Abstract
Background
There are a number of studies regarding to the susceptibility of A20 SNPs in rheumatoid arthritis (RA); however, a few of these studies have shown an association between polymorphisms in the A20 gene and RA risk in the Chinese population. The aim of this study was to investigate the characteristics of A20 gene polymorphisms, the association between polymorphisms and clinical significance in Chinese RA patients.
Methods
PCR and sequencing were used to identify A20 gene polymorphisms in peripheral blood mononuclear cells (PBMCs) (50 cases), synovial fluid (11 cases) from RA patients and PBMCs from 30 healthy individuals. Quantitative Real-time PCR (qRT-PCR) was used to analyze the A20 mRNA expression in 38 RA patients and 40 healthy individuals. Pearson’s Chi square test and two independent-samples Wilcoxon tests were used for statistical analysis.
Results
Eight single nucleotide polymorphisms (SNPs) (rs5029937, rs3799491, rs598493, rs2307859, rs146534657, rs2230926, rs661561, and rs582757) were identified in PBMCs of RA patients. One new mutation (14284 T > A) was identified in synovial fluid mononuclear cells from one RA case. rs146534657 was identified for the first time in two RA cases. Patients with rs146534657 (12411 A > G, Asn102Ser) AG genotype or rs2230926 (12486 T > G, Phe127Cys) TG genotype had poor outcome. Significantly lower A20 mRNA expression was found in PBMCs from RA patients compared with healthy individuals (p < 0.001). There was a higher A20 mRNA expression in RA patients with rs2230926 TG genotype and rs146534657 AG genotype (11.56 ± 7.39) than patients with rs2230926 TT genotype and rs146534657 AA genotype (5.63 ± 4.37) (p = 0.031).
Conclusion
Significantly lower A20 expression was found in RA patients. The polymorphisms of A20 were characterized in RA patients. We detected rs146534657 for the first time and identified a new A20 mutation (14284 T > A). A20 rs2230926 TG genotype and rs146534657 AG genotype may be related to poor outcome in RA patients.
Electronic supplementary material
The online version of this article (doi:10.1186/s12967-015-0566-1) contains supplementary material, which is available to authorized users.
Keywords: A20 gene, Polymorphism, Mutation, Rheumatoid arthritis
Background
Rheumatoid arthritis (RA) is a systemic autoimmune inflammatory disease characterized by proliferative synovitis, the infiltration of inflammatory cells into synovial tissue, progressive joint destruction and disability. Several changes in the T cell compartment have been described in immune-mediated diseases [1]. Abnormal T cell immunity plays a critical role in the development of RA. Inflammatory mediators, such as interleukin-6 (IL-6), interleukin-1 (IL-1) and tumor necrosis factor alpha (TNF-α), are significantly over expressed in RA. Recently, RA treatment has been transformed by the development of biologics targeting TNF-α and IL-6 [2]. Moreover, many inflammatory mediators involved in the pathology of RA are regulated by nuclear factor kappa B (NF-κB) transcription factors [3]. Overexpression of NF-κB is a common characteristic of RA. Although the RA etiology is not fully understood, it is known that a strong genetic component plays a major role in this disease [4, 5].
A20, also known as TNFAIP3, was first identified as a TNF primary response transcript encoding a 790 amino acid protein with a unique zinc finger motif, and it is a ubiquitin-editing enzyme that is an essential negative regulator of inflammation via its zinc finger domains in C-terminus and OUT (ovarian tumor) domain in N-terminus. A20 acts as a negative-feedback regulator of NF-κB activation in response to multiple stimuli, including TNF, IL-1, TLR (Toll-like receptor) and NLR [Nod (nucleotide-binding oligomerization domain)-like receptor] ligands [6, 7].
Genome-wide association studies have implicated the A20 locus in susceptibility to multiple autoimmune diseases in different cohorts, including RA, systemic lupus erythematosus (SLE), psoriasis, celiac disease, type 1 diabetes, inflammatory bowel disease, and coronary artery disease. Alterations in the activity or expression of A20 may influence the pathogenesis of RA [8–10]. Many A20 single nucleotide polymorphisms (SNPs) were found to be associated with the susceptibility to autoimmune disease [11–13].
There are a number of studies regarding the susceptibility of A20 SNPs in RA [11]. However, only a few of these studies have found an association between A20 gene polymorphisms and the risk of RA in Chinese population [14]. Moreover, there are no data comparing the characteristics of A20 polymorphisms and mutations in different fluids and tissues of RA. Thus, in this study, we analyzed the distribution of A20 gene polymorphisms in peripheral blood and synovial fluid as well as the A20 expression level, and evaluated the potential association of A20 polymorphisms with clinical characteristics of RA in Chinese population.
