Abstract
Background
Porphyromonas gingivalis inhibits oral epithelial wound healing in vitro more strongly than other oral bacteria, but it is unknown why P. gingivalis is such a potent inhibitor of wound healing.
Objective
Therefore, the aim of this study was to investigate the influence of major virulence factors of P. gingivalis on wound healing in an in vitro wound-healing model. The influence of the capsular polysaccharide, the Arg- and Lys- gingipains, the major fimbriae and lipopolysaccharide (LPS) was investigated.
Design
A standardized scratch was made in a confluent layer of human oral epithelial cells HO-1-N-1. The epithelial cells were then challenged with different concentrations of several P. gingivalis wild-type strains and knockout mutants. Closure of the scratch was determined after 17 h and compared to control conditions without bacteria.
Results
The P. gingivalis strains ATCC 33277, W83, and W50 significantly inhibited wound healing. The presence of a capsular polysaccharide lowered significantly the inhibition of epithelial cell migration, while gingipain activity significantly increased the inhibition of cell migration. LPS and the major fimbriae did not influence epithelial cell migration. None of the tested P. gingivalis strains completely prevented the inhibition of cell migration, suggesting that other characteristics of P. gingivalis also play a role in the inhibition of wound healing, and that further research is needed.
Conclusions
The capsular polysaccharide and the Arg- and Lys- gingipains of P. gingivalis influenced the capacity of P. gingivalis to hinder wound healing, while LPS and the major fimbriae had no effect.
Keywords: gingipains, capsular polysaccharide, P. gingivalis, LPS, cell migration essay, oral epithelial cells, re-epithelialization
In hematopoietic stem cell transplant patients, the oral microbiota is traditionally assumed to colonize existing ulcerative lesions rather than to cause them (1). However, recently Laheij and de Soet (2) and Stringer and Logan (3) suggested a more important role for oral microorganisms in oral mucositis. Oral microorganisms are likely able to contribute to the development of oral mucositis by a shared pathway and may prolong the existence of established ulcerations by delaying their healing.
Recently, two studies described the negative influence of oral bacteria on the healing of wounds, using an in vitro scratch assay (4, 5). Laheij et al. (5) studied the influence of Porphyromonas gingivalis, Prevotella nigrescens, Prevotella intermedia, Tannerella forsythia, and Streptococcus mitis in different concentrations on the closure of a scratch in epithelial cells from an oral epithelial cell line. Bhattacharya et al. (4) studied the effect of P. gingivalis and Fusobacterium nucleatum on wound closure and on epithelial cell migration, proliferation, and apoptosis in primary epithelial cells. In both studies, P. gingivalis was a potent inhibitor of re-epithelialization of the scratch.
The mechanisms by which P. gingivalis acts as a strong inhibitor of wound closure have not been studied so far. It is known that P. gingivalis has a range of virulence factors that enable it to invade into host tissues and to deregulate the immune system to promote survival inside the host (6). Important virulence factors of P. gingivalis are lipopolysaccharide (LPS), gingipain proteinases, the capsular polysaccharide, and fimbriae (7). These virulence factors are involved in the host–pathogen recognition (LPS) (8), they trigger intracellular signaling events and an immune response (LPS, gingipains) (6), they are involved in binding of P. gingivalis to host cells (gingipains, fimbriae) (6, 9), and they are involved in invasive growth (capsular polysaccharide) (10).
It was hypothesized that these virulence factors could account for the inhibiting effect that P. gingivalis had on wound healing. Therefore, the aim of this study was to investigate the role of these virulence factors of P. gingivalis on wound closure by using natural variants and mutant strains in an in vitro scratch assay using oral epithelial cells.
Materials and methods
Cell culture
A human oral buccal epithelial cell line HO-1-N-1 was provided by the Japanese Collection of Research Bioresources (Osaka, Japan). HO-1-N-1 is an immortalized epithelial cell-like cell line that originated from a squamous cell carcinoma located on the buccal mucosa. The epithelial cells were grown in tissue culture flasks with Dulbecco's modified Eagle's medium-F12 (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal calf serum (Hyclone, Logan, UT, USA), 2% antibiotics (penicillin, streptomycin and fungizone, Sigma, St. Louis, MO, USA) in a humidified atmosphere at 37°C with 5% CO2. Cell culture medium was refreshed twice a week.
Bacterial strains and cultures
The P. gingivalis strains are listed in Table 1. All P. gingivalis strains were grown anaerobically in Brain–Heart-Infusion (BD Difco, Le Pont de Claix, France) broth enriched with hemin (5 mg/L) and menadione (1 mg/L) until mid-log phase. In case of mutant selection and growth, 50 µg/ml of erythromycin (Em) or 1 µg/ml tetracycline was added to the medium. The purity of the cultures was checked by Gram staining and culturing. Bacterial cultures were washed twice with Dulbecco's phosphate-buffered saline (DPBS, Invitrogen) and resuspended in keratinocyte serum-free medium (SFM; Invitrogen) at OD690 0.1 (5×108 CFU/ml). Experiments were performed with either viable or heat-inactivated bacteria, in which case the bacterial cultures were heated at 60°C for 60 min. The absence of growth on blood agar plates confirmed the killing of the bacteria.
Table 1.
