Abstract
The mucosal gene expression in rectosigmoid mucosa (RSM) in irritable bowel syndrome with diarrhea (IBS-D) is unknown. Our objectives were, first, to study mRNA expression [by RT2 PCR of 19 genes pertaining to tight junctions, immune activation, intestinal ion transport and bile acid (BA) homeostasis] in RSM in IBS-D patients (n = 47) and healthy controls (n = 17) and study expression of a selected protein (PDZD3) in 10 IBS-D patients and 4 healthy controls; second, to assess RSM mRNA expression according to genotype and fecal BA excretion (high ≥2,337 μmol/48 h); and third, to determine whether genotype or mucosal mRNA expression is associated with colonic transit or BA parameters. Fold changes were corrected for false detection rate for 19 genes studied (P < 0.00263). In RSM in IBS-D patients compared with controls, mRNA expression of GUC2AB, PDZD3, and PR2Y4 was increased, whereas CLDN1 and FN1 were decreased. One immune-related gene was upregulated (C4BP4) and one downregulated (CCL20). There was increased expression of a selected ion transport protein (PDZD3) on immunohistochemistry and Western blot in IBS-D compared with controls (P = 0.02). There were no significant differences in mucosal mRNA in 20 IBS-D patients with high compared with 27 IBS-D patients with normal BA excretion. GPBAR1 (P < 0.05) was associated with colonic transit. We concluded that mucosal ion transport mRNA (for several genes and PDZD3 protein) is upregulated and barrier protein mRNA downregulated in IBS-D compared with healthy controls, independent of genotype. There are no differences in gene expression in IBS-D with high compared with normal fecal BA excretion.
Keywords: neurotransmitters, ion channels, cytokines, barrier, immune, secretion, PDZD3, GUCA2B
the cause of loose bowel movements in patients with irritable bowel syndrome with diarrhea (IBS-D) is partly attributed to acceleration of colonic transit, documented in ∼45% of patients with IBS-D (15), and to intestinal secretory mechanisms (reviewed in Refs. 7 and 8). Documented secretory mechanisms include increased duodenal and rectosigmoid expression of secretory transmitters (e.g., 5-HT), reduced expression of the serotonin reuptake protein, and fecal excretion of secretogranins or chromogranins (25, 26, 46). There is also evidence of reduced expression of proabsorption mechanisms (e.g., mucosal PYY, somatostatin, NPY).
IBS has also been associated with changes in RSM expression of immune factors, barrier function, and mucus secretion (1, 4, 5, 9, 22, 33, 51, 56, 60, 63, 65, 68). In a few instances, altered mucosal gene expression in tissues was associated with the inherited genotype, such as TNFSF15 gene, which is associated with IBS and has been linked with functional alterations of mucosal immune and protective functions (51, 68).
Differences in jejunal mucosal expression (at gene and protein levels) and distribution of apical junction complex proteins between IBS patients and controls support the observed alterations in barrier function in colonic mucosa in patients with IBS-D (36, 37). We also demonstrated a borderline difference in the zonula occludens 1 (ZO-1) intensity score in the small bowel mucosa (P = 0.06) of patients with IBS-D compared with healthy controls, with lower intensity in HLA-DQ2/8-positive relative to HLA-DQ2/8-negative patients with IBS-D (56).
In a prior study, based on next-generation RNA sequencing and confirmation by reverse transcriptase polymerase chain reaction (RT-PCR), we examined RSM from nine patients with IBS-D and nine healthy controls and identified differential expression of secretory and barrier genes, suggesting that the transcriptome is different in IBS-D compared with controls (11). The upregulated mechanisms associated with changes in ion transport included PDZD3. PDZ adapter proteins are involved in multiple ion transport functions in the intestine, including sodium absorption through sodium-hydrogen exchange (NHE3), as well as guanylyl cyclase C receptor (GC-C, or GUCY2C)-induced chloride and water secretion through cGMP signaling that leads to cystic fibrosis transmembrane conductance regulator phosphorylation and chloride-bicarbonate exchange through the SLC26 anion exchanger (35). These ion-exchange mechanisms also result in alterations in intestinal fluid transport.
Another upregulated ion transport mechanism identified in our prior study was increased GUCA2B mRNA. GUCA2B encodes uroguanylin, an endogenous ligand for GC-C receptor, increasing cyclic GMP, chloride, and water secretion.
The present study hypotheses were as follows: first, there is upregulation of genes and proteins associated with intestinal secretion in the colonic mucosa of patients with IBS-D, particularly in patients with high fecal bile acid (BA) excretion; second, variations in inherited genes are associated with the mRNA expression of the same genes in the colonic mucosa; and third, genetic variation and/or RSM expression of the genes are associated with alterations in intermediate phenotype in IBS-D, specifically colonic transit and parameters of BA homeostasis.
The aims of this study were, first, to replicate the prior pilot study by targeted mRNA analysis using quantitative RT-PCR of RSM biopsies from 47 IBS-D patients and 17 healthy controls and to analyze the protein expression of one of the proteins associated with ion transport, PDZD3, in 10 IBS-D patients and 4 healthy controls; second, to determine whether there are differences in mucosal mRNA expression in subgroups of IBS-D with high compared with normal fecal BA excretion; third, to determine whether genotype was associated with level of expression of mRNA in RSM, colonic transit, or BA parameters; and fourth, to assess whether the level of expression of mRNA in RSM was associated with colonic transit.
METHODS
Ethical Approval
The study was approved by Mayo Clinic Institutional Review Board on October 28, 2011. Written, informed consent was received from participants prior to inclusion in the study.
Study Design
We appraised bowel functions, total fecal BA excretion over 48 h, fasting serum C4 (7α-hydroxy-4-cholesten-3-one) and FGF19, colonic transit, genotype, and RSM mRNA expression in 47 patients with IBS-D (by Rome III criteria). Fecal total BA excretion was used to differentiate patients with high or normal BA excretion, suggestive of BA diarrhea.