Methods
Study population
This study included 50 cases with untreated RA (7 males and 43 females, age: 13–74 years, median age: 55.6 years), and 40 healthy individuals (6 males and 34 females, age: 18–70 years, median age: 55.0 years) served as controls. (Among them, DNA samples were isolated from 50 RA patients and 30 healthy for SNPs analysis, while RNA samples were isolated from 38 RA patients and 40 healthy individuals for A20 gene expression level detection). There was no significant difference in the distribution of the genders or ages between the cases and controls. RA diagnoses were based on the American College of Rheumatology criteria and expert opinion (1987 ACR criteria) [15]. All RA patients were assessed for clinical disease activity by a trained rheumatologist using disease activity score 28 (DAS 28), and their erythrocyte sedimentation rate (ESR), C reactive protein (CRP), rheumatoid factor (RF), and anti-cyclic citrullinated peptide antibody (CCP) were collected [16]. RF and CRP were measured by immune nephelometry, and anti-CCP antibody was measured by enzyme-linked immunoabsorbent assay (ELISA). In the active phase, all of the parameters were abnormal, including the ESR: 69.61 ± 34.63 mm/h, CRP: 35.67 ± 28.68 mg/l, RF: 247.89 ± 377.42 IU/ml, DAS28: 7.50 ± 1.28, and anti-CCP antibody positivity was found for 29 patients, while anti-CCP antibody negativity was found for 16 patients, anti-CCP antibody was not detected in 5 patients. The healthy individuals were in healthy status without any cancer, type 2 diabetes, hypertension, or autoimmune inflammatory disease. Neither RA patients nor healthy individuals smoked. Peripheral blood mononuclear cells (PBMCs) were isolated from heparinized venous blood by Ficoll-Paque gradient centrifugation. The synovial fluid was collected from RA patients whose knees were examined for therapeutic purpose. RNA and DNA extraction and cDNA synthesis from PBMCs were performed according to the manufacturer’s instructions (Trizol, Invitrogen, USA, and Superscript III Kit, Gibco, Gaithersburg, MD, USA) [17, 18]. All procedures were conducted according to the guidelines of the Medical Ethics Committee of the Health Bureau of the Guangdong Province in China.
PCR and sequencing
To amplify different domains of genomic DNA that cover A20 exons 2–9 (coding region) and exon/intron junctions of the A20 gene according to the structure of the A20 gene, 11 pairs of primers were purchased (Additional file 1: Table S1). PCR was performed as described in our previous study [19]. The PCR products were used for mutation analysis of the A20 coding sequence by direct sequencing using the Big Dye Terminator v3.1 Cycle Sequencing Kit (Perkin Elmer, ABI) and the ABI PRISM 3100-Avant genetic analyzer. Direct sequencing was performed by Invitrogen Biotechnology Company. Sequences of different samples from patients with RA and healthy individuals were analyzed with BLAST software (http://blast.ncbi.nlm.nih.gov/Blast.cgi) to identify polymorphisms or mutations in the A20 gene.
Quantitative Real-time PCR (qRT-PCR)
To compare the A20 mRNA expression level, qRT-PCR was performed using specific A20 primers (Additional file 1: Table S1), and the relative amount of the genes of interest and the β2M reference gene was measured in two independent assays. Specific amplification of the PCR products was analyzed by melting curve analysis. The data were presented as the relative expression of the genes of interest compared with the internal control gene as determined by the method. qRT-PCR was performed as previously described [20–22].
Statistical analysis
All statistical analyses were performed with SPSS (v. 13.0) software. A P value of less than 0.05 was considered statistically significant. Pearson’s Chi square test was used to compare the distribution of genotypes and alleles between RA group and healthy control group. In cases in which the genotype had a frequency of less than 1, Fisher’s exact test was applied. Two independent-samples Wilcoxon tests were performed to compare the median A20 mRNA expression level of RA group and controls group.
Results
The distribution and frequency of A20 SNPs in RA
We used 11 primer pairs, which cover the A20 coding region, and several introns to amplify segments of A20 genomic DNA, and positive PCR products were confirmed by sequencing. DNA samples from 50 cases with RA and 30 healthy individuals were selected for this part of the study. We detected eight SNPs of A20 gene: rs5029937 (position 11571 with a nucleotide substitution of G to T), rs3799491 (position 11812, G > A), rs598493 (position 11822, T > C), rs2307859 (position 12387–12389 delCCT), rs146534657 (position 12411, A > G, resulting in an amino acid substitution at position 102 from asparagine to serine, Asn 102 Ser), rs2230926 (position 12486, T > G, resulting in an amino acid substitution at position 127 from phenylalanine to cysteine, Phe127Cys), rs661561 (position 13751, A > C) and rs582757 (position 14244, C > T).
Most SNPs are located in intron 2 and 5, while two SNPs (rs146534657 and rs2230926) are located in exon 3 (Table 1; Figures 1, 2). However, the SNP frequencies appeared to be lower in the detected samples; the highest frequency was 24% (12/50 cases) for rs3799491 GA/AA genotype, and the lowest was 4.0% (2/50 case) for rs5029937 GT genotype, rs146534657 AG genotype, and without rs2307859 genotype respectively. In addition, the genotypes of four SNPs, rs2307859 (CCT deletion), rs661561 (homozygous), rs582757 (homozygous) and rs598493 (homozygous), appeared to be common genetic alterations in RA as they were identified in 41 cases, while the remaining 9 cases demonstrated different genotypic characteristics. However, 6 of the 8 SNPs were also identified in the healthy control group with similar frequency, and rs146534657 AG genotype and rs2307859 wild genotype were not identified in the healthy controls. However, there was no statistically significant difference in the proportion of genotypic data between the RA group and healthy group (Table 2). Moreover, genetic alterations in RA and healthy samples with the rs2230926 TG genotype and/or rs146534657 AG genotype were different from those lacking both SNPs (Table 3).
Table 1.
Identified SNPs of the A20 gene in PBMCs and synovial fluid from RA
| SNP number | Chromosome position | Gene position | Major/minor | Function |
|---|---|---|---|---|
| rs5029937 | 137874014 | Intron 2 | G/T | – |
| rs3799491 | 137874255 | Intron 2 | G/A | – |
| rs598493 | 137874265 | Intron 2 | C/T | – |
| rs2307859 | 137874830 | Intron 2 | 12387_12389 delCCT | – |
| rs146534657 | 137874854 | Exon 3 | A/G | Asn102Ser |
| rs2230926 | 137874929 | Exon 3 | T/G | Phe127Cys |
| rs661561 | 137876194 | Intron 5 | C/A | – |
| rs582757 | 137876687 | Intron 5 | T/C | – |
| NEW1 (14284) | 137876727 | Intron 5 | T/A | – |
Figure 1.
