Abstract
An explosion of knowledge regarding the genetic and genomic basis for rare and common diseases has provided a framework for revolutionizing the practice of medicine. Achieving the reality of a genomic medicine era requires that basic discoveries are effectively translated into clinical practice through implementation of genetic and genomic testing. Clinical genetic tests have become routine for many inherited disorders and can be regarded as the standard-of-care in many circumstances including disorders affecting the cardiovascular system. New, high-throughput methods for determining the DNA sequence of all coding exons or complete genomes are being adopted for clinical use to expand the speed and breadth of genetic testing. Along with these extraordinary advances have emerged new challenges to practicing physicians for understanding when and how to use genetic testing along with how to appropriately interpret test results. This review will acquaint readers with general principles of genetic testing including newer technologies, test interpretation and pitfalls. The focus will be on testing genes responsible for monogenic disorders and on other emerging applications such as pharmacogenomic profiling. The discussion will be extended to the new paradigm of direct-to-consumer genetic testing and the value of assessing genomic risk for common diseases.
Keywords: genetic testing, genes, mutation, polymorphism, pharmacogenomics
Logistical Considerations for Genetic Testing
Genetic testing is a specialized diagnostic procedure that can be performed by commercial and research laboratories. However, in the United States, clinical genetic testing laboratories must meet stringent criteria for quality standards that conform to the federal Clinical Laboratory Improvement Amendments (CLIA).1 Most research laboratories operate without CLIA certification, and data generated in this setting are not strictly appropriate for inclusion in patient medical records or for making clinical decisions. Discoveries made by research laboratories should be confirmed by a CLIA-certified clinical genetics laboratory if the results are meaningful to patient care.
Unlike commonly used laboratory assays, genetic testing should be performed after informing the patient about the potential risks, benefits and limitations. Involvement of a genetic counselor is ideal in circumstances where either physician time or knowledge is limited. The potential negative impact of learning the results of a genetic test must be anticipated. Patients should be properly educated and carefully counseled about their long-term risks of having a genetic condition without inciting excessive apprehension by implying that genotype is an absolute predictor of disease. Physicians should also be sensitive to the potential socioeconomic fallout (e.g., insurability) from a genetic diagnosis and vigorously guard confidentiality of test results. In the United States, the Genetic Information Non-discrimination Act (GINA) prohibits workplace and health insurance discrimination based on genetic predisposition, but this sensitive information should be protected nonetheless.
Indications for Genetic Testing
Genetic testing is performed under a variety of circumstances and applied at various time points from before life begins to after death occurs (Table 1). The most common scenarios for clinical genetic testing for cardiovascular disorders occur beyond the perinatal period and include diagnostic, presymptomatic and postmortem testing. Diagnostic testing undertakes the primary discovery of genetic defects by screening a panel of genes previously associated with a specific disease. Directed genetic testing (e.g., detection of specific mutation or genomic variant) is performed when there is a known familial risk for a disease and a previously identified mutation in the parents or other first degree relative. Postmortem genetic testing is increasingly performed in the evaluation of sudden unexplained death especially in the young and yields positive findings in up to a quarter of autopsies.2,3 Preimplantation genetic testing is performed on in vitro fertilized embyros to guide selection of unaffected embryos for transfer to a recipient uterus.4 Although risk for cardiovascular disorders is not typically determined, there have been successful uses of preimplantation genetic diagnosis in cases of Holt-Oram syndrome5,6 and familial cardiomyopathy.7 Direct-to-consumer genetic testing is discussed later in the article.
Table 1.
Uses of Genetic Testing in Cardiovascular Disorders
| Pre-implantation testing: Performed on human embryos derived by in vitro fertilization to guide selection of unaffected embryos for implantation. |
| Diagnostic testing: Performed at any age to diagnose specific genetic disorders. |
| Carrier testing: Performed on individuals with a family history of a genetic disorder to guide reproductive counseling about the risks of having an affected child. |
| Presymptomatic testing: Performed individuals who are at-risk for a genetic disorder at an age before the disorder typically develops. This is also called predictive testing. |
| Postmortem testing: Performed on deceased individuals as part of an evaluation for the cause of death especially when sudden cardiac death was evident. |
| Pharmacogenomic testing: Performed to determine risk for unfavorable or adverse effects of drug therapy, or to determine the most appropriate dose. |
| Risk prediction testing: Testing genomic loci with known population-based disease associations to assess risk for the condition in an individual. |
| Direct-to-consumer testing: Commercial genetic testing offered to individuals without involvement of a physician. This type of testing is not designed to make specific genetic diagnoses. |
A freely available internet resource, GeneTestsTM, provides a searchable database of clinical genetic testing laboratories, specialty clinics and other relevant information (Table 2).8 The current database consists of entries from more than 600 laboratories worldwide offering tests for an aggregate of more than 3,000 genetic conditions. A variety of test types are indexed including molecular (e.g., DNA sequence), cytogenetic and biochemical assays. The clinic directory has information for more than 1,000 genetics clinics internationally. GeneTests™ can be searched using disease or gene names to retrieve a listing of laboratories performing specific tests along with their contact information. The database can also be searched to find laboratories and specialty clinics within a specific region.
Table 2.
Internet resources and databases
| Genetic Testing Resources |
| GeneTests™ |
| http://www.genetests.org/ |
| Genetic Testing Registry |
| http:/www.ncbi.nlm.nih.gov/gtr/ |
| GeneReviews® |
| http://www.ncbi.nlm.nih.gov/books/NBK1116/ |
| United Kingdom Gene Testing Network |
| http://ukgtn.nhs.uk/ |
| Genetic Variant Databases |
| Single Nucleotide Polymorphism Database (dbSNP) |
| http://www.ncbi.nlm.nih.gov/projects/SNP/ |
| NIH ClinVar |
| https://www.ncbi.nlm.nih.gov/clinvar/ |
| The Human Gene Mutation Database (HGMD) |
| http://www.hgmd.cf.ac.uk/ac/ |
| 1000 Genomes Project |
| http://www.1000genomes.org/ |
| Exome Variant Server |
| http://evs.gs.washington.edu/EVS/ |
| Human Genetic Variation Browser (HGVD) |
| http://www.genome.med.kyoto-u.ac.jp/SnpDB |
| Pharmacogenomic Resources |
| Pharmacogenomics Knowledgebase (PharmGKB) |
| https://www.pharmgkb.org/ |
| Clinical Pharmacogenetics Implementation Consortium (CPIC) |
| http://www.pharmgkb.org/page/cpic |
Information in GeneTests™ has recently transitioned to a resource hosted by the National Center for Biotechnology Information (NCBI), the Genetic Testing Registry, which is a repository for testing information provided by laboratories. Both GeneTests™ and the Genetic Testing Registry provide gene and disease specific information derived in part from another resource, GeneReviews®, a collection of expertly authored and peer-reviewed articles on genes and genetic disorders. This site is also hosted by NCBI. A similar searchable database, the United Kingdom Gene Testing Network, provides online access to a catalog of genetic tests provided by clinically accredited laboratories in the UK.
There are several monogenic diseases primarily or secondarily affecting the cardiovascular system (Table 3). For many cardiovascular disorders in which Mendelian inheritance has already been established, genetic testing may already be indicated, and there have been published guidelines for the use and interpretation of genetic tests for many specific disorders including channelopathies and familial cardiomyopathies.9,10 In other circumstances, an important step in determining which specific patients will benefit most from a genetic work-up is distinguishing cases that arise from a single gene mutation or definable genomic defect from those with a more complex etiology (e.g., combined impact of multiple genetic, developmental and environmental factors). Clinical evidence suggesting a single genetic locus etiology may include: 1) familial segregation of the trait in a pattern consistent with Mendelian inheritance (e.g., autosomal dominant, autosomal recessive or X-linked), 2) extreme phenotype with either unusual severity or early age of onset, and 3) associated clinical features that suggest a specific syndrome.