Patient Selection
Patients were recruited by public advertisement or by invitation to participate from a database of ∼1,200 patients with IBS living in communities within ∼120 miles of Mayo Clinic in Rochester, MN. Inclusion criteria were based on symptoms by use of a validated diary questionnaire that characterized IBS symptoms and, particularly, bowel functions (53). Participants also completed the Hospital Anxiety and Depression Inventory (67). These patients had been evaluated at Mayo Clinic, and alternative diagnoses such as inflammatory bowel disease, cancer, and celiac disease were excluded. The main exclusion criteria were intake of medications that could interfere with the study tests, bleeding diathesis (owing to the need for rectosigmoid biopsies), and alanine aminotransferase or aspartate aminotransferase greater than twice the upper normal limit to avoid interference with the assessment of BA parameters.
Stored Biospecimens
We used stored samples from patients who had consented to the use of biospecimens for future research in prior studies (1, 10, 57) conducted at Mayo Clinic in Rochester, MN. These samples were obtained in 47 patients with IBS-D and 17 healthy controls. To appraise specificity of the observations in IBS-D, we analyzed RSM samples from 10 patients with irritable bowel syndrome with constipation (IBS-C) previously studied in our laboratory (1).
Participants' rectosigmoid biopsies were preserved in a solution of RNAlater, stored at −80°C. For all the mucosal biopsies, the RNA concentration was 130.3 ± 6.2 (mean ± SE) ng/μl and the RNA integrity number (RIN) ranged from 9.2 to 10 (mean 9.9 ± 0.02).
DNA was similarly stored at −80°C after prior extraction from peripheral venous blood.
Measurement of Colonic Transit by Scintigraphy
Overall colonic transit was measured by validated scintigraphy with 111In-labeled activated charcoal particles delivered to the ileocolonic junction in a methacrylate-coated gelatin capsule. The primary transit end points were colonic geometric center at 24 h and 48 h; in addition, ascending colonic emptying T1/2 was measured by linear interpolation of radioisotopic content in the ascending colon at 4, 6, 24, and 48 h (23).
Identification of Subgroups of IBS-D Patients Based on Fecal BA Excretion
Total and main fecal BA excretions (per 48 h on 100 g fat diet) were measured by HPLC/tandem mass spectrometry (48, 57, 64). This assay was adapted from a method used with serum samples (52).
IBS-D subgroups were identified by fecal BA excretion >2,337 μmol/48 h based on 90th percentile of 45 healthy volunteers studied in our laboratory (64). The 90th percentile was used to define the upper limit of normal range, consistent with the observation that a normal distribution (estimated from 5th and 95th percentiles) requires sampling of 500 normal people (2).
Other BA Parameters
Fasting serum C4 (a measure of hepatic synthesis rate of BAs sampled in the morning) was measured by HPLC/tandem mass spectrometry (16); serum C4 is a validated method for detecting BA malabsorption (6). Similarly, fasting levels of fibroblast growth factor-19 (FGF19) were measured by a commercial enzyme-linked immunosorbent assay (FGF19 Quantikine Enzyme-Linked Immunosorbent Assay Kit; R&D Systems, Minneapolis, MN) as in prior studies (43, 64).
Gene Expression Method by RT2 PCR Array
Selection of genes of interest.
We developed a custom profile including 19 genes to assess the effect of IBS-D on the expression of tight junction proteins, chemokines, markers of innate immunity, ion channels, and transmitters that have been demonstrated to be differentially expressed in RSM of patients with IBS (11). The custom profile included two housekeeping genes for normalization and three control genes that check for sample quality and reaction quality (Table 1).
Table 1.
Genes of interest included in the RT-PCR analysis of rectosigmoid mucosa
| Gene Symbol | Refseq No. | Official Full Name |
|---|---|---|
| C4BPA | NM_000715 | Complement component 4 binding protein, alpha |
| CCL20 | NM_004591 | Chemokine (C-C motif) ligand 20 |
| CLDN1 | NM_021101 | Claudin 1 |
| FGFR4 | NM_002011 | Fibroblast growth factor receptor 4 |
| FN1 | NM_002026 | Fibronectin 1 |
| GPBAR1 | NM_170699 | G protein-coupled bile acid receptor 1 (syn. TGR5) |
| GUCA2B | NM_007102 | Guanylate cyclase activator 2B (uroguanylin) |
| IFIT3 | NM_001549 | Interferon-induced protein with tetratricopeptide repeats 3 |
| NR1H4 | NM_005123 | Nuclear receptor subfamily 1, group H, member 4 (syn. Farnesoid X receptor) |
| OCLN | NM_002538 | Occludin |
| P2RY4 | NM_002565 | Pyrimidinergic receptor P2Y, G-protein coupled, 4 |
| PDZD3 | NM_024791 | PDZ domain containing 3 |
| RBP2 | NM_004164 | Retinol binding protein 2, cellular |
| SLC10A2 | NM_000452 | Solute carrier family 10 (sodium/bile acid cotransporter family), member 2 (syn. Apical Sodium-coupled Bile acid transporter) |
| SLC6A4 | NM_001045 | Solute carrier family 6 (neurotransmitter transporter, serotonin), member 4 (syn. Serotonin transporter) |
| TFF1 | NM_003225 | Trefoil factor 1 |
| TJP1 | NM_175610 | Tight junction protein 1 (syn. zonula occludens 1) |
| TNFSF15 | NM_005118 | Tumor necrosis factor (ligand) superfamily, member 15 |
| VIP | NM_003381 | Vasoactive intestinal peptide |
| ACTB | NM_001101 | Actin, beta (housekeeping gene) |
| GAPDH | NM_002046 | Glyceraldehyde-3-phosphate dehydrogenase (housekeeping gene) |
| HGDC | SA_00105 | Human Genomic DNA Contamination |
| RTC | SA_00104 | Reverse Transcription Control |
| PPC | SA_00103 | Positive PCR Control |
Syn., synonym.