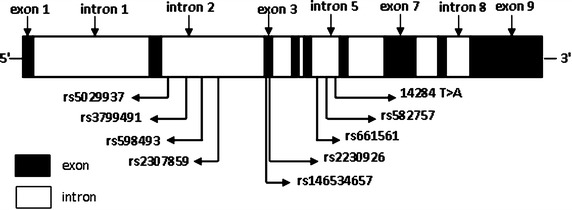
Location of SNPs in the A20 gene locus.
Figure 2.
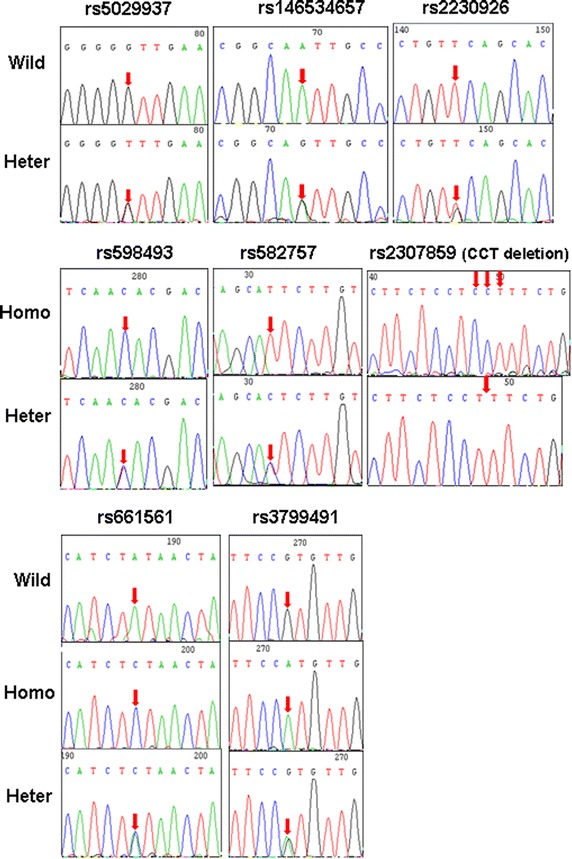
Identification of A20 SNPs in RA patients. Arrows indicate sites of nucleotide changes. Wild wild-type, Homo homozygous, Heter heterozygous.
Table 2.
Frequencies of the alleles and genotypes of the A20 polymorphisms in RA patients (case) and healthy individuals (control)
| SNPs | Genotype frequency n (%) | Allele frequency n (%) | ||||
|---|---|---|---|---|---|---|
| Major homozygote | Heterozygote | Minor homozygote | Major allele | Minor allele | ||
| rs5029937 | GG | GT | TT | G | T | |
| Case | 48 (96.0) | 2 (4.0) | 0 (0) | 98 (98.0) | 2 (2.0) | |
| Control | 28 (93.3) | 2 (6.7) | 0 (0) | 58 (96.7) | 2 (3.3) | |
| p-value | – | 1.000 | – | 1.000 | ||
| rs3799491 | GG | GA | AA | GA/AA | G | A |
| Case | 38 (76.0) | 11 (22.0) | 1 (2.0) | 12 (24) | 87 (87.0) | 13 (13.0) |
| Control | 21 (70.0) | 9 (30.0) | 0 (0) | 9 | 51 (85.0) | 9 (15.0) |
| p-value | – | 0.454 | 1.000* | 0.555 | – | 0.722 |
| rs598493 | CC | TC | TT | C | T | |
| Case | 44 (88.0) | 6 (12.0) | 0 (0) | 94 (94.0) | 6 (6.0) | |
| Control | 26 (86.7) | 4 (13.3) | 0 (0) | 56 (93.3) | 4 (6.7) | |
| p-value | – | 1.000 | – | 1.000 | ||
| rs2307859 | –/CCT | Wild type | ||||
| Case | 48 (96.0) | 2 (4.0) | ||||
| Control | 30 (100) | 0 (0) | ||||
| p-value | – | 0.525* | ||||
| rs146534657 | AA | AG | GG | A | G | |
| Case | 48 (96.0) | 2 (4.0) | 0 (0) | 98 (98.0) | 2 (2.0) | |
| Control | 30 (100) | 0 (0) | 0 (0) | 60 (100) | 0 (0) | |
| p-value | – | 0.525* | – | 0.528* | ||
| rs2230926 | TT | TG | GG | T | G | |
| Case | 47 (94.0) | 3 (6.0) | 0 (0) | 97 (97.0) | 3 (3.0) | |
| Control | 28 (93.3) | 2 (6.7) | 0 (0) | 58 (96.7) | 2 (3.3) | |
| p-value | – | 1.000 | – | 1.000 | ||
| rs661561 | CC | AC | AA | AC/AA | C | A |
| Case | 41 (82.0) | 8 (16.0) | 1 (2.0) | 9 (18) | 90 (90.0) | 10 (10.0) |
| Control | 24 (80) | 6 (20) | 0 (0) | 6 (20) | 54 (90.0) | 6 (10.0) |
| p-value | – | 0.678 | 1.000* | 0.824 | – | 1.000 |
| rs582757 | TT | CT | CC | T | C | |
| Case | 44 (88.0) | 6 (12.0) | 0 (0) | 94 (94.0) | 6 (6.0) | |
| Control | 26 (86.7) | 4 (13.3) | 0 (0) | 56 (93.3) | 4 (6.7) | |
| p-value | – | 1.000 | – | 1.000 | ||
n number of participants.
* Fisher’s exact p value.
