Abstract
Postranslational modifications of histones have been correlated with virtually all chromatintemplated processes, including gene expression regulation, DNA replication, mitosis and meiosis, and DNA repair. In order to better understand the mechanistic basis by which histone modifications participate in the control of cellular processes, it is essential to identify and characterize downstream effector proteins, or “readers”, that are responsible for recognizing different marks and translating them into specific biological outcomes. Ideally, identification of potential histone-binding effectors should occur in an unbiased fashion. Although in the recent years much progress has been made in identifying readers of histone modifications, in particular methylation, recognition of the majority of known histone marks is still poorly understood. Here I describe a simple and unbiased biochemical pull-down assay that allows for the identification of novel histone effector proteins and utilizes biotinylated histone peptides modified at various residues. I provide detailed protocols and suggestions for troubleshooting.
Keywords: histone modifications, peptide pull-down, chromatin, effector protein, histone methylation, histone acetylation, histone phosphorylation, biotinylated peptide
1. Introduction and overview of the assay
Histone proteins are extensively posttranslationally modified at specific residues by a variety of mechanisms including acetylation, methylation, phosphorylation, ADP-ribosylation, citrullination, ubiquitinylation and sumoylation (for recent reviews on histone modifications and histone modifying enzymes see: [1, 2, 3, 4, 5]). Although specific modifications have been well correlated with distinct biological processes, precise mechanisms by which histone modifications transmit their biological signals into meaningful biological readout are, in general, poorly understood. There is mounting evidence, however, that modifications act through the recruitment of downstream molecules, or “readers”, which specifically recognize a particular modification in the context of the histone molecule. Association of the downstream effectors may then lead to changes in accessibility of the DNA template to the transcriptional machinery, recruitment of enzymatic activities, e.g. ATP-dependent chromatin remodeling complexes, or changes in the higher order structure of chromatin, which would, in turn, dictate specific regulatory outcomes. In addition to bromodomain-containing proteins, which have been shown to recognize acetylated lysine residues, many readers of histone methylation marks have been characterized recently, including chromodomain, Tudor domain, MBT-repeats, WD40-repeats, and PHD finger proteins (for reviews on effector proteins see: [6, 7]).
Histones are most heavily decorated at the N- and C-terminal tails, and known effectors do not appear to recognize more than 10 amino acids of the histone sequence. Therefore assays with modified peptides corresponding in sequence to the histone tails are particularly useful for studying readers of histone modifications. There are many excellent biochemical and biophysical assays suitable for analysis of peptide-protein interactions. However, most of these assays are restricted to “candidate” approaches in which a limited, pre-determined set of proteins can be tested for binding to a particular modified or unmodified peptide. One significant advantage of using a peptide pull-down assay from nuclear extracts, in addition to its simplicity, is the fact that it is an unbiased approach, and therefore allows for identification of factors that would otherwise not have been likely candidates (see for example [8, 9]). The assay also has its caveats: it is not quantitative and, due to extensive washes needed to minimize background, the association off-rate significantly impacts on observed binding.
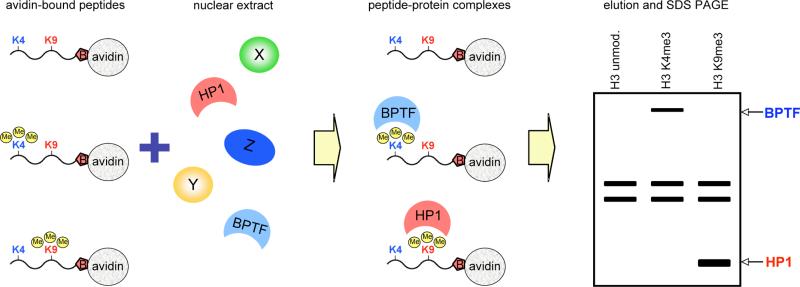
The principle of the peptide pull-down assay is illustrated in Figure 1. Biotinylated histone peptides that are either unmodified or modified at specific residues are immobilized on avidin beads and incubated with nuclear extract. After incubation, beads are extensively washed, allowing for separation of bound and un-bound proteins. Bound proteins are then eluted from the beads and resolved by SDS PAGE. Proteins present in the modified, but not unmodified, peptide pull-down lane are identified by mass spectrometry, and represent candidate readers of a specific histone modification.
