Abstract
Seven acidophilic actinobacteria isolated from humus and mineral layers of a spruce forest soil were examined using a polyphasic approach. Chemotaxonomic properties of the isolates were found to be consistent with their classification in the genus Actinospica. The strains formed a distinct phyletic line in the Actinospica 16S rRNA gene tree being most closely related to Actinospica robiniae DSM 44927T (98.7–99.3 % similarity). DNA:DNA relatedness between isolate CSCA57T and the type strain of A. robiniae was found to be low at 40.8 (±6.6) %. The isolates were shown to have many phenotypic properties in common and were distinguished readily from the type strains of Actinospica acidiphila and A. robiniae using a range of phenotypic features. On the basis of these data the seven isolates were considered to represent a new species for which the name Actinospica durhamensis sp. nov. is proposed. The type strain of the species is CSCA 57T (=DSM 46820T = NCIMB 14953T).
Electronic supplementary material
The online version of this article (doi:10.1007/s10482-015-0496-1) contains supplementary material, which is available to authorized users.
Keywords: Actinobacteria, Polyphasic taxonomy, Actinospica durhamensis sp. nov., Spruce forest soil
Introduction
The actinobacterial genus Actinospica is the sole representative of the family Actinospicaceae of the order Catenulisporales (Donadio et al. 2010a, b). The genus was proposed by Cavaletti et al. (2006) to accommodate two strains isolated from a soil sample collected from a wooded area in Gerenzano, Italy, namely Actinospica acidiphila GE134766T and Actinospica robiniae GE134769T. The strains were found to be obligate acidophiles (pH range 4.2–6.2) that shared a combination of chemotaxonomic and morphological properties, notably the presence of 3-hydroxydiaminopimelic acid in whole-organism hydrolysates, and were distinguished using a battery of cultural and chemical markers (Cavaletti et al. 2006; Donadio et al. 2010c).
In a continuation of our bioprospecting studies on acidophilic and acidotolerant filamentous actinobacteria isolated from a spruce forest soil (Golinska et al. 2013a, b, c) many slow-growing obligate acidophilic strains were found to have colonial properties consistent with their assignment to the genus Actinospica. A polyphasic taxonomic study was carried out on seven of these isolates in order to establish their taxonomic status. The resultant data showed that the isolates form a new Actinospica species for which we propose the name Actinospica durhamensis sp. nov.
Materials and methods
Organisms, maintenance and biomass preparation
The organisms, strains HGG19, HSCA45, A1GG17, A1GG29, A1SCA6, CGG1, CSCA57T, were isolated from the humus (H), A1 and C mineral layers of a spruce soil at Hamsterley Forest, County Durham; the site and the dilution plate procedures used to isolate the strains have been described elsewhere (Golinska et al. 2013a, c). They were isolated from acidified starch-casein plates (Kűster and Williams 1964) using either agar (SCA) or gellan gum (GG) as gelling agents. The isolates were maintained on acidified (pH 5.5) yeast extract-malt extract ISP medium 2 (Shirling and Gottlieb 1966) plates at 4 °C and as hyphal fragments in glycerol (20 % v/v) at −80 °C, as were A. acidiphila DSM 44926T and A. robiniae DSM 44927T.
Biomass for the chemotaxonomic and molecular systematic studies was prepared by growing the isolates in shake flasks of acidified ISP 2 broth (pH 5.5) at 150 revolutions per minute for 3 weeks at 28 °C. Cells were harvested by centrifugation and washed twice in distilled water; biomass for the chemotaxonomic analyses was freeze-dried and that for the molecular work stored at −20 °C. Cells for the phenotypic tests was washed ten times with distilled water then diluted to give a turbidity of 5 on the McFarland scale and used to inoculate the test media.