Table 3.
A20 genetic alterations with rs230926 and rs146534657 in PBMCs of RA patients and healthy individuals
| Cases | rs5029937 | rs3799491 | rs598493 | rs2307859 | rs146534657 | rs2230926 | rs661561 | rs582757 |
|---|---|---|---|---|---|---|---|---|
| PHLa | Wild | Wild | Homo | Wild | Heter | Heter | Wild | Homo |
| LXSa | Wild | Heter | Homo | Del | Heter | Wild | Heter | Homo |
| WYFa | Heter | Wild | Homo | Wild | Wild | Heter | Heter | Homo |
| LWLa | Heter | Wild | Homo | Del | Wild | Heter | Homo | Homo |
| HI1 | Heter | Wild | Homo | Del | Wild | Heter | Heter | Homo |
| HI2 | Heter | Heter | Homo | Del | Wild | Heter | Heter | Homo |
HI healthy individuals, Wild wild-type, Homo homozygous, Heter heterozygous, Del deletion.
aRA cases.
Characteristics of A20 SNPs in synovial fluid from RA patients
We further analyzed the characteristics of A20 SNPs in cells from the synovial fluid and PBMCs from 11 cases with RA whose knees were examined for therapeutic purpose at the same time as peripheral blood collection. A total of seven A20 SNPs (rs5029937, rs3799491, rs598493, rs2307859, rs2230926, rs661561 and rs582757) were identified. The identical SNPs in the synovial fluid and peripheral blood were identified in 10 RA cases (Table 4), and a new mutation (14284 T > A) was identified in synovial fluid from only one RA case whose peripheral blood sample did not contain this mutation (Table 4; Figure 3).
Table 4.
A20 gene polymorphism characteristics in peripheral blood and synovial fluid from RA patients
| Case no. | Samples | rs5029937 | rs3799491 | rs598493 | rs2307859 | rs2230926 | rs661561 | rs582757 | 14284 T > A |
|---|---|---|---|---|---|---|---|---|---|
| 1 | S | Wild | Wild | Homo | Del | Wild | Homo | Homo | Wild |
| B | Wild | Wild | Homo | Del | Wild | Homo | Homo | Wild | |
| 2 | S | Wild | Wild | Homo | Del | Wild | Homo | Homo | Wild |
| B | Wild | Wild | Homo | Del | Wild | Homo | Homo | Wild | |
| 3 | S | Wild | Wild | Heter | Del | Wild | Heter | Heter | Wild |
| B | Wild | Wild | Heter | Del | Wild | Heter | Heter | Wild | |
| 4 | S | Wild | Wild | Homo | Del | Wild | Homo | Homo | Heter |
| B | Wild | Wild | Homo | Del | Wild | Homo | Homo | Wild | |
| 5 | S | Wild | Wild | Homo | Del | Wild | Homo | Homo | Wild |
| B | Wild | Wild | Homo | Del | Wild | Homo | Homo | Wild | |
| 6 | S | Heter | Wild | Homo | Wild | Heter | Heter | Homo | Wild |
| B | Heter | Wild | Homo | Wild | Heter | Heter | Homo | Wild | |
| 7 | S | Wild | Homo | Homo | Del | Wild | Homo | Homo | Wild |
| B | Wild | Homo | Homo | Del | Wild | Homo | Homo | Wild | |
| 8 | S | Wild | Wild | Homo | Del | Wild | Homo | Homo | Wild |
| B | Wild | Wild | Homo | Del | Wild | Homo | Homo | Wild | |
| 9 | S | Wild | Wild | Homo | Del | Wild | Homo | Homo | Wild |
| B | Wild | Wild | Homo | Del | Wild | Homo | Homo | Wild | |
| 10 | S | Wild | Heter | Homo | Del | Wild | Homo | Homo | Wild |
| B | Wild | Heter | Homo | Del | Wild | Homo | Homo | Wild | |
| 11 | S | Wild | Wild | Homo | Del | Wild | Homo | Homo | Wild |
| B | Wild | Wild | Homo | Del | Wild | Homo | Homo | Wild |
B peripheral blood, S synovial fluid, Wild wild-type, Homo homozygous, Heter heterozygous, Del deletion.
Figure 3.
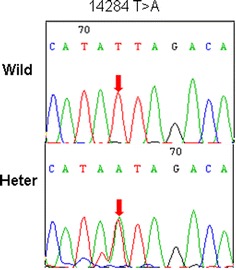
New mutation identified in A20 in synovial fluid from a patient with RA. Arrows indicate sites of nucleotide changes. Wild wild-type, Heter heterozygous.
The association between A20 SNPs and A20 expression level
We analyzed the A20 expression level in cDNA samples from 38 RA patients and 40 healthy individuals by real-time PCR, and a significantly lower A20 expression level (5.96 ± 4.82) was found compared with that in healthy individuals (34.54 ± 26.89) (p < 0.001) (Figure 4). We also compared the A20 expression level of different SNPs, it appeared that there was a higher A20 expression level in RA patients with rs2230926 TG genotype and rs146534657 AG genotype (11.56 ± 7.39) than patients with rs2230926 TT genotype and rs146534657 AA genotype (5.63 ± 4.37) (P = 0.031) (Figure 5). In addition, there was no significant difference of A20 expression level between RA patients with rs3799491 AA/GA genotypes (6.75 ± 5.25) and with rs3799491 GG genotype (5.61 ± 4.79) (P = 0.560) (Figure 6a). The A20 expression level in RA patients with rs661561 AA/AC genotypes (7.23 ± 6.82) appeared to be higher than with rs661561 CC genotype (5.59 ± 4.47); however, there was also no significant difference (p = 0.452) (Figure 6b).