P. gingivalis strains used in this study
| Strains | Properties | Source |
|---|---|---|
| W83 (wild type) ΔEpSC |
Capsular serotype 1 (K1, thick capsule) Non-encapsulated (K−) |
(12) (11) |
| ATCC 33277 (wild type) KDP 129 KDP 133 FimA |
Non-encapsulated (K−) Lacking lysine gingipain (Δkgp) Lacking arginine gingipain (Δrgp ArgpB) Lacking major fimbriae (ΔfimA) |
(13) (13) This paper, S1 |
| W50 (wild type) K1A E8 |
Capsular serotype 1 (K1, thick capsule) Lacking lysine gingipain (Δkgp) Lacking arginine gingipain (Δrgp ArgpB) |
(12)
(14) (14) |
| HG91 | Non-encapsulated (K−) | (12) |
| ATCC49417 | Capsular serotype 4 (K4, thin capsule) | |
| HG1691 | Capsular serotype (K6, thick capsule) | Own isolate |
Lipopolysaccharide
The influence of LPS from P. gingivalis was tested by adding pure LPS from P. gingivalis to the epithelial cells or by blocking LPS in the cell wall of P. gingivalis. Ultrapure LPS from P. gingivalis was purchased (InvivoGen, Toulouse, France) and used in the assay in a concentration of 10 µg/ml. To exclude the influence of LPS, polymyxin B (Sigma–Aldrich, St. Louis, MO), an LPS inhibitor, was added to 5×108 CFU/ml heat-inactivated P. gingivalis ATCC 33277 in a concentration of 50 µg/ml.
Gingipains
Besides using gingipain mutants from two wild-type strains, the influence of gingipain activity was also studied by using gingipain inhibitors. KYT-1 (an arginine gingipain inhibitor) and KYT-36 (a lysine gingipain inhibitor) peptides (15) (Peptide Institute, Osaka, Japan) were added in the assay to viable P. gingivalis W50 at a concentration of 1×10−6 mol/ml.
In vitro wound closure assay
Scratch assays were performed as previously described (5). Cell cultures were trypsinized and seeded in 24-well plates at 3×105 to 5×105 cells/ml. When confluency was reached, a scratch with a sterile 1-ml pipet point was made in each well. The wells were washed three times with DPBS and challenged with viable or heat-inactivated bacteria or LPS from P. gingivalis. In control wells, only SFM was added. Bacteria were added at a multiplicity of infection (MOI) of 1,000 (1,000 bacteria vs. 1 epithelial cell), 100, and 10. Most P. gingivalis cells remained viable and metabolically active until the end of the experiment. Images were taken of the middle of the well immediately after adding the bacteria and 17 h later with an inverted digital phase-contrast microscope (Advanced Microscopy Group, Evos, FL). The surface of the scratch was calculated with Photoshop CS4 (version 11.0.1, Adobe). After 17 h, closure of the wound under control conditions varied between 60 and 80%. In addition, several images were taken to analyze migration of epithelial cells over time and to construct a time-lapse movie (Supplemental file S2).
The percentage of closure of the scratch was calculated as: 100 – [(surface area of the scratch at time 17 h/surface area of the scratch at time 0)×100]. To account for differences in wound closure between experiments, the closure of the scratch relative to control (relative closure) was calculated by dividing the percentage of closure of the scratch with the average percentage of closure of the scratch under control conditions (SFM)×100%. Relative closure between 1 and 100% indicated inhibition of wound closure, with 0% indicating complete inhibition. Each treatment was performed in five different wells per experiment, and each experiment was completed on three separate occasions.
Statistics
Non-parametric tests were performed since not all assumptions for parametric testing were met. Differences in relative closure between the different MOIs of specific bacterial strains and control were tested for significance with the Kruskal–Wallis test (α=0.05). Mann–Whitney U tests were performed as post hoc tests, and α was corrected for multiple testing. Since three treatments were compared, and the number of pairwise comparisons was three, α for the Mann–Whitney U-tests was set at 0.05/3=0.0167. Differences between different MOIs of the same strain were tested with the Wilcoxon signed rank test (α=0.05). SPSS version 21.0 (IBM SPSS, Chicago, IL) was used to perform the analyses. Generally, a p-value<0.05 was considered statistically significant.