Table 3.
Genetic disorders affecting the cardiovascular system.
| Cardiomyopathies |
| Hypertrophic cardiomyopathy |
| Familial dilated cardiomyopathy |
| Arrhythmogenic cardiomyopathy |
| Restrictive cardiomyopathy |
| Left ventricular noncompaction |
| Arrhythmia susceptibility |
| Congenital long QT syndrome |
| Brugada syndrome |
| Catecholamineric polymorphic ventricular tachycardia |
| Atrial fibrillation |
| Short QT syndrome |
| Disorders of lipid metabolism |
| Familial hypercholesterolemia |
| Vascular disorders |
| Primary pulmonary hypertension |
| Hereditary hemorrhagic telangiectasia |
| Disorders of blood pressure regulation |
| Liddle syndrome |
| Glucocorticoid remediable aldosteronism |
| Bartter syndrome |
| Gitelman syndrome |
| Pseudohypoaldosteronism |
| Congenital heart malformations |
| Multi-system and developmental disorders with cardiovascular manifestations |
| Marfan syndrome |
| Duchenne muscular dystrophy |
| Noonan syndrome |
| LEOPARD syndrome |
| Holt-Oram syndrome |
| CHARGE syndrome |
Recognizing Mendelian inheritance patterns requires the ascertainment of a thorough and reliable family history with construction of a multi-generation pedigree. However, this task requires considerable time and may not be practical for busy clinicians. Therefore, alternative strategies for acquiring family history data that involve properly trained allied health professionals (e.g., genetic counselors, nurse practitioners, physician assistants) who are familiar with the phenotype, computer or internet resources developed for collecting family health history, or carefully compiled disease-specific survey tools should be used when physician time is limited. Nuances of Mendelian inheritance such as incomplete penetrance (e.g., presence of a mutation does not correlate with disease in all cases) and subclinical disease expression may confound interpretation of pedigree data. For these reasons, involvement of genetic counselors or referral to a medical geneticist is highly recommended.
Extreme phenotypes may suggest a monogenic disorder even in the absence of a clear family history, such as with de novo mutations. Unusually early onset of phenotypes that are more typically adult-onset such as hypertensive stroke, coronary artery disease, heart failure and sudden cardiac death should raise suspicion of a monogenic condition. Additional clinical features, which may affect organs or tissues outside the cardiovascular system, may herald the presence of a specific genetic or genomic syndrome.
Genetic testing may have value beyond establishing or confirming a particular diagnosis in just the primary patient. Demonstrating a specific mutation or genomic defect provides an opportunity to offer targeted testing to relatives who may be affected by the same disease or are at-risk based on their shared genetic makeup. Collateral testing of first degree relatives (e.g., siblings, parents, offspring), referred to as ‘cascade’ screening, can identify pre-symptomatic individuals who may benefit from additional diagnostic procedures or prophylactic therapy. Positive genetic test results may also help tailor therapy and provide a basis for reproductive genetic counseling.
Next-generation Clinical Genetic Testing
An extensive technological repertoire exists to test DNA for a range of medically relevant genetic and genomic defects ranging from single nucleotide variants to chromosomal aberrations. Recent advances in DNA sequencing technology have greatly expanded the scope and capabilities of clinical genetic testing laboratories to discovery medically actionable mutations in cardiovascular disorders.11,12 Until the past few years, the standard sequencing platforms used the Sanger method and this approach served as the ‘workhorse’ for sequencing the human genome. The advent of next-generation sequencing brings a paradigm-shift in the scale and complexity of clinical genetic testing enabling laboratories to perform testing on large panels of disease-relevant genes, all coding exons (exome) or whole genomes. This has allowed genetic diagnoses in small families with rare disorders and discovery of novel disease-causing genes at much lower cost per nucleotide. In 2013, the Food and Drug Administration (FDA) granted marketing authorization for the first next-generation sequencer to be used for clinical genetic testing.13 During the past 3 years, there has been an explosion in the number of reports demonstrating successful uses of exome and genome sequencing to uncover the genetic basis for several disorders with cardiovascular manifestations in the research setting (Supplemental Table S1). Accompanying these advances are new challenges for handling and interpreting massive quantities of data as well as managing discovery of medically actionable incidental findings.
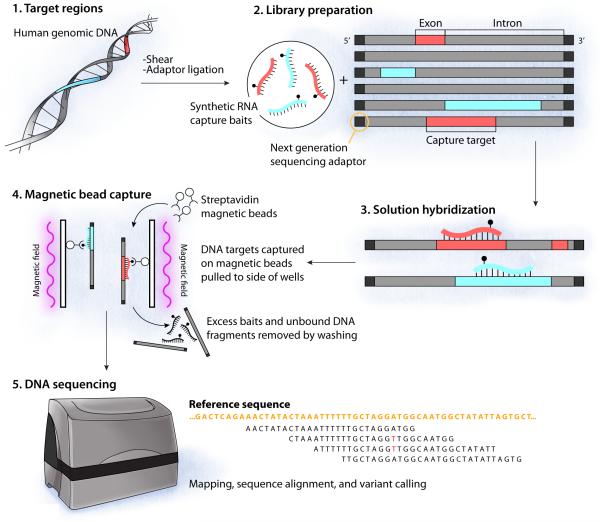
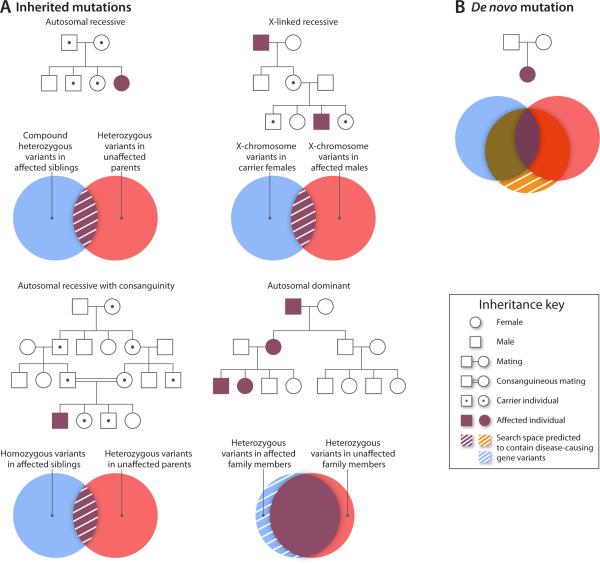
Sequence capture technology coupled with next-generation sequencing has enabled two new genetic testing paradigms: simultaneous sequencing of multiple genes, and whole-exome sequencing.14 Figure 1 illustrates the general workflow of these approaches. Initially, genomic DNA is sheared into small fragments (typically 200-500 bp), then these fragments of DNA are mixed with a capture reagent. The capture reagent is a solution mixture of thousands of synthetic DNA or RNA molecules designed with base pair complementarity to all sequences within the target of interest (e.g., all exons of a candidate gene panel, or all coding exons in the genome).12 For exome sequencing, the capture reagent will generally target approximately 180,000 exons and ~50 Mb or ~1% of the entire human genome. The captured patient DNA is then sequenced on a next-generation sequencer and an intensive bioinformatics analysis is performed to read the sequence (base calling), assess data quality, align sequences to a reference genome, call and annotate variants. Additional details regarding the technical aspects and limitations of exome sequencing have been described elsewhere.12,14 An adequate ‘depth’ of coverage (e.g., redundancy with which each nucleotide is sequenced) is necessary to assure reliability and accuracy. Certain technical limitations can affect the analytical quality of exome data, and Sanger sequencing is often used to validate results and to eliminate false positives. Analysis of exome data to identify disease associated variants is aided greatly by knowledge of family structure and by collecting sequence data from first degree relatives who are either affected or unaffected (Fig. 2).
Figure 1.