Figure.
Schematics of the peptide pull-down assay. Biotinylated histone peptides either unmodified, or modified at specific residues are immobilized on avidin beads and incubated with extract. To illustrate the principle of the assay, H3 peptide, either unmodified, or trimethylated at lysine 4 (K4), or trimethylated at lysine 9 (K9) is shown. Specific effector proteins bind to histone peptides in a modification dependent manner. This is illustrated by specific recognition of the H3 K4me3 peptide by BPTF (shown in blue), and of the H3 K9me3 peptide by HP1 (shown in red). Bound proteins are then eluted from the avidin beads, and resolved by SDS PAGE. Proteins present in the modified, but not unmodified peptide pull-down lane represent candidate readers of a specific histone modification. For example, BPTF is specifically present in the H3 K4me3 pull-down, whereas HP1 is recovered in the H3 K9me3 pull-down.
2. Method
2.1. Design and synthesis of the biotinylated peptides
Histone peptides with residues variously modified (e.g. methylated, acetylated and phosphorylated) and conjugated to biotin, can be synthesized chemically. I recommend synthesis of peptides about 20 amino acids long, with biotin conjugated through a linker on the C-terminus for N-terminal histone peptides, and on the N-terminus for the C-terminal histone peptides. Modification should be positioned close to the center of the peptide (with the exception of modifications located very close to the terminus, e.g. H3 K4 methylation), allowing for 6-8 amino acid overhang on each side, which may be necessary for recognition by the effector protein.
For each modified peptide, corresponding unmodified peptide should be synthesized to use as a negative control for a pull-down. As a positive control, include a peptide binding a known effector protein. I recommend using H3 K9me3 amino acids 1-20 peptide, which very robustly associates with three isoforms of HP1, a protein that is highly expressed in all cell types tested.
Alternatively, peptides can be synthesized with a terminal cysteine and then conjugated to a resin using SulfoLink (Pierce). Although this approach has been successfully used by several laboratories (see for example: [10, 11]), in our hands pull-downs with SulfoLink conjugated peptides tend to give much higher unspecific backgrounds compared with pull-downs with avidin-bound peptides.
Synthesis of the biotinylated histone peptides containing various modifications can be requested from a university peptide facility or a commercial source (e.g. Rockefeller University Proteomics Resource Center, Sigma, GL Biochem). Some of the ready-made modified and unmodified biotinylated histone peptides are also available from Upstate Biotech. I recommend using peptides that have been HPLC purified to purity of 80% or higher and analyzed by mass spectrometry. After synthesis peptides should be aliquoted, lyophilized and stored dry at −80° C.
2.2. Preparation of the peptide-bound resin
Reagents
Chemically synthesized biotinylated peptides (described in 2.1)
Immobilized avidin (Pierce)
PBS (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.47 mM KH2PO4, pH 7.4)
TritonX-100 (20% v/v stock solution)
Sodium azide (10% w/v stock solution)
Protocol
1. Resuspend 100 μg of each lyophilized peptide in 400 μl of PBS. Use corresponding set of unmodified and appropriately modified peptides. Typically, I use 100 μg of peptide per 400 μl of immobilized avidin beads (Pierce); however, if beads of different biotin binding capacity are used, the peptide-resin ratio should be adjusted accordingly. 2. Pre-bind biotinylated peptides to the immobilized avidin beads. Wash 400 μl of avidin beads three times in 1 ml of PBS/0.1% TritonX-100, remove excess of buffer and add peptide resuspended in PBS. Incubate for 3h at room temperature (RT) with rotation, then separate avidin resin from the unbound peptide by washing 3 times in PBS/0.1% TritonX-100.
3. Resuspend avidin-bound peptides in 400 μl of PBS to prepare a 50% slurry and add sodium azide to a final concentration of 0.1% for long-term storage; this amount of immobilized peptide is typically sufficient for 20 pull-down assays as described below. Avidin-bound peptides can be stored at 4°C for at least a month; that said, methylated peptides are usually very stable, whereas phosphorylated peptides may be more labile due to hydrolysis.