Results
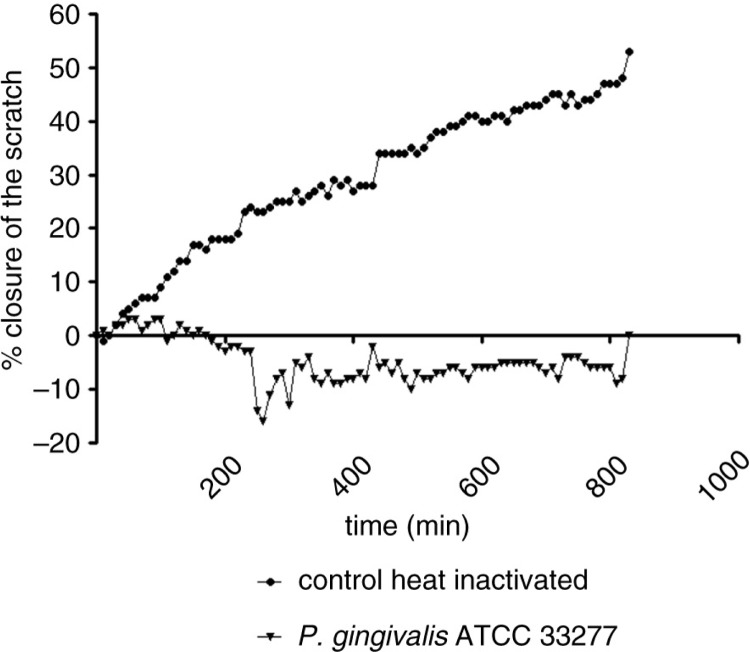
Figure 1 shows the percentage of closure of a scratch in oral epithelial cells challenged with a high concentration of P. gingivalis ATCC 33277 (MOI 1,000) or with medium only (control) in time. Closure of the scratch under control conditions starts almost immediately, and closure in time follows a linear pattern. A time-lapse movie of that experiment can be found in Supplemental file S2.
Fig. 1.
Percentage closure of a scratch in oral epithelial cells challenged with P. gingivalis ATCC 33277 and medium only.
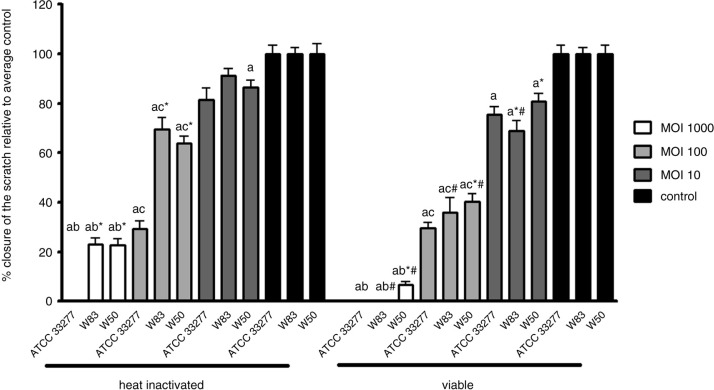
The heat-inactivated P. gingivalis strains ATCC 33277, W83, and W50 significantly inhibited the migration of oral epithelial cells at all tested MOIs compared to control in a dose-responsive manner, although the inhibition by the heat-inactivated W83 at MOI 10 did not reach statistical significance (Fig. 2). Viable P. gingivalis ATCC 33277, W83, and W50 significantly inhibited the migration of oral epithelial cells at all tested MOIs compared to control in a dose-responsive manner as well (Fig. 2). Viable W83 and W50 inhibited cell migration more than their heat-inactivated variants (not statistically significant for W50 at MOI 10); however, no differences were found between heat-inactivated and viable P. gingivalis ATCC 33277 (p>0.05).
Fig. 2.
Mean relative closure (±SEM) from all biological replicates of scratches in oral epithelial cells challenged with heat-inactivated and viable P. gingivalis strains ATCC 33277, W83, and W50. a=significantly different from control (p<0.0083). b=significantly different from MOI 100 of the same strain (p<0.0083). c=significantly different from MOI 1,000 of the same strain (p<0.0083). *=significantly different from ATCC 33277 of the same MOI (p<0.05). #=significantly different from heat-inactivated P. gingivalis of the same MOI and strain (p<0.05).
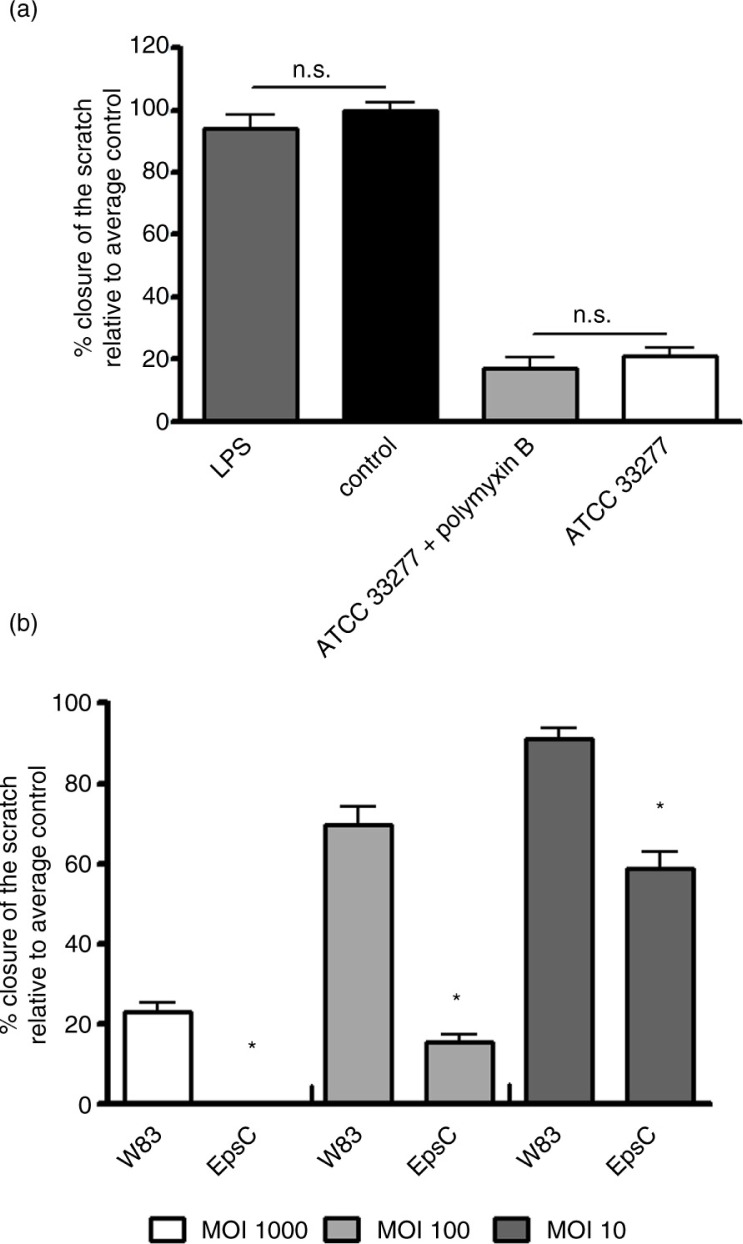
Adding ultrapure LPS from P. gingivalis to the oral epithelial cells did not result in the inhibition of the migration of the epithelial cells (p=0.39; Fig. 3a). When P. gingivalis LPS was blocked by the addition of polymyxin B, it did not affect relative closure of the scratch in epithelial cells compared to P. gingivalis alone (p=0.38; Fig. 3a).