Figure 4.
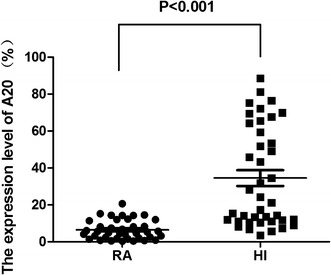
A20 expression level in RA patients and healthy individuals (HI).
Figure 5.
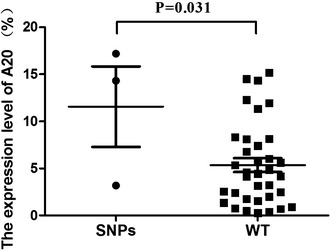
Comparison of A20 expression level between SNPs (RA patients with rs2230926 TG genotype and rs146534657 AG genotype) and WT (RA patients with rs2230926 TT genotype and rs146534657 AA genotype).
Figure 6.
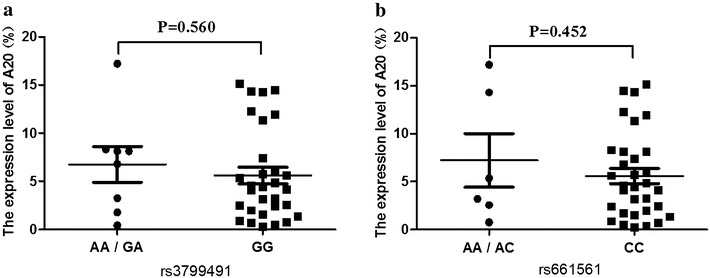
Comparison of the A20 expression level between samples with rs3799491 or rs661561 SNPs. a A20 expression level in patients with rs3799491 AA/GA genotypes and with rs3799491 GG genotype. b A20 expression level in patients with rs661561 AA/AC genotypes (heterozygous) and with rs661561 CC genotype (homozygous).
The association between A20 alteration and RA clinical characteristics
First, we compared the difference of SNPs and the status in RA patients with or without anti-CCP antibody positive, we found there were no correlation between the eight SNPs and anti-CCP antibody. We also found that there was no significant difference of A20 expression level between anti-CCP antibody positive and anti-CCP antibody negative RA groups (P > 0.05).
Among the 50 RA patients, rs2230926 TG genotype was identified in 3 RA patients, one patient both with rs2230926 TG genotype and rs146534657 AG genotype was poor response to immune suppression therapy, such as methotrexate, infliximab and anti-TNF-α antibody; the second and the third patients both with rs2230926 TG genotype and rs5029937 GT genotype suffered severe disease with systemic joint destruction. rs146534657 AG genotype was identified in two RA patients, the second patient suffered obvious joint deformity in multiple joints of the hands and the knee. Overall, the SNPs rs2230926 and/or rs146534657, which located in exon 3, appeared to be related to poor clinical outcome in RA. Moreover, there were no significantly different clinical characteristics for the patient with the newly detected mutation (14284 T > A).
Discussion
A20 plays a central role in the control of NF-kB activation, A20 also negative regulates NLRP3 inflammasome to protect against arthritis [23]. It is known to be associated with susceptibility to multiple autoimmune diseases [6]. Polymorphisms and mutations in the A20 gene are linked to various autoimmune diseases. Recent studies from different countries have reported that A20 is frequently inactivated by deletions and/or mutations in autoimmune diseases, including RA [11]. Previous studies of A20 in RA have demonstrated an association with several disease risk SNPs, including rs2230926, rs6920220 and rs10499194 [24]. However, the frequencies of the reported A20 abnormalities in different reports are dissimilar. Moreover, our previous study suggested that the A20 polymorphisms associated with disease susceptibility may be different for different ethnic groups [19]. Little is known about the genetic alteration characteristics of A20 gene in Chinese population. In this study, we evaluated the role of the polymorphisms in the A20 gene on the risk for RA.
A number of studies have demonstrated a strong association between rs2230926 and RA, and this SNP plays a functional role in the development of RA. rs2230926 is located in exon 3 of A20, and results in an amino acid (at 127 position) substitution from phenylalanine to cysteine (Phe127Cys). This risk allele (Cys127) leads to reduced inhibition of NF-kB activation or reduced A20 mRNA levels [25]. There is a significant association between rs2230926 and increased risk for SLE and RA in the Japanese population [26]. Moreover, the rs2230926 polymorphism is associated with RA susceptibility in Europeans and Asians [24]. In this study, we found only three cases with rs2230926 TG genotype in 50 RA samples; however, we also found two cases with rs2230926 TG genotype among 30 healthy individuals. Therefore, this preliminary result may indicate a lower incidence of rs2230926 TG/GG genotype in Chinese RA patients, as least in our study group. Although patients with rs2230926 TG genotype demonstrated refractory and poorer outcome, there was no significant association between RA patients and healthy individuals, which was similar to findings in our previous study on T cell acute lymphoblastic leukemia (T-ALL) [19]. Analogous results were reported by Orozco et al. who found that rs2230926 is not independently associated with RA [27]. In addition, findings from a Korean group showed that rs2230926 in A20 is not associated with RA susceptibility in the Korean population [28]. However, results from Zhang et al. showed that rs2230926 is associated with RA risk in the northern Chinese population [29]. Because the RA patients and healthy individuals in this study were all from southern China, whether there are different genotypes of A20 gene in northern and southern Chinese people remains an open question. An alternative may be that, due to the limited samples in this study, the association between rs2230926 and RA in Chinese population requires further investigation with a greater number of samples.
