Abstract
We focus on the identification of complete and recombined ribosomal DNA-bearing chromosomes, and the dynamics of chromosomal number and position of ribosomal DNA (rDNA) loci in the F2-F4 generations derived from the F1 hybrid of Festuca pratensis Huds. (2n = 4x = 28) × Lolium perenne L. (2n = 4x = 28). Lolium genomic DNA and rRNA genes were mapped by means of genomic and fluorescence in situ hybridization (GISH and FISH). The results revealed that plants of the three generations share various rDNA loci profiles with chromosome structural changes, possibly as a result of chromosomal inter- and intra-rearrangements. We observed an asymmetrical variation in the number of recombinant arms with and without rDNA loci between parental genomes. The Lolium genome was more affected by rearrangements in arms with rDNA loci, while Festuca was more affected in arms without them. Statistically significant differences between L. perenne and F. pratensis genomes concerned the number of recombined chromosomes without rDNA, and the number of recombined rDNA-bearing chromosomal arms of marked chromosomes, showing a tendency of F. pratensis genome-like chromosomes to be less stable, compared with L. perenne. We postulate a novel genome-dependent range and type of chromosome variation in plants of the F2-F4 generations derived from F. pratensis × L. perenne hybrid.
Electronic supplementary material
The online version of this article (doi:10.1007/s00709-014-0734-9) contains supplementary material, which is available to authorized users.
Keywords: Chromosomal rearrangements, Festuca × Lolium hybrid, GISH, rDNA-FISH, 5S rDNA, 35S rDNA
In the study of different plant genomes, including cultivars and inter-generic hybrids of the Festuca-Lolium complex, the possibility to identify mitotic and meiotic chromosomes is of major importance. Modern cytogenetic analyses, such as fluorescence and genomic in situ hybridization (FISH and GISH) techniques, have been widely used to resolve many processes of chromosome evolution, including structural rearrangements (Levin 2004), as well as extensive studies on phylogenetic and genomic relationships (Robledo et al. 2009), and to enhance our knowledge of plant genome structure and differentiation (D’Hont 2005; Maluszynska and Hasterok 2005; Cai et al. 2006; Zwierzykowski et al. 2008; Wolny et al. 2011; Wan et al. 2012; Chacón et al. 2012). Using a combination of double FISH with 5S and 35S ribosomal DNA (rDNA) probes, chromosome morphology and rDNA loci patterns have been described in Festuca pratensis (Thomas et al. 1997; Harper et al. 2004; Książczyk et al. 2010) and Lolium perenne (Thomas et al. 1996; Książczyk et al. 2010; Rocha et al. 2014), as well as in various amphiploid and introgression forms of Festulolium (Kopecký et al. 2006; Kosmala et al. 2006; Książczyk et al. 2010, 2012; Harper et al. 2011).
Contrasting patterns of genome organization in Festuca and Lolium species, involving chromosome substitution and homoeologous chromosome pairing, numerical and structural chromosome instability have already been observed in several generations of inter-generic Festuca × Lolium hybrids (Zwierzykowski et al. 1998, 2006, 2012). Studies of genome instability in the Festuca-Lolium complex has differentiated genetically close genomes such as Lolium multiflorum and F. pratensis (Thomas et al. 1994; Zwierzykowski et al. 1998; Kopecký et al. 2006; Kosmala et al. 2006), L. perenne and F. pratensis (King et al. 1998; Zwierzykowski et al. 2006, 2012), and L. multiflorum and Festuca arundinacea (Humphreys and Pašakinskienė 1996). The application of GISH/FISH in F1 hybrids of F. pratensis × L. perenne allowed the identification of L. perenne chromosome 3 and F. pratensis chromosomes 2 and 3 (Lolium and Festuca chromosomes were numbered according to Thomas 1981). This approach revealed variation in the number and position of rDNA sites, which can be easily monitored in these hybrids (Książczyk et al. 2010).
Numerous arrangements of rDNA chromosomal patterns within species were revealed in cultivars of Festuca spp. and Lolium spp. (Thomas et al. 1996, 1997, 2001; Harper et al. 2004). Moreover, it was shown by Książczyk et al. (2010) that variation in the number of 5S rDNA sites (gain/loss) occurred even among individuals derived from the same cultivar of F. pratensis. Pedrosa-Harand et al. (2006) also showed variation in the number of 35S rDNA loci among individuals of the same Phaseolus vulgaris accession. In F1 hybrids of F. pratensis × L. perenne, a new distally and interstitially located locus of 5S rDNA was observed (Książczyk et al. 2010; T. Książczyk and K. Molik, unpublished data), while in the tetraploid BC1 plants obtained from crosses of F1 hybrid F. pratensis × L. perenne into L. perenne, only a distally located new locus of 5S rDNA was found (Książczyk et al. 2012). The appearance of a distally located 5S rDNA site in hexaploid Festuca gigantea (Thomas et al. 1997), diploid Festuca drymeja (Harper et al. 2004), and diploid and tetraploid F. pratensis (Książczyk et al. 2010) may suggest its common distribution within Festuca species; however, the origin and extent of such a variation still remains unclear.
The present study aimed at characterizing the chromosomal number and position of rDNA sites, as well as mitotic chromosome behavior, in three successive generations, F2-F4, derived from F1 hybrid of F. pratensis (4x) × L. perenne (4x). As reported before, a cytogenetic examination of synthetic allotetraploid F1 hybrid of F. pratensis × L. perenne revealed various numbers of 5S and 35S rDNA sites (Książczyk et al. 2010). It was later showed by Książczyk et al. (2012) that the L. perenne chromosome 3 (5S + 35S rDNA) and F. pratensis chromosome 2 (35S rDNA) and 3 (5S rDNA) are involved in recombination, showing rearrangements in the BC1 plants. To the best of our knowledge, however, little is known about parental chromosome identification in the Festuca-Lolium complex, or any precise monitoring of recognized and unrecognized rearranged chromosomes of both parental genomes. We deal with this novel aspect in the present paper.
Materials and methods
Plant material
Tetraploid hybrids of F. pratensis (Fp) × L. perenne (Lp) (2n = 4x = 28, described here as Fp × Lp) were generated by inter-crossing autotetraploid forms of both species. F. pratensis Huds. (2n = 4x = 28) spontaneous tetraploid plants, obtained from twin seedlings of diploid cultivars, were used as the female parent, and L. perenne L. (2n = 4x = 28) as the male parent (Zwierzykowski et al. 2006). Four partially fertile female and male F1 hybrids were inter-crossed under controlled conditions, and the F2 progeny was generated. Generations F3-F4 were obtained by inter-crossing of 150 genotypes in control conditions. In this work, 30 randomly chosen plants (ten per each generation) with the tetraploid (2n = 4x = 28) number of chromosomes were used for cytogenetic analyses. In general, tetraploids comprised 66.7 % of plants studied (Z. Zwierzykowski, unpublished data). The cultivars of F. pratensis (4x), L. perenne (4x), and F1 hybrids of F. pratensis (4x) × L. perenne (4x) were previously studied in order to determine the number and position of rDNA sites (Książczyk et al. 2010; Online Resource S1). Due to the inter-cross of four F1 hybrids to produce the F2 progeny, a hypothetical model of F1 karyotypes was presented, considering a theoretical rDNA loci pattern (Online Resource S1). All four F1 plants used for hybridization in situ experiments had various rDNA loci patterns; hence, based on the number and position of their rDNA loci (four to five sites of 5S rDNA and seven to nine sites of 35S rDNA), we expected to observe seven to nine sites of 35S rDNA, consisting of both homologues of Fp chromosome 2 with large 35S rDNA sites, and 7 undifferentiated Lp chromosomes (besides both homologues of chromosome 3) with 35S rDNA, 2 large interstitial 5S rDNA sites in both Fp homologues of chromosome 3, 2 large interstitial 5S rDNA sites in the short arms of both Lp homologues of chromosome 3 (Online Resource S1). In addition, we expected zero to one small 5S rDNA locus in a distal region of unrecognized Fp chromosomes. All changes from each generation studied, in F. pratensis × L. perenne hybrids with equal number of Festuca and Lolium chromosomes, compared with the expected number and position of rDNA sites, were treated as possible variations in the rDNA loci pattern.
