3 enrichment at genes associated with early embryonic lineages (e.g., Msx2, Fgf5, Enc1, and Bmp3), resulting in modest “ectopic” DNA methylation and subsequent reduction in gene expression. With that said, we do not observe clear evidence of transcriptional silencing in highly expressed housekeeping genes, indicating that a modest level of newly acquired promoter hypermethylation may not be sufficient to repress genes that are maintained in a strongly expressed state.
Interestingly, we found that WWD-mediated hypermethylation of CpG at H3K4me3/CGI loci is generally less efficient (≤ 30%) than at intergenic regions (≥ 60%). This result suggests that H3K4me3/CGI loci are refractory to de novo Dnmt3a2 action, which is probably the result of a number of contributing factors including DNA demethylation (e.g., methyl-cytosine oxidation; Figure S4D) (Wu and Zhang, 2014), competition for binding with other regulatory proteins/complexes, and other Dnmts such as Dnmt3L. Dnmt3L, a catalytically inactive Dnmt3, is abundantly expressed in TKO and the established ESC lines. Dnmt3L is known to interact with Dnmt3a/3b (Jia et al., 2007; Jurkowska et al., 2011) as well as Dnmt3a2 (Ooi et al., 2007). Given that ADD3l binding to the H3 N-terminus is disrupted by H3K4me3 (Ooi et al., 2007) (Figure 1C) and Dnmt3L incorporation stimulates Dnmt3a2 enzymatic activity (Gowher et al., 2005; Kareta et al., 2006), Dnmt3L itself may facilitate WT-, WWD-, R-Dnmt3a2 binding/activity to unmodified H3, limiting WWD-Dnmt3a2 access to H3K4 methylation. Dnmt3L has also been reported to exist in a complex with polycomb repressive complex 2 (PRC2) at bivalent promoters enriched with PRC2, H3K27me3, and H3K4me3 (Neri et al., 2013). This may further limit access of the WWD-Dnmt3a2 at these bivalent loci.
ADD3a repels both H3K4 methylation and H3T3 phosphorylation using two separate surface regions, suggesting that these modifications act independently to influence DNA methylation patterns. Our genome-wide and functional results with engineered mutants provide strong evidence that these two H3 modifications instruct Dnmt3a2 during different cellular processes. H3T3ph exclusively occurs during mitosis and removes multiple reading modules that bind to the H3 N-terminus and H3K4me3 (Garske et al., 2010). H3T3ph attracts the chromosome passenger complex to mitotic centromeres (Kelly et al., 2010; Wang et al., 2010). Mitotic H3T3ph, as with other well studied mitotic phosphorylation sites (e.g., H3S10ph), serves as a general “off switch” for the ejection of key downstream chromatin enzymes and complexes (Oliver and Denu, 2011) that contribute to proper kinetochore function and faithful chromosomal segregation. Our results suggest that Dnmt3a2 should be added to the list of ejected proteins during mitosis, particularly at centromeric regions. In the H3T3ph-insensitive Dnmt3a2 mutant (R-Dnmt3a2), we report chromosomal abnormalities not exhibited by WT nor WWD (Figure 6D, E). We suggest that R-Dnmt3a2 is not properly “ejected” by a H3T3ph-driven mechanism, and that this removal is crucial for proper chromatin segregation. Interestingly, H3S10ph, which also abundantly occurs during mitosis, does not disrupt ADD (and PWWP-ADD) binding whereas H3T6ph did (not studied here due to our inability to design a binding mutant). H3K9me3 does not affect ADD binding (Ooi et al., 2007; Otani et al., 2009), so it is likely that ADD3a acts primarily to sense “front-end” H3T3 and H3K4 modifications.
In summary, guided by a structure-based approach, our engineering of the ADD domain of Dnmt3a provides informative insights into the role that histone modifications play in establishing downstream patterns of DNA methylation in ESCs. We demonstrate the biological significance of exquisite DNA methylation-histone links that are essential for proper differentiation and development, as well as chromosome stability and segregation. We anticipate that this approach will be broadly applicable for the elucidation of epigenetic networks altered by “reader” mutations, whether introduced artificially or naturally in human diseases.