Interestingly, we found two RA patients with rs2230926 TG genotype did not accompany rs2307859 (CCT deletion) which was a common alteration in 48 RA patients and 30 healthy individuals, it may be possible that absence of rs2307859 genotype is a risk factor to RA when rs2230926 TG genotype is present. Moreover, the genetic alteration of A20 in one patient with both rs2230926 TG genotype and rs146534657 AG genotype and poor outcome appeared to be different from most RA patients because it appeared with rs661561 AA genotype, which was found in almost all of the RA patients and healthy individuals with homozygous (CC genotype) or heterozygous (AC genotype) [30]. Whether such genetic alterations in A20 leads to high risk for RA needs further investigation. In addition, similar genetic alterations in A20 were identified in T-ALL [19].
In this study, we detected rs146534657 for the first time in two RA patients. This SNP which was described in GenBank but not identified for any disease, is a nonsynonymous variant located in exon 3 of A20 and results in an amino acid (at position 102) substitution from asparagine to serine (Asn 102 Ser). According to the clinical information of the patients, those who had both rs2230926 TG genotype and rs146534657 AG genotype had poor outcome, and whether rs2230926 and rs146534657 are associated with refractory RA requires further investigation with more samples to demonstrate.
In this study, we also analyzed the alteration of SNPs in A20 that were reported in different autoimmune diseases, such as rs5029937, which was strongly associated with RA in a large European RA cohort [27, 31] and significantly associated with SLE in southwestern Chinese population [32]; rs582757, which was reported to be associated with rheumatic heart disease (RHD) in the Chinese Han population [33] and weakly associated with reduced risk for RA in Caucasians [34], also associated with psoriasis [35]; rs598493, which was significantly increased in the GG genotype in Graves’ disease (GD) [30]; and rs3799491, which has not been reported in RA or other diseases. However, in this study, rs5029937 GT genotype was identified only in two RA patients who with rs2230926 TG genotype and suffered severe disease with systemic joint destruction, and whether rs5029937 is associated with the destruction of bone and cartilage in RA need further investigation. Moreover, rs582757 and rs598493 were identified in all samples including RA patients and healthy individuals as having a homozygous or heterozygous genotype; thus, it could be concluded that these are a common genotype in the Chinese population. In addition, rs3799491 GA/AA genotype was found in 12 out of 50 RA patients and 9 out of 30 healthy individuals, and we found no significant difference in these alterations between RA patients and healthy individuals. These results may indicate that the A20 polymorphisms associated with RA susceptibility are different in different populations.
To define the A20 gene polymorphism characteristics of synovial fluid from RA patients, we compared A20 polymorphisms in samples from peripheral blood and synovial fluid from 11 RA patients. Interestingly, one new mutation (14284 T > A) was identified in synovial fluid from one RA patient. This result may indicate the possibility of inconsistent genetic alterations in different tissues from the same patient. However, the significance and frequency of this new mutation need further investigation.
As a NF-kB negative regulator, A20 is critical for limiting inflammation by terminating TNF-induced NF-kB responses in vivo [36]. We also analyzed the A20 expression level in PBMCs of RA patients, and a significantly lower A20 expression level may be related to enhanced NF-kB and RA pathogenesis [22, 37]. There was a negative relationship between the A20 mRNA expression level and RA activity. Interestingly, PBMCs from RA patients with the rs2230926 TG genotype and rs146534657 AG genotype had a higher A20 expression level compared with RA patients with rs2230926 TT genotype and rs146534657 AA genotype, and a similar finding was reported for T-ALL in our previous study [19]. This result may imply that such a rs2230926 TG genotype may be related to the maintenance of A20 expression level. However, this hypothesis is based only on results from a limited case analysis, and further research involving more samples is needed to determine representative results. Moreover, this finding appeared to be inconsistent with the disease status of patients; thus, remaining an open question.
Conclusion
In conclusion, we characterized the A20 gene polymorphisms of RA, detected the SNP rs146534657 for the first time in RA, and found a new mutation (14284 T > A) in synovial fluid of RA. Our data suggested that the rs2230926 TG genotype and rs146534657 AG genotype might be related to RA poor outcome. Significantly lower A20 mRNA expression was found in RA patients. However, further research involving more samples is needed to determine representative results.
Authors’ contribution
YQL contributed to the concept development and study design. LHZ and LW performed PCR and sequencing, XW performed real-time PCR, LLZ, ZWL and LX prepared the figures, SHC, LJY, BL and XLW prepared PBMCs, DNA, RNA and cDNA and helped establish the techniques, LHZ, YZ, HXW, JR and ZXL were responsible for clinical diagnoses and performed clinical data acquisition. YQL and LHZ coordinated the study and helped draft the manuscript. All authors read and approved the final manuscript.
Acknowledgements
This study was supported by Grants from the National Natural Science Foundation of China (No. 91129720), the Collaborated Grant for HK-Macao-TW of the Ministry of Science and Technology (2012DFH30060) and the Medical Science Foundation of Guangdong Province (B2014223).
Compliance with ethical guidelines
Competing interests The authors declare that they have no competing interests.
Additional files
Table S1. Details of primers used in the PCR and real-time PCR.
Footnotes
Lihua Zhu and Liang Wang have equally contributed this work
Contributor Information
Lihua Zhu, Email: 125741799@qq.com.
Liang Wang, Email: 15857266@qq.com.
Xu Wang, Email: 742630368@qq.com.
Lingling Zhou, Email: 251582936@qq.com.
Ziwei Liao, Email: lzw122623@aliyun.com.
Ling Xu, Email: lingxu114@163.com.
Huixia Wu, Email: 56085299@qq.com.
Jie Ren, Email: 41607951@qq.com.
Zhaoxia Li, Email: 547932486@qq.com.
Lijian Yang, Email: jnyanglijian@163.com.