Illustration of steps involved in DNA capture and next-generation sequencing (adapted by permission from Macmillan Publishers Ltd: Nature Genetics Reviews, Bamshad, et al., 2011).12 Intact human genomic DNA is sheared into random, small fragments then synthetic adaptor sequences are added to the fragment ends. Next, the pool of adaptor-ligated DNA fragments are incubated with a complex mixture of biotinylated RNA ‘baits’ designed to hybridize to all coding exons by complementary nucleic acid base pairing. Following hybridization, targeted DNA regions are captured using streptavidin coated magnetic beads, which selectively bind the biotinylated ‘bait’ strands and simultaneously immobilize any bound DNA fragments. After washing to remove excess bait and unbound DNA, a library of captured DNA is prepared for sequencing. Finally, all captured DNA is sequenced a several fold redundancy using a next-generation sequencer and the data output are analyzed.
Figure 2.
Approaches for gene discovery using next-generation sequencing data (adapted by permission from Macmillan Publishers Ltd: Nature Genetics Reviews, Boycott, et al., 2013).83 Pedigrees illustrating various inheritance models are presented in which symbols represent males (squares), females (circles), affected (shaded), unaffected (unshaded) and obligatory mutation carriers (symbols with a central dot). (A) Mendelian inheritance patterns (autosomal recessive, autosomal dominant and X-linked) are illustrated. (B) Pedigree illustrating a parent-child trio in which a de novo (e.g., non-inherited) mutation is present in the affected child. Venn diagrams beneath each representative pedigree indicate how variants can be shared among family members. The hatched regions within these diagrams denote the search space (e.g., regions of overlap or non-overlap) predicted to contain disease-causing gene variants.
Whole genome sequencing may soon be sufficiently cost effective and time efficient to warrant its use as the mainstay in next-generation clinical genetic testing. Although the costs of sequencing an entire human genome are falling rapidly, there remain substantial challenges to data analysis that will slow widespread clinical implementation.15 However, there is evidence that these challenges may be surmountable sooner than expected. A recent report demonstrated the feasibility of a rapid turnaround of genome sequencing results for determining the molecular diagnosis of a severe case of neonatal long-QT syndrome type 2 (KCNH2 mutation) in a clinical setting.16
Classifying Variants and Interpreting Test Results
Genetic variants identified by clinical genetic testing laboratories are classified according to defined schema to enable a concise language for describing the best estimate of the clinical significance of a reported sequence variation (Table 4). In 2008, the American College of Medical Genetic (ACMG) Laboratory Quality Assurance Committee recommended six interpretative categories of sequence variants to standardize laboratory reporting of genetic test results.17 Two of these categories are intended for variants with the strongest supporting evidence for pathogenicity (‘Disease causing’) or the lack of pathogenicity (‘Not disease causing’). Other categories provide descriptors for variants with less certain clinical significance. A similar five-tiered scheme was proposed by the International Agency for Research on Cancer (IARC) to classify variants detected in subjects at-risk for hereditary cancer syndromes.18 The IARC scheme additionally assigns quantitative probability ranges for each category. Commercial genetic testing laboratories may deploy additional derivatives of these classification schemes for standardized reporting. Importantly, evidence for or against pathogenicity of a given variant may evolve with new information and/or experimental data. Certain variants once deemed pathogenic may require reclassification based on new findings.19,20
Table 4.
Classification of Genetic Variant Pathogenicity.
| American College of Medical Genetics 17 |
| • Disease causing |
| Sequence variation has been reported previously and is a recognized cause of the disorder |
| • Likely disease causing |
| Sequence variation has not been reported previously but is a type (e.g., nonsense mutation, frameshift) expected to cause the disorder |
| • Possibly disease causing |
| Sequence variation has not been reported previously and is a type that may or may not be causative of the disorder (e.g., nonsynonymous variant) |
| • Likely not disease causing |
| Sequence variation has not been reported previously and is probably not causative of the disease |
| • Not disease causing |
| Sequence variation has previously been reported and is a recognized neutral (e.g., benign) variant |
| • Variant of unknown clinical significance (VUS) |
| Sequence variation is not known or expected to be causative of disease but is associated with a clinical presentation |
| International Agency for Research on Cancer18 |
| • Definitely pathogenic |
| >0.99 probability of being pathogenic |
| • Likely Pathogenic |
| 0.95-0.99 probability of being pathogenic |
| • Uncertain |
| 0.05-0.949 probability of being pathogenic |
| • Likely not pathogenic |
| 0.001-0.049 probability of being pathogenic |
| • Not pathogenic |
| <0.001 probability of being pathogenic |
Allele frequency in reference populations is typically used to distinguish common genetic variants from rare, candidate mutations. Resources for ascertaining population-based allele frequencies are listed in Table 2. The Single Nucleotide Polymorphism Database (dbSNP) is a freely available catalog of genetic variation within different species. Another resource, ClinVar, archives evidence-based reports of the relationships among genetic variation and phenotypes. The Human Gene Mutation Database (HGMD) maintains a collection of data on mutations in genes underlying or associated with human inherited disease.
More recently, databases of human genome and exome sequences have been curated to determine variant frequencies. The 1000 Genomes Project archives low coverage genome sequences on >1,000 individuals without disclosed phenotypic information.21 Similarly, the Exome Sequencing Project funded by a Grand Opportunity (GO) grant from the National Institute of Heart, Lung and Blood Diseases (NHLBI) has populated the Exome Variant Server (EVS) with variant data deduced from exome sequencing of more than 6,500 individuals.22 The Human Genetic Variation Browser [HGVD] database includes exome data obtained from 1208 Japanese subjects. These resources can be used to determine if a discovered variant is novel and therefore likely disease-causing by virtue of its absence or rarity in the general population. Certain variants with known association with specific genetic disorders may be captured by these large scale projects but the significance of these findings is uncertain. The presence of a genetic variant in a reference database does not necessarily exclude its potential pathogenicity, especially when the observed allele frequency is below the estimated population frequency of the disease. For example, the congenital long-QT syndrome has been estimated to affect 1 in 2500 live births based on a large scale neonatal ECG screening study coupled with candidate gene mutation discovery in nearly 45,000 Italian neonates.23 By contrast, other variants reported as disease-associated in familial cardiomyopathy and certain arrhythmic disorders are present in reference databases at frequencies higher than expected based on the population prevalence of the respective phenotypes.24-27 These findings suggest the need for caution in interpreting incidentally discovered variants without stringent criteria for assigning pathogenicity (e.g., segregation with phenotype in more than one family, functional evidence of deleterious consequences).
Variants of Unknown Clinical Significance
Results from clinical genetic testing may be confounded by the discovery of ‘variants of unknown significance’ (VUS) for which there is insufficient data to establish whether or not a particular variant predisposes to a disease. With the expanded use of exome/genome data in clinical medicine, interpreting VUS will become a larger challenge especially when variants in genes associated with monogenic disorders are incidentally discovered and reported. This problem is particularly vexing for most genetic disorders that have a high level of allelic heterogeneity and a preponderance of ‘private’ mutations. Additional evidence should be sought when possible to more firmly establish the relationship with disease risk in a family tested positive for a VUS. Segregation of the VUS among affected and unaffected family members may provide additional support for disease association, although this may be difficult to ascertain in small families, disorders with recessive inheritance or with incomplete penetrance. Laboratory research to establish whether a VUS has deleterious consequences may offer additional clues to pathogenicity but these are not standardized assays that yield results suitable for clinical decision making. Nonetheless, collaboration between clinicians and researchers provides an avenue to decrypt the growing burden of VUS.