EXPERIMENTAL PROCEDURES
Protein Preparation
The ADD domains encompassing residues 479-610 or 476-611 of human Dnmt3a were cloned into pGEX-6p-1 vector with an N-terminal GST tag. All mutant ADD3a were generated by the QuickChange site-directed mutagenesis kit (Stratagene) in the frame of Dnmt3a(476-611) and were verified by sequencing. The purification procedures of these mutant proteins were essentially the same as that of the wild type protein. The constructs were overexpressed in E. coli BL21 (DE3) strain (Novagen). After overnight induction with 0.2 mM isopropyl β-D-thiogalactoside at 16 °C in LB media, cells were harvested and disrupted by the EmulsiFlex-C3 homogenizer (Avestin). After centrifugation, the supernatant was loaded into GST affinity column. Bound proteins were eluted with 20 mM glutathione and cleaved using PreScission protease. The GST tag was removed by anion exchange chromatography on Q column, and the ADD3a domain protein was finally purified by size-exclusion chromatography on Hiload 16/60 Superdex 75 column (GE Healthcare) in 20 mM HEPES-Na pH 7.5, 100 mM NaCl. The peak fractions were concentrated to about 10-15 mg ml−1 for crystallization.
Peptide pull-down assays
Biotin-conjugated histone peptides were incubated overnight at 4 °C with High Capacity Streptavidin Agarose (Pierce) and washed five times to remove unbound peptides. For the pull-downs of the wild type and mutant Dnmt3a-ADD proteins, 10 μl of packed peptide-Streptavidin resins were incubated with 50 μg protein for 4 hours at 4 °C in buffer containing 20 mM Hepes pH 7.9, 150 mM KCl, 0.01% NP-40, 10% glycerol, 5mM BME, 0.4mM PMSF, and protease inhibitors (Roche). After extensive washing, proteins bound to the beads were eluted with buffer at pH 2.5, separated by SDS-PAGE, and examined by Coomassie blue staining.
WT/mutant Dnmt3a2 ESC generation and NPC differentiation
The coding sequence of Dnmt3a2 was amplified from pCMV6-Dnmt3a (MC200622, OriGene) and subcloned into pBluescriptKS. Plasmids encoding mutant Dnmt3a2 were made from pBluescriptKS-Dnmt3a2 by site-directed mutagenesis. Each plasmid construct was verified by DNA sequencing. WT and mutant Dnmt3a2 cDNAs were amplified by PCR from pBluescriptKS-Dnmt3a2 with the sense primer introducing a 3× FLAG and a XbaI site at its 5′end and the antisense primer a BamHI site. The reaction products were digested with the corresponding enzymes and ligated into the PiggyBac-EF1-ires-GFP vector (System Biosciences). To generate WT/mutant Dnmt3a2 ESCs, TKO ESCs were seeded on a 6-well plate at a density of 1×105 cells and the next day the cells were transfected using FugeneHD (Roche) with 2 μg of PiggyBac-EF1-Dnmt3a2-ires-GFP and 0.8 μg of PiggyBac transpose vector. GFP positive cells were selected after 3 days with BD FACSAriaII-2. After single cell or bulk sorting, ESCs were immediately plated onto feeders and expanded for further studies. To perform the subsequent experiments, feeders were removed and ESCs were cultured for at least two passages on gelatin-coated plates. ESCs were differentiated into the NPC through embryoid body formation for 4 days, followed by treatment with 5 μM retinoic acid for additional 4 days.
Detailed materials, crystallization, data collection and structure analysis, Isothermal titration calorimetry measurements, ChIP-seq, RRBS and RNA-seq data analysis and validation experiments are described in Supplementary Information.
Supplementary Material
Highlights.
A structure of the ADD3a reveals an amino acid sensitive to H3T3 phosphorylation.
Two ADD3a domain mutants allow binding to methylated H3K4 or phosphorylated H3T3.
The H3K4 methylation-binding mutant perturbs the ESC differentiation program.
The H3T3 phosphorylation-binding mutant leads to chromosomal instability.