Assay method.
For mRNA expression, RNA was purified from human RSM biopsies by using the Qiagen RNeasy Kit (Qiagen, Valencia, CA), including on-column DNase treatment to remove genomic DNA. RNA quality was assessed on the Agilent Bioanalyzer. The resulting RNA (RIN > 7) was reverse transcribed by use of the RT2 First Strand Kit (Qiagen), and samples were analyzed for expression by a Custom Profiler RT2 PCR Array (Qiagen).
Genotyping Method
DNA was extracted from venous blood, and candidate genotype analysis was conducted by established PCR-based methods, as previously detailed in prior publications: rs4795541 [SLC6A4 (34)]; rs4263839 [TNFSF15 (68)]; rs11554825 [GPBAR1 (18)]; rs434434 [FGFR4 (13)]; rs188096 [SLC10A2 (18)]; rs17618244 [KLB (18)]; rs1966265 [FGFR4 (18)]; and rs351855 [FGFR4 (18)]. Farnesoid X receptor (FXR) single-nucleotide polymorphisms (SNPs) rs17030285 and rs4764980 were also assessed by TaqMan assay (catalog no. C_34126156_10; catalog C_3127933_20). Gene names are given according to Hugo Gene Nomenclature.
Briefly, genomic DNA was isolated from whole blood by use of the QIAamp DNA Blood Maxi Kit (Qiagen, Valencia, CA) and stored at −80°C until genotyping. The serotonin transporter protein promoter polymorphism (SLC6A4 rs4795541), also referred to as 5-HTTLPR, was determined by PCR-based fragment length. The remaining eight SNPs were analyzed by TaqMan SNP Genotyping Assays (Applied Biosystems, Foster City, CA) per the manufacturer's instructions. Following polymerase chain reaction amplification, end reactions were analyzed by using ABI 7300 Real-Time PCR System by Sequence Detection Software (Applied Biosystems).
Confocal Immunofluorescence Microscopy of PDZD3
We chose to study expression of PDZD3 because of its involvement in both sodium absorptive and chloride secretory mechanisms.
Confocal immunofluorescence microscopy was performed as previously described (44). Briefly, unstained RSM sections were deparaffinized and rehydrated, boiled in antigen unmasking solution (Vector Laboratories, Burlingame, CA), quenched with Image-iT FX signal enhancer (Life Technologies, Grand Island, NY), and blocked with 1% BSA/10% FBS/0.1% Triton X-100 in PBS. Slides were then incubated overnight at 4°C with primary antibodies PDZD3 (1:1,000, LifeSpan BioSciences, Seattle, WA) and cytokeratin 8/18 (1:50, Santa Cruz, Dallas, TX). Fluorescent-labeled goat anti-rabbit IgG H+L (Alexa Fluor 568, Abcam, Cambridge, MA) and donkey anti-mouse IgG H+L (Alexa Fluor 488, Life Technologies) were applied to the slides; the slides were then rinsed and mounted with ProLong Gold with DAPI (Molecular Probes, Life Technologies) and were visualized on a Zeiss LSM 510 confocal microscope with a ×63 magnification oil objective.
PDZD3 Protein Measurements by Western Blots
Whole cell lysates were isolated from human colonic biopsies with the RIPA Lysis Buffer System (Santa Cruz), and concentrations were determined by BCA quantification (Pierce, Rockford, IL). Proteins were separated by use of 4–15% Mini-PROTEAN TGX gels (Bio-Rad, Hercules, CA) and blotted onto nitrocellulose membranes. The membranes were blocked with 5% milk in PBS/0.2% Tween, after which PDZD3 primary antibody (1:4,000, LSBio Systems, Seattle, WA) was applied overnight at 40°C.
Vinculin (1:500, Santa Cruz) was used for normalization of protein loading. Membranes were washed and incubated with horseradish peroxidase-conjugated secondary antibody (goat anti-rabbit or donkey anti-goat, Santa Cruz) and visualized with Amersham ECL Prime Western Blotting Detection Reagent (GE Healthcare Life Sciences, Pittsburgh, PA), and autoradiography. Band densities were quantified with the Gel Pro Analyzer 6.0.0.349 (Media Cybernetics, Rockville, MD).
Statistical Analysis
All statistical analyses were conducted with SigmaPlot (Systat Software, San Jose, CA 95110). For mRNA expression analysis, the RT2 Profiler PCR Array software package was used. This package uses 2−ΔΔCT-based fold-change calculations and the Student's t-test to calculate two-tail, equal-variance P values. P values for comparison of the whole IBS-D group vs. the healthy control group were corrected for false detection rate (FDR; Bonferroni correction) so that P < 0.00263 was statistically significant.
Comparison of the protein expression of PDZD3 based on Western blot analysis was performed based on two-tailed Student's t-test.
We used the general genetic model (that is, comparison of the three genotypes at each gene SNP) to assess the effect of each genotype separately on the expression of the related gene in terms of mRNA expression in RSM; three-group comparisons were conducted by ANOVA on ranks test. The mRNA expression data were also analyzed using the dominant genetic model (major allele homozygous vs. combined heterozygotes and minor allele homozygous groups) for consistency with the rest of the study analysis.
The dominant genetic model was used to assess the associations of genotype with mRNA expression and quantitative traits of interest, such as colonic transit or BA parameters. Group comparisons were performed by the Mann-Whitney rank-sum test.
We used Spearman correlations to explore the association of mRNA expression with colonic transit, focusing on ascending colon emptying T1/2, which is a continuous variable and has no upper and lower bounds, whereas colonic transit, based on geometric center at 24 and 48 h, is limited by values of 1 (all isotope in ascending colon) and 5 (all isotope in stool).