Shaohua Chen, Email: jnshaohuachen@163.com.
Bo Li, Email: jnlibo517@163.com.
Xiuli Wu, Email: siulier@163.com.
Yi Zhou, Email: zhou21yi@163.com.
Yangqiu Li, Email: yangqiuli@hotmail.com.
References
- 1.Ji X, Zhang L, Peng J, Hou M. T cell immune abnormalities in immune thrombocytopenia. J Hematol Oncol. 2014;7:72. doi: 10.1186/s13045-014-0072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moore AR, Allden S, Bourne T, Denis MC, Kranidioti K, Okoye R, et al. Collagen II antibody-induced arthritis in Tg1278TNFko mice: optimization of a novel model to assess treatments targeting human TNFα in rheumatoid arthritis. J Transl Med. 2014;12:285. doi: 10.1186/s12967-014-0285-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Okamoto H, Kobayashi A. Tyrosine kinases in rheumatoid arthritis. J Inflamm (Lond) 2011;8:21. doi: 10.1186/1476-9255-8-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dhaouadi T, Sfar I, Haouami Y, Abdelmoula L, Turki S, Hassine LB, et al. Polymorphisms of Toll-like receptor-4 and CD14 in systemic lupus erythematosus and rheumatoid arthritis. Biomark Res. 2013;1:20. doi: 10.1186/2050-7771-1-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.MacGregor AJ, Snieder H, Rigby AS, Koskenvuo M, Kaprio J, Aho K, et al. Characterizing the quantitative genetic contribution to rheumatoid arthritis using data from twins. Arthritis Rheum. 2000;43:30–37. doi: 10.1002/1529-0131(200001)43:1<30::AID-ANR5>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 6.Vereecke L, Beyaert R, van Loo G. Genetic relationships between A20/TNFAIP3, chronic inflammation and autoimmune disease. Biochem Soc Trans. 2011;39:1086–1091. doi: 10.1042/BST0391086. [DOI] [PubMed] [Google Scholar]
- 7.Hymowitz SG, Wertz IE. A20: from ubiquitin editing to tumour suppression. Nat Rev Cancer. 2010;10:332–341. doi: 10.1038/nrc2775. [DOI] [PubMed] [Google Scholar]
- 8.Tavares RM, Turer EE, Liu CL, Advincula R, Scapini P, Rhee L, et al. The ubiquitin modifying enzyme A20 restricts B cell survival and prevents autoimmunity. Immunity. 2010;33:181–191. doi: 10.1016/j.immuni.2010.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elsby LM, Orozco G, Denton J, Worthington J, Ray DW, Donn RP. Functional evaluation of TNFAIP3 (A20) in rheumatoid arthritis. Clin Exp Rheumatol. 2010;28:708–714. [PMC free article] [PubMed] [Google Scholar]
- 10.Scherer HU, van der Linden MP, Kurreeman FA, Stoeken-Rijsbergen G, Cessie SL, Huizinga TW, et al. Association of the 6q23 region with the rate of joint destruction in rheumatoid arthritis. Ann Rheum Dis. 2010;69:567–570. doi: 10.1136/ard.2008.106161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lodolce JP, Kolodziej LE, Rhee L, Kariuki SN, Franek BS, McGreal NM, et al. African-derived genetic polymorphisms in TNFAIP3 mediate risk for autoimmunity. J Immunol. 2010;184:7001–7009. doi: 10.4049/jimmunol.1000324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coornaert B, Carpentier I, Beyaert R. A20: central gatekeeper in inflammation and Immunity. J Biol Chem. 2009;284:8217–8221. doi: 10.1074/jbc.R800032200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Musone SL, Taylor KE, Nititham J, Chu C, Poon A, Liao W, et al. Sequencing of TNFAIP3 and association of variants with multiple autoimmune diseases. Genes Immun. 2011;12:176–182. doi: 10.1038/gene.2010.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang X, Li W, Zhang X, Zhao L, Zhang X, Jiang L, et al. Single nucleotide polymorphisms in TNFAIP3 were associated with the risks of rheumatoid arthritis in northern Chinese Han population. BMC Med Genet. 2014;15:56. doi: 10.1186/1471-2350-15-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arnett FC, Edworthy SM, Bloch DA, McShane DJ, Fries JF, Cooper NS, et al. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 1988;31:315–324. doi: 10.1002/art.1780310302. [DOI] [PubMed] [Google Scholar]
- 16.Turunen S, Koivula MK, Melkko J, Alasaarela E, Lehenkari P, Risteli J. Different amounts of protein-bound citrulline and homocitrulline in foot joint tissues of a patient with anti-citrullinated protein antibody positive erosive rheumatoid arthritis. J Transl Med. 2013;11:224. doi: 10.1186/1479-5876-11-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rechsteiner M, Müller R, Reineke T, Goede J, Bohnert A, Zhong Q, et al. Modelling of a genetically diverse evolution of systemic mastocytosis with chronic myelomonocytic leukemia (SM-CMML) by next generation sequencing. Exp Hematol Oncol. 2014;3:18. doi: 10.1186/2162-3619-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang L, Xu M, Wang C, Zhu L, Hu J, Chen S, et al. The feature of distribution and clonality of TCR γ/δ subfamilies T cells in patients with B-cell non-Hodgkin’s lymphoma. J Immunol Res. 2014;2014:241246. doi: 10.1155/2014/241246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu L, Zhang F, Shen Q, Chen S, Wang X, Wang L, et al. Characteristics of A20 gene polymorphisms in T-cell acute lymphocytic leukemia. Hematology. 2014;19:448–454. doi: 10.1179/1607845414Y.0000000160. [DOI] [PubMed] [Google Scholar]