Phylogenetic analyses
Extraction of genomic DNA, PCR-mediated amplification of the 16S rRNA genes of the isolates and direct sequencing of the purified PCR products were carried out as described by Golinska et al. (2013a, c). The closest phylogenetic neighbours based on 16S rRNA gene similarities were found using the EzTaxon server (http://eztaxon-e.ezbiocloud.net/: Kim et al. 2012). The resultant 16S rRNA gene sequences were aligned with corresponding sequences of the type strains of A. acidiphila and A. robiniae and those of Catenulispora species, using ClustalW. Phylogenetic analyses were carried out using the MEGA5 (Tamura et al. 2011) and PHYML (Guindon and Gascuel 2003) software packages. Phylogenetic trees were inferred by using the maximum-likelihood (Felsenstein 1981), maximum-parsimony (Fitch 1971) and neighbour-joining (Saitou and Nei 1987) tree-making algorithms, and evolutionary distances generated using the distance model described by Jukes and Cantor (1969). The tree topologies were evaluated by a bootstrap analysis (Felsenstein 1985) of the neighbour-joining data based on 1000 resamplings using MEGA5 software. The root position of the unrooted trees were estimated using the 16S rRNA gene sequence of Bifidobacterium bifidum NBRC 14252T (GenBank accession number NR 113873).
DNA:DNA relatedness
Reciprocal DNA:DNA pairing assays were carried out between isolate CSCA 57T and A. robiniae DSM 44927T at the DSMZ (Braunschweig, Germany). Cells were disrupted by using a Constant System TS 0.75 KW machine (IUL Instruments, Germany) and the crude DNA lysates purified by chromatography on hydroxyapatite after Cashion et al. (1977). DNA–DNA hybridization was carried out as described by De Ley et al. (1970) using the modifications described by Huss et al. (1983) and a model Cary 100 Bio UV/VIS-spectrophotometer equipped with a Peltier-thermostatted 6 × 6 multicell changer and a temperature controller with an in situ temperature probe (Varian) at 70 °C.
Chemotaxonomy
Standard chromatographic procedures were used to determine the isoprenoid quinone (Collins 1985), polar lipid (Minnikin et al. 1984) and whole-organism sugar (Hasegawa et al. 1983), composition of the seven isolates, using the type strain of A. robiniae DSM 44927T as control. Cellular fatty acids of isolate CSCA57T were extracted, methylated and determined by gas chromatography (Hewlett Packard instrument 6890) and analysed using the standard Sherlock Microbial Identification (MIDI) system, version 5 (Sasser 1990). The isomers of diaminopimelic acid of two representative strains, namely isolates A1SCA17 and CSCA57T, were determined by the identification service of the DSMZ (Braunschweig, Germany), as described by Schumann (2011). The G+C mol% of the DNA of strain CSCA57T was determined following the procedure described by Gonzalez and Saiz-Jimenez (2002).
Cultural and morphological properties
All of the isolates were examined for cultural and morphological properties following growth on acidified tryptone–yeast extract, yeast extract–malt extract, oatmeal, inorganic salts–starch, glycerol–asparagine, peptone–yeast extract–iron and tyrosine agars; ISP media 1–7, respectively (Shirling and Gottlieb 1966).
Phenotypic tests
A broad range of phenotypic tests were carried out on the isolates and the type strains of A. acidiphila and A. robiniae using acidified yeast carbon base (Sigma) without amino acids and acidified yeast nitrogen base (Sigma) media for the nitrogen and carbon utilization tests, respectively. The isolates were also examined for their ability to degrade a range of organic compounds, as described by Williams et al. (1983) and to grow at various temperatures (4, 10, 15, 20, 25, 30, 35 and 40 °C), pH values (4.0, 4.5, 5.0, 5.5, 6.0, 6.5, 7.0 and 7.5) and sodium chloride concentrations (1, 3, 5, 7 and 10 %, w/v) using acidified ISP2 agar (Shirling and Gottlieb 1966); apart from the temperature tests all of the media were incubated at 28 °C for 3 weeks.