2.3. Peptide pull-down assay from nuclear extracts
Reagents
Avidin bound peptides prepared as described in 2.2
Immobilized avidin (Pierce)
Buffer A (10 mM HEPES pH 7.9, 1.5 mM MgCl2, 10 mM KCl, freshly added protease inhibitors and DTT to 1mM)
Buffer C (20 mM HEPES pH 7.9, 25% v/v glycerol, 420 mM KCl, 1.5 mM MgCl2, 0.2 mM EDTA, freshly added protease inhibitors and DTT to 1mM)
Buffer D (20 mM HEPES pH 7.9, 20% v/v glycerol, 0.2 mM EDTA, 0.2% TritonX-100, freshly added protease inhibitors, and KCl at the indicated concentration)
Low-HEPES buffer (4mM HEPES pH7.9, 10mM NaCl)
TritonX-100 (20% v/v stock solution)
100 mM glycine pH 2.8
1 M Tris pH 8
4X Laemmli sample buffer (0.2 M Tris-HCl, pH 6.8, 8% SDS, 40% glycerol, 0.04% bromophenol blue); freshly added DTT to 40 mM
SilverQuest silver staining kit (Invitrogen)
Protease inhibitor coctail (Roche)
PMSF (100 mM stock solution in ethanol)
Protocol
1. Prepare nuclear extract from HEK293 cells, or other cell line of choice, using standard high salt extraction protocol [12]. Briefly, lyse cells by Dounce homogenizing in hypotonic buffer A, separate nuclear and cytoplasmic fractions, and extract nuclear pellet with buffer C. Typically, I use extract from 108 cells per peptide, but the amount of necessary input material may vary depending on the effector protein abundance. 2. Lower the salt concentration in the extract to 150mM by dialyzing against buffer D/150mM KCl or diluting with buffer D/no salt. Adjust the extraction buffer volume such that 1ml of nuclear extract corresponds to 108 cell equivalents (a total protein concentration of about 2-5 μg/μl).
Add protease inhibitors at every step of extract preparation on, and if you are planning to work with phosphorylated peptides, include phosphatase inhibitors as well. Extract can be flash-frozen in liquid nitrogen and kept at –80°C or used directly for the pull-down assay. Using freshly prepared extracts can be advantageous for identification of proteins that are particularly susceptible to degradation. 3. Add TritonX-100 to the nuclear extract to a final concentration of 0.1% and spin at 16 000g/10 min/4°C, to remove any precipitate that may have formed. Pre-clear the nuclear extract with avidin beads by adding 80 μl of 50% immobilized avidin slurry, pre-washed with buffer D/150mM KCl /0.1% TritonX-100. Incubate for 1h at 4°C with rotation, spin at low speed and collect the supernatant.
4. For each peptide, add 40 μl of 50% peptide-bound slurry prepared in step 2.2 to a fresh tube and wash once with buffer D/150mM KCl /0.1% TritonX-100. Remove supernatant and add pre-cleared nuclear extract (108 cell equivalent) to each avidin-bound peptide. Incubate with rotation for 3h to overnight at 4°C. Spin at low speed, remove supernatant and flash-freeze it for further analysis.
5. Transfer beads to a fresh tube using wash buffer. Wash eight times with 1 ml of buffer D/300mM KCl/0.1% TritonX-100 (see also Suggestions for troubleshooting).
6. Perform the final wash with low-HEPES buffer. This step is to ensure you can change pH readily during elution with glycine.
7. Elute twice for 10 minutes at RT with 100 mM glycine pH 2.8, using 1-2 equivalents of the bead volume (e.g. 20 μl) for each elution. Carefully remove each eluate using capillary pipette tip, combine the eluates and neutralize pH by adding 1/10 volume of 1M Tris pH 8. Centrifuge briefly to remove any residual beads, transfer the supernatant to a fresh tube, and add Laemmli buffer. Often, it is helpful to analyze in parallel proteins that remained on the beads after glycine treatment. To do so, after collecting glycine eluates, resuspend the beads in 100 mM Tris pH 8, add Laemmli buffer and boil (see also Suggestions for troubleshooting).