Chromosome preparations
Root tips of all 30 F2-F4 plants were collected in ice water, refrigerated for 24 h, fixed in ethanol with glacial acetic acid (3:1, v/v), and then stored at −20 °C until use. Further treatment was performed according to Zwierzykowski et al. (1998). Chromosome analysis was carried out on 3-5 well-spread metaphases. Each chromosomal preparation was derived from a different single root tip, so that each preparation corresponded to one individual.
DNA probes
Three kinds of probes were used: (i) total genomic DNA from F. pratensis and L. perenne extracted from young leaves using Novabeads Plant DNA Maxi Kit according to the manufacturer’s procedure (Novazym Poland; after modifications), and further treatment of extracted DNA was carried out as described by Książczyk et al. (2010); (ii) the 5S rDNA probe was generated by PCR amplification of a 410-bp BamHI sub-clone of the 5S rDNA from the wheat clone pTa794 (Gerlach and Dyer 1980) and also labeled by PCR with tetramethyl-rhodamine-5-dUTP (Roche) by using universal M13 “forward” (5′-CAG GGT TTT CCC AGT CAC GA-3′) and “reverse” (5′-CGG ATA ACA ATT TCA CAC AGG A-3′) sequencing primers. The thermal cycling program was as follows: 94 °C for 1 min, 39 cycles of 94 °C for 40 s, 55 °C for 40 s, and 72 °C for 90 s, and finally, 72 °C for 5 min; (iii) the 26S rDNA probe, used for detection of 35S rDNA loci, was made by nick translation of a 2.3-kb ClaI sub-clone of the 26S rDNA coding region of Arabidopsis thaliana (Unfried and Gruendler 1990) with digoxigenin-11-dUTP (Roche). The conditions for this reaction were as follows: 15 °C for 95 min and 65 °C for 10 min.
In situ hybridization (FISH and GISH)
The FISH procedure was performed as described by Książczyk et al. (2010). The Lp and Fp chromosomes identified by rDNA-FISH were numbered according to Thomas (1981). For distinguishing the two subgenomes of the hybrids, GISH was done using the total genomic DNA of Lp and Fp as a probe and block, respectively. Before GISH, incubation of slides previously subjected to FISH experiments was carried out as described by Książczyk et al. (2010). The GISH procedure was adapted from Kosmala et al. (2006), with minor modifications (Książczyk et al. 2010). The following observations were made for each plant studied: (i) the total number of complete Lp and Fp rDNA-bearing chromosomes, (ii) the total number of complete Lp and Fp non-rDNA-bearing chromosomes, (iii) the total number of recombinant Lp and Fp rDNA-bearing chromosomes, (iv) the total number of recombinant Lp and Fp non-rDNA-bearing chromosomes, (v) the total number of complete and recombinant Lp and Fp chromosomal arms with rDNA site, (vi) the total number of complete and recombinant Lp and Fp chromosomal arms without rDNA site, and (vii) frequency of rDNA-bearing chromosomes (3L as well as 2F and 3F) involved in recombination.
Image capturing and processing
All images were acquired using either an Olympus XM10 CCD camera attached to an Olympus BX 61 automatic epifluorescence microscope, or an F-View II CCD camera attached to an Olympus BX 60 epifluorescence microscope. Image processing and superimpositions were done using Olympus Cell-F imaging software and Micrographx Picture Publisher software.
Statistical analysis
To evaluate the influence of three generations on Lp and Fp chromosome changes, and to study difference between both genomes, cytogenetic data were statistically processed by the Pearson’s chi-squared test at P ≤ 0.05, according to standard procedures within GenStat® version 15.1 (Payne et al. 2012).
Results
rDNA loci pattern versus recombination in the F2-F4 generations
Among plants of the three generations the number of 5S rDNA sites ranged from 4 to 6, with a predominant number of five signals, although six signals of 5S rDNA loci were observed only in plants of the F3 generation (Table 1). There were two to three sites in Lp of the F2, one to three in Lp of the F3, and two to four in Lp of the F4 (chromosome no. 3). In addition, there were one to two sites in Fp (chromosome no. 3; F2 and F3) and zero to two sites in Fp (chromosome no. 3; F4) large (main) 5S rDNA sites interstitially located, while a small 5S rDNA locus was found in a distal region of one (F2 and F4) or two (F3) unrecognized Fp chromosomes. In one F2 plant, the additional small 5S rDNA locus was distally located in the recombinant undifferentiated Lp chromosome (data not presented), but the recombinant Lp chromosome was apparently lost in later generations. The number of 35S rDNA sites ranged from 8 to 11 in the F2, 8 to 10 in the F3, and 7 to 12 in the F4 (Table 1). The number of clearly identifiable Lp homologues of chromosome 3 ranged 2–3 (F2), 1–3 (F3), and 2–4 (F4), although plants with both Lp homologues of chromosome 3 were “dominant” over the generations (24/30 plants). Through the generations studied, 4 to 7 (F2), 3 to 6 (F3), 1 and 4 to 6 (F4) signals of 35S rDNA sites were always located at secondary constrictions on Lp homologues of cytologically undifferentiated chromosomes 1 and 2, as well as proximally located close to the centromere of chromosome 7. Signals of 35S rDNA sites were also located at the secondary constriction on Lp and Fp homologues of chromosomes 3 and 2, respectively, showing the differentiation in their patterns as follows: 2–3 Lp and 1–2 Fp (F2), 1–3 Lp, and 2 Fp (F3), and also 2–4 Lp and 1–4 Fp (F4).
Table 1.
Number and chromosomal position of rDNA sites in plants of F2-F4 generations derived from the allotetraploid F. pratensis × L. perenne hybrid
| Generation/plant no. | 2n | Chromosome ratio Lp/Fp | No. of rDNA sites (positiona) | No. of chromosomes with both rDNA sites | ||||
|---|---|---|---|---|---|---|---|---|
| 5S rDNA | 35S rDNA | |||||||
| Lp (is) | Fp (is) | Fpb (d) | Lp (sc/p) | Fp (sc/p) | ||||
| F2-7 | 28 | 14:14 | 2 | 2 | 1 | 6 | 2 | 2 |
| F2-9 | 28 | 14:14 | 2 | 2 | 1 | 7 | 2 | 2 |
| F2-13 | 28 | 14:14 | 2 | 2 | 0 | 7 | 2 | 2 |
| F2-15 | 28 | 14:14 | 2 | 2 | 1 | 6 | 2 | 2 |
| F2-80 | 28 | 14:14 | 2 | 2 | 1 | 9 | 2 | 2 |
| F2-126 | 28 | 14:14 | 2 | 2 | 0 | 9 | 2 | 2 |
| F2-11 | 28 | 15:13 | 2 | 2 | 1 | 8 | 2 | 2 |
| F2-28 | 28 | 15:13 | 2 | 2 | 1 | 8 | 2 | 2 |
| F2-122 | 28 | 15:13 | 3 | 1 | 1 | 7 | 2 | 3 |
| F2-79 | 28 | 17:11 | 2 | 2 | 1 | 9 | 1 | 2 |
| F3-28 | 28 | 14:14 | 2 | 2 | 2 | 8 | 2 | 2 |
| F3-34 | 28 | 14:14 | 2 | 2 | 0 | 8 | 2 | 2 |
| F3-150 | 28 | 14:14 | 2 | 2 | 0 | 7 | 2 | 2 |
| F3-18 | 28 | 13:15 | 2 | 2 | 2 | 6 | 2 | 2 |
| F3-57 | 28 | 13:15 | 2 | 2 | 1 | 8 | 2 | 2 |
| F3-123 | 28 | 13:15 | 2 | 2 | 0 | 7 | 2 | 2 |
| F3-1 | 28 | 15:13 | 2 | 2 | 1 | 6 | 2 | 2 |
| F3-96 | 28 | 15:13 | 3 | 1 | 1 | 6 | 2 | 3 |
| F3-106 | 28 | 16:12 | 1 | 2 | 1 | 7 | 2 | 1 |
| F3-139 | 28 | 16:12 | 2 | 2 | 0 | 7 | 2 | 2 |
| F4-28 | 28 | 14:14 | 2 | 2 | 1 | 8 | 2 | 2 |
| F4-33 | 28 | 14:14 | 2 | 2 | 0 | 8 | 2 | 2 |
| F4-53 | 28 | 14:14 | 2 | 2 | 1 | 8 | 1 | 2 |
| F4-83 | 28 | 14:14 | 2 | 2 | 0 | 8 | 2 | 2 |
| F4-6 | 28 | 12:16 | 2 | 2 | 0 | 3 | 4 | 2 |
| F4-10 | 28 | 15:13 | 2 | 2 | 0 | 6 | 3 | 2 |
| F4-104 | 28 | 15:13 | 2 | 2 | 1 | 6 | 3 | 2 |
| F4-109 | 28 | 16:12 | 3 | 1 | 1 | 8 | 2 | 3 |
| F4-135 | 28 | 17:11 | 3 | 1 | 1 | 9 | 2 | 3 |
| F4-25 | 28 | 18:10 | 4 | 0 | 0 | 9 | 3 | 4 |
| Test resultc | n.s. | n.s. | n.s. | n.s. | n.s. | P = 0.022 | n.s. | |
Lp L. perenne chromosomes, Fp F. pratensis chromosomes
aPosition of rDNA sequences is shown in brackets: interstitial (is), distal (d), secondary constriction (sc), proximal (p)
bUnrecognized F. pratensis chromosome with an additional 5S rDNA locus
cThe distribution of values for 5S and 35S rDNA loci between generations was compared, and significant differences between distributions for Lp and Fp were assessed using Pearson’s chi-squared test (P ≤ 0.05); statistically significant difference for Fp genome-like 35S rDNA between generations was found at P = 0.022 (n.s. means P > 0.05).