Fig. 3.
Mean relative closure (±SEM) from all biological replicates of scratches in oral epithelial cells challenged with LPS from P. gingivalis and LPS inactivated P. gingivalis by polymyxin B (a) and heat-inactivated P. gingivalis strains W83 and EpsC mutant (b). *p<0.01.
The capsule lacking (non-encapsulated) P. gingivalis mutant EpsC inhibited cell migration significantly more than the encapsulated wild-type strain W83 (Fig. 3b). Relative closure of the scratch in oral epithelial cells challenged with heat-inactivated P. gingivalis strains of different capsular serotypes is shown in Table 2. Not all capsular serotypes inhibited wound healing to the same extent (all MOIs, p<0.002). The non-encapsulated strains (ATCC 33277 and HG91) inhibited epithelial cell migration significantly more than the K1 and K6 capsular serotypes at all MOIs (p<0.002), except for MOI 10 of strain HG91 versus ATCC 33277. The relative closure of the K4 capsular serotype was close to that of the K− strain ATCC 33277. The K4 strain inhibited migration of oral epithelial cells significantly more than the K1 and K6 capsular serotypes at all MOIs (p<0.001).
Table 2.
Relative closure (mean±SEM) of scratch in oral epithelial cells challenged with heat-inactivated P. gingivalis strains with several capsular serotypes and a fimbriae-lacking mutant
| Strains | MOI 1,000 | MOI 100 | MOI 10 |
|---|---|---|---|
| ATCC 33277 (K−) | 0% | 29% (±3) | 81% (±5) |
| HG91 (K−) | 0% | 11% (±1) | 69% (±3) |
| W83 (K1) | 23% (±3) | 70% (±5) | 91% (±3) |
| ATCC49417 (K4) | 0% | 30% (±5) | 72% (±6) |
| HG1691 (K6) | 17% (±2) | 48% (±4) | |
| Heat-inactivated FimA mutant | 7% (±6) | 38% (±10) | 85% (±6) |
| Viable FimA mutant | 0% | 23% (±2) | 75% (±5) |
K−, K1, K4, and K6 strains all significantly inhibited epithelial cell migration (p=0.001). ATCC 33277 and HG91 inhibited epithelial cell migration significantly more than K1 and K6 (p<0.002). K4 inhibited epithelial cell migration significantly more than K1 and K6 (p<0.001), K4 inhibited epithelial cell migration significantly less than HG91 but not ATCC 33277.