There have been several computational strategies developed to help predict the potential effects of genetic variants on protein function in the research setting. Two of the more widely used methods are PolyPhen-2 and SIFT (Sorting Intolerant From Tolerant). SIFT uses protein sequence homology to assess the likelihood that a position-specific amino acid substitution will be damaging based on the premise that important residues will be conserved in the protein family throughout evolution.28,29 SIFT was originally developed using prokaryotic gene mutation data, but was later tested on a large set of annotated human mutation data. PolyPhen-2 uses protein sequence-based and structure-based features to make predictions.30 Another approach (Evolutionary Diagnosis [EvoD]) featuring statistical models based on evolutionarily weighted training data has been suggested to offer improved predictive power.31 Newer, purportedly better methods have emerged recently,32,33 but no particular algorithm appears superior to all others.34 Disease specific models may have better performance.35 Importantly, there have been few attempts to experimentally validate these in silico prediction models.34,36
Managing Negative Results and Incidental Findings
Interpretation of a ‘negative’ genetic test in a symptomatic person is a significant challenge. A true negative result (e.g., technically successful, but no pathogenic findings) may occur because the test does not target the causative gene, possibly because a previously unknown genetic culprit is involved or the test panel was not comprehensive. Theoretically, this phenomenon will occur less frequently when exome sequencing is used as the testing platform. False negative results (e.g., no pathogenic findings reported even when one exists in the targeted gene) have other causes such as location of a mutation outside the region interrogated by the test37 and existence of types of mutations (e.g., multi-exon deletion, duplication) missed by the most commonly used testing strategies that are designed for finding single nucleotide changes.38-40 Repeat testing may sometimes overcome false negative results especially when there is a high level of clinical suspicion.41
The use of exome or genome sequencing may uncover medically actionable variants unrelated to the primary disorder that prompted the test. This point was illustrated by an examination of 1,000 participants randomly selected from the 6,500 subjects studied by the NHLBI Exome Sequencing Project.42 In a survey of pathogenic variants in 114 genes selected because of associations with medically actionable genetic conditions, the frequency of highly penetrant and actionable variants was 1.2% for individuals of African descent and 3.4% for subjects with European ancestry. Among the pathogenic variants identified were several in genes associated with familial cardiomyopathy and congenital arrhythmia susceptibility. This demonstration of the prevalence of incidental findings in exome data in a research setting has prompted considerable debate regarding the best practice for reporting and managing such information.
Recently, the American College of Medical Genetics (ACMG) Working Group on Incidental Findings in Clinical Exome and Genome Sequencing recommended that laboratories performing clinical sequencing seek and report mutations in 57 genes including 30 genes responsible for cardiovascular phenotypes (Table 5).43,44 These recommendations were designed to establish an initial reporting standard for laboratories engaged in exome/genome sequencing for genetic diagnosis. Inherent in the ethical framework within which these recommendations were based was the assumption that the ordering clinician would bear responsibility for obtaining informed consent from the patient including pretest and posttest genetic counseling about the potential risks and benefits of testing.45 The ACMG recommendations have been challenged both on scientific and ethical grounds,46,47 but defended by emphasizing that incidental findings provide an opportunity for patient education and collaboration between patient and provider to define the best course of action.48 A recent amendment to these recommendations by ACMG suggests that patients should be given an opportunity to ‘opt out’ of the analysis of medically actionable genetic variants at the time samples are sent for initial testing.49 Genetic counselors may have special value in helping patients make such decisions.
Table 5.
Genes involved with cardiovascular disorders recommended for automatic reporting of incidental variants from exome and genome testing.
| Phenotype | Gene | Variants to Report* |
|---|---|---|
| Vascular disorders | ||
| FBN1 | KP, EP | |
| TGFBR1 | KP, EP | |
| Marfan syndrome, Loeys-Dietz syndrome, familial aortic aneurysm | TGFBR2 | KP, EP |
| SMAD3 | KP, EP | |
| ACTA2 | KP, EP | |
| MYLK | KP, EP | |
| MYH11 | KP, EP | |
| Cardiomyopathies | ||
| MYBPC3 | KP, EP | |
| MYH7 | KP | |
| TNNT2 | KP, EP | |
| TNNI3 | KP | |
| TPM1 | KP | |
| Hypertrophic cardiomyopathy, dilated cardiomyopathy | MYL3 | KP |
| ACTC1 | KP | |
| PRKAG2 | KP | |
| GLA | KP, EP | |
| MYL2 | KP | |
| LMNA | KP, EP | |
| Arrhythmic disorders | ||
| Catecholaminergic polymorphic ventricular tachycardia | RYR2 | KP |
| PKP2 | KP, EP | |
| DSP | KP, EP | |
| Arrythmogenic cardiomyopathy | DSC2 | KP, EP |
| TMEM43 | KP | |
| DSG2 | KP, EP | |
| KCNQ1 | KP, EP | |
| LQTS, Brugada syndrome | KCNH2 | KP, EP |
| SCN5A | KP, EP | |
| Dyslipidemias | ||
| LDLR | KP, EP | |
| Familial hypercholesterolemia | APOB | KP |
| PCSK9 | KP |
KP, known pathogenic (previously reported and is a recognized cause of the disease); EP, expected pathogenic (unreported variant but of a type such as truncating that is expected to cause the disorder).
Pharmacogenomic Profiling
Clinical genetic testing can be applied to reveal genomic variants associated with inter-individual differences in drug responses (e.g., variable therapeutic efficacy or adverse effects). The term pharmacogenetics was originally coined as the study of unusual drug response traits exhibiting Mendelian inheritance in families (e.g., glucose-6-phosphate dehydrogenase deficiency [G6PD, Xq28], pseudocholinesterase deficiency [CHE1, 3q25], malignant hyperthermia susceptibility [RYR1, 19q13]). By contrast, pharmacogenomics has been used to describe mainly population-based studies defining genes or loci associated with differences in drug responses among groups of unrelated individuals. Drug response variability is often explainable by differences in either pharmacokinetics (e.g., drug metabolism for biotransformation or elimination) or pharmacodynamics (e.g., response of the target molecule). More than 125 FDA-approved drugs have pharmacogenomic information in their labeling including some with ‘boxed warnings’ advising physicians to acquire specific genomic data on patients for whom the drug is being considered. The Clinical Pharmacogenetics Implementation Consortium (CPIC) formed by the NIH-funded Pharmacogenomics Research Network50 has developed a series of evidence-based, consensus guidelines to enable the translation of clinical genetic test results into actionable prescribing decisions for specific drugs.51
The emergence of pharmacogenomics has offered new opportunities for achieving the goal of personalized medicine by utilizing clinical testing for medically actionable variants including drugs commonly prescribed for cardiovascular disorders.52-54 Genetic testing may have value in predicting efficacy of specific drug therapy (e.g., CYP2C19 genotyping in the setting of antiplatelet therapy with clopidogrel),55 identifying persons what are at risk for specific adverse reactions (e.g., SLCO1B1 genotyping to assess risk of simvastatin-induced muscle toxicity),56,57 or for determining initial dosage (VKORC1 and CYP2C9 genotyping for warfarin dosing).58 Strategies for implementing pharmacogenomic testing in clinical settings have either adopted a one gene at a time approach59,60 or advocated for preemptive testing of multiple variants.61-63 Decision support is essential for educating providers about interpretation of test results and for presenting specific prescribing actions. Physician adoption and utilization of pharmacogenomics testing can be high in settings where point-of-care decision support is provided,64 but somewhat less effective when results are merely faxed to providers several days later.65 Further research including randomized clinical trials, such as those recently reported for oral anticoagulants,66-68 are needed to determine value of pharmacogenomic testing in clinical practice.
Testing for Complex Genetic Traits
Whereas genetic diagnostics have become routine and standard-of-care for many monogenic disorders, laboratory assessments of inherited risk for more common and genetically complex diseases are seldom performed in medical practice.69 During the past decade, genome-wide association studies (GWAS) have mapped more than a thousand disease susceptibility loci based on the “common disease-common variant” hypothesis that posits a major portion of risk for a common disease in populations is conferred by a limited number of common genetic variants.70 A frequent observation made by GWAS is that common variants account for only a small proportion of population attributable risk (estimated by an odds ratio with reported values often less than 1.5), and results from these population-based genomic studies have been difficult to translate into risk predictions for individuals.