Generally, 30 F2-F4 plants showed up to 11 various rDNA loci patterns, of which 6 patterns were repeated two to six times (Table 1). Thirteen F2-F4 plants had an equal number of chromosomes, 14Lp and 14Fp, showing up to eight various rDNA loci patterns, of which four patterns were repeated two to three times (Table 1). The hypothetical model of F1 Fp × Lp karyotype (14Lp:14Fp) (Online Resource S1) assumes a presence of four signals (2Lp + 2Fp) of 5S rDNA and nine signals (7Lp + 2Fp) of 35S rDNA, which was only observed in one plant of the F3 and F4 and in two plants of the F2. However, the expected four to five signals of 5S rDNA and seven to nine signals of 35S rDNA were observed in 6 out of 13 plants (14Lp:14Fp) of the F2-F4 generations. In seven remaining plants, six had the expected number of 5S rDNA loci (four to five), but an unexpected number of 35S rDNA (10–11); the seventh plant had six unexpected signals of 5S rDNA and ten signals of 35S rDNA (Table 1). In two out of four Fp × Lp plants with a lower number of Lp chromosomes (12–13 Lp chromosomes; Table 1), the hypothetical (5S/35S: 4/9) and expected (4/7) number of rDNA loci was observed (13Lp:15Fp and 12Lp:16Fp), while the other two plants (13Lp:15Fp) had the expected number of 5S or 35S rDNA loci. Among 13 Fp × Lp plants with the higher number of Lp chromosomes (15–18 Lp chromosomes; Table 1), the hypothetical (4/9) and expected (4/9, 5/8, 5/9) number of rDNA loci was observed in seven plants (five plants with 15Lp:13Fp and two plants with 16Lp:12Fp; Table 1), although the distribution of rDNA loci was consistent with the hypothetical model of rDNA loci pattern only in one plant (16Lp:12Fp) (Online Resource S1). Six other plants had unexpected 5S and 35S rDNA loci patterns. It is worth mentioning that 4/12 (5S/35S) rDNA loci pattern occurred in the Fp × Lp plant with 18Lp and 10Fp chromosomes (Table 1), and the number and position of rDNA loci were also not consistent with the hypothetical model of rDNA loci pattern (Online Resource S1), showing three homologues of Fp chromosome 2 (instead of two) and a lack of two homologues of Fp chromosome 3, which might be absent due to a lower number of Fp chromosomes in this plant (Table 1).
Sixty-five Lp homologues of chromosome 3 were observed in Fp × Lp plants (Table 1), and 48 of these chromosomes did not undergo numerical changes, while among 157 Lp homologues of chromosomes 1, 2, and 7, only 16 Lp rDNA-carrying ones were stable, and their number was consistent with the hypothetical model (Online Resource S1). In the F2-F4 plants studied, among 63 homologues of Fp chromosome 2, and 54 homologues of Fp chromosome 3, no numerical changes were noted in 48 and 50 homologues, respectively. In the F2 generation, Lp homologue(s) of chromosome 3 was rarely involved in recombination, showing rearrangement (only one case; Figs. 1a, b and 2), while no Fp homologues of chromosomes 2 and 3 were found to be rearranged. In the F3 generation, the Lp homologue of chromosome 3 and Fp homologues of chromosomes 2 and 3 were involved in recombination, showing rearrangements (14 cases; Fig. 2), and, in turn, in the F4 generation, the Lp homologue of chromosome 3 and Fp homologues of chromosomes 2 and 3 (Fig. 1e, f) were also recombined (21 cases; Fig. 2). Over the generations, the variation in the number of rDNA-carrying chromosomes of both parental genomes seemed to be asymmetrical and genome-dependent (Table 1); statistically significant difference for rDNA-bearing chromosomes between generations was found for Fp chromosome 2 (35S rDNA) at P = 0.022.
Fig. 1.
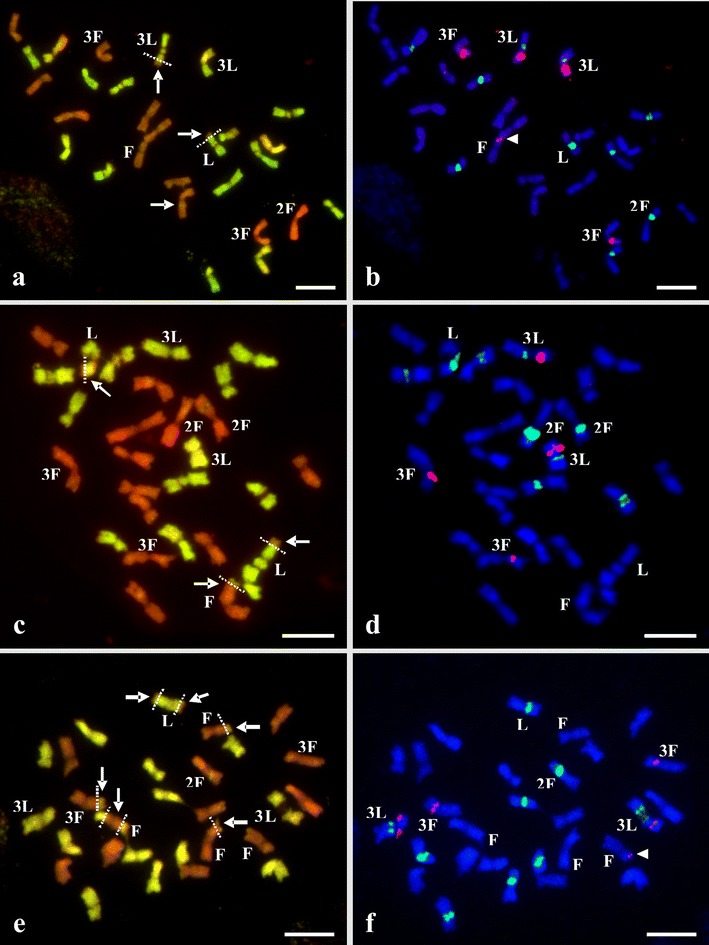
Chromosome identification of parental species in plants of the F2-F4 generations derived from F. pratensis (4x) × L. perenne (4x) hybrid using GISH (a, c, e) and FISH (b, d, f). GISH images (a, c, e) were created after FISH hybridization using total genomic DNA from Lp as a probe labeled with digoxigenin and detected by anti-digoxigenin conjugated with fluorescein (green/yellow), with blocking genomic DNA of Fp (orange/red); chromosomes were counterstained with propidium iodide. FISH images (b, d, f) were created using probes as follows: (i) 5S rDNA labeled with rhodamine (red) and (ii) 26S rDNA labeled with digoxigenin and detected by anti-digoxigenin conjugated with FITC (green); chromosomes were counterstained with DAPI (blue). GISH and FISH images are marked by white arrows indicating Lp and Fp recombinant chromosomes (R), by white arrowheads indicating additional location of 5S rDNA locus, and by the white lines with intervals indicating recombination breakpoints. a, b F2 plant [17Lp (2R) +11Fp (1R)]. c, d F3 plant [14Lp (2R) +14Fp (1R)]. e, f F4 plant [14Lp (1R) +14Fp (4R)]. The nomenclature of rDNA-bearing chromosomes (Arabic numerals) follows the system of Thomas (1981). Uppercase letters denote the genomic origin of tagged chromosomes. Scale bars represent 5 μm
Fig. 2.