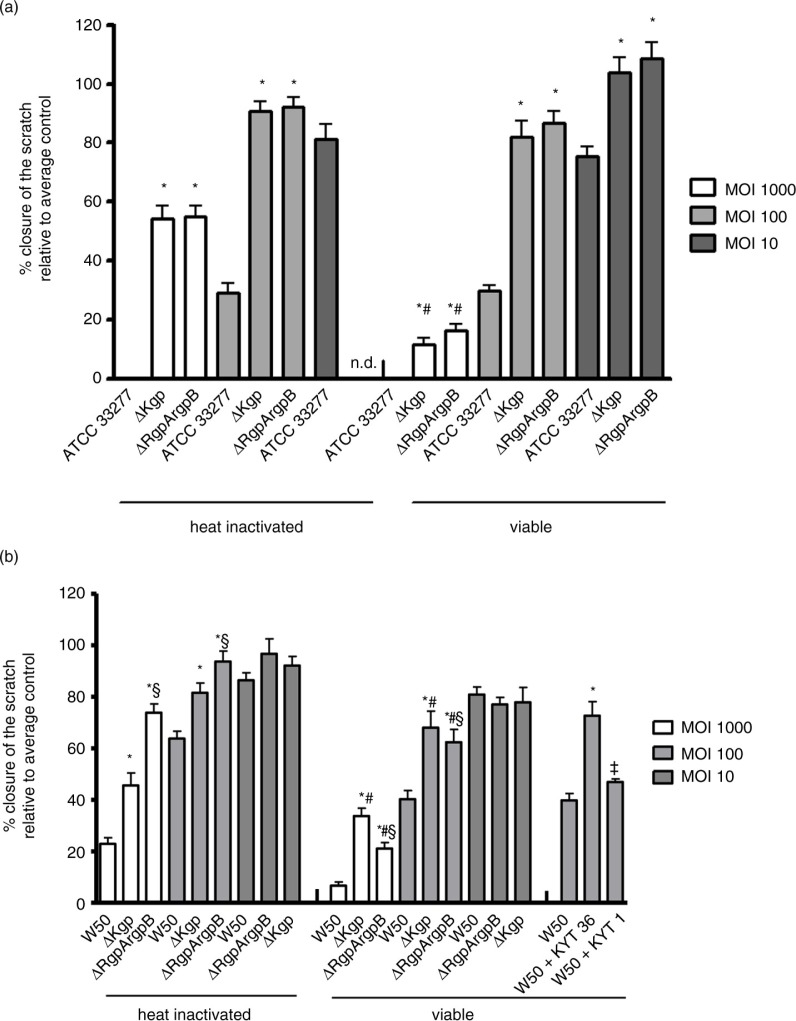
The influence of protease activity was tested using gingipain mutants constructed from both the ATCC 33277 and the W50 strains. For strain ATCC 33277, migration of oral epithelial cells was significantly more inhibited by the wild-type strain than by the both gingipains mutants (Fig. 4a). There was no significant difference in relative closure between the kgp and rgpArgpB mutants. The viable gingipain mutants inhibited wound closure more strongly than the heat-inactivated variants at MOI 1,000 (Fig. 4a). For the W50 strain, it was also found that the gingipain mutants, either viable or heat inactivated, inhibited the epithelial cell migration significantly less than the wild-type strain (Fig. 4b). The viable gingipain mutants significantly inhibited wound closure more than the heat-inactivated variants at MOI 1,000 and MOI 100 (Fig. 3b). The viable rgpArgpB mutant led to more inhibition of wound closure than the kgp mutant at MOI 1,000 and MOI 100 (Fig. 4b). Adding the gingipain inhibitors KYT-1 or KYT-36 to viable P. gingivalis W50 led to significantly less inhibition of wound closure compared with the wild type (Fig. 4b). Wound closure was more inhibited by the rgpArgpB gene compared with the Kgp gene, as shown by the results from the mutant strain and the KYT inhibitors. Moreover, heat inactivation of the gingipain mutants resulted in a higher inhibition by the kgp mutant compared with the rgpArgpB mutant.
Fig. 4.
Mean relative closure (±SEM) from all biological replicates of scratches in oral epithelial cells challenged with heat inactivated and viable P. gingivalis wild type strain ATCC 33277, ΔKgp, and ΔRgpArgpB mutants (a) and heat-inactivated and viable P. gingivalis wild-type strain W50, ΔKgp, and ΔRgpArgpB mutants and with addition of KYT-36 and KYT-1 peptides (b). *=significantly different from wild-type strain of the same MOI (p<0.001). ‡=significantly different from wild-type strain of the same MOI (p<0.05). #=significantly different from heat-inactivated P. gingivalis of the same MOI and strain (p<0.05). §=significantly different from the ΔKgp mutant (p<0.05).
Viable FimA mutants, lacking the major fimbriae, did not differ in inhibiting epithelial cell migration compared to the wild-type strain (MOI 100: p=0.10, MOI 10: p=0.627). The heat-inactivated FimA mutant did inhibit epithelial cell migration significantly less than the wild type at high concentrations (Table 2; MOI 1,000: p<0.01, MOI 100: p<0.05, MOI 10: p=0.575), although the difference was very small.
Discussion
Previous research described the effect of P. gingivalis on cell migration and wound healing, finding a significant inhibiting effect of P. gingivalis. The responsible virulence factors, however, were not determined (4, 5, 16, 17). By comparing different naturally occurring variants and mutant strains of P. gingivalis, major virulence factors of P. gingivalis were studied in an in vitro wound-healing model to find out which of these affect wound healing. In the in vitro wound-healing model that was used, the migration of cells is studied, which is an important step in wound healing (18, 19).
Epithelial cell migration under control conditions was a linear process, and the model used was successfully applied by Oudhoff et al. (20) among others to show other wound healing processes. Others also found that in vivo wound healing was a linear process (21). Measuring extra time points could, however, give extra information about the relationship between bacteria and bacterial products and epithelial cell migration. These measurements could be incorporated in future experiments.
All of the P. gingivalis strains that we used in this study inhibited migration of oral epithelial cells. The difference between viable and heat-inactivated P. gingivalis W83 and W50 suggests that there were heat-resistant and heat-sensitive factors of P. gingivalis that influenced migration of oral epithelial cells. The heat-resistant virulence factors of P. gingivalis that were expected to account for this effect were LPS, the capsular polysaccharide, and the fimbriae, whereas the heat-sensitive factors could be the gingipains (22). We found that the capsular polysaccharide and the kpg and rgpArgpB genes influenced migration of oral epithelial cells significantly, whereas LPS and the major fimbriae did not influence migration of oral epithelial cells at all.