The Center for Disease Control (CDC) has recently launched the Evaluation of Genomic Applications in Practice and Prevention (EGAPP) initiative to establish and evaluate an evidence-based process for assessing applications of genomic technologies in the clinical setting.71 Using this systematic approach, an evaluation of specific cardiovascular disease genomic risk variants in 29 candidate genes found only weak to moderate evidence supporting clinical validity. A possible exception was with the association of 9p21 variants with heart disease, which seemed more robust although the estimated additional benefit of knowing genotype at this locus was judged as negligible.72,73 Further studies including clinical trials are needed to determine the impact of genomic risk testing on clinical outcomes, physician practice, patient behavior and health care costs.
Despite the uncertain benefits to individuals undergoing genomic risk profiling, a few for-profit business ventures have capitalized on direct-to-consumer marketing of genomic testing. The leader in this emerging industry has been 23 and Me, Inc. (Mountain View, CA). Such companies market laboratory genetic testing along with interpretive services for assessing genomic risk for specific diseases, pharmacogenomic profiling and analysis of ancestry directly to consumers without requiring input from healthcare providers.74 The intent of these services is to empower individuals with personal genomic information that might help improve health through lifestyle changes and more informed medical decision-making. Collections of population-based genomic data also has enabled research opportunities.75-77 Initial research on the impact of direct-to-consumer genomic testing suggests there is little harm or benefit.78,79
The advent of direct-to-consumer genetic testing ignited considerable debate related to the medical value of the genomic risk discoveries on which these proprietary tests are based and on the accuracy of the data provided to consumers.74,80 Unlike other genetic diagnostics, which are classified as medical devices requiring FDA approval, direct-to-consumer tests have not been subject to regulation. In late 2013, following the launch of an aggressive campaign to sell its product, the FDA ordered 23 and Me to discontinue marketing its primary genomic profiling service.81,82 New regulatory regimes stimulated by these actions could impact other emerging genomic testing paradigms including whole-genome sequencing.
Summary
Technical advances have ushered in a new era of genetically-informed diagnosis that will impact both rare and common cardiovascular disorders. Understanding the capabilities and limitations of genetic testing in various clinical settings is vital to effectively translate the tsunami of medical breakthroughs generated during this genomic era into clinical practice. Accomplishing these goals will require changes in medical education, health care delivery systems, medical informatics, and more informed patients and providers.
Supplementary Material
Acknowledgments
Funding Sources: This work was supported in part by NIH grant HL083374.
Footnotes
Conflict of Interest Disclosures: The author has no financial disclosures related to this work
References
- 1.Schwartz MK. Genetic testing and the clinical laboratory improvement amendments of 1988: present and future. Clin Chem. 1999;45:739–745. [PubMed] [Google Scholar]
- 2.Tester DJ, Ackerman MJ. Postmortem long QT syndrome genetic testing for sudden unexplained death in the young. J Am Coll Cardiol. 2007;49:240–246. doi: 10.1016/j.jacc.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 3.Tester DJ, Medeiros-Domingo A, Will ML, Haglund CM, Ackerman MJ. Cardiac channel molecular autopsy: insights from 173 consecutive cases of autopsy-negative sudden unexplained death referred for postmortem genetic testing. Mayo Clin Proc. 2012;87:524–539. doi: 10.1016/j.mayocp.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bodurtha J, Strauss JF., III. Genomics and perinatal care. N Engl J Med. 2012;366:64–73. doi: 10.1056/NEJMra1105043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He J, McDermott DA, Song Y, Gilbert F, Kligman I, Basson CT. Preimplantation genetic diagnosis of human congenital heart malformation and Holt-Oram syndrome. Am J Med Genet A. 2004;126A:93–98. doi: 10.1002/ajmg.a.20487. [DOI] [PubMed] [Google Scholar]
- 6.McDermott DA, He J, Song YS, Kligman I, Basson CT. Update: PGD and Holt-Oram syndrome. Am J Med Genet A. 2005;136:223. doi: 10.1002/ajmg.a.30804. [DOI] [PubMed] [Google Scholar]
- 7.Kuliev A, Pomerantseva E, Polling D, Verlinsky O, Rechitsky S. PGD for inherited cardiac diseases. Reprod Biomed Online. 2012;24:443–453. doi: 10.1016/j.rbmo.2011.12.009. [DOI] [PubMed] [Google Scholar]
- 8.Pagon RA, Tarczy-Hornoch P, Baskin PK, Edwards JE, Covington ML, Espeseth M, Beahler C, Bird TD, Popovich B, Nesbitt C, Dolan C, Marymee K, Hanson NB, Neufeld-Kaiser W, Grohs GM, Kicklighter T, Abair C, Malmin A, Barclay M, Palepu RD. GeneTests-GeneClinics: genetic testing information for a growing audience. Hum Mutat. 2002;19:501–509. doi: 10.1002/humu.10069. [DOI] [PubMed] [Google Scholar]
- 9.Ackerman MJ, Priori SG, Willems S, Berul C, Brugada R, Calkins H, Camm AJ, Ellinor PT, Gollob M, Hamilton R, Hershberger RE, Judge DP, Le MH, McKenna WJ, Schulze-Bahr E, Semsarian C, Towbin JA, Watkins H, Wilde A, Wolpert C, Zipes DP. HRS/EHRA expert consensus statement on the state of genetic testing for the channelopathies and cardiomyopathies: this document was developed as a partnership between the Heart Rhythm Society (HRS) and the European Heart Rhythm Association (EHRA). Europace. 2011;13:1077–1109. doi: 10.1093/europace/eur245. [DOI] [PubMed] [Google Scholar]
- 10.Hershberger RE, Lindenfeld J, Mestroni L, Seidman CE, Taylor MR, Towbin JA. Genetic evaluation of cardiomyopathy--a Heart Failure Society of America practice guideline. J Card Fail. 2009;15:83–97. doi: 10.1016/j.cardfail.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 11.Shendure J, Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 12.Bamshad MJ, Ng SB, Bigham AW, Tabor HK, Emond MJ, Nickerson DA, Shendure J. Exome sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet. 2011;12:745–755. doi: 10.1038/nrg3031. [DOI] [PubMed] [Google Scholar]
- 13.Collins FS, Hamburg MA. First FDA authorization for next-generation sequencer. N Engl J Med. 2013;369:2369–2371. doi: 10.1056/NEJMp1314561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mamanova L, Coffey AJ, Scott CE, Kozarewa I, Turner EH, Kumar A, Howard E, Shendure J, Turner DJ. Target-enrichment strategies for next-generation sequencing. Nat Methods. 2010;7:111–118. doi: 10.1038/nmeth.1419. [DOI] [PubMed] [Google Scholar]
- 15.Dewey FE, Grove ME, Pan C, Goldstein BA, Bernstein JA, Chaib H, Merker JD, Goldfeder RL, Enns GM, David SP, Pakdaman N, Ormond KE, Caleshu C, Kingham K, Klein TE, Whirl- Carrillo M, Sakamoto K, Wheeler MT, Butte AJ, Ford JM, Boxer L, Ioannidis JP, Yeung AC, Altman RB, Assimes TL, Snyder M, Ashley EA, Quertermous T. Clinical interpretation and implications of whole-genome sequencing. JAMA. 2014;311:1035–1045. doi: 10.1001/jama.2014.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Priest JR, Ceresnak SR, Dewey FE, Malloy-Walton LE, Dunn K, Grove ME, Perez MV, Maeda K, Dubin AM, Ashley EA. Molecular diagnosis of long-QT syndrome at 10 days of life by rapid whole genome sequencing. Heart Rhythm. 2014;11:1707–13. doi: 10.1016/j.hrthm.2014.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richards CS, Bale S, Bellissimo DB, Das S, Grody WW, Hegde MR, Lyon E, Ward BE. ACMG recommendations for standards for interpretation and reporting of sequence variations: Revisions 2007. Genet Med. 2008;10:294–300. doi: 10.1097/GIM.0b013e31816b5cae. [DOI] [PubMed] [Google Scholar]