Results and discussion
Relatively little attention has been paid to acidophilic filamentous actinobacteria even though they were discovered many years ago (Jensen 1928). It is now known that these organisms are common in acidic habitats, notably coniferous soils (Williams et al. 1971; Khan and Williams 1975; Goodfellow and Dawson 1978; Goodfellow and Simpson 1987) and are considered to have a role in the turnover of organic matter (Goodfellow and Williams 1983; Williams et al. 1984). It is important to clarify the taxonomy of acidophilic filamentous actinobacteria, not least because they are a source of antifungal agents (Williams and Khan 1974) and acid stable enzymes (Williams and Flowers 1978; Williams and Robinson 1981).
Chemotaxonomic, cultural and phenotypic properties
All of the strains isolated from the H, A1 and C horizons of the Hamsterley Forest spruce soil were found to have menaquinone, polar lipid and whole-cell sugar patterns as well as phenotypic properties consistent with their classification in the genus Actinospica (Cavaletti et al. 2006; Donadio et al. 2010c). The isolates were shown to be aerobic, Gram-positive, non-acid-alcohol fast, nonmotile actinobacteria that grew from pH 4.0–6.0, produced extensively branched substrate mycelia, whole-organism hydrolysates that contained arabinose, galactose, mannose and rhamnose (trace), contained di, tetra-, hexa- and octa-hydrogenated menaquinones in the ratio 5, 22, 40 and 30 %, respectively, and major amounts of diphosphatidylglycerol, phosphatidylethanolamine (taxonomically significant component) and phosphatidylglycerol (phospholipid pattern 2 sensu Lechevalier et al. 1977; Online supplementary Fig. 1). The whole-organism hydrolysates of isolates A1SCA17 and CSCA57T were found to contain 2, 6-diamino-3-hydroxy and meso-diaminopimelic acids in a ratio 1:2, respectively. The fatty acid profile of isolate CSCA57T was shown to contain major proportions (>10 %) of iso-C15:0, (22.2 %), anteiso-C15:0 (14.2 %), iso-C16:0 (27.7 %) and anteiso-C17:0 (16.7 %), minor proportions (>1.1 %) of iso-C14 (1.65 %), C16:0 (3.5 %), anteiso-C17:1ω9c (1.7 %), iso-C17:0 (3.9 %), C17:1ω8c (1.1 %), C17:0 (1.1 %) and summed feature iso-C17:1ω9c (2.9 %) and trace amounts (<0.7 %) of a few components. This isolate was also found to have a DNA G+C ratio of 68.0 mol% and to grow to varying degrees on most of the ISP media producing substrate mycelial pigments that distinguished them from the type strains of A. acidiphila and A. robiniae (Table 1). None of the isolates formed aerial hyphae on the ISP media.
Table 1.
Growth and cultural characteristics of the isolates and the type strains of A. acidiphila and A. robiniae on acidified ISP media after incubation for 4 weeks at 28 °C
| Isolates | A1GG17, A1GG29, A1SCA6, CGG1, CSCA57T, HGG19, HSCA45 | A. acidiphila DSM 449226T (GE 134766T) | A. robiniae DSM 44927T (GE 134769T) | |||||
|---|---|---|---|---|---|---|---|---|
| Growth | Colour of substrate mycelia | Growth | Colour of: aerial spore mass | Substrate mycelium | Growth | Colour of: aerial spore mass | Substrate mycelium | |
| Yeast extract–malt extract agar (ISP 2) | ++++ | Medium yellow | +++ | White | Pale greenish yellow | +++ | Whitish | Light greenish yellow |
| Oatmeal agar (ISP 3) | +++ | Light yellowish brown | +++ | White | Grayish yellow | +++ | Whitish | Dark grayish yellow |
| Inorganic salts-starch agar (ISP 4) | +++ | Greenish white | +++ | Absent | White | + | Absent | White |
| Glycerol-asparagine agar (ISP 5) | ++ | White | ++ | White | Greenish white | + | Absent | White |
| Peptone-yeast extract iron agar (ISP 6) | + | Medium yellow | + | White | White | – | Absent | Absent |