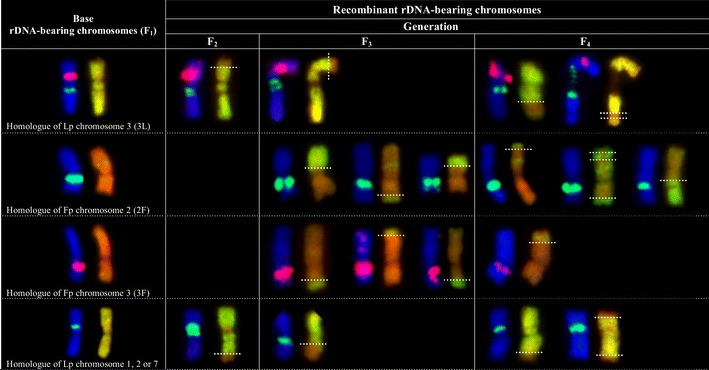
rDNA-FISH/GISH of known Lp and Fp rDNA-bearing chromosomes in plants of the F2-F4 generations derived from F. pratensis (4x) × L. perenne (4x) hybrid. Data of FISH/GISH analyses of base rDNA-bearing chromosomes from the F1 generation were published previously (Książczyk et al. 2010). The color of the chromosome band label indicates the fluorochrome used in each experiment (pink for rhodamine; 5S rDNA, green and green/yellow for FITC; 35S rDNA and Lp genomic DNA, respectively). FISH painted chromosomes were counterstained with DAPI (blue), while GISH ones were counterstained with propidium iodide (red)
Structural dynamics of rDNA loci patterns in the F2-F4 generations
The frequency profile of complete and recombinant rDNA-bearing and non-rDNA-bearing chromosomes of both genomes in the hybrids of the three generations is given in Online Resource S2. Recombinant rDNA-bearing chromosomes were only observed in the Lp genome of the F2 (Figs. 1a, b and 2) (mean 0.8/genotype), in both Lp (mean 0.9) and Fp (mean 1.1) of the F3, as well as in both Lp (mean 0.9) and Fp (mean 1.8) of the F4 (data not presented). Over the generations, 26 (F2), 47 (F3), and 69 (F4) recombined Lp and Fp chromosomes were observed (Table 2). Among 26 recombined chromosomes in the F2 generation, 6 Lp and 12 Fp were non-rDNA-bearing ones. No Fp, but eight Lp were rDNA-bearing (Table 2), of which one Lp had both 5S and 35S rDNA loci (chromosomes 3, Figs. 1a, b and 2), six had Lp 35S rDNA locus (chromosomes 1, 2, or 7, Figs. 1a, b and 2), and one unknown Lp had 5S rDNA locus (data not showed). Among 47 recombined chromosomes of the F3, 9 Lp and 18 Fp were non-rDNA-bearing ones, while 9 Lp and 11 Fp were rDNA-bearing, of which 4 Lp had both 5S and 35S rDNA loci (chromosome 3, Fig. 2), 5 had Lp 35S rDNA locus (chromosomes 1, 2, or 7, Figs. 1c, d and 2), 6 had Fp 35S rDNA locus (chromosome 2), 4 had Fp large 5S rDNA locus (chromosome 3), and 1 had Fp small 5S rDNA locus (unknown chromosome). It should be pointed out that three types of Fp rDNA-bearing chromosomes (two known and one unrecognized; Table 1) were found to be more frequently recombined (11 cases) based on five known types of Lp rDNA-bearing ones (nine cases). Among 69 recombined chromosomes in the F4, 12 Lp and 30 Fp were non-rDNA-bearing ones, 9 Lp and 18 Fp were rDNA-bearing ones, of which 5 Lp had both 5S and 35S rDNA loci (chromosome 3, Figs. 1e, f and 2), 4 had Lp 35S rDNA locus (chromosomes 1, 2, or 7, Fig. 1e, f), 11 had Fp 35S rDNA locus (chromosome 2), 6 had Fp large 5S rDNA locus (chromosome 3, Fig. 1e, f), and 1 had Fp small 5S rDNA locus (unknown chromosome, Fig. 1e, f). Again, Fp rDNA-bearing chromosomes were found to be the most frequently recombined in the F4. It is worth mentioning that the number of recombined rDNA-bearing chromosomes was doubled for Fp ones, comparing both genomes (Table 2, Online Resource S2). Over the generations, the variation in the number of recombinant Lp and Fp arms with (m+) and without (m−) rDNA loci also seemed to be genome-dependent (Table 2).
Table 2.
Number of non-recombined and recombined non-rDNA- and rDNA-bearing chromosomes and their genome assignment in plants of F2-F4 generations derived from the allotetraploid F. pratensis × L. perenne hybrid
| Generation/plant no. | Chromosome ratio M+/M− | No. of non-recombined chromosomes | No. of recombined chromosomes | No. of recombined arms | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M+ | M− | M+ | M− | m+ | m− | ||||||||
| Lp | Fp | Lp | Fp | Lp | Fp | Lp | Fp | Lp | Fp | Lp | Fp | ||
| F2-7 | 11:17 | 6 | 5 | 7 | 8 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 |
| F2-9 | 12:16 | 6 | 5 | 6 | 7 | 1 | 0 | 1 | 2 | 1 | 0 | 0 | 0 |
| F2-13 | 11:17 | 7 | 4 | 5 | 10 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| F2-15 | 11:17 | 6 | 5 | 7 | 8 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 |
| F2-80 | 14:14 | 9 | 5 | 5 | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| F2-126 | 13:15 | 9 | 4 | 5 | 9 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| F2-11 | 13:15 | 6 | 5 | 7 | 7 | 2 | 0 | 0 | 1 | 0 | 0 | 2 | 0 |
| F2-28 | 13:15 | 6 | 5 | 7 | 6 | 2 | 0 | 0 | 2 | 2 | 0 | 0 | 0 |
| F2-122 | 11:17 | 6 | 4 | 7 | 7 | 1 | 0 | 1 | 3 | 1 | 0 | 0 | 0 |
| F2-79 | 13:15 | 7 | 4 | 8 | 6 | 2 | 0 | 0 | 1 | 1 | 0 | 1 | 0 |
| F3-28 | 14:14 | 7 | 6 | 6 | 5 | 1 | 0 | 0 | 3 | 1 | 0 | 0 | 0 |
| F3-34 | 12:16 | 7 | 3 | 5 | 7 | 1 | 1 | 1 | 3 | 1 | 0 | 0 | 1 |
| F3-150 | 11:17 | 6 | 4 | 6 | 9 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 0 |
| F3-18 | 12:16 | 6 | 6 | 6 | 8 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 |
| F3-57 | 13:15 | 6 | 5 | 5 | 8 | 2 | 0 | 0 | 2 | 2 | 0 | 0 | 0 |
| F3-123 | 11:17 | 6 | 1 | 7 | 8 | 1 | 3 | 0 | 2 | 1 | 1 | 0 | 2 |
| F3-1 | 11:17 | 4 | 4 | 8 | 7 | 2 | 1 | 1 | 1 | 2 | 0 | 0 | 1 |
| F3-96 | 11:17 | 6 | 3 | 8 | 6 | 0 | 1 | 1 | 3 | 0 | 0 | 0 | 1 |
| F3-106 | 12:16 | 7 | 3 | 6 | 6 | 0 | 2 | 3 | 1 | 0 | 1 | 0 | 1 |
| F3-139 | 11:17 | 6 | 1 | 8 | 8 | 1 | 3 | 1 | 1 | 1 | 1 | 0 | 2 |
| F4-28 | 13:15 | 8 | 4 | 4 | 5 | 0 | 1 | 2 | 4 | 0 | 0 | 0 | 1 |
| F4-33 | 12:16 | 6 | 2 | 6 | 7 | 2 | 2 | 0 | 3 | 2 | 2 | 0 | 0 |
| F4-53 | 12:16 | 7 | 3 | 5 | 7 | 1 | 1 | 1 | 3 | 1 | 0 | 0 | 1 |
| F4-83 | 12:16 | 6 | 4 | 6 | 4 | 2 | 0 | 0 | 6 | 1 | 0 | 1 | 0 |
| F4-6 | 9:19 | 2 | 1 | 9 | 9 | 1 | 5 | 0 | 1 | 1 | 1 | 0 | 4 |
| F4-10 | 10:18 | 6 | 4 | 8 | 6 | 0 | 1 | 1 | 2 | 0 | 1 | 0 | 0 |
| F4-104 | 12:16 | 6 | 3 | 7 | 6 | 0 | 3 | 2 | 1 | 0 | 0 | 0 | 3 |
| F4-109 | 12:16 | 7 | 2 | 7 | 5 | 1 | 2 | 1 | 3 | 1 | 1 | 0 | 1 |
| F4-135 | 13:15 | 7 | 2 | 7 | 5 | 2 | 2 | 1 | 2 | 1 | 0 | 1 | 2 |
| F4-25 | 12:16 | 9 | 2 | 5 | 2 | 0 | 1 | 4 | 5 | 0 | 0 | 0 | 1 |
| Test resulta | n.s. | n.s. | P = 0.001 | n.s. | n.s. | n.s. | P = 0.001 | n.s. | n.s. | n.s. | n.s. | n.s. | P = 0.013 |
| Test resultb | n.a. | n.a. | n.a. | n.s. | P = 0.008 | P = 0.035 | n.s. | ||||||
Lp L. perenne chromosomes, Fp F. pratensis chromosomes, M+ Lp and Fp marked chromosomes, M− Lp and Fp non-marked chromosomes, m+ Lp and Fp chromosomal arms with rDNA locus, m− Lp and Fp chromosomal arms without rDNA locus, n.a. means not analyzed
n.s. P > 0.05
aThe distribution of M+/m+ and M−/m− values for non-recombined and recombined chromosomes as well as recombined arms between generations was compared, and significant differences between distributions for Lp and Fp were assessed using Pearson’s chi-squared test (P ≤ 0.05); statistically significant differences between generations were found both for Fp rDNA-carrying non-recombined and recombined chromosomes (at P = 0.001) and for Fp non-rDNA-carrying recombined arms (at P = 0.013)