- 18.Plon SE, Eccles DM, Easton D, Foulkes WD, Genuardi M, Greenblatt MS, Hogervorst FB, Hoogerbrugge N, Spurdle AB, Tavtigian SV. Sequence variant classification and reporting: recommendations for improving the interpretation of cancer susceptibility genetic test results. Hum Mutat. 2008;29:1282–1291. doi: 10.1002/humu.20880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tester DJ, Valdivia C, Harris-Kerr C, Alders M, Salisbury BA, Wilde AA, Makielski JC, Ackerman MJ. Epidemiological, Molecular, and functional evidence suggest A572D-SCN5A should not be considered an independent LQT3-susceptibility mutation. Heart Rhythm. 2010;7:912–919. doi: 10.1016/j.hrthm.2010.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Das KJ, Ingles J, Bagnall RD, Semsarian C. Determining pathogenicity of genetic variants in hypertrophic cardiomyopathy: importance of periodic reassessment. Genet Med. 2014;16:286–293. doi: 10.1038/gim.2013.138. [DOI] [PubMed] [Google Scholar]
- 21.Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu W, O'Connor TD, Jun G, Kang HM, Abecasis G, Leal SM, Gabriel S, Rieder MJ, Altshuler D, Shendure J, Nickerson DA, Bamshad MJ, Akey JM. Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature. 2013;493:216–220. doi: 10.1038/nature11690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwartz PJ, Stramba-Badiale M, Crotti L, Pedrazzini M, Besana A, Bosi G, Gabbarini F, Goulene K, Insolia R, Mannarino S, Mosca F, Nespoli L, Rimini A, Rosati E, Salice P, Spazzolini C. Prevalence of the Congenital Long-QT Syndrome. Circulation. 2009;120:1781–1767. doi: 10.1161/CIRCULATIONAHA.109.863209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Refsgaard L, Holst AG, Sadjadieh G, Haunso S, Nielsen JB, Olesen MS. High prevalence of genetic variants previously associated with LQT syndrome in new exome data. Eur J Hum Genet. 2012;20:905–908. doi: 10.1038/ejhg.2012.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jabbari J, Jabbari R, Nielsen MW, Holst AG, Nielsen JB, Haunso S, Tfelt-Hansen J, Svendsen JH, Olesen MS. New exome data question the pathogenicity of genetic variants previously associated with catecholaminergic polymorphic ventricular tachycardia. Circ Cardiovasc Genet. 2013;6:481–489. doi: 10.1161/CIRCGENETICS.113.000118. [DOI] [PubMed] [Google Scholar]
- 26.Andreasen C, Refsgaard L, Nielsen JB, Sajadieh A, Winkel BG, Tfelt-Hansen J, Haunso S, Holst AG, Svendsen JH, Olesen MS. Mutations in genes encoding cardiac ion channels previously associated with sudden infant death syndrome (SIDS) are present with high frequency in new exome data. Can J Cardiol. 2013;29:1104–1109. doi: 10.1016/j.cjca.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 27.Risgaard B, Jabbari R, Refsgaard L, Holst A, Haunso S, Sadjadieh A, Winkel B, Olesen M, Tfelt-Hansen J. High prevalence of genetic variants previously associated with Brugada syndrome in new exome data. Clin Genet. 2013;84:489–495. doi: 10.1111/cge.12126. [DOI] [PubMed] [Google Scholar]
- 28.Ng PC, Henikoff S. Accounting for human polymorphisms predicted to affect protein function. Genome Res. 2002;12:436–446. doi: 10.1101/gr.212802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu L, Kumar S. Evolutionary Balancing is Critical for Correctly Forecasting Disease Associated Amino Acid Variants. Mol Biol Evol. 2013;30:1252–1257. doi: 10.1093/molbev/mst037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu H, Huff CD, Moore B, Flygare S, Reese MG, Yandell M. VAAST 2.0: improved variant classification and disease-gene identification using a conservation-controlled amino acid substitution matrix. Genet Epidemiol. 2013;37:622–634. doi: 10.1002/gepi.21743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bendl J, Stourac J, Salanda O, Pavelka A, Wieben ED, Zendulka J, Brezovsky J, Damborsky J. PredictSNP: robust and accurate consensus classifier for prediction of disease-related mutations. PLoS Comput Biol. 2014;10:e1003440. doi: 10.1371/journal.pcbi.1003440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thusberg J, Olatubosun A, Vihinen M. Performance of mutation pathogenicity prediction methods on missense variants. Hum Mutat. 2011;32:358–368. doi: 10.1002/humu.21445. [DOI] [PubMed] [Google Scholar]
- 35.Jordan DM, Kiezun A, Baxter SM, Agarwala V, Green RC, Murray MF, Pugh T, Lebo MS, Rehm HL, Funke BH, Sunyaev SR. Development and validation of a computational method for assessment of missense variants in hypertrophic cardiomyopathy. Am J Hum Genet. 2011;88:183–192. doi: 10.1016/j.ajhg.2011.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Galehdari H, Saki N, Mohammadi-Asl J, Rahim F. Meta-analysis diagnostic accuracy of SNP-based pathogenicity detection tools: a case of UTG1A1 gene mutations. Int J Mol Epidemiol Genet. 2013;4:77–85. [PMC free article] [PubMed] [Google Scholar]
- 37.Crotti L, Lewandowska MA, Schwartz PJ, Insolia R, Pedrazzini M, Bussani E, Dagradi F, George AL, Jr., Pagani F. A KCNH2 branch point mutation causing aberrant splicing contributes to an explanation of genotype-negative long QT syndrome. Heart Rhythm. 2009;6:212–218. doi: 10.1016/j.hrthm.2008.10.044. [DOI] [PubMed] [Google Scholar]
- 38.Tester DJ, Benton AJ, Train L, Deal B, Baudhuin LM, Ackerman MJ. Prevalence and spectrum of large deletions or duplications in the major long QT syndrome-susceptibility genes and implications for long QT syndrome genetic testing. Am J Cardiol. 2010;106:1124–1128. doi: 10.1016/j.amjcard.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koopmann TT, Alders M, Jongbloed RJ, Guerrero S, Mannens MM, Wilde AA, Bezzina CR. Long QT syndrome caused by a large duplication in the KCNH2 (HERG) gene undetectable by current polymerase chain reaction-based exon-scanning methodologies. Heart Rhythm. 2006;3:52–55. doi: 10.1016/j.hrthm.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 40.Eddy CA, Maccormick JM, Chung SK, Crawford JR, Love DR, Rees MI, Skinner JR, Shelling AN. Identification of large gene deletions and duplications in KCNQ1 and KCNH2 in patients with long QT syndrome. Heart Rhythm. 2008;5:1275–1281. doi: 10.1016/j.hrthm.2008.05.033. [DOI] [PubMed] [Google Scholar]
- 41.Medlock MM, Tester DJ, Will ML, Bos JM, Ackerman MJ. Repeat long QT syndrome genetic testing of phenotype-positive cases: prevalence and etiology of detection misses. Heart Rhythm. 2012;9:1977–1982. doi: 10.1016/j.hrthm.2012.08.010. [DOI] [PubMed] [Google Scholar]
- 42.Dorschner MO, Amendola LM, Turner EH, Robertson PD, Shirts BH, Gallego CJ, Bennett RL, Jones KL, Tokita MJ, Bennett JT, Kim JH, Rosenthal EA, Kim DS, Tabor HK, Bamshad MJ, Motulsky AG, Scott CR, Pritchard CC, Walsh T, Burke W, Raskind WH, Byers P, Hisama FM, Nickerson DA, Jarvik GP. Actionable, pathogenic incidental findings in 1,000 participants’ exomes. Am J Hum Genet. 2013;93:631–640. doi: 10.1016/j.ajhg.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Green RC, Berg JS, Grody WW, Kalia SS, Korf BR, Martin CL, McGuire AL, Nussbaum RL, O'Daniel JM, Ormond KE, Rehm HL, Watson MS, Williams MS, Biesecker LG. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med. 2013;15:565–574. doi: 10.1038/gim.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Couzin-Frankel J. Return of unexpected DNA results urged. Science. 2013;339:1507–1508. doi: 10.1126/science.339.6127.1507. [DOI] [PubMed] [Google Scholar]
- 45.McGuire AL, Joffe S, Koenig BA, Biesecker BB, McCullough LB, Blumenthal-Barby JS, Caulfield T, Terry SF, Green RC. Point-counterpoint. Ethics and genomic incidental findings. Science. 2013;340:1047–1048. doi: 10.1126/science.1240156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Holtzman NA. ACMG recommendations on incidental findings are flawed scientifically and ethically. Genet Med. 2013;15:750–751. doi: 10.1038/gim.2013.96. [DOI] [PubMed] [Google Scholar]
- 47.Townsend A, Adam S, Birch PH, Friedman JM. Paternalism and the ACMG recommendations on genomic incidental findings: patients seen but not heard. Genet Med. 2013;15:751–752. doi: 10.1038/gim.2013.105. [DOI] [PubMed] [Google Scholar]
- 48.Green RC, Lupski JR, Biesecker LG. Reporting genomic sequencing results to ordering clinicians: incidental, but not exceptional. JAMA. 2013;310:365–366. doi: 10.1001/jama.2013.41703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.ACMG Updates Recommendation on “Opt Out” for Genome Sequencing Return of Results. 2014 https://www.acmg.net/docs/Release_ACMGUpdatesRecommendations_final.pdf.