| Tyrosine agar (ISP 7) | ++ | White | +++ | White | Greenish white | ++ | Absent | White |
The isolates either did not grow or showed scant growth on acidified tryptone-yeast extract agar. None of the strains formed diffusible pigments or aerial spore mass
++++ abundant, +++ good, ++ poor, + scant growth, – no growth
16S rRNA gene sequencing and DNA:DNA relatedness studies
Near complete 16S rRNA gene sequences of isolates A1GG17, A1GG29, A1SCA6, CGG1, CSCA57T, HGG19 and HSCA45 (GenBank accession numbers; KJ 445730, KJ 445737, KJ 445731, KJ 445733, KJ 445732, KJ 445735 and KJ 445736, respectively) were generated. The isolates were found to have identical or almost identical 16S rRNA gene sequences (99.9–100 % nucleotide [nt] similarity) and were shown to form a branch in the Actinospica gene tree that was supported by all of the tree-making algorithms and by a 100 % bootstrap value (Fig. 1). In turn, the isolates were found to be most closely related to the type strain of A. robiniae DSM 44927T, a relationship that was underpinned by a 100 % bootstrap value and by all of the tree-making methods. The isolates shared 16S rRNA gene similarities with A. robiniae DSM 44927T that fell within the range 98.7 and 99.3 %, values equivalent to 10–18 nt differences at 1399–1421 locations. Corresponding 16S rRNA gene sequence similarities between the isolates and the type strain of A. acidiphila lay within the range 96.6–97.2 %, values that corresponded to between 40 and 49 nt differences at between 1397 and 1421 locations. The Actinospica 16S rRNA gene clade was distinguished readily from a second clade accommodating the type strains of Catenulispora species (Fig. 1). The isolates were also shown to exhibit the pattern of 16S rRNA gene signatures characteristic of members of the family Actinospicaceae (Donadio et al. 2010b).
Fig. 1.
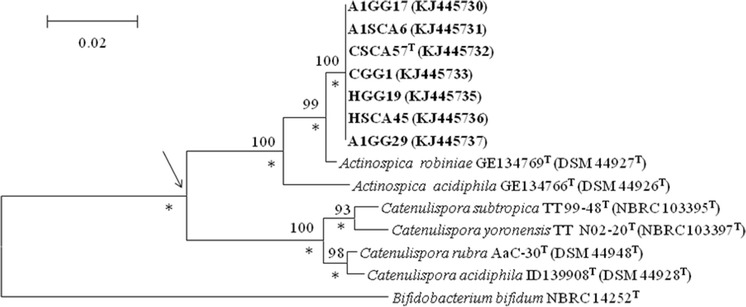
Neighbour-joining tree based on nearly complete 16S rRNA gene sequences (1399–1532 nucleotides) showing relationships between the isolates and between them and the type strains of Actinospica species. Asterisks indicate branches of the tree that were also found using the maximum-likelihood and maximum-parsimony tree-making algorithms. Numbers at the nodes indicate the percentage bootstrap values based on 1000 re-sampled datasets, only values above 50 % are given. T type strain. Bar 0.02 substitutions per nucleotide position. The root position of the tree was determined by using Bifidobacterium bifidum NBRC 14252T as outgroup
The DNA:DNA similarity value between isolate CSCA57T and A. robiniae DSM 44927T was shown to be 40.8 (±6.6) %, a result that is well below the 70 % cut-off point recommended for assigning bacterial strains to the same genomic species (Wayne et al. 1987).
Phenotypic tests
The isolates had many phenotypic features in common, some of which were shown to distinguish them from the type strains of A. acidiphila and A. robiniae (Table 2). The isolates, unlike A. robiniae DSM 44927T, their nearest phylogenetic neighbour, grew on d-trehalose as a sole carbon source, on l-alanine, l-isoleucine, l-phenylalamine and l-valine as sole nitrogen sources and in the presence of 1 %, w/v sodium chloride. Conversely, the A. robiniae strain, unlike the isolates grew on d-lactose and d-raffinose as sole carbon sources. Some, but not all, of the isolates grew at pH 6.5 and used l-histidine as a sole nitrogen source. It can also be seen from Table 2 that the isolates were found to have sugar and polar lipid patterns that distinguish them from the A. acidiphila and A. robiniae strains. The source of the isolates and features that distinguish them are shown in Table 3.