bThe distribution of M+/m+ and M−/m− values for recombined chromosomes and arms between both genomes was compared, and significant differences between distributions for Lp and Fp were assessed using Pearson’s chi-squared test (P ≤ 0.05); statistically significant differences between Lp and Fp genomes were found with respect to the number of recombined chromosomes without marker (M−) (at P = 0.008), and also with respect to the number of recombined arms with marker (m+) (at P = 0.035)
The distribution of values for rDNA- (M+) and non-rDNA-bearing (M−) chromosomes between generations was compared (Table 2), and statistically significant structural differences for rDNA-bearing chromosomes between generations were found for non-recombined (P = 0.001) and recombined (P = 0.001) Fp rDNA-bearing ones, and also for Fp non-rDNA-bearing arms of recombined chromosomes with the marker (P = 0.013). In the case of the remaining structural characters given in Table 2, no statistically significant differences for Lp and Fp chromosomes between generations were found at the P < 0.05. The distribution of M+/m+ and M−/m− values for recombined chromosomes and arms between both genomes was also compared (Table 2 and Online Resource S3), and statistically significant differences between distributions for Lp and Fp genomes were found for P = 0.008 with respect to the number of recombined chromosomes without marker (M−), and also for P = 0.035 with respect to the number of recombined arms with marker (m+).
Discussion
Our results show a Festuca-like and Lolium-like dynamic pattern of chromosome variation in the F. pratensis × L. perenne hybrids, occurring during early, F2-F4, generations following hybridization. The numerous changes, which seemed to occur independently within the Fp and Lp genomes, often altered non-3L, non-2F, and non-3F chromosomes. The presence of rearrangements in rDNA-bearing chromosomes concerns chromosomal arms with or without rDNA loci. The comparison of rDNA profiles in plants observed in the F1, and then in F2-F4 generations revealed, as it was expected, further differentiation in number and position of rDNA loci and parental split of rDNA loci patterns in Fp × Lp hybrids.
Over the F2-F4, the proportion of rearranged rDNA-bearing and non-rDNA-bearing chromosomes of parental genomes increased from generation to generation, and the frequency was higher for non-marked chromosomes than for rDNA-carrying ones. The number of recombinant and non-recombinant Lp and Fp rDNA-bearing chromosomes, and the frequency of structural rearrangements in rDNA-bearing ones, also increased from generation to generation, although the respective value of these characters was always higher for Fp chromosomes with rDNAs. The hypothesis was that over the generations the variation of parental genomes is asymmetrical, as borne out in the present work, and significant differences in this variation were always biased in favor of the Fp chromosomes. The recombination pattern was consistent with previous observations recorded for Fp chromosomes, in which there was an increase in the recombination profile in plants from the F2 to the F8 generation in selected population of Fp × Lp hybrids (Zwierzykowski et al. 2006, 2011). The present work shows that the non-recombinant and recombinant Lp rDNA-bearing chromosomes remained on a comparable level over the generations, while the non-recombinant and recombinant Fp rDNA-bearing ones were much increased, and exceeded double the Lp value for Fp in the F4. This observation does not suggest, however, a greater capacity of the Lp genome to be structurally more stable than the Fp one, but the profile of Fp chromosomes to recombine more often than those of Lp ones has been already proved to some extent (Zwierzykowski et al. 2006, 2011). In the F2-F4 generations of the hybrids, we observed an asymmetrical pattern of rDNA-carrying chromosome variation in the number of recombinant Lp and Fp arms with and without rDNA loci. In the Lp and Fp recombination pattern of arms with and without rDNA locus, the distribution of values increased over the generations, but statistically significant difference between generations was found in the Fp genome for arms without rDNA locus. This indicates that in the case of marked chromosomes, the Fp genome was more affected by changes in arms without any rDNA locus. On the other hand, it is showed that statistically significant differences between Lp and Fp genomes were found in respect to the number of recombined chromosomes without rDNA locus (M−) and recombined arms with rDNA one (m+) (Online Resource S3b, c). This means that in the case of the number of recombined M− chromosomes, no recombination event is more frequent for Lp chromosomes, but two or three recombination events are more frequent for Fp ones (Online Resource S3b), although no statistically significant differences were found between distribution of these characters over the generations separately for Lp and Fp genomes. Similarly, no statistically significant differences were found between distribution of m+ character over the generations separately for Lp and Fp genomes, but the comparison of this character between Lp and Fp genomes showed significant difference; no recombination event is more frequent for Fp m+ chromosomes, but one or two recombination events are more frequent for Lp m+ ones (Online Resource S3c). This confirms our observations found in present work that statistically significant difference in the recombination profile was found for Fp-genome-like m− arms. On the contrary, statistically significant differences between distributions of M+ character were found over the generations separately for Lp and Fp genomes, but no difference was found between distributions of this character for Lp and Fp genomes, when compared. Again, such a general tendency of recombination profiles in Lp and Fp chromosomes seems to be in agreement with the previous data found in plants of the F2-F4 generations (Zwierzykowski et al. 2012), confirming a balance of chromatin, which progressively to favors the dominant Lolium genome and higher predisposition of Festuca chromosomes to be structurally more often modified (Zwierzykowski et al. 2006).