- 50.Giacomini KM, Brett CM, Altman RB, Benowitz NL, Dolan ME, Flockhart DA, Johnson JA, Hayes DF, Klein T, Krauss RM, Kroetz DL, McLeod HL, Nguyen AT, Ratain MJ, Relling MV, Reus V, Roden DM, Schaefer CA, Shuldiner AR, Skaar T, Tantisira K, Tyndale RF, Wang L, Weinshilboum RM, Weiss ST, Zineh I. The pharmacogenetics research network: from SNP discovery to clinical drug response. Clin Pharmacol Ther. 2007;81:328–345. doi: 10.1038/sj.clpt.6100087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Caudle KE, Klein TE, Hoffman JM, Muller DJ, Whirl-Carrillo M, Gong L, McDonagh EM, Sangkuhl K, Thorn CF, Schwab M, Agundez JA, Freimuth RR, Huser V, Lee MT, Iwuchukwu OF, Crews KR, Scott SA, Wadelius M, Swen JJ, Tyndale RF, Stein CM, Roden D, Relling MV, Williams MS, Johnson SG. Incorporation of Pharmacogenomics into Routine Clinical Practice: the Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline Development Process. Curr Drug Metab. 2014;15:209–217. doi: 10.2174/1389200215666140130124910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weeke P, Roden DM. Applied pharmacogenomics in cardiovascular medicine. Annu Rev Med. 2014;65:81–94. doi: 10.1146/annurev-med-101712-122545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Turner RM, Pirmohamed M. Cardiovascular pharmacogenomics: expectations and practical benefits. Clin Pharmacol Ther. 2014;95:281–293. doi: 10.1038/clpt.2013.234. [DOI] [PubMed] [Google Scholar]
- 54.Johnson JA, Cavallari LH. Pharmacogenetics and cardiovascular disease--implications for personalized medicine. Pharmacol Rev. 2013;65:987–1009. doi: 10.1124/pr.112.007252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scott SA, Sangkuhl K, Stein CM, Hulot JS, Mega JL, Roden DM, Klein TE, Sabatine MS, Johnson JA, Shuldiner AR. Clinical Pharmacogenetics Implementation Consortium guidelines for CYP2C19 genotype and clopidogrel therapy: 2013 update. Clin Pharmacol Ther. 2013;94:317–323. doi: 10.1038/clpt.2013.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wilke RA, Ramsey LB, Johnson SG, Maxwell WD, McLeod HL, Voora D, Krauss RM, Roden DM, Feng Q, Cooper-Dehoff RM, Gong L, Klein TE, Wadelius M, Niemi M. The clinical pharmacogenomics implementation consortium: CPIC guideline for SLCO1B1 and simvastatin-induced myopathy. Clin Pharmacol Ther. 2012;92:112–117. doi: 10.1038/clpt.2012.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ramsey LB, Johnson SG, Caudle KE, Haidar CE, Voora D, Wilke RA, Maxwell WD, McLeod HL, Krauss RM, Roden DM, Feng Q, Cooper-Dehoff RM, Gong L, Klein TE, Wadelius M, Niemi M. The Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for SLCO1B1 and simvastatin-induced myopathy: 2014 update. Clin Pharmacol Ther. 2014;96:423–428. doi: 10.1038/clpt.2014.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Johnson JA, Gong L, Whirl-Carrillo M, Gage BF, Scott SA, Stein CM, Anderson JL, Kimmel SE, Lee MT, Pirmohamed M, Wadelius M, Klein TE, Altman RB. Clinical Pharmacogenetics Implementation Consortium Guidelines for CYP2C9 and VKORC1 genotypes and warfarin dosing. Clin Pharmacol Ther. 2011;90:625–629. doi: 10.1038/clpt.2011.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weitzel KW, Elsey AR, Langaee TY, Burkley B, Nessl DR, Obeng AO, Staley BJ, Dong HJ, Allan RW, Liu JF, Cooper-Dehoff RM, Anderson RD, Conlon M, Clare-Salzler MJ, Nelson DR, Johnson JA. Clinical pharmacogenetics implementation: Approaches, successes, and challenges. Am J Med Genet C Semin Med Genet. 2014;166:56–67. doi: 10.1002/ajmg.c.31390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shuldiner AR, Palmer K, Pakyz RE, Alestock TD, Maloney KA, O'Neill C, Bhatty S, Schub J, Overby CL, Horenstein RB, Pollin TI, Kelemen MD, Beitelshees AL, Robinson SW, Blitzer MG, McArdle PF, Brown L, Jeng LJ, Zhao RY, Ambulos N, Vesely MR. Implementation of pharmacogenetics: The University of Maryland personalized anti-platelet pharmacogenetics program. Am J Med Genet C Semin Med Genet. 2014;166:76–84. doi: 10.1002/ajmg.c.31396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.O'Donnell PH, Bush A, Spitz J, Danahey K, Saner D, Das S, Cox NJ, Ratain MJ. The 1200 patients project: creating a new medical model system for clinical implementation of pharmacogenomics. Clin Pharmacol Ther. 2012;92:446–449. doi: 10.1038/clpt.2012.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pulley JM, Denny JC, Peterson JF, Bernard GR, Vnencak-Jones CL, Ramirez AH, Delaney JT, Bowton E, Brothers K, Johnson K, Crawford DC, Schildcrout J, Masys DR, Dilks HH, Wilke RA, Clayton EW, Shultz E, Laposata M, McPherson J, Jirjis JN, Roden DM. Operational implementation of prospective genotyping for personalized medicine: the design of the Vanderbilt PREDICT project. Clin Pharmacol Ther. 2012;92:87–95. doi: 10.1038/clpt.2011.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hoffman JM, Haidar CE, Wilkinson MR, Crews KR, Baker DK, Kornegay NM, Yang W, Pui CH, Reiss UM, Gaur AH, Howard SC, Evans WE, Broeckel U, Relling MV. PG4KDS: A model for the clinical implementation of pre-emptive pharmacogenetics. Am J Med Genet C Semin Med Genet. 2014;166:45–55. doi: 10.1002/ajmg.c.31391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.O'Donnell PH, Danahey K, Jacobs M, Wadhwa NR, Yuen S, Bush A, Sacro Y, Sorrentino MJ, Siegler M, Harper W, Warrick A, Das S, Saner D, Corless CL, Ratain MJ. Adoption of a clinical pharmacogenomics implementation program during outpatient care-initial results of the University of Chicago “1,200 Patients Project”. Am J Med Genet C Semin Med Genet. 2014;166:68–75. doi: 10.1002/ajmg.c.31385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Desai NR, Canestaro WJ, Kyrychenko P, Chaplin D, Martell LA, Brennan T, Matlin OS, Choudhry NK. Impact of CYP2C19 genetic testing on provider prescribing patterns for antiplatelet therapy after acute coronary syndromes and percutaneous coronary intervention. Circ Cardiovasc Qual Outcomes. 2013;6:694–699. doi: 10.1161/CIRCOUTCOMES.113.000321. [DOI] [PubMed] [Google Scholar]