RESULTS
Demographics and Quantitative BA and Transit Characteristics
Patients with high fecal BA excretion had faster overall colonic transit compared with patients with normal fecal BA excretion (Table 2). Overall, in IBS-D subjects, there were no significant differences in the serum concentrations of FGF19 or C4 in the two groups of IBS-D patients (Table 2); however, across all the groups studied, there was a significant inverse relationship between serum FGF19 and serum C4 (r = −0.297, P = 0.045), as has been previously documented in the literature, including by our group (58). The lack of significant differences in serum FGF19 or C4 in the two IBS-D groups and controls likely represents a type 2 error, since only four of the healthy controls had serum FGF19 and C4 measurements.
Table 2.
Demographics and bile acid and colonic transit characteristics
| Parameter | Healthy Controls | IBS-C | Whole IBS-D Group | IBS-D + High BA Excretion | IBS-D + Normal BA Excretion | P Value High vs. Normal BA Excretiona |
|---|---|---|---|---|---|---|
| N | 17 | 10 | 47 | 20 | 27 | |
| Age, yr | 38.1 ± 2.7 | 48.3 ± 4.3 | 40.7 ± 1.7 | 40.0 ± 2.4 | 41.4 ± 2.5 | |
| BMI, kg/m2 | 27.3 ± 1.5 | 26.4 ± 2.1 | 30.1 ± 1.2 | 32.3 ± 1.8 | 28.5 ± 1.6 | 0.051 |
| Total fecal BA, μmol/48 h | 1,322.4 ± 656.8b | 2787.6 ± 480.7 | 5167.2 ± 874.9 | 1024.9 ± 129.0 | <0.001 | |
| Mean % fecal LCA/CDCA/DCA/CA/UDCA | 38/1/58/1/3 | 28/6/54/8/4 | 24/8/51/13/4 | 32/5/55/5/3 | Not tested | |
| Serum C4, ng/ml | 35.7 ± 5.7b | 34.4 ± 3.8 | 34.5 ± 4.7 | 34.3 ± 5.7 | NS | |
| Serum FGF19, pg/ml | 162.0 ± 39.4c | 118.8 ± 10.8 | 126.7 ± 22.3 | 117.2 ± 13.9 | NS | |
| CT GC24 hd | 2.2 ± 0.2 | 2.0 ± 0.3e | 2.9 ± 0.2 | 3.3 ± 0.3 | 2.5 ± 0.2 | 0.017 |
| CT GC48 h | 3.7 ± 0.5b | 2.9 ± 0.4e | 4.2 ± 0.1 | 4.5 ± 0.2 | 4.0 ± 0.2 | 0.022 |
| Asc. colon T1/2, h | 13.0 ± 2.8b | 15.0 ± 1.2 | 13.1 ± 1.9 | 16.4 ± 1.6 | 0.124 |
Values are means±SE.
Mann-Whitney rank-sum test;
based on analysis in 3 among these healthy participants;
based on analysis in 4 among these healthy participants;
based on data from 12 healthy controls;
based on 7 patients with irritable bowel syndrome (IBS) with constipation (IBS-C). IBS-D, irritable bowel syndrome with diarrhea; BA, bile acid; BMI, body mass index; CT, colonic transit; Asc., ascending; NS, not significant. CA, cholic acid; CDCA, chenodeoxycholic acid; DCA, deoxycholic acid; UDCA, ursodeoxycholic acid; LCA, lithocholic acid.
The percentages of primary (cholic acid, CA; chenodeoxycholic acid, CDCA) and secondary BA (deoxycholic acid, DCA; ursodeoxycholic acid, UDCA; and lithocholic acid, LCA) of the combined secretory BA (DCA and CDCA) in the three groups were not different (Table 2). However, given the higher total fecal BA in the IBS-D subgroup with high fecal BA excretion, there was an approximately eightfold higher mass of CDCA in stool in the IBS-D group with high BA excretion, compared with the IBS-D group with normal BA excretion. Thus, on average, there was 8% of mean BA excretion of 5,167 μmol per 48 h or ∼0.41 mmol as CDCA in stool of IBS-D with high fecal BA excretion, compared with average 5% of mean BA excretion of 1,025 μmol per 48 h or ∼0.05 mmol as CDCA in stool of IBS-D with normal fecal BA excretion.
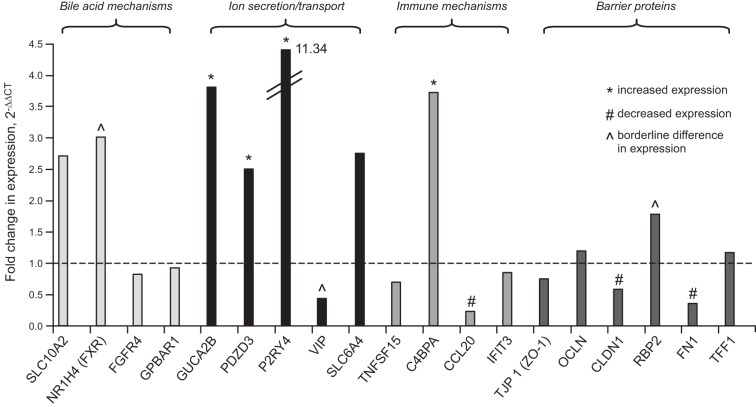
mRNA Fold Change in RSM from IBS Relative to Healthy Controls
The mean (and 95% confidence interval) fold changes (based on 2−ΔΔCT) in IBS-D and subgroups of IBS-D patients relative to healthy controls are illustrated in Table 3 and Fig. 1. By using FDR for 19 gene comparisons, these observations suggest, first, no significant fold changes in expression of genes associated with absorption [SLC10A2 (ASBT)], effects [GPBAR1(TGR5)] or feedback regulation of synthesis (FGFR4) of BAs; however, there were borderline changes in expression of FXR (P = 0.0081); second, increased expression of GUCA2B, PDZD3, and P2RY4, reflecting ion transport mechanisms; there was borderline reduction in expression of vasoactive intestinal peptide (VIP) (P = 0.0030); third, increased expression of C4BPA and reduced expression of CCL20, reflecting immune functions; and fourth, decreased expression of several barrier proteins, claudin-1 and fibronectin-1, and borderline increased expression of retinol binding protein (RBP-2, P = 0.0031).