Recombination of chromosomes with arms carrying rDNA loci has also been found in wheat/rye translocations (Lukaszewski and Gustafson 1983), in Triticum × Dasypyrum hybrids (Minelli et al. 2005) and in allotetraploid Secale × Dasypyrum forms (Książczyk et al. 2011b). A similar approach using FISH/GISH has already been used in F1 plants of F. pratensis × L. perenne hybrids (Książczyk et al. 2010), in which existing cytological landmarks showed some vulnerability of particular Lp and Fp rDNA loci to change their position, especially when those sequences were located at a secondary constriction, which is relatively unstable chromosomal region (Schubert and Wobus 1985). Significantly, rDNAs may be targets of rearrangements, as was shown in newly synthesized allotetraploids of Brassica species (Książczyk et al. 2011a; Xiong et al. 2011). Chromosomal rearrangements may involve many processes, e.g., activation of transposable elements or epigenetic regulation (Soltis and Soltis 1993), as well as structural rearrangements such as inversions and translocations (Levin 2004). The variation in the number and location of 35S rDNA signals found in L. perenne can be due to the formation of breaks and/or gaps in 35S rDNA sites and can randomly fragment the 35S rDNA regions (Huang et al. 2008; Rocha et al. 2014). The incidence of fragile sites may be involved in the process of chromosomal variation, including rearrangements and amplifications constituting a potential mechanism for speciation (Brown and O’Neill 2010) and its role in evolution, by asserting that fragile sites may generate chromosomal instability as representing fragile regions of the genome and are able to undergo recombination events (Ruiz-Herrera and Robinson 2007). Thus, the variation of rDNAs has led to the hypothesis that rDNA clusters are mobile (Schubert and Wobus 1985) and that some rDNA changes in chromosomal location may be activated by transposons enabling the traveling of (r)DNA to a new site (Raskina et al. 2004). A transposase-mediated transposition of rDNA might be postulated as the key mechanism in chromosome evolution (Raskina et al. 2004; Datson and Murray 2006; Pedrosa-Harand et al. 2006), and such a model could be responsible for the presence of distally located new loci of 5S rDNA within F. pratensis cultivars (Książczyk et al. 2010). It has also been found by many authors that retroelements play a major role in shaping and remodeling genomes during evolution by their influence on chromosome stability (Feuillet and Keller 2002). Langdon et al. (2000) have shown that a single ancestral family of retrotransposons related to the Ty3-gypsy family is the source of all Poaceae centromere-specific retroelement sequences. In solanaceous species, maize, rice and Arabidopsis, terminal-repeat retrotransposons in miniature (TRIM), the smallest known LTR retrotransposons, can be mobilized by other retroelements and are found to be actively involved in the reshaping of their genomes (Witte et al. 2001). It is anticipated that TRIM-like elements might exist in forage grasses, such as L. perenne and F. pratensis, and could be involved in some rDNA mobility. In the Fp genome, the main 5S rDNA loci are closely embedded in pericentromeric heterochromatin that is typically rich in transposable elements, so a transposon-mediated rearrangement could contribute to a loss or transposition of 5S rDNA sequences observed in Fp-genome-like chromosomes of Fp × Lp hybrids, suggesting extensive chromosome rearrangements resulting from genome imbalance during polyploid formation, as it was recently shown in Tragopogon allotetraploids (Malinska et al. 2010). The question is whether a similar model of (5S) rDNA mobility is present in the tetraploid F. pratensis × L. perenne hybrids? Thus, various proportions of centromere-specific retroelements between the two syntenic 3F and 3L chromosomes carrying rDNA loci in the proximal region might account for differences found in the recombination of arms with and without rDNA loci between both genomes.
Changes of rDNA sites, e.g., gaining of 5S ribosomal RNA (rRNA) genes, appears to have occurred more frequently in four tetraploid F1 plants of F. pratensis × L. perenne hybrid (Książczyk et al. 2010), being used to obtain the F2 generation. In the present work, the Fp genome-like 5S rDNA loci found in unrecognized chromosomes were more affected by numerical changes than the Fp and Lp genome-like 5S rDNA ones found in homologues of chromosome 3. This postulates the existence of genome-dependent dynamics of 5S rDNA loci pattern. In the majority of studied Fp × Lp hybrids, the amplification of 5S rRNA gene loci in Fp genome-like chromosomes was observed, and such additional and near-terminally located 5S rDNA sites were also found in F1 plants previously studied (Książczyk et al. 2010). The origin and uniparental extent of 5S rDNA variation remains unclear, although in some plants, “novel” or migrated loci were positioned at near-terminal and terminal regions of the chromosomes (Li and Zhang 2002), suggesting that the loci might change position through dispersion of minor loci without chromosome rearrangements (Dubcovsky and Dvorák 1995). We think that this mechanism could explain the appearance of novel 5S rDNA sites in Fp chromosomes. Interchromosomal exchanges might be facilitated by the terminal or near-terminal location of the rDNA loci, and the terminal location of the rDNA locus and the loss of its interstitial site suggests that concerted evolution of the particular rDNA locus (and sequence homogeneity) has probably occurred (Li and Zhang 2002), and inter-locus unequal crossing over could be proposed to play a role in concerted evolution (Wendel et al. 1995; Raskina et al. 2004; Pedrosa-Harand et al. 2006). The most intriguing aspect in the 5S rDNA variation observed in Fp-genome-like chromosomes in the hybrids is the appearance of novel 5S rDNA loci, which may result from transposon activity and may not be associated with the loss of major 5S rDNA sites on Fp homologues of chromosome 3 (the loss of 5S rDNA site resulted rather from an incomplete number of Fp chromosomes in some plants of the F2-F4 generations), and it is likely that the Fp genome does not address this hypothesis in a similar way as proposed for concerted evolution in other plants. Further analyses are necessary to prove or reject this hypothesis as well as for deeper understanding of the mechanisms responsible for the genome-dependent rDNA dynamics in genomes of the Festuca-Lolium complex.