8. Analyze eluates from each pull-down on SDS PAGE gel and visualize proteins by silver staining using a mass spectrometry-compatible protocol (e.g. SilverQuest silver staining kit from Invitrogen). Handle gel with caution to prevent contamination with keratin. Excise protein bands present in the modified, but not unmodified, peptide pull-down gel lane, and identify entrapped proteins by mass spectrometry.
9. Confirm specific association of the identified protein with the modified peptide by immunoblotting eluates with specific antibodies, if available, and by performing pull-down assays using purified recombinant protein as input (for examples, see: [8, 9]).
3. Suggestions for troubleshooting
A major challenge in using the aforementioned protocol to identify novel proteins that recognize specific histone modifications is optimizing the signal to noise ratio; namely, retaining proteins bound in the modification-specific manner and, at the same time, reducing non-specific binding, which may obscure analysis and identification of specific effectors. Note that some proteins will specifically bind to histone tails regardless of its modification status. For example, I repeatedly recover a histone chaperone with modified and unmodified H3 tail peptide pull-downs.
However, if positive controls in your pull-down assay are working (e.g. you specifically recover HP1 in a H3 K9me pull-down), but proteins binding a modification of interest cannot be detected, than it is worthwhile to optimize the conditions for your particular peptide set. Below are several suggestions on how the assay may be optimized.
3.1. Using the “right” input extract
Success in identifying a histone modification specific reader is highly dependent on its abundance in the input extract. However, while searching for previously unidentified effector proteins, the abundance of the protein in the extract cannot be tested directly. Nevertheless, if initial analysis does not yield positive results, it is important to use extracts from different cell lines, as some factors may not be expressed well in a particular cell type. For example, while searching for H3 K4me3 binding proteins, I identified BPTF in pull-downs from HEK293, but not HeLa nuclear extracts.
Many histone-bound factors will not efficiently extract with high salt (420-500 mM) during nuclear extract preparation. This is not a problem for abundant proteins present in nucleus in excess (e.g. HP1). However, if the vast majority of the protein of interest is tightly bound to chromatin, it may be advantageous to use an alternative extract preparation method, which utilizes sonication (see, for example: [13]).
Finally, it is helpful to consider a cellular process in which a histone modification of interest is involved. For example, if the modification is associated with mitosis, synchronized mitotic extracts would be an appropriate input for pull-down; if it is associated with DNA damage repair, extracts from cells induced for the DNA damage response should be used.
3.2. Optimizing washing conditions
The protocol described here calls for washes with buffer containing 300mM salt, which usually result in relatively low backgrounds in pull-downs and do not disrupt association of HP1, WDR5, BPTF or CBX7 with H3 peptides [8, 9, 14]. However, salt concentration of 250 mM disrupts association of NuRD complex with H3 peptides [10]. Therefore 300 mM salt washes may be too stringent to retain some interactions, and washing with buffer containing lower salt may result in better recovery of the protein of interest. Conversely, for interactions that are less salt-sensitive, increasing salt and detergent concentration in washing buffer may be beneficial, as it will result in lower background.
3.3. Optimizing elution conditions
Some proteins do not efficiently elute with 100 mM glycine pH 2.8. As an alternative, base elution can be used (with 0.5 N NH4OH/0.5 mM EDTA), or bound proteins can be eluted by competition with excess of free peptide. The latter approach is the most specific, and can eliminate the majority of the non-specifically bound proteins. However, it requires large amounts of peptide (typically, I elute with 0.5 mg/ml peptide in PBS), and in some cases the elution efficiency is very poor. Elutions can be performed with the same peptide that was used for pull-down, or, alternatively, to increase specificity, first with the unmodified peptide followed by the modified peptide.