Table 3.
mRNA fold change in rectosigmoid mucosa from IBS-D patients relative to healthy controls
| Parameter Data: Mean (95% CI) | IBS-C Group | P IBS-C vs. Control | Whole IBS-D Group | P IBS-D vs. Control | IBS-D + high BA Excretion (n = 20) | IBS-D + Normal BA Excretion (n = 27) | P, high vs. Normal BA Excretion |
|---|---|---|---|---|---|---|---|
| Bile acid absorption and effects | |||||||
| SLC10A2 (ASBT) | 1.86 (0.49, 3.22) | 0.0759 | 2.71 (1.37, 4.06) | 0.0298 | 3.27 (0.92, 5.63) | 2.36 (1.06, 3.66) | |
| NR1H4 (FXR) | 1.64 (0.86, 2.41) | 0.0420 | 3.02 (1.95, 4.08) | 0.0081 | 2.98 (1.50, 4.46) | 3.04 (1.90, 4.19) | |
| FGFR4 | 1.14 (0.86, 1.41) | 0.2030 | 0.82 (0.69, 0.95) | 0.0463 | 0.82 (0.67, 0.95) | 0.82 (0.67, 0.98) | |
| GPBAR1 | 1.45 (1.02, 1.88) | 0.0178 | 0.93 (0.74, 1.12) | 0.2298 | 0.90 (0.70, 1.11) | 0.95 (0.75, 1.16) | |
| Ion transport mechanisms | |||||||
| GUCA2B | 1.45 (0.56, 2.34) | 0.3359 | 3.81 (2.25, 5.36) | 0.00016 | 3.78 (1.99, 5.56) | 3.83 (2.23, 5.42) | |
| PDZD3 | 1.14 (0.00, 2.48) | 0.0099 | 2.51 (1.88, 3.14) | 0.000001 | 2.49 (1.72, 3.17) | 2.56 (1.88, 3.23) | |
| P2RY4 | 4.27 (1.45, 7.09) | 0.0016 | 11.34 (4.11, 18.58) | 0.000001 | 9.30 (2.05, 16.56) | 13.14 (5.01, 21.26) | |
| VIP | 3.21 (1.61, 4.80) | 0.0008 | 0.44 (0.24, 0.63) | 0.0030 | 0.46 (0.20, 0.73) | 0.42 (0.20, 0.63) | |
| SLC6A4 | 1.97 (0.73, 3.21) | 0.0726 | 2.75 (1.30, 4.21) | 0.0104 | 2.44 (0.71, 4.17) | 3.01 (1.38, 4.64) | |
| Immune functions | |||||||
| TNFSF15 | 1.17 (0.50, 1.84) | 0.1901 | 0.69 (0.48, 0.90) | 0.0185 | 0.55 (0.33, 0.77) | 0.81 (0.56, 1.06) | 0.10 |
| C4BPA | 1.97 (0.76, 3.17) | 0.0189 | 3.73 (2.37, 5.08) | 0.00003 | 4.14 (2.27, 6.00) | 3.45 (2.02, 4.88) | |
| CCL20 | 1.43 (0.35, 2.51) | 0.7339 | 0.23 (0.10, 0.36) | 0.000001 | 0.22 (0.08, 0.35) | 0.24 (0.10, 0.38) | |
| IFIT3 | 0.92 (0.33, 1.50) | 0.3555 | 0.84 (0.38, 1.30) | 0.0279 | 0.90 (0.39, 1.42) | 0.79 (0.35, 1.23) | |
| Barrier functions | |||||||
| TJP 1 (ZO-1) | 1.64 (1.14, 2.14) | 0.0070 | 0.76 (0.57, 0.96) | 0.0457 | 0.76 (0.57, 0.96) | 0.92 (0.72, 1.11) | 0.06 |
| OCLN | 1.77 (1.25, 2.30) | 0.0017 | 1.19 (0.87, 1.51) | 0.2900 | 1.12 (0.79, 1.45) | 1.25 (0.91, 1.59) | |
| CLDN1 | 1.20 (0.69, 1.70) | 0.7099 | 0.59 (0.39, 0.80) | 0.0004 | 0.63 (0.40, 0.86) | 0.57 (0.35, 0.78) | |
| RBP2 | 1.14 (0.04, 2.25) | 0.1005 | 1.78 (1.16, 2.40) | 0.0031 | 1.72 (1.08, 2.36) | 1.83 (1.13, 2.52) | |
| FN1 | 1.33 (0.77, 1.90) | 0.4134 | 0.35 (0.23, 0.47) | 0.00001 | 0.28 (0.16, 0.40) | 0.40 (0.26, 0.54) | 0.10 |
| TFF1 | 3.34 (0.00, 7.22) | 0.1918 | 1.17 (0.72, 1.63) | 0.8739 | 1.12 (0.63, 1.61) | 1.21 (0.72, 1.70) | |
Bolded data show univariately significant fold changes (without correction for false detection resulting from multiple comparisons) relative to healthy controls. The decreased expression is indicated by italics. Bolded P values reflect the comparison of whole IBS-D group vs. control corrected for FDR (Bonferroni correction) so that P < 0.00263 is statistically significant. There were no significant differences (with only few P values ≤0.10, without FDR correction) in mRNA fold changes in IBS-D patients with high vs. low fecal BA excretion; all other P values were >0.20.