Table 2.
Phenotypic properties that distinguish the isolates from the type strains of A. acidiphila and A. robiniae
| Characteristic | Isolates A1GG17, A1GG29, A1SCA6, CGG1, CSCA57T, HGG19, HSCA45 | A. acidiphila DSM 44926T (GE 134766T) | A. robiniae DSM 44927T (GE 134769T) |
|---|---|---|---|
| Degradation of Tween 20 | – | – | + |
| Growth on sole carbon sources (1 %, w/v) | |||
| d-Lactose | – | – | + |
| d-Raffinose | – | + | + |
| d-Trehalose | + | + | – |
| Growth on sole nitrogen sources (0.1 %, w/v) | |||
| Acetamide | + | + | – |
| l-Alanine | + | + | – |
| l-Isoleucine | + | + | – |
| l-Phenylalanine | + | – | – |
| l-Valine | + | + | – |
| Growth at 15 °C | + | – | – |
| pH 4.0 | + | – | – |
| Growth in presence of 1 %, w/v NaCl | + | + | – |
| Chemotaxonomy | |||
| Diagnostic sugars in whole-organism hydrolysates | Ara, gal, man, rham (trace) | Ara, man, rham, xyl | Gal, man, rham |
| Predominantphospholipids | DPG, PE, PG, PI | DPG, PE, methyl-PE, PI | DPG, PE, methyl PE, PI |
| GC content of DNA (mol%) | 68.0 | 69.2 | 70.8 |
The chemotaxonomic data on the type strains of A. acidiphila and A. robiniae was taken from Cavaletti et al. (2006)
+ positive, − negative, Ara arabinose, gal galactose, man mannose, rham rhamnose, xyl xylose, DPG diphosphatidylglycerol, PE phosphatidylethanolamine, methyl-PE methyl phophatidylethanolamine, PG phosphatidylglycerol, PI phophatidylinositol
Table 3.
Properties that distinguish between isolates
| Characteristics | HGG19 | HSCA45 | A1GG17 | A1GG29 | A1SCA6 | CGG1 | CSCA57T |
|---|---|---|---|---|---|---|---|
| Source | Humus horizon H | Humus horizon H | Mineral horizon A1 | Mineral horizon A1 | Mineral horizon A1 | Mineral horizon C | Mineral horizon C |
| Growth at pH 6.5 | − | − | + | − | + | − | − |
| Growth on l-histidine as a sole nitrogen source | + | − | + | + | − | − | + |
| H2S production | + | + | + | + | − | + | + |
+ positive, − negative
Conclusion
The genotypic and phenotypic profiles of the isolates show that they form a well-circumscribed taxon within the genus Actinospica. It is, therefore, proposed that they represent a novel species, Actinospica durhamensis sp. nov.
Description of Actinospica durhamensis sp. nov.
Actinospica durhamensis (dur. ham. en’ sis. N.L. fem. adj. durhamensis, belonging to Durham, a county in the North East of England, the source of the isolates).