Conclusions
Our results show that the chromosome variation in plants of the F2-F4 generations derived from the F1 hybrid of F. pratensis × L. perenne seem to argue for genome-dependent dynamics of chromosome changes. They also show the independent character of rDNA loci patterns within L. perenne and F. pratensis genomes. A statistically significant difference between distributions of values for 35S rDNA loci over the generation was found for F. pratensis genome-like chromosome 2, and F. pratensis genome-like chromosomes were more affected by rDNA loci changes, showing the presence of an additional 5S rDNA locus found in unrecognized F. pratensis chromosomes. Moreover, statistically significant differences between parental genomes were found both for non-recombined and recombined F. pratensis rDNA-bearing chromosomes (M+) and for recombined F. pratensis non-rDNA-carrying arms (m−) of marked chromosomes. Statistically significant differences between L. perenne and F. pratensis genomes were also found for recombined F. pratensis chromosomes without marker (M−) and recombined F. pratensis chromosomal arms of marked chromosomes (m+), indicating a tendency of F. pratensis genome-like chromosomes to be less stable in plants of the F2-F4 generations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
(DOC 44 kb)
Ideograms of the rDNA loci number and position in L. perenne (A), F. pratensis (B) and a hypothetical model of F1 karyotypes illustrating rDNA-bearing chromosome complement in F. pratensis (4x) × L. perenne (4x) hybrid (C). The rDNA loci pattern of L. perenne and F. pratensis is taken from Książczyk et al. (2010). (PDF 103 kb)
Frequency profile of complete and recombinant L. perenne and F. pratensis rDNA-bearing and non-rDNA-bearing chromosomes in plants of the F2-F4 generations derived from F. pratensis (4x) × L. perenne (4x) hybrid. M+ Lp and Fp rDNA-bearing chromosomes, M- Lp and Fp non-rDNA-bearing chromosomes, M+(L) Lp and M+(F) Fp rDNA-bearing chromosomes, M-(L) Lp and M-(F) Fp non-rDNA-bearing chromosomes, M+(L)R recombinant Lp rDNA-bearing chromosomes, M+(F)R recombinant Fp rDNA-bearing chromosomes, M+(L + F)R total number of recombinant Lp and Fp rDNA-bearing chromosomes, M-(L + F)R total number of recombinant Lp and Fp non-rDNA-bearing chromosomes. (PDF 51 kb)
Acknowledgments
This work was supported by the Polish Ministry of Science and Higher Education (grant no. N N310 090736). We are grateful to Mr. Włodzimierz Zwierzykowski (Institute of Plant Genetics PAS, Poznań, Poland) for excellent technical assistance. We also thank Prof. R. Neil Jones (Aberystwyth University, Aberystwyth, UK) for critical reading and English revision of the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Brown JD, O’Neill RJ. Chromosomes, conflict, and epigenetics: chromosomal speciation revisited. Annu Rev Genom Hum G. 2010;11:291–316. doi: 10.1146/annurev-genom-082509-141554. [DOI] [PubMed] [Google Scholar]
- Cai Q, Zhang D, Liu Z, Wang XR. Chromosomal localization of 5S and 18S rDNA in five species of subgenus Strobus and their implications for genome evolution of Pinus. Ann Bot. 2006;97:715–722. doi: 10.1093/aob/mcl030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacón J, Sousa A, Baeza CM, Renner SS. Ribosomal DNA distribution and a genus-wide phylogeny reveal patterns of chromosomal evolution in Alstroemeria (Alstroemeriaceae) Am J Bot. 2012;99(9):1501–1512. doi: 10.3732/ajb.1200104. [DOI] [PubMed] [Google Scholar]
- Datson PM, Murray BG. Ribosomal DNA locus evolution in Nemesia: transposition rather than structural rearrangement as the key mechanism. Chromosome Res. 2006;14:845–857. doi: 10.1007/s10577-006-1092-z. [DOI] [PubMed] [Google Scholar]
- D’Hont A. Unraveling the genome structure of polyploids using FISH and GISH; examples of sugarcane and banana. Cytogenet Genome Res. 2005;109(1–3):27–33. doi: 10.1159/000082378. [DOI] [PubMed] [Google Scholar]
- Dubcovsky J, Dvorák J. Ribosomal RNA multigene loci: nomads of the Triticeae genomes. Genetics. 1995;140:1367–1377. doi: 10.1093/genetics/140.4.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuillet C, Keller B. Comparative genomics in the grass family: molecular characterization of grass genome structure and evolution. Ann Bot. 2002;89(1):3–10. doi: 10.1093/aob/mcf008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach WL, Dyer TA. Sequence organisation of the repeating units in the nucleus of wheat which contain 5S rRNA genes. Nucleic Acids Res. 1980;8:4851–4865. doi: 10.1093/nar/8.21.4851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper J, Armstead I, Thomas A, James C, Gasior D, Bisaga M, Roberts L, King I, King J. Alien introgression in the grasses Lolium perenne (perennial ryegrass) and Festuca pratensis (meadow fescue): the development of seven monosomic substitution lines and their molecular and cytological characterization. Ann Bot. 2011;107(8):1313–1321. doi: 10.1093/aob/mcr083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JA, Thomas ID, Lovatt JA, Thomas HM. Physical mapping of rDNA sites in possible diploid progenitors of polyploid Festuca species. Plant Syst Evol. 2004;245:163–168. doi: 10.1007/s00606-003-0110-2. [DOI] [Google Scholar]
- Huang J, Ma L, Yang F, Fei Y, Fei S, Li L. 45S rDNA regions are chromosome fragile sites expressed as gaps in vitro on metaphase chromosomes of root-tip meristematic cells in Lolium spp. PLoS ONE. 2008;3:1–7. doi: 10.1371/journal.pone.0002167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys MW, Pašakinskienė I. Chromosome painting to locate genes for drought resistance transferred from Festuca arundinacea into Lolium multiflorum. Heredity. 1996;77:530–534. doi: 10.1038/hdy.1996.180. [DOI] [Google Scholar]
- King IP, Morgan WG, Armstead IP, Harper JA, Hayward MD, Bollard A, Nash JV, Forster JW, Thomas HM. Introgression mapping in the grasses I Introgression of Festuca pratensis chromosomes and chromosome segments into Lolium perenne. Heredity. 1998;81:462–467. doi: 10.1046/j.1365-2540.1998.00437.x. [DOI] [Google Scholar]
- Kopecký D, Loureiro J, Zwierzykowski Z, Ghesquière M, Doležel J. Genome constitution and evolution in Lolium × Festuca hybrid cultivars (Festulolium) Theor Appl Genet. 2006;113:731–742. doi: 10.1007/s00122-006-0341-z. [DOI] [PubMed] [Google Scholar]
- Kosmala A, Zwierzykowski Z, Gąsior D, Rapacz M, Zwierzykowska E, Humphreys MW. GISH/FISH mapping of genes for freezing tolerance transferred from Festuca pratensis to Lolium multiflorum. Heredity. 2006;96:243–251. doi: 10.1038/sj.hdy.6800787. [DOI] [PubMed] [Google Scholar]
- Książczyk T, Apolinarska B, Kulak-Książczyk S, Wiśniewska H, Stojałowski S, Łapiński M. Identification of the chromosome complement and the spontaneous 1R/1V translocations in allotetraploid Secale cereale × Dasypyrum villosum hybrids through cytogenetic approaches. J Appl Genet. 2011;52:305–311. doi: 10.1007/s13353-011-0048-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Książczyk T, Kovarik A, Eber F, Huteau V, Khaitova L, Tesarikova Z, Coriton O, Chèvre AM. Immediate unidirectional epigenetic reprogramming of NORs occurs independently of rDNA rearrangements in synthetic and natural form of a polyploid species Brassica napus. Chromosoma. 2011;120(6):557–571. doi: 10.1007/s00412-011-0331-z. [DOI] [PubMed] [Google Scholar]
- Książczyk T, Taciak M, Zwierzykowski Z. Variability of ribosomal DNA sites in Festuca pratensis, Lolium perenne, and their intergeneric hybrids, revealed by FISH and GISH. J Appl Genet. 2010;51(4):449–460. doi: 10.1007/BF03208874. [DOI] [PubMed] [Google Scholar]
- Książczyk T, Zwierzykowski Z, Zwierzykowska E (2012) Chromosomal rearrangements in BC1 progeny obtained from crosses of tetraploid F. pratensis × L. perenne hybrids with tetraploid L. perenne. In: Barth S, Milbourne D (eds) Breeding strategies for sustainable forage and turf grass improvement. Springer, Netherlands, pp 97–101
- Langdon T, Seago C, Mende M, Leggett M, Thomas H, Forster JW, Jones RN, Jenkins G. Retrotransposon evolution in diverse plant genomes. Genetics. 2000;156(1):313–325. doi: 10.1093/genetics/156.1.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin DA. The role of chromosome change in plant evolution. Syst Botany. 2004;29(2):460–461. doi: 10.1600/036364404774195449. [DOI] [Google Scholar]