Until now, only the effect of the capsule on the immune system and on human cells has been described. Both stimulating and inhibiting effects were reported (11, 23, 24). In the present experiments, the non-encapsulated bacteria inhibited wound closure more strongly than the encapsulated bacteria. Not only was the presence of the capsule important, but the thickness of the capsule as well. The K4 serotype has a relatively thin capsule, while the K1 and the K6 serotypes have significantly thicker capsules (25). The K4 P. gingivalis strain had a relative closure rate that was in between that of the K− and the K1–K6 serotypes. These findings suggest that a capsule may more or less depend on its thickness to hide cell-wall bound factors that might influence cell migration such as protease activity of the gingipains or other, yet unknown antigens on the cell wall of P. gingivalis.
Gingipains are important for the adherence of P. gingivalis to epithelial cells (9). The absence of specific protease activity of the gingipain mutants used in this study was confirmed using the method described by Kaman et al. (26) (data not shown). Moreover, the inhibition of protease activity by specific inhibitors gave similar results. The protease activity of the gingipains of P. gingivalis was an important determinant of epithelial cell migration. Knocking out and inhibiting gingipain activity led to improved wound closure, suggesting that gingipain activity inhibited cell migration.
The gingipains account for 85% of the protease activity of P. gingivalis (27). Furthermore, in the RgpArgpB mutant the kgp gene is still functional, and in the Kgp mutant the RgpArgpB gene is still functional. Thus, these viable gingipain mutants still possessed some protease activity, while the heat treatment inactivated almost all protease activities (22). Furthermore, gingipains have a function in the modification of several cell wall components (28). Therefore, P. gingivalis that lacks arginine or lysine gingipain activity may have a slightly different cell wall composition compared to the wild type, which may lead to differences in attachment to or recognition by the epithelial cells. This effect would have been more pronounced in the experiments with the viable bacteria, which may contribute to the differences in relative closure between experiments with heat-inactivated and viable bacteria found in this study.
LPS did not influence epithelial cell migration, and our results are in agreement with those of Kraus et al. (29). However, many previous studies have shown that LPS could stimulate cell migration through activation of the TLR4/NF-κB signal pathway. We noticed that in most of those studies, Escherichia coli LPS was tested. Kocgozlu et al. (30) showed that LPS from P. gingivalis was different from many other Gram-negative bacteria. It used TLR2 to activate epithelial cells and TLR4 to activate endothelial cells. This finding may explain Kraus’ observations and ours. Moreover, LPS from P. gingivalis is known to contain numerous lipid A species (31, 32). The different lipid A structures could differentially modulate host innate immune responses in human gingival epithelia (32) and perhaps wound-healing processes. It is worthwhile for studying further, even though LPS did not appear to be a major virulence factor in this study.
The fimbriae are important for the adherence of P. gingivalis to epithelial cells (6), and the inhibition of the fimbriae on wound healing was described by Nakagawa et al. (33). Therefore, our hypothesis was that the absence of major fimbriae would lead to less inhibition of wound healing. In our experiments, however, we proved that the fimA gene was not involved in migration of oral epithelial cells because we found no difference in relative closure between the viable wild-type strain and the fimA mutant. When epithelial cells were challenged with heat-inactivated bacteria, we did find significantly less inhibition of wound healing of the fimA mutant strain compared to the wild type. Because the difference was small, it may not be biologically relevant. We could not confirm the importance of fimbriae in the inhibition of migration of oral epithelial cells as described by Nakagawa et al. (33). This might be explained by the difference in the cell lines that were used. Nakagawa et al. used HeLa cells of cervical epithelial origin. HeLa cells may react differently on a challenge with P. gingivalis than our oral epithelial cell line. Moreover, the amount of wound closure Nakagawa et al. reported was generally lower.
Because there was an inhibition in migration of oral epithelial cells even in the presence of a capsule and the absence of either the Arg- or Lys- gingipains, we hypothesize that there must be other, yet unknown, antigens on the cell wall that are responsible for the inhibiting effect of P. gingivalis. This is supported by the finding that there is no difference in the percentage of wound closure between heat-inactivated and viable P. gingivalis ATCC 33277. In this strain, it must be a heat-resistant factor that exceeds the influence of the gingipains. Another possibility is that a combination of virulence factors, the ones that were tested in this research or other yet unknown factors, have a synergistic effect in explaining the inhibiting effect on the migration of oral epithelial cells.
In conclusion, the P. gingivalis strains ATCC 33277, W83, and W50 strongly inhibited the migration of oral epithelial cells. Part of that effect was accounted for by the absence of a capsular polysaccharide exposing cell wall bound virulence factors and Arg- and Lys- gingipain activity. LPS and the major fimbriae did not influence migration of oral epithelial cells. Because not all inhibition could be explained by the absence of capsular polysaccharide or the Arg- and Lys- gingipains, other yet unknown characteristics of P. gingivalis must have contributed to this effect, making further research necessary.
Supplementary Material
Acknowledgements
We thank Dr. F. Bikker for providing the FRET assay for testing gingipain activity and Dr. J. Aduse-Opoku for providing the P. gingivalis E8 and K1A strains.