Aerobic, Gram-positive, non-acid-alcohol-fast, acidophilic actinobacteria which form an extensively branched substrate mycelium, but do not form aerial hyphae on ISP media. Strains grow at 15–33 °C, optimally ~28 °C, from pH 4.0 to 6.0, optimally at pH 5.5 and in the presence of 1 % w/v, but not 3 %, w/v sodium chloride. Tweens 40 and 60 are metabolised, but not adenine, casein, chitin, elastin, gelatin, guanine, hypoxanthine, starch, Tweens 20 and 80, tyrosine, uric acid, xanthine or xylan. l-arabinose, d-cellobiose, dextran, d-fructose, d-galactose, d-glucosamine, d-glucose, glycogen, d-melibiose, β-methyl-α-glucoside, l-rhamnose and d-ribose are used as sole carbon sources for energy and growth, but not adonitol, amygdalin, d- or l-arabitol, meso-erythritol, d-glucuronic acid, glycerol, meso-inositol, inulin, d-lactose, d-mannitol, d-melezitose, α-methyl-β-glucoside, d-raffinose, d-salicin, d-sucrose or xylitol (all at 1 %, w/v). Acetate, adipate, benzoate, butyrate, citrate, fumarate, hippurate, oxalate, propionate, pyruvate and succinate (sodium salts) or p-hydroxybenzoic acid (all at 0.1 %, w/v) are not used as sole sources of carbon. l-arginine, l-asparagine, l-aspartic acid, l-cysteine, l-glutamic acid, l-hydroxyproline, l-methionine, l-serine and l-threonine are used as a sole nitrogen sources (all at 0.1 %, w/v). Additional phenotypic properties are cited in the text and in Tables 1 and 2. The major fatty acids are iso-C15:0, anteiso-C15:0, iso-C16:0 and anteiso-C17:0. The G+C content of the DNA of the type strain is 68.0 mol%. Additional chemotaxonomic properties are given in the text and in Table 2.
The species contains the type strain CSCA57T (=DSM 46820T = NCIMB 14953T) and isolates HGG19, HSCA45, A1SCA6, A1GG29, A1GG17, CGG1, all of which were isolated from spruce soil from Hamsterley Forest, County Durham, England. The Genbank accession number of the 16S rRNA gene sequence of strain CSCA57T is KJ445732.
Electronic supplementary material
Supplementary Fig. 1 Two dimensional thin-layer chromatography of polar lipids of isolate CSCA57T stained with molybdenum blue (Sigma). Chloroform : methanol : water (32.5 : 12.5 : 2.0 v/v) were used in the first direction and chloroform : acetic acid : methanol : water (40 : 7.5 : 6 : 2 v/v) in the second direction. DPG, diphosphatidylglycerol; PE, phosphatidylethanolamine; PI, phosphatidylinositol; PIMS, phosphatidylinositol mannosides (DOC 805 kb)
Acknowledgments
This study was supported by a research fellowship from Nicolaus Copernicus University (Toruń, Poland) “Enhancing Educational Potential of Nicolaus Copernicus University in the Disciplines of Mathematical and Natural Sciences” (Project No. POKL.04.01.01-00-081/10) and Grant Symphony 1, No. 2013/08/W/NZ8/00701 from The Polish National Science Centre.
Conflict of interest
The authors declare that they have no conflict of interest.
Compliance with ethical standards
This article does not contain any studies with human participants or animals performed by any of the authors.
References
- Cashion P, Hodler-Franklin MA, McCully J, Franklin M. A rapid method for base ratio determination of bacterial DNA. Anal Biochem. 1977;81:461–466. doi: 10.1016/0003-2697(77)90720-5. [DOI] [PubMed] [Google Scholar]
- Cavaletti L, Monciardini P, Schumann P, Rohde M, Bamonte R, Busti E, Sosio M, Donadio S. Actinospica robiniae gen. nov., sp. nov. and Actinospica acidiphila sp. nov.: proposal for Actinospicaceae fam. nov. and Catenulisporinae subord. nov. in the order Actinomycetales. Int J Syst Evol Microbiol. 2006;56:1747–1753. doi: 10.1099/ijs.0.63859-0. [DOI] [PubMed] [Google Scholar]