Fig. 1.
Pictorial summary of mean fold changes in mRNA expression of candidate mechanisms in irritable bowel syndrome with diarrhea (IBS-D) patients (n = 47) compared with healthy controls (n = 17), plotted from data in Table 3 (which includes 95% confidence interval for all fold changes in expression).
There were no differences in the mucosal expression of any of the genes of interest among IBS-D patients with high compared with normal fecal BA excretion; however, there were numerical (univariate, FDR-uncorrected values, P ≤ 0.10) reductions in expression of the mRNA of tight junction proteins [ZO-1 (TJP1), and FN-1] and decreased expression of TNFSF15 (immune marker).
The mean (and 95% confidence interval) fold changes (based on 2−ΔΔCT) in IBS-D, and subgroups of IBS-D and IBS-C patients relative to healthy controls are illustrated in Table 3. The only significant change, corrected for FDR, was increased expression of PR2Y4, VIP, and occludin (OCLN).
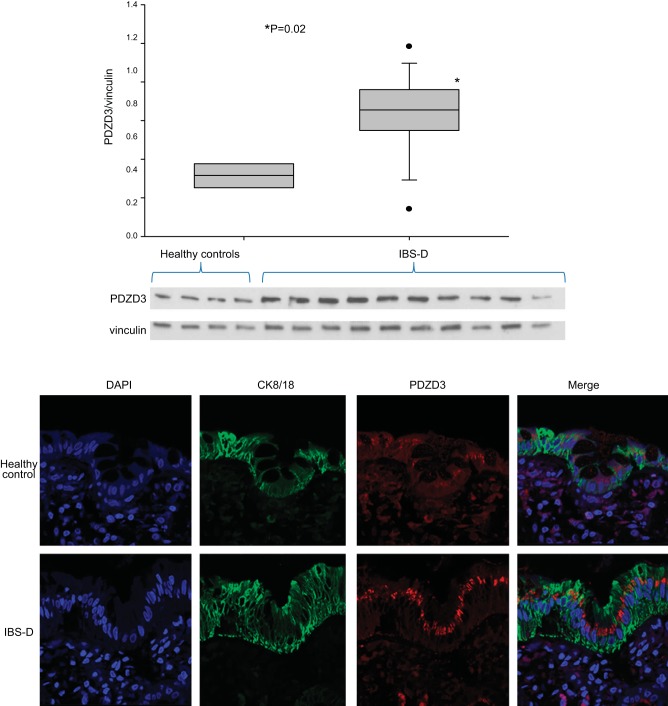
PDZD3 Protein Expression by Immunohistochemistry and Western Blot
We used immunofluorescence to assess protein expression of the regulatory protein, PDZD3. Figure 2 shows the increased expression of this ion transport protein in RSM of patients with IBS-D compared with normal healthy volunteers by Western blot and immunohistochemistry. Western blots quantitating PDZD3 protein (with vinculin as control) showed increased expression of PDZD3 (P = 0.02 by Student's t-test) in IBS-D patients (n = 10) compared with healthy controls (n = 4).
Fig. 2.
Top: Western blots quantitating PDZD3 protein (with vinculin as control). Note the increased expression (P = 0.02) of PDZD3 in IBS-D patients (n = 10) compared with healthy controls (n = 4). Bottom: immunofluorescence for PDZD3 in rectosigmoid colon mucosa of a patient with IBS-D compared with a normal, healthy control (×63 magnification). Stains show nuclear staining (DAPI), cytokeratin (C8/18, marker of epithelial cells), PDZD3, and merge. Note the localization of increased PDZD3 protein in epithelial cells in mucosa from the patient with IBS-D compared with the healthy control.
Relationship of Genotype and mRNA Expression (corrected for β-Actin) in IBS
Using general genetic model.
mRNA expressions (corrected for the expression of β-actin) showed no significant associations with genotypes except for a trend in the expression of GPBAR1 (Table 4); however, the observation that homozygous genotypes were both associated with higher expression than the heterozygotes questions any biological significance of this finding.
Table 4.
Relationship of genotype and mRNA expression (median, IQR; corrected for β-actin) by general genetic model (comparing expression in 3 genotypes: AA, Ab, bb) in IBS (analysis used ANOVA on ranks to compare 3 genotypes)
| Genotype | MAF | N | mRNA Expression | P |
|---|---|---|---|---|
| Serotonin transporter | ||||
| SLC6A4 rs4795541LL | 0.447 (L) | 7 | 16.59 (15.47, 17.44) | 0.463 |
| SLC6A4 rs4795541LS | 28 | 16.49 (15.19, 17.95) | ||
| SLC6A4 rs4795541SS | 12 | 15.72 (14.87, 16.75) | ||
| Immune function | ||||
| TNFSF15 rs4263839AA | 0.38 (A) | 6 | 12.94 (12.11, 13.85) | 0.27 |
| TNFSF15 rs4263839AG | 24 | 12.55 (12.11, 13.54) | ||
| TNFSF15 rs4263839GG | 17 | 12.28 (12.07, 12.96) | ||
| Bile acid absorption, homeostasis, or receptor | ||||
| GPBAR1 rs11554825CC | 0.436 (C) | 10 | 13.05 (12.53, 13.33) | 0.082 |
| GPBAR1 rs11554825CT | 21 | 12.86 (12.52, 13.03) | ||
| GPBAR1 rs11554825TT | 16 | 13.18 (12.72, 13.59) | ||
| NR1H4 rs17030285 CC | 0.128 (G) | 35 | 12.80 (11.59, 13.04) | 0.30 |
| NR1H4 rs17030285 CG | 12 | 11.83 (11.15, 12.46) | ||
| NR1H4 rs4764980 AA | 0.49 (G) | 12 | 12.30 (11.60, 12.75) | 0.98 |
| NR1H4 rs4764980 AG | 23 | 12.03 (11.09, 13.22) | ||
| NR1H4 rs4764980 GG | 12 | 12.18 (11.15, 12.93) | ||
| SLC10A2 rs188096 AC | 0.106 (A) | 10 | 16.65 (15.83, 17.80) | 0.82 |
| SLC10A2 rs188096 CC | 37 | 17.16 (15.11, 17.87) | ||
| FGFR4 rs1966265 AA | 0.213 (A) | 2 | 10.15 (9.55, 10.75) | 0.18 |
| FGFR4 rs1966265 AG | 16 | 11.06 (10.79, 11.32) | ||
| FGFR4 rs1966265 GG | 29 | 10.85 (10.46, 11.52) | ||
| FGFR4 rs434434 AG | 0.191 (A) | 18 | 10.80 (10.52, 11.11) | 0.28 |
| FGFR4 rs434434 GG | 29 | 11.03 (10.54, 11.52) | ||
| FGFR4 rs351855 AA | 0.287 (A) | 2 | 11.17 (10.51, 11.83) | 0.73 |
| FGFR4 rs351855 AG | 25 | 11.03 (10.51, 11.52) | ||
| FGFR4 rs351855 GG | 20 | 10.83 (10.62, 11.17) | ||
MAF, minor allele frequency.