Conflict of interest and funding
All authors declare no conflict of interest.
References
- 1.Sonis ST. Mucositis: the impact, biology and therapeutic opportunities of oral mucositis. Oral Oncol. 2009;45:1015–20. doi: 10.1016/j.oraloncology.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 2.Laheij AM, de Soet JJ. Can the oral microflora affect oral ulcerative mucositis? Curr Opin Support Palliat Care. 2014;8:180–7. doi: 10.1097/SPC.0000000000000053. [DOI] [PubMed] [Google Scholar]
- 3.Stringer AM, Logan RM. The role of oral flora in the development of chemotherapy-induced oral mucositis. J Oral Pathol Med. 2014;44:81–7. doi: 10.1111/jop.12152. [DOI] [PubMed] [Google Scholar]
- 4.Bhattacharya R, Xu F, Dong G, Li S, Tian C, Ponugoti B, et al. Effect of bacteria on the wound healing behavior of oral epithelial cells. PLoS One. 2014;9:e89475. doi: 10.1371/journal.pone.0089475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laheij AM, de Soet JJ, Veerman EC, Bolscher JG, van Loveren C. The influence of oral bacteria on epithelial cell migration in vitro . Mediators Inflamm. 2013;2013 doi: 10.1155/2013/154532. 154532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bostanci N, Belibasakis GN. Porphyromonas gingivalis: an invasive and evasive opportunistic oral pathogen. FEMS Microbiol Lett. 2012;333:1–9. doi: 10.1111/j.1574-6968.2012.02579.x. [DOI] [PubMed] [Google Scholar]
- 7.Lamont RJ, Jenkinson HF. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis . Microbiol Mol Biol Rev. 1998;62:1244–63. doi: 10.1128/mmbr.62.4.1244-1263.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pathirana RD, O'Brien-Simpson NM, Reynolds EC. Host immune responses to Porphyromonas gingivalis antigens. Periodontol 2000. 2010;52:218–37. doi: 10.1111/j.1600-0757.2009.00330.x. [DOI] [PubMed] [Google Scholar]
- 9.Chen T, Nakayama K, Belliveau L, Duncan MJ. Porphyromonas gingivalis gingipains and adhesion to epithelial cells. Infect Immun. 2001;69:3048–56. doi: 10.1128/IAI.69.5.3048-3056.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laine ML, van Winkelhoff AJ. Virulence of six capsular serotypes of Porphyromonas gingivalis in a mouse model. Oral Microbiol Immunol. 1998;13:322–5. doi: 10.1111/j.1399-302x.1998.tb00714.x. [DOI] [PubMed] [Google Scholar]
- 11.Brunner J, Scheres N, El Idrissi NB, Deng DM, Laine ML, van Winkelhoff AJ, et al. The capsule of Porphyromonas gingivalis reduces the immune response of human gingival fibroblasts. BMC Microbiol. 2010;10:5. doi: 10.1186/1471-2180-10-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Steenbergen TJ, de Soet JJ, de Graaff J. Genetic relationship between different subspecies of Bacteroides melaninogenicus . Antonie Van Leeuwenhoek. 1979;45:513. doi: 10.1007/BF00443288. [DOI] [PubMed] [Google Scholar]
- 13.Okamoto K, Nakayama K, Kadowaki T, Abe N, Ratnayake DB, Yamamoto K. Involvement of a lysine-specific cysteine proteinase in hemoglobin adsorption and heme accumulation by Porphyromonas gingivalis . J Biol Chem. 1998;273:21225–31. doi: 10.1074/jbc.273.33.21225. [DOI] [PubMed] [Google Scholar]
- 14.Aduse-Opoku J, Davies NN, Gallagher A, Hashim A, Evans HE, Rangarajan M, et al. Generation of lys-gingipain protease activity in Porphyromonas gingivalis W50 is independent of Arg-gingipain protease activities. Microbiology. 2000;146:1933–40. doi: 10.1099/00221287-146-8-1933. [DOI] [PubMed] [Google Scholar]
- 15.Kadowaki T, Baba A, Abe N, Takii R, Hashimoto M, Tsukuba T, et al. Suppression of pathogenicity of Porphyromonas gingivalis by newly developed gingipain inhibitors. Mol Pharmacol. 2004;66:1599–606. doi: 10.1124/mol.104.004366. [DOI] [PubMed] [Google Scholar]
- 16.Nakayama K, Kadowaki T, Okamoto K, Yamamoto K. Construction and characterization of arginine-specific cysteine proteinase (Arg-gingipain)-deficient mutants of Porphyromonas gingivalis. Evidence for significant contribution of Arg-gingipain to virulence. J Biol Chem. 1995;270:23619–26. doi: 10.1074/jbc.270.40.23619. [DOI] [PubMed] [Google Scholar]
- 17.Hintermann E, Haake SK, Christen U, Sharabi A, Quaranta V. Discrete proteolysis of focal contact and adherens junction components in Porphyromonas gingivalis-infected oral keratinocytes: a strategy for cell adhesion and migration disabling. Infect Immun. 2002;70:5846–56. doi: 10.1128/IAI.70.10.5846-5856.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matthay MA, Thiery JP, Lafont F, Stampfer F, Boyer B. Transient effect of epidermal growth factor on the motility of an immortalized mammary epithelial cell line. J Cell Sci. 1993;106:869–78. doi: 10.1242/jcs.106.3.869. [DOI] [PubMed] [Google Scholar]