4. Concluding remarks
Peptide pull-down assay provides a straightforward and unbiased approach to discovery of novel proteins reading histone modifications. The simplicity of the assay makes it easy to perform in any laboratory. In principle, the approach described here can be used to identify proteins recognizing any posttranslational modification on histone tails, provided that appropriately modified biotinylated peptides can be synthesized chemically. Precursor amino acids for modifications like acetylation, phosphorylation of serine and threonine residues, citrullination and different forms of methylation (mono-, di- and trimethylation of lysines, symmetric and asymmetric dimethylation of arginines) are commonly available. Chemical synthesis of peptides containing large modifications, like ubiquitinylation and sumoylation, represent a more significant challenge, but ubiquitin and sumo moieties, for example, can be added to the histone peptide using chemical ligation methods.
The peptide pull-down assay is particularly useful for discovery of histone tail modifications readers, given that histone tails are generally unstructured (therefore biotinylated peptide mimicks the native epitope well), and currently known effectors do not appear to recognize more than 10 amino acids of the histone tail sequence. Nevertheless, this assay can also be used for discovery of proteins that recognize modifications on histone globular domains or proteins other than histones. In this case, however, the caveat is that the modified peptide may not be presented to the reader protein in the right conformation, or that additional molecular contacts are required to stabilize the interaction. Finally, I point out that principals underlying the peptdide pull-down assay described here may be modified to screen immobilized peptide libraries for histone peptide readers in a high throughput, more automated format. A reverse approach with protein array containing immobilized GST fusion proteins probed with different methylated peptides has already been described [15].
Acknowledgements
I thank David Allis for his support and encouragement, and David Allis, Lindsey Baker, Elizabeth Duncan and Tomek Swigut for critical readings of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nightingale KP, O'Neill LP, Turner BM. Curr Opin Genet Dev. 2006;16:125–136. doi: 10.1016/j.gde.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 2.Shilatifard A. Annu Rev Biochem. 2006 [Google Scholar]
- 3.Lin W, Dent SYR. Curr Opin Genet Dev. 2006;16:137–142. doi: 10.1016/j.gde.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 4.Martin C, Zhang Y. Nat Rev Mol Cell Biol. 2005;2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- 5.van Attikum H, Gasser SM. Nat Rev Mol Cell Biol. 6(10):757–765. doi: 10.1038/nrm1737. [DOI] [PubMed] [Google Scholar]
- 6.Daniel JA, Pray-Grant MG, Grant PA. Cell Cycle. 2005;4:919–926. doi: 10.4161/cc.4.7.1824. [DOI] [PubMed] [Google Scholar]
- 7.de la Cruz X, Lois S, Snchez-Molina S, Martnez-Balbs MA. Bioessays. 2005;27:164–175. doi: 10.1002/bies.20176. [DOI] [PubMed] [Google Scholar]
- 8.Wysocka J, Swigut T, Milne TA, Dou Y, Zhang X, Burlingame AL, Roeder RG, Brivanlou AH, Allis CD. Cell. 2005;121:859–872. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 9.Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, Wu C, Allis CD. Nature, AOP. 2006 doi: 10.1038/nature04815. http://dx.doi.org/10.1038/nature04815. [DOI] [PubMed]
- 10.Zegerman P, Canas B, Pappin D, Kouzarides T. J Biol Chem. 2002;277:11621–11624. doi: 10.1074/jbc.C200045200. [DOI] [PubMed] [Google Scholar]
- 11.Sims RJ, Chen C-F, Santos-Rosa H, Kouzarides T, Patel SS, Reinberg D. J Biol Chem. 2005;280:41789–41792. doi: 10.1074/jbc.C500395200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dignam JD, Lebovitz RM, Roeder RG. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roeder RG. J Biol Chem. 1974;249:241–248. [PubMed] [Google Scholar]
- 14.Bernstein E, Duncan EM, Masui O, Gil J, Heard E, Allis CD. Mol Cell Biol. 2006;26:2560–2569. doi: 10.1128/MCB.26.7.2560-2569.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim J, Daniel J, Espejo A, Lake A, Krishna M, Xia L, Zhang Y, Bedford MT. EMBO Rep. 2006;7:397–403. doi: 10.1038/sj.embor.7400625. [DOI] [PMC free article] [PubMed] [Google Scholar]