- 66.Kimmel SE, French B, Kasner SE, Johnson JA, Anderson JL, Gage BF, Rosenberg YD, Eby CS, Madigan RA, McBane RB, bdel-Rahman SZ, Stevens SM, Yale S, Mohler ER, III, Fang MC, Shah V, Horenstein RB, Limdi NA, Muldowney JA, III, Gujral J, Delafontaine P, Desnick RJ, Ortel TL, Billett HH, Pendleton RC, Geller NL, Halperin JL, Goldhaber SZ, Caldwell MD, Califf RM, Ellenberg JH. A pharmacogenetic versus a clinical algorithm for warfarin dosing. N Engl J Med. 2013;369:2283–2293. doi: 10.1056/NEJMoa1310669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Verhoef TI, Ragia G, de BA, Barallon R, Kolovou G, Kolovou V, Konstantinides S, Le CS, Maltezos E, van der Meer FJ, Redekop WK, Remkes M, Rosendaal FR, van Schie RM, Tavridou A, Tziakas D, Wadelius M, Manolopoulos VG, Maitland-van der Zee AH. A randomized trial of genotype-guided dosing of acenocoumarol and phenprocoumon. N Engl J Med. 2013;369:2304–2312. doi: 10.1056/NEJMoa1311388. [DOI] [PubMed] [Google Scholar]
- 68.Pirmohamed M, Burnside G, Eriksson N, Jorgensen AL, Toh CH, Nicholson T, Kesteven P, Christersson C, Wahlstrom B, Stafberg C, Zhang JE, Leathart JB, Kohnke H, Maitland-van der Zee AH, Williamson PR, Daly AK, Avery P, Kamali F, Wadelius M. A randomized trial of genotype-guided dosing of warfarin. N Engl J Med. 2013;369:2294–2303. doi: 10.1056/NEJMoa1311386. [DOI] [PubMed] [Google Scholar]
- 69.Ramos EM, Din-Lovinescu C, Berg JS, Brooks LD, Duncanson A, Dunn M, Good P, Hubbard TJ, Jarvik GP, O'Donnell C, Sherry ST, Aronson N, Biesecker LG, Blumberg B, Calonge N, Colhoun HM, Epstein RS, Flicek P, Gordon ES, Green ED, Green RC, Hurles M, Kawamoto K, Knaus W, Ledbetter DH, Levy HP, Lyon E, Maglott D, McLeod HL, Rahman N, Randhawa G, Wicklund C, Manolio TA, Chisholm RL, Williams MS. Characterizing genetic variants for clinical action. Am J Med Genet C Semin Med Genet. 2014;166:93–104. doi: 10.1002/ajmg.c.31386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H, Klemm A, Flicek P, Manolio T, Hindorff L, Parkinson H. The NHGRI GWAS Catalog, a curated resource of SNP- trait associations. Nucleic Acids Res. 2014;42:D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Teutsch SM, Bradley LA, Palomaki GE, Haddow JE, Piper M, Calonge N, Dotson WD, Douglas MP, Berg AO. The Evaluation of Genomic Applications in Practice and Prevention (EGAPP) Initiative: methods of the EGAPP Working Group. Genet Med. 2009;11:3–14. doi: 10.1097/GIM.0b013e318184137c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Palomaki GE, Melillo S, Neveux L, Douglas MP, Dotson WD, Janssens AC, Balkite EA, Bradley LA. Use of genomic profiling to assess risk for cardiovascular disease and identify individualized prevention strategies--a targeted evidence-based review. Genet Med. 2010;12:772–784. doi: 10.1097/GIM.0b013e3181f8728d. [DOI] [PubMed] [Google Scholar]
- 73.Recommendations from the EGAPP Working Group: genomic profiling to assess cardiovascular risk to improve cardiovascular health. Genet Med. 2010;12:839–843. doi: 10.1097/GIM.0b013e3181f872c0. [DOI] [PubMed] [Google Scholar]
- 74.Caulfield T, McGuire AL. Direct-to-consumer genetic testing: perceptions, problems, and policy responses. Annu Rev Med. 2012;63:23–33. doi: 10.1146/annurev-med-062110-123753. [DOI] [PubMed] [Google Scholar]
- 75.Eriksson N, Macpherson JM, Tung JY, Hon LS, Naughton B, Saxonov S, Avey L, Wojcicki A, Pe'er I, Mountain J. Web-based, participant-driven studies yield novel genetic associations for common traits. PLoS Genet. 2010;6:e1000993. doi: 10.1371/journal.pgen.1000993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tung JY, Do CB, Hinds DA, Kiefer AK, Macpherson JM, Chowdry AB, Francke U, Naughton BT, Mountain JL, Wojcicki A, Eriksson N. Efficient replication of over 180 genetic associations with self-reported medical data. PLoS ONE. 2011;6:e23473. doi: 10.1371/journal.pone.0023473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Do CB, Tung JY, Dorfman E, Kiefer AK, Drabant EM, Francke U, Mountain JL, Goldman SM, Tanner CM, Langston JW, Wojcicki A, Eriksson N. Web-based genome-wide association study identifies two novel loci and a substantial genetic component for Parkinson's disease. PLoS Genet. 2011;7:e1002141. doi: 10.1371/journal.pgen.1002141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bloss CS, Schork NJ, Topol EJ. Effect of direct-to-consumer genomewide profiling to assess disease risk. N Engl J Med. 2011;364:524–534. doi: 10.1056/NEJMoa1011893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bloss CS, Darst BF, Topol EJ, Schork NJ. Direct-to-consumer personalized genomic testing. Hum Mol Genet. 2011;20:R132–R141. doi: 10.1093/hmg/ddr349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fears R, ter M V. The perspective from EASAC and FEAM on direct-to-consumer genetic testing for health-related purposes. Eur J Hum Genet. 2013;21:703–707. doi: 10.1038/ejhg.2012.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Downing NS, Ross JS. Innovation, risk, and patient empowerment: the FDA-mandated withdrawal of 23andMe's Personal Genome Service. JAMA. 2014;311:793–794. doi: 10.1001/jama.2014.148. [DOI] [PubMed] [Google Scholar]
- 82.Annas GJ, Elias S. 23andMe and the FDA. N Engl J Med. 2014;370:985–988. doi: 10.1056/NEJMp1316367. [DOI] [PubMed] [Google Scholar]
- 83.Boycott KM, Vanstone MR, Bulman DE, MacKenzie AE. Rare-disease genetics in the era of next-generation sequencing: discovery to translation. Nat Rev Genet. 2013;14:681–691. doi: 10.1038/nrg3555. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.