- 20.Lin C, Zheng H, Wang C, Yang L, Chen S, Li B, et al. Mutations increased overexpression of Notch1 in T-cell acute lymphoblastic leukemia. Cancer Cell Int. 2012;12:13. doi: 10.1186/1475-2867-12-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Y, Liu S, Shen Q, Zha X, Zheng H, Yang L, et al. Differential gene expression profiles of PPP2R5C-siRNA treated malignant T cells. DNA Cell Biol. 2013;32:573–581. doi: 10.1089/dna.2013.2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang X, Zhu L, Liao Z, Zhang F, Xu L, Xu Y, et al. Alternative expression pattern of MALT1-A20-NF-kB in patients with rheumatoid arthritis. J Immunol Res. 2014;2014:492872. doi: 10.1155/2014/492872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vande Walle L, Van Opdenbosch N, Jacques P, Fossoul A, Verheugen E, Vogel P, et al. Negative regulation of the NLRP3 inflammasome by A20 protects against arthritis. Nature. 2014;512:69–73. doi: 10.1038/nature13322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee YH, Bae SC, Choi SJ, Ji JD, Song GG. Associations between TNFAIP3 gene polymorphisms and rheumatoid arthritis: a meta-analysis. Inflamm Res. 2012;61:635–641. doi: 10.1007/s00011-012-0455-5. [DOI] [PubMed] [Google Scholar]
- 25.Musone SL, Taylor KE, Lu TT, Nititham J, Ferreira RC, Ortmann W, et al. Multiple polymorphisms in the TNFAIP3 region are independently associated with systemic lupus erythematosus. Nat Genet. 2008;40:1062–1064. doi: 10.1038/ng.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shimane K, Kochi Y, Horita T, Ikari K, Amano H, Hirakata M, et al. The association of a nonsynonymous single-nucleotide polymorphism in TNFAIP3 with systemic lupus erythematosus and rheumatoid arthritis in the Japanese population. Arthritis Rheum. 2010;62:574–579. doi: 10.1002/acr.20194. [DOI] [PubMed] [Google Scholar]
- 27.Orozco G, Hinks A, Eyre S, Ke X, Gibbons LJ, Bowes J, et al. Combined effects of three independent SNPs greatly increase the risk estimate for RA at 6q23. Hum Mol Genet. 2009;18:2693–2699. doi: 10.1093/hmg/ddp193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim SK, Choe JY, Bae J, Chae SC, Park DJ, Kwak SG, et al. TNFAIP3 gene polymorphisms associated with differential susceptibility to rheumatoid arthritis and systemic lupus erythematosus in the Korean population. Rheumatology (Oxf) 2014;53:1009–1013. doi: 10.1093/rheumatology/ket473. [DOI] [PubMed] [Google Scholar]
- 29.Zhang X, Li W, Zhang X, Zhang X, Jiang L, Guo Y, et al. The association of single nucleotide polymorphisms in TNFAIP3 with rheumatoid arthritis in the Chinese population. Eur J Immunol. 2013;38:214–220. doi: 10.5114/ceji.2013.36066. [DOI] [Google Scholar]
- 30.Song RH, Yu ZY, Wang Q, Muhali FS, Jiang WJ, Xiao L, et al. Polymorphisms of the TNFAIP3 region and Graves’ disease. Autoimmunity. 2014;47:459–465. doi: 10.3109/08916934.2014.914504. [DOI] [PubMed] [Google Scholar]
- 31.Plant D, Flynn E, Mbarek H, Dieudé P, Cornelis F, Arlestig L, et al. Investigation of potential non-HLA rheumatoid arthritis susceptibility loci in a European cohort increases the evidence for nine markers. Ann Rheum Dis. 2010;69:1548–1553. doi: 10.1136/ard.2009.121020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhong H, Li XL, Li M, Hao LX, Chen RW, Xiang K, et al. Replicated associations of TNFAIP3, TNIP1 and ETS1 with systemic lupus erythematosus in a southwestern Chinese population. Arthritis Res Ther. 2011;13:R186. doi: 10.1186/ar3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hua R, Xu JB, Wang JC, Zhu L, Li B, Liu Y, et al. Association of TNFAIP3 polymorphism with rheumatic heart disease in Chinese Han population. Immunogenetics. 2009;61:739–744. doi: 10.1007/s00251-009-0405-8. [DOI] [PubMed] [Google Scholar]
- 34.Dieguez-Gonzalez R, Calaza M, Perez-Pampin E, Balsa A, Blanco FJ, Cañete JD, et al. Analysis of TNFAIP3, a feedback inhibitor of nuclear factor-kappa B and the neighbor intergenic 6q23 region in rheumatoid arthritis susceptibility. Arthritis Res Ther. 2009;11:R42. doi: 10.1186/ar2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nititham J, Taylor KE, Gupta R, Chen H, Ahn R, Liu J, et al. Meta-analysis of the TNFAIP3 region in psoriasis reveals a risk haplotype that is distinct from other autoimmune diseases. Genes Immun. 2015;16:120–126. doi: 10.1038/gene.2014.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee EG, Boone DL, Chai S, Libby SL, Chien M, Lodolce JP, et al. Failure to regulate TNF-induced NF-kappaB and cell death responses in A20-deficient mice. Science. 2000;289:2350–2354. doi: 10.1126/science.289.5488.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoon HK, Byun HS, Lee H, Jeon J, Lee Y, Li Y, et al. Intron-derived aberrant splicing of A20 transcript in rheumatoid arthritis. Rheumatology (Oxf) 2013;52:347–427. doi: 10.1093/rheumatology/kes292. [DOI] [PubMed] [Google Scholar]