ACKNOWLEDGMENTS
We thank M. Okano for Dnmt TKO ESCs. We thank members of the Allis laboratory for technical advice and discussion. We thank the staff at beamlines BL17U of the Shanghai Synchrotron Radiation Facility and 24ID-E of the Advanced Photon Source at the Argonne National Laboratory for their assistance in data collection. We also thank the staff at the Epigenomics Core Facility of Weill Cornell Medical College, the Rockefeller University Genomics Resource Center, the Bio-Imaging Resource Center, and the Flow Cytometry Resource Center. We also thank the staff at the Molecular Cytogenetics, the Molecular Cytology, and The Center of Comparative Medicine and Pathology of Memorial Sloan Kettering Cancer Center. This work is supported by a Women & Science Postdoctoral Fellowship (to KMN), a NIMH grant (PHS MH 094698, to CDA), a Leukemia Lymphoma Society Program Project Award (NORTHWESTERN-LLS 7006-13, to AMM, DJP and CDA), STARR Foundation Awards (I5-A554 and I5-A558 to DJP), program grants (The General Program of National Natural Science Foundation of China, 31270763, The Major State Basic Research Development Program in China, 2011CB965300 and Program for New Century Excellent Talents in University, to HL), and funds from the National Cancer Institute (SH). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does the mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
Structure data are deposited in the Protein Data Bank with the accession numbers of 4QBQ, 4QBR, and 4QBS for ADD3a(479-610)-H31-15, ADD3a(476-611)(G550D)-H31-10, and ADD3a(476-611)(E545R)-H31-10T3ph complexes, respectively. The complete genome-wide data are deposited in the Gene Expression Omnibus with the accession number of GSE57577.
AUTHOR CONTRIBUTIONS
K.M.N., H.L., and C.D.A. conceived of the project. K.M.N. and H.W. designed the experiments. K.M.N., H.W., and W.W. collected and analyzed the data. K.M.N., H.R.K., F.F., C.H.L., and S.D. performed the bioinformatics analyses. S.H.H., A.M.M., and D.J.P. provided essential resources and reagents. K.M.N., H.L., and C.D.A. wrote the manuscript.
REFERENCES
- Bibel M, Richter J, Lacroix E, Barde YA. Generation of a defined and uniform population of CNS progenitors and neurons from mouse embryonic stem cells. Nat. Protoc. 2007;2:1034–1043. doi: 10.1038/nprot.2007.147. [DOI] [PubMed] [Google Scholar]
- Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat. Rev. Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- Dai J, Sultan S, Taylor SS, Higgins JM. The kinase haspin is required for mitotic histone H3 Thr 3 phosphorylation and normal metaphase chromosome alignment. Genes Dev. 2005;19:472–488. doi: 10.1101/gad.1267105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards JR, O’Donnell AH, Rollins RA, Peckham HE, Lee C, Milekic MH, Chanrion B, Fu Y, Su T, Hibshoosh H, Gingrich JA, Haghighi F, Nutter R, Bestor TH. Chromatin and sequence features that define the fine and gross structure of genomic methylation patterns. Genome Res. 2010;20:972–980. doi: 10.1101/gr.101535.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle W, Tseng BS, Dormann HL, Ueberheide BM, Garcia BA, Shabanowitz J, Hunt DF, Funabiki H, Allis CD. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature. 2005;438:1116–1122. doi: 10.1038/nature04219. [DOI] [PubMed] [Google Scholar]
- Fischle W, Wang Y, Allis CD. Binary switches and modification cassettes in histone biology and beyond. Nature. 2003;425:475–479. doi: 10.1038/nature02017. [DOI] [PubMed] [Google Scholar]
- Garske AL, Oliver SS, Wagner EK, Musselman CA, LeRoy G, Garcia BA, Kutateladze TG, Denu JM. Combinatorial profiling of chromatin binding modules reveals multisite discrimination. Nat. Chem. Biol. 2010;6:283–290. doi: 10.1038/nchembio.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford CA, Ziller MJ, Gu H, Trapnell C, Donaghey J, Tsankov A, Shalek AK, Kelley DR, Shishkin AA, Issner R, Zhang X, Coyne M, Fostel JL, Holmes L, Meldrim J, Guttman M, Epstein C, Park H, Kohlbacher O, Rinn J, Gnirke A, Lander ES, Bernstein BE, Meissner A. Transcriptional and Epigenetic Dynamics during Specification of Human Embryonic Stem Cells. Cell. 2013;153:1149–1163. doi: 10.1016/j.cell.2013.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowher H, Liebert K, Hermann A, Xu G, Jeltsch A. Mechanism of stimulation of catalytic activity of Dnmt3A and Dnmt3B DNA-(cytosine-C5)-methyltransferases by Dnmt3L. J. Biol. Chem. 2005;280:13341–13348. doi: 10.1074/jbc.M413412200. [DOI] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh CL. In vivo activity of murine de novo methyltransferases, Dnmt3a and Dnmt3b. Mol. Cell Biol. 1999;19:8211–8218. doi: 10.1128/mcb.19.12.8211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson JP, Lindroth AM, Cao X, Jacobsen SE. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature. 2002;416:556–560. doi: 10.1038/nature731. [DOI] [PubMed] [Google Scholar]