Using dominant genetic model.
There was a significant association of GPBAR1 genotype and mRNA expression (P = 0.043) in RSM, based on analysis using the dominant genetic model (Table 5).
Table 5.
Relationship of genotype and mRNA expression (median, IQR, corrected for ß-actin) by dominant genetic model (comparing expression in homozygous major allele genotype vs. combining heterozygotes and minor allele homozygous) in IBS (comparisons with Mann-Whitney test)
| Genotype | MAF | N | mRNA Expression | P |
|---|---|---|---|---|
| Serotonin transporter | ||||
| SLC6A4 rs4795541 LL | 0.447 (L) | 7 | 16.59 (15.47, 17.44) | 0.70 |
| SLC6A4 rs4795541 LS/SS | 37 | 16.22 (14.95, 17.24) | ||
| Immune function | ||||
| TNFSF15 rs4263839 GG | 0.38 (A) | 17 | 12.28 (12.07, 12.96) | 0.12 |
| TNFSF15 rs4263839 AA/AG | 30 | 12.58 (12.14, 13.62) | ||
| Bile acid absorption, homeostasis, or receptor | ||||
| GPBAR1 rs11554825TT | 0.436 (C) | 16 | 13.18 (12.72, 13.59) | 0.043 |
| GPBAR1 rs11554825 CC/CT | 31 | 12.86 (12.54, 13.17) | ||
| NR1H4 rs17030285 CC | 0.128 (G) | 35 | 12.80 (11.59, 13.04) | 0.30 |
| NR1H4 rs 17030285 CG | 12 | 11.83 (11.15, 12.46) | ||
| NR1H4 rs4764980 AA | 0.49 (G) | 12 | 12.30 (11.60, 12.75) | 0.86 |
| NR1H4 rs4764980 AG/GG | 35 | 12.11 (11.09, 13.12) | ||
| SLC10A2 rs188096AC | 0.106 (A) | 10 | 16.65 (15.83, 17.80) | 0.821 |
| SLC10A2 rs188096CC | 37 | 17.16 (15.11, 17.87) | ||
| FGFR4 rs1966265 GG | 0.213 (A) | 29 | 10.85 (10.46, 11.52) | 0.50 |
| FGFR4 rs1966265 AG/AA | 18 | 11.01 (10.74, 11.25) | ||
| FGFR4 rs434434 AG | 0.191 (A) | 18 | 10.80 (10.52, 11.11) | 0.28 |
| FGFR4 rs434434 GG | 29 | 11.03 (10.54, 11.52) | ||
| FGFR4 rs351855 GG | 0.287 (A) | 20 | 10.83 (10.62, 11.17) | 0.48 |
| FGFR4 rs351855 AA/AG | 27 | 11.03 (10.51, 11.52) | ||
S is minor allele for 5-HTTLPR; T is minor allele for GPBAR1.
Spearman Correlation of mRNA Expression with Ascending Colon Emptying T1/2
We sought the association of mRNA expression (corrected for β-actin) in RSM with ascending colon emptying T1/2. There were no significant relationships between mRNA expression of FGFR4 (r = −0.06, P = 0.71), GPBAR1 (r = −0.08, P = 0.63), GUCA2B (r = 0.22, P = 0.13), P2RY4 (r = 0.16, P = 0.27), NR1H4 (r = −0.11, P = 0.48), TNFSF15 (r = −0.63, P = 0.673), SLC10A2 (r = −0.08, P = 0.62), and SLC6A4 (r = 0.147, P = 0.39) and ascending colon emptying T1/2.
Relationship of Genotype with Colonic Transit in IBS-D (using Dominant Genetic Model)
GPBAR1 was significantly associated with colonic transit at 24 and 48 h, as well as ascending colon emptying T1/2 (Table 6); a borderline association of 5-HTTLPR with colonic transit at 24 and 48 h was also observed (P ≤ 0.081).
Table 6.