- 19.Singer AJ, Clark RA. Cutaneous wound healing. N Engl J Med. 1999;341:738–46. doi: 10.1056/NEJM199909023411006. [DOI] [PubMed] [Google Scholar]
- 20.Oudhoff MJ, Bolscher JG, Nazmi K, Kalay H, van't Hof W, Amerongen AV, et al. Histatins are the major wound-closure stimulating factors in human saliva as identified in a cell culture assay. FASEB J. 2008;22:3805–12. doi: 10.1096/fj.08-112003. [DOI] [PubMed] [Google Scholar]
- 21.Ghoghawala SY, Mannis MJ, Murphy CJ, Rosenblatt MI, Isseroff RR. Economical LED based, real-time, in vivo imaging of murine corneal wound healing. Exp Eye Res. 2007;84:1031–8. doi: 10.1016/j.exer.2007.01.021. [DOI] [PubMed] [Google Scholar]
- 22.Abe N, Kadowaki T, Okamoto K, Nakayama K, Ohishi M, Yamamoto K. Biochemical and functional properties of lysine-specific cysteine proteinase (Lys-gingipain) as a virulence factor of Porphyromonas gingivalis in periodontal disease. J Biochem. 1998;123:305–12. doi: 10.1093/oxfordjournals.jbchem.a021937. [DOI] [PubMed] [Google Scholar]
- 23.Singh A, Wyant T, Anaya-Bergman C, Aduse-Opoku J, Brunner J, Laine ML, et al. The capsule of Porphyromonas gingivalis leads to a reduction in the host inflammatory response, evasion of phagocytosis, and increase in virulence. Infect Immun. 2011;79:4533–42. doi: 10.1128/IAI.05016-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vernal R, Leon R, Silva A, van Winkelhoff AJ, Garcia-Sanz JA, Sanz M. Differential cytokine expression by human dendritic cells in response to different Porphyromonas gingivalis capsular serotypes. J Clin Periodontol. 2009;36:823–9. doi: 10.1111/j.1600-051X.2009.01462.x. [DOI] [PubMed] [Google Scholar]
- 25.Laine ML, Appelmelk BJ, van Winkelhoff AJ. Novel polysaccharide capsular serotypes in Porphyromonas gingivalis . J Periodontal Res. 1996;31:278–84. doi: 10.1111/j.1600-0765.1996.tb00494.x. [DOI] [PubMed] [Google Scholar]
- 26.Kaman WE, Galassi F, de Soet JJ, Bizzarro S, Loos BG, Veerman EC, et al. Highly specific protease-based approach for detection of Porphyromonas gingivalis in diagnosis of periodontitis. J Clin Microbiol. 2012;50:104–12. doi: 10.1128/JCM.05313-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Potempa J, Pike R, Travis J. Titration and mapping of the active site of cysteine proteinases from Porphyromonas gingivalis (gingipains) using peptidyl chloromethanes. Biol Chem. 1997;378:223–30. doi: 10.1515/bchm.1997.378.3-4.223. [DOI] [PubMed] [Google Scholar]
- 28.Fitzpatrick RE, Wijeyewickrema LC, Pike RN. The gingipains: scissors and glue of the periodontal pathogen, Porphyromonas gingivalis . Future Microbiol. 2009;4:471–87. doi: 10.2217/fmb.09.18. [DOI] [PubMed] [Google Scholar]
- 29.Kraus D, Winter J, Jepsen S, Jager A, Meyer R, Deschner J. Interactions of adiponectin and lipopolysaccharide from Porphyromonas gingivalis on human oral epithelial cells. PLoS One. 2012;7:e30716. doi: 10.1371/journal.pone.0030716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kocgozlu L, Elkaim R, Tenenbaum H, Werner S. Variable cell responses to P. gingivalis lipopolysaccharide. J Dent Res. 2009;88:741–5. doi: 10.1177/0022034509341166. [DOI] [PubMed] [Google Scholar]
- 31.Darveau RP, Pham TT, Lemley K, Reife RA, Bainbridge BW, Coats SR, et al. Porphyromonas gingivalis lipopolysaccharide contains multiple lipid A species that functionally interact with both toll-like receptors 2 and 4. Infect Immun. 2004;72:5041–51. doi: 10.1128/IAI.72.9.5041-5051.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu Q, Darveau RP, Samaranayake LP, Wang CY, Jin L. Differential modulation of human {beta}-defensins expression in human gingival epithelia by Porphyromonas gingivalis lipopolysaccharide with tetra- and penta-acylated lipid A structures. Innate Immun. 2009;15:325–35. doi: 10.1177/1753425909104899. [DOI] [PubMed] [Google Scholar]
- 33.Nakagawa I, Inaba H, Yamamura T, Kato T, Kawai S, Ooshima T, et al. Invasion of epithelial cells and proteolysis of cellular focal adhesion components by distinct types of Porphyromonas gingivalis fimbriae. Infect Immun. 2006;74:3773–82. doi: 10.1128/IAI.01902-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.