- Collins MD. Isoprenoid quinone analysis in bacterial classification and identification. In: Goodfellow M, Minnikin DE, editors. Chemical methods in bacterial systematics. London: Academic Press; 1985. pp. 267–287. [Google Scholar]
- De Ley J, Cattoir H, Reynaerts A. The quantitative measurement of DNA hybridization from renaturation rates. Eur J Biochem. 1970;12:133–142. doi: 10.1111/j.1432-1033.1970.tb00830.x. [DOI] [PubMed] [Google Scholar]
- Donadio S, Cavaletti L, Monciardini P (2010a) Family 11. Actinospicaceae Cavaletti, Monciardini, Schumann, Rohde, Bamote, Busti, Sosio and Donadio 2006, 1751VP. In: Goodfellow M, Kämpfer P, Busse H-J, Trujillo ME, Suzuki K-I, Ludwig W, Whitman WB (eds) Bergey’s manual of systematic bacteriology, vol. 5, 2nd edn. The Actinobacteria, Part A. Springer, New York, p 232
- Donadio S, Cavaletti L, Monciardini P (2010b) Order IV. Catenulisperales ord. nov. In: Goodfellow M, Kämpfer P, Busse H-J, Trujillo ME, Suzuki K-I, Ludwig W, Whitman WB (eds) Bergey’s manual of systematic bacteriology, vol. 5, 2nd edn. The Actinobacteria, Part A, Springer, New York, p 225
- Donadio S, Cavaletti L, Monciardini P (2010c) Genus 1. Actinospica Cavaletti, Monciardini, Schumann, Rohde, Bamote, Busti, Sosio and Donadio 2006, 1751VP. In: Goodfellow M, Kämpfer P, Busse H-J, Trujillo ME, Suzuki K-I, Ludwig W, Whitman WB (eds) Bergey’s manual of systematic bacteriology, vol. 5, 2nd edn. The Actinobacteria, Part A, Springer, New York, pp 232–234
- Felsenstein J. Evolutionary trees from DNA sequences: maximum-likelihood approach. J Mol Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
- Fitch WM. Towards defining the course of evolution: minimum change for a specific tree topology. Syst Zool. 1971;20:406–416. doi: 10.2307/2412116. [DOI] [Google Scholar]
- Golinska P, Ahmed L, Wang D, Goodfellow M. Streptacidiphilus durhamensis sp. nov., isolated from a spruce forest soil. Antonie Van Leeuwenhoek. 2013;104:199–206. doi: 10.1007/s10482-013-9938-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golinska P, Kim B-Y, Dahm H, Goodfellow M. Streptacidiphilus hamsterleyensis sp. nov., isolated from a spruce forest soil. Antonie Van Leeuwenhoek. 2013;104:965–972. doi: 10.1007/s10482-013-0015-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golinska P, Wang D, Goodfellow M. Nocardia aciditolerans sp. nov., isolated from a spruce forest soil. Antonie Van Leeuwenhoek. 2013;103:1079–1088. doi: 10.1007/s10482-013-9887-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez JM, Saiz-Jimenez C. A fluorimetric method for the estimation of G+C mol% content in microorganisms by thermal denaturation temperature. Environ Microbiol. 2002;4:770–773. doi: 10.1046/j.1462-2920.2002.00362.x. [DOI] [PubMed] [Google Scholar]
- Goodfellow M, Dawson D. Qualitative and quantitative studies of bacteria colonizing Picea sitchensis litter. Soil Biol Biochem. 1978;10:303–307. doi: 10.1016/0038-0717(78)90027-5. [DOI] [Google Scholar]
- Goodfellow M, Simpson KE. Ecology of streptomycetes. Front Appl Microbiol. 1987;2:97–125. [Google Scholar]
- Goodfellow M, Williams ST. Ecology of actinomycetes. Ann Rev Microbiol. 1983;37:189–216. doi: 10.1146/annurev.mi.37.100183.001201. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hasegawa T, Takizawa M, Tanida S. A rapid analysis for chemical grouping of aerobic actinomycetes. J Gen Appl Microbiol. 1983;29:319–322. doi: 10.2323/jgam.29.319. [DOI] [Google Scholar]
- Huss VAR, Festl H, Schleifer KH. Studies on the spectrophotometric determination of DNA hybridization from renaturation rates. Syst Appl Microbiol. 1983;4:184–192. doi: 10.1016/S0723-2020(83)80048-4. [DOI] [PubMed] [Google Scholar]
- Jensen HL. Actinomyces acidophilus n. sp., a group of acidophilic actinomycetes isolated from the soil. Soil Sci. 1928;25:225–233. doi: 10.1097/00010694-192803000-00005. [DOI] [Google Scholar]