- Li D, Zhang X. Physical localization of the 18S-5.8S-26S rDNA and sequence analysis of ITS regions in Thinopyrum ponticum (Poaceae: Triticeae): Implications for concerted evolution. Ann Bot. 2002;90(4):445–452. doi: 10.1093/aob/mcf213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukaszewski AJ, Gustafson JP. Translocations and modifications of chromosomes in Triticale × wheat hybrids. Theor Appl Genet. 1983;64:239–248. doi: 10.1007/BF00303771. [DOI] [PubMed] [Google Scholar]
- Malinska H, Tate JA, Matyasek R, Leitch AR, Soltis DE, Soltis PS, Kovarik A. Similar patterns of rDNA evolution in synthetic and recently formed natural populations of Tragopogon (Asteraceae) allotetraploids. BMC Evol Biol. 2010;10(1):291. doi: 10.1186/1471-2148-10-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maluszynska J, Hasterok R. Identification of individual chromosomes and parental genomes in Brassica juncea using GISH and FISH. Cytogenet Genome Res. 2005;109:310–314. doi: 10.1159/000082414. [DOI] [PubMed] [Google Scholar]
- Minelli S, Ceccarelli M, Mariani M, De Pace C, Cionini PG. Cytogenetics of Triticum × Dasypyrum hybrids and derived lines. Cytogenet Genome Res. 2005;109:385–392. doi: 10.1159/000082424. [DOI] [PubMed] [Google Scholar]
- Payne RW, Murray DM, Harding SA, Baird DB, Soutar DM. An introduction to GenStat for Windows (15th Edition) Hemel Hempstead, UK: VSN International; 2012. [Google Scholar]
- Pedrosa-Harand A, de Almeida CCS, Mosiolek M, Blair MW, Schweizer D, Guerro M. Extensive ribosomal DNA amplification during Andean common bean (Phaseolus vulgaris L.) evolution. Theor Appl Genet. 2006;112:924–933. doi: 10.1007/s00122-005-0196-8. [DOI] [PubMed] [Google Scholar]
- Raskina O, Belyayev A, Nevo E. Activity of the En/Spm-like transposons in meiosis as a base for chromosome repatterning in a small, isolated, peripheral population of Aegilops speltoides Tausch. Chromosome Res. 2004;12:153–161. doi: 10.1023/B:CHRO.0000013168.61359.43. [DOI] [PubMed] [Google Scholar]
- Robledo G, Lavia GI, Seijo G. Species relations among wild Arachis species with the A genome as revealed by FISH mapping of rDNA loci and heterochromatin detection. Theor Appl Genet. 2009;118:1295–1307. doi: 10.1007/s00122-009-0981-x. [DOI] [PubMed] [Google Scholar]
- Rocha LC, de Oliveira Bustamante F, Silveira RAD, Torres GA, Mittelmann A, Techio VH. Functional repetitive sequences and fragile sites in chromosomes of Lolium perenne L. Protoplasma. 2014 doi: 10.1007/s00709-014-0690-4. [DOI] [PubMed] [Google Scholar]
- Ruiz-Herrera A, Robinson TJ. Afrotherian fragile sites, evolutionary breakpoints and phylogenetic inference from genomic assemblies. BMC Evol Biol. 2007;7:1–15. doi: 10.1186/1471-2148-7-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert I, Wobus U. In situ hybridization confirms jumping nucleolus organizing regions in Allium. Chromosoma. 1985;92:143–148. doi: 10.1007/BF00328466. [DOI] [Google Scholar]
- Soltis DE, Soltis PS. Molecular data and the dynamic nature of polyploidy. Crit Rev Plant Sci. 1993;12:243–273. doi: 10.1080/07352689309701903. [DOI] [Google Scholar]
- Thomas HM. The Giemsa C-band karyotypes of six Lolium species. Heredity. 1981;46:263–267. doi: 10.1038/hdy.1981.33. [DOI] [Google Scholar]
- Thomas HM, Harper JA, Morgan WG. Gross chromosome rearrangements are occurring in an accession of the grass Lolium rigidum. Chromosome Res. 2001;9:585–590. doi: 10.1023/A:1012499303514. [DOI] [PubMed] [Google Scholar]
- Thomas HM, Harper JA, Meredith MR, Morgan WG, King IP. Physical mapping of ribosomal DNA sites in Festuca arundinacea and related species by in situ hybridization. Genome. 1997;40:406–410. doi: 10.1139/g97-054. [DOI] [PubMed] [Google Scholar]
- Thomas HM, Harper JA, Meredith MR, Morgan WG, Thomas ID, Timms E, King IP. Comparison of ribosomal DNA sites in Lolium species by fluorescence in situ hybridization. Chromosome Res. 1996;4:486–490. doi: 10.1007/BF02261775. [DOI] [PubMed] [Google Scholar]
- Thomas HM, Morgan WG, Meredith MR, Humphreys MW, Thomas H, Leggett JM. Identification of parental and recombined chromosomes in hybrid derivatives of Lolium multiforum × Festuca pratensis by genomic in situ hybridization. Theor Appl Genet. 1994;88:909–913. doi: 10.1007/BF00220795. [DOI] [PubMed] [Google Scholar]
- Unfried I, Gruendler P. Nucleotide sequence of the 5.8S and 25S rRNA genes and the internal transcribed spacers from Arabidopsis thaliana. Nucleic Acids Res. 1990;18:4011. doi: 10.1093/nar/18.13.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan T, Zhang XL, Gregan J, Zhang Y, Guo P, Guo JH. A dynamic evolution of chromosome in subgenus Potamogeton revealed by physical mapping of rDNA loci detection. Plant Syst Evol. 2012 [Google Scholar]
- Wendel JF, Schnabel A, Seelanan T (1995) Bidirectional interlocus concerted evolution following allopolyploid speciation in cotton (Gossypium). Proc Natl Acad Sci USA 92: 280–284 [DOI] [PMC free article] [PubMed]
- Witte CP, He QH, Bureau T, Kumar A. Terminal-repeat retrotransposons in miniature (TRIM) are involved in restructuring plant genomes. Proc Natl Acad Sci U S A. 2001;98(24):13778–13783. doi: 10.1073/pnas.241341898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolny E, Lesniewska K, Hasterok R, Langdon T. Compact genomes and complex evolution in the genus Brachypodium. Chromosoma. 2011;120(2):199–212. doi: 10.1007/s00412-010-0303-8. [DOI] [PubMed] [Google Scholar]
- Xiong Z, Gaeta R, Pires C. Homoeologous shuffling and chromosome compensation maintain genome balance in resynthesized allopolyploid Brassica napus. Proc Natl Acad Sci U S A. 2011;108:7908–7913. doi: 10.1073/pnas.1014138108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwierzykowski Z, Kosmala A, Zwierzykowska E, Jones N, Jokś W, Bocianowski J. Genome balance in six successive generations of the allotetraploid Festuca pratensis × Lolium perenne. Theor Appl Genet. 2006;113:539–547. doi: 10.1007/s00122-006-0322-2. [DOI] [PubMed] [Google Scholar]
- Zwierzykowski Z, Książczyk T, Taciak M, Zwierzykowska E, Jones N, Kosmala A (2012) Genome constitution in selected and unselected plants of F2-F4 generations derived from an allotetraploid Festuca pratensis × Lolium perenne hybrid. In: Barth S, Milbourne D (eds) Breeding strategies for sustainable forage and turf grass improvement. Springer, Netherlands, pp 97–101
- Zwierzykowski Z, Tayyar R, Brunell M, Lukaszewski AJ. Genome recombination in intergeneric hybrids between tetraploid Festuca pratensis and Lolium multiflorum. J Hered. 1998;89(4):324–328. doi: 10.1093/jhered/89.4.324. [DOI] [Google Scholar]
- Zwierzykowski Z, Zwierzykowska E, Taciak T, Jones N, Kosmala A, Krajewski P. Chromosome pairing in allotetraploid hybrids of Festuca pratensis × Lolium perenne revealed by genomic in situ hybridization (GISH) Chromosome Res. 2008;16:575–585. doi: 10.1007/s10577-008-1198-6. [DOI] [PubMed] [Google Scholar]
- Zwierzykowski Z, Zwierzykowska E, Taciak M, Kosmala A, Jones RN, Zwierzykowski W, Książczyk T, Krajewski P. Genomic structure and fertility in advanced breeding populations derived from an allotetraploid Festuca pratensis × Lolium perenne cross. Plant Breed. 2011;130:476–480. doi: 10.1111/j.1439-0523.2010.01839.x. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC 44 kb)
Ideograms of the rDNA loci number and position in L. perenne (A), F. pratensis (B) and a hypothetical model of F1 karyotypes illustrating rDNA-bearing chromosome complement in F. pratensis (4x) × L. perenne (4x) hybrid (C). The rDNA loci pattern of L. perenne and F. pratensis is taken from Książczyk et al. (2010). (PDF 103 kb)
Frequency profile of complete and recombinant L. perenne and F. pratensis rDNA-bearing and non-rDNA-bearing chromosomes in plants of the F2-F4 generations derived from F. pratensis (4x) × L. perenne (4x) hybrid. M+ Lp and Fp rDNA-bearing chromosomes, M- Lp and Fp non-rDNA-bearing chromosomes, M+(L) Lp and M+(F) Fp rDNA-bearing chromosomes, M-(L) Lp and M-(F) Fp non-rDNA-bearing chromosomes, M+(L)R recombinant Lp rDNA-bearing chromosomes, M+(F)R recombinant Fp rDNA-bearing chromosomes, M+(L + F)R total number of recombinant Lp and Fp rDNA-bearing chromosomes, M-(L + F)R total number of recombinant Lp and Fp non-rDNA-bearing chromosomes. (PDF 51 kb)