- Jia D, Jurkowska RZ, Zhang X, Jeltsch A, Cheng X. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature. 2007;449:248–251. doi: 10.1038/nature06146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, Strouboulis J, Wolffe AP. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat. Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- Jurkowska RZ, Rajavelu A, Anspach N, Urbanke C, Jankevicius G, Ragozin S, Nellen W, Jeltsch A. Oligomerization and binding of the Dnmt3a DNA methyltransferase to parallel DNA molecules: heterochromatic localization and role of Dnmt3L. J. Biol. Chem. 2011;286:24200–24207. doi: 10.1074/jbc.M111.254987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kareta MS, Botello ZM, Ennis JJ, Chou C, Chedin F. Reconstitution and mechanism of the stimulation of de novo methylation by human DNMT3L. J. Biol. Chem. 2006;281:25893–25902. doi: 10.1074/jbc.M603140200. [DOI] [PubMed] [Google Scholar]
- Kelly AE, Ghenoiu C, Xue JZ, Zierhut C, Kimura H, Funabiki H. Survivin reads phosphorylated histone H3 threonine 3 to activate the mitotic kinase Aurora B. Science. 2010;330:235–239. doi: 10.1126/science.1189505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnertz B, Ueda Y, Derijck AA, Braunschweig U, Perez-Burgos L, Kubicek S, Chen T, Li E, Jenuwein T, Peters AH. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 2003;13:1192–1200. doi: 10.1016/s0960-9822(03)00432-9. [DOI] [PubMed] [Google Scholar]
- Lei H, Oh SP, Okano M, Juttermann R, Goss KA, Jaenisch R, Li E. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development. 1996;122:3195–3205. doi: 10.1242/dev.122.10.3195. [DOI] [PubMed] [Google Scholar]
- Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, Patel DJ. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442:91–95. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, Edsall L, Antosiewicz-Bourget J, Stewart R, Ruotti V, Millar AH, Thomson JA, Ren B, Ecker JR. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markaki Y, Christogianni A, Politou AS, Georgatos SD. Phosphorylation of histone H3 at Thr3 is part of a combinatorial pattern that marks and configures mitotic chromatin. J. Cell Sci. 2009;122:2809–2819. doi: 10.1242/jcs.043810. [DOI] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, Gnirke A, Jaenisch R, Lander ES. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger E, Imhof A, Patel D, Kahl P, Hoffmeyer K, Friedrichs N, Muller JM, Greschik H, Kirfel J, Ji S, Kunowska N, Beisenherz-Huss C, Gunther T, Buettner R, Schule R. Phosphorylation of histone H3T6 by PKCbeta(I) controls demethylation at histone H3K4. Nature. 2010;464:792–796. doi: 10.1038/nature08839. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, Lee W, Mendenhall E, O’Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C, Lander ES, Bernstein BE. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- Neri F, Krepelova A, Incarnato D, Maldotti M, Parlato C, Galvagni F, Matarese F, Stunnenberg HG, Oliviero S. Dnmt3L antagonizes DNA methylation at bivalent promoters and favors DNA methylation at gene bodies in ESCs. Cell. 2013;155:121–134. doi: 10.1016/j.cell.2013.08.056. [DOI] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Oliver SS, Denu JM. Dynamic interplay between histone H3 modifications and protein interpreters: emerging evidence for a “histone language”. Chembiochem. 2011;12:299–307. doi: 10.1002/cbic.201000474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, Cheng X, Bestor TH. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otani J, Nankumo T, Arita K, Inamoto S, Ariyoshi M, Shirakawa M. Structural basis for recognition of H3K4 methylation status by the DNA methyltransferase 3A ATRX-DNMT3-DNMT3L domain. EMBO Rep. 2009;10:1235–1241. doi: 10.1038/embor.2009.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schug J, Schuller WP, Kappen C, Salbaum JM, Bucan M, Stoeckert CJ., Jr. Promoter features related to tissue specificity as measured by Shannon entropy. Genome Biol. 2005;6:R33. doi: 10.1186/gb-2005-6-4-r33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, Ren B. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat. Rev. Genet. 2013;14:204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- Tamaru H, Selker EU. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature. 2001;414:277–283. doi: 10.1038/35104508. [DOI] [PubMed] [Google Scholar]