- Jukes TH, Cantor CR. Evolution of protein molecules. In: Munro HN, editor. Mammalian protein metabolism. New York: Academic Press; 1969. pp. 21–132. [Google Scholar]
- Khan MR, Williams ST. Studies on the ecology of actinomycetes in soil. VIII. Distribution and characteristics of acidophilic actinomycetes. Soil Biol Biochem. 1975;7:345–348. doi: 10.1016/0038-0717(75)90047-4. [DOI] [Google Scholar]
- Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S, Chun J. Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol. 2012;62:716–721. doi: 10.1099/ijs.0.038075-0. [DOI] [PubMed] [Google Scholar]
- Kűster E, Williams ST. Selection of media for isolation of streptomycetes. Nature. 1964;202:928–929. doi: 10.1038/202928a0. [DOI] [PubMed] [Google Scholar]
- Lechevalier MP, De Biévre C, Lechevalier HA. Chemotaxonomy of aerobic actinomycetes: phospholipid composition. Biochem Syst Ecol. 1977;5:249–260. doi: 10.1016/0305-1978(77)90021-7. [DOI] [Google Scholar]
- Minnikin DE, O’Donnell AG, Goodfellow M, Alderson G, Athalye M, Schaal A, Parlett JH. An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods. 1984;2:233–241. doi: 10.1016/0167-7012(84)90018-6. [DOI] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for constructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids. MIDI Technical Note 101, MIDI Inc. Newark
- Schumann P. Peptidoglycan structure. Methods Microbiol. 2011;38:101–129. doi: 10.1016/B978-0-12-387730-7.00005-X. [DOI] [Google Scholar]
- Shirling EB, Gottlieb D. Methods for characterization of Streptomyces species. Int J Syst Bacteriol. 1966;16:313–340. doi: 10.1099/00207713-16-3-313. [DOI] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. Mega 5: molecular evolutionary genetics analysis using maximum likelihood evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne LG, Brenner DJ, Colwell RR, et al. International Committee on Systematic Bacteriology. Report on the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol. 1987;37:463–465. doi: 10.1099/00207713-37-4-463. [DOI] [Google Scholar]
- Williams ST, Flowers TH. The influence of pH on starch hydrolysis by neutrophilic and acidophilic actinomycetes. Microbios. 1978;20:99–106. [PubMed] [Google Scholar]
- Williams ST, Khan MR. Antibiotics—a soil microbiologist’s viewpoint. Post Hig Med Dosw. 1974;28:395–408. [PubMed] [Google Scholar]
- Williams ST, Robinson CS. The role of streptomycetes in decomposition of chitin in acidic soils. J Gen Microbiol. 1981;127:55–63. [Google Scholar]
- Williams ST, Davies FL, Mayfield CI, Khan MR. Studies on the ecology of actinomycetes in soil. II. The pH requirements of streptomycetes from two acid soils. Soil Biol Biochem. 1971;3:187–195. doi: 10.1016/0038-0717(71)90014-9. [DOI] [Google Scholar]
- Williams ST, Goodfellow M, Alderson G, Wellington EMH, Sneath PHA, Sackin MJ. Numerical classification of Streptomyces and related genera. J Gen Microbiol. 1983;129:1743–1813. doi: 10.1099/00221287-129-6-1743. [DOI] [PubMed] [Google Scholar]
- Williams ST, Lanning S, Wellington EMH. Ecology of actinomycetes. In: Goodfellow M, Mordarski M, Williams ST, editors. The biology of actinomycetes. London: Academic Press; 1984. pp. 481–528. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1 Two dimensional thin-layer chromatography of polar lipids of isolate CSCA57T stained with molybdenum blue (Sigma). Chloroform : methanol : water (32.5 : 12.5 : 2.0 v/v) were used in the first direction and chloroform : acetic acid : methanol : water (40 : 7.5 : 6 : 2 v/v) in the second direction. DPG, diphosphatidylglycerol; PE, phosphatidylethanolamine; PI, phosphatidylinositol; PIMS, phosphatidylinositol mannosides (DOC 805 kb)