- Tatton-Brown K, Seal S, Ruark E, Harmer J, Ramsay E, Del Vecchio DS, Zachariou A, Hanks S, O’Brien E, Aksglaede L, Baralle D, Dabir T, Gener B, Goudie D, Homfray T, Kumar A, Pilz DT, Selicorni A, Temple IK, Van ML, Yachelevich N, van MR, Rahman N. Mutations in the DNA methyltransferase gene DNMT3A cause an overgrowth syndrome with intellectual disability. Nat. Genet. 2014;46:385–388. doi: 10.1038/ng.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JP, Skene PJ, Selfridge J, Clouaire T, Guy J, Webb S, Kerr AR, Deaton A, Andrews R, James KD, Turner DJ, Illingworth R, Bird A. CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature. 2010;464:1082–1086. doi: 10.1038/nature08924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsumura A, Hayakawa T, Kumaki Y, Takebayashi S, Sakaue M, Matsuoka C, Shimotohno K, Ishikawa F, Li E, Ueda HR, Nakayama J, Okano M. Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes Cells. 2006;11:805–814. doi: 10.1111/j.1365-2443.2006.00984.x. [DOI] [PubMed] [Google Scholar]
- Wang F, Dai J, Daum JR, Niedzialkowska E, Banerjee B, Stukenberg PT, Gorbsky GJ, Higgins JM. Histone H3 Thr-3 phosphorylation by Haspin positions Aurora B at centromeres in mitosis. Science. 2010;330:231–235. doi: 10.1126/science.1189435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Lin C, Lu D, Ning Z, Cox T, Melvin D, Wang X, Bradley A, Liu P. Chromosomal transposition of PiggyBac in mouse embryonic stem cells. Proc. Natl. Acad. Sci. U. S. A. 2008;105:9290–9295. doi: 10.1073/pnas.0801017105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Coskun V, Tao J, Xie W, Ge W, Yoshikawa K, Li E, Zhang Y, Sun YE. Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science. 2010;329:444–448. doi: 10.1126/science.1190485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Zhang Y. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell. 2014;156:45–68. doi: 10.1016/j.cell.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, Wu C, Allis CD. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- Wyszynski MW, Gabbara S, Bhagwat AS. Substitutions of a cysteine conserved among DNA cytosine methylases result in a variety of phenotypes. Nucleic Acids Res. 1992;20:319–326. doi: 10.1093/nar/20.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Schultz MD, Lister R, Hou Z, Rajagopal N, Ray P, Whitaker JW, Tian S, Hawkins RD, Leung D, Yang H, Wang T, Lee AY, Swanson SA, Zhang J, Zhu Y, Kim A, Nery JR, Urich MA, Kuan S, Yen CA, Klugman S, Yu P, Suknuntha K, Propson NE, Chen H, Edsall LE, Wagner U, Li Y, Ye Z, Kulkarni A, Xuan Z, Chung WY, Chi NC, Antosiewicz-Bourget JE, Slukvin I, Stewart R, Zhang MQ, Wang W, Thomson JA, Ecker JR, Ren B. Epigenomic Analysis of Multilineage Differentiation of Human Embryonic Stem Cells. Cell. 2013;153:1134–1148. doi: 10.1016/j.cell.2013.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi Y, Honda T, Tanno Y, Watanabe Y. Two histone marks establish the inner centromere and chromosome bi-orientation. Science. 2010;330:239–243. doi: 10.1126/science.1194498. [DOI] [PubMed] [Google Scholar]
- Yan XJ, Xu J, Gu ZH, Pan CM, Lu G, Shen Y, Shi JY, Zhu YM, Tang L, Zhang XW, Liang WX, Mi JQ, Song HD, Li KQ, Chen Z, Chen SJ. Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nat. Genet. 2011;43:309–315. doi: 10.1038/ng.788. [DOI] [PubMed] [Google Scholar]
- Young RA. Control of the embryonic stem cell state. Cell. 2011;144:940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Jurkowska R, Soeroes S, Rajavelu A, Dhayalan A, Bock I, Rathert P, Brandt O, Reinhardt R, Fischle W, Jeltsch A. Chromatin methylation activity of Dnmt3a and Dnmt3a/3L is guided by interaction of the ADD domain with the histone H3 tail. Nucleic Acids Res. 2010;38:4246–4253. doi: 10.1093/nar/gkq147. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.