Relationship of genotype with CT and ascending colon emptying T1/2 in IBS-D
| Genotype | N | Colonic GC24 | P | Colonic GC48 | P | AC Emptying T1/2 h | P |
|---|---|---|---|---|---|---|---|
| Serotonin transporter | |||||||
| 5-HTTLPR rs4795541 LL | 7 | 3.91 (3.01, 4.42) | 0.079 | 5.00 (4.17, 5.00) | 0.081 | 15.14 (5.41, 17.41) | 0.362 |
| 5-HTTLPR rs4795541 LS/SS | 40 | 2.43 (1.78, 3.89 | 4.57 (3.33, 4.99) | 16.39 (10.46, 19.47) | |||
| Immune function | |||||||
| TNFSF15 rs4263839 GG | 17 | 2.97 (1.70,4.56) | 0.79 | 4.67 (3.99, 5.00) | 0.40 | 15.57 (10.67, 18.57) | 0.825 |
| TNFSF15 rs4263839 AA/AG | 30 | 2.72 (1.90,3.90) | 4.58 (3.16, 5.00) | 16.60 (5.30, 20.15) | |||
| Bile acid absorption, homeostasis, or receptor | |||||||
| GPBAR1 rs11554825 CC | 10 | 3.9 (3.33, 4.49) | 0.010 | 5.00 (4.72, 5.00) | 0.005 | 9.18 (4.79, 15.58) | 0.034 |
| GPBAR1 rs11554825CT/TT | 37 | 2.26 (1.78, 3.38) | 4.24 (3.02, 4.93) | 16.60 (10.49, 19.59) | |||
| KLB rs17618244 AG* | 17 | 2.83 (2.03,4.32) | 0.42 | 4.86 (3.79, 5.00) | 0.237 | 13.1 (7.33, 17.64) | 0.514 |
| KLB rs17618244 GG | 30 | 2.26 (1.77,3.90) | 4.41 (3.04, 4.93) | 16.67 (7.08, 19.59) | |||
| FGFR4 rs1966265 GG | 29 | 2.53 (1.80,4.32) | 0.947 | 4.83 (2.91, 5.00) | 0.418 | 15.57 (6.83, 20.61) | 0.897 |
| FGFR4 rs1966265 AG/AA | 18 | 2.78 (1.88,3.89) | 4.50 (3.68, 4.95) | 16.27 (7.33, 18.67) | |||
| FGFR4 rs 434434 GA | 18 | 2.53 (1.84,4.07) | 0.973 | 4.64 (2.91, 5.00) | 0.920 | 16.30 (9.11, 20.00) | 0.622 |
| FGFR4 rs 434434 GG | 29 | 2.78 (1.82,4.20) | 4.65 (3.72, 4.98) | 14.65 (6.95, 18.39) | |||
| FGFR4 rs 351855 GG | 20 | 2.53 (1.80,4.32) | 0.947 | 4.83 (2.91, 5.00) | 0.418 | 15.57 (6.83, 20.61) | 0.897 |
| FGFR4 rs 351855 AA/AG | 27 | 2.78 (1.89,3.89) | 4.50 (3.68, 4.95) | 16.27 (7.33, 18.67) | |||
| SLC10A2 rs188096 AC | 10 | 2.41 (1.73, 4.47) | 0.88 | 4.66 (2.91, 5.00) | 0.95 | 14.75 (5.12, 19.2) | 0.795 |
| SLC10A2 rs188096 CC | 37 | 2.78 (1.94, 3.89) | 4.65 (3.64, 5.00) | 16.00 (7.49, 19.0) | |||
| FXR rs4764980 AA | 12 | 2.26 (1.87, 3.89) | 0.80 | 4.52 (3.69, 4.93) | 0.88 | 15.62 (6.79, 19.9) | 0.96 |
| FXR rs4764980 AG/GG | 35 | 2.80 (1.79, 4.31) | 4.66 (3.20, 5.00) | 16.00 (7.33, 19.6) | |||
Data show median and interquartile range; analysis is by Mann-Whitney rank-sum test. Note the significant association with GPBAR1 genotype and the borderline association with SLC6A4 (5-HTTLPR polymorphism). S is minor allele for 5-HTTLPR; T is minor allele for GPBAR1.
Number of AA KLB genotype identified.
AC, ascending colon.
Association of BA Homeostasis Genes (SLC10A2, KLB and FGFR4 Genotypes) and BA Measurements in Patients with IBS-D (using Dominant Genetic Model)
In these 47 patients with IBS-D, the variants in the genes associated with BA homeostasis were not significantly associated with serum FGF19, serum C4, and 48-h fecal BA excretion (Table 7).
Table 7.
Associations of variants in genes (associated with BA absorption and feedback regulation of synthesis) with serum FGF-19, serum C4, and 48-h fecal BA excretion in IBS-D
| Genotype | N | Fasting Serum FGF-19, pg/ml | P | Fasting Serum C4, ng/ml | P | Total Fecal BA, μmol/48 h | P |
|---|---|---|---|---|---|---|---|
| SLC10A2 rs188096 AC | 10 | 117 (63, 246) | 0.13 | 28 (16, 60) | 0.73 | 2723 (1037, 7766) | 0.25 |
| SLC10A2 rs188096 CC | 37 | 86 (47, 135) | 26 (14, 41) | 1827 (737, 2887) | |||
| KLB rs17618244 AG | 17 | 110 (55, 173) | 0.38 | 26 (13,54) | 0.99 | 2127 (1493, 4056) | 0.30 |
| KLB rs17618244 GG | 30 | 81 (48, 138) | 26 (15.5, 46) | 1559 (694, 3055) | |||
| FGFR4 rs351855GG | 20 | 74.2 (46.5, 157.8) | 0.45 | 39 (13,61) | 0.13 | 1850 (1073, 2916) | 0.87 |
| FGFR4 rs351855 AG/AA | 27 | 99.8 (57.7, 144.7) | 23 (16, 36) | 1829 (733, 3442) | |||
| FGFR4 rs1966265 GG | 29 | 94.3 (49.3, 154.2) | 1.00 | 27 (16, 59) | 0.37 | 2127 (903, 4543) | 0.16 |
| FGFR4 rs1966265AG/AA | 18 | 101.7 (49.1,140.1) | 23 (14, 38) | 1559 (666, 2741) | |||
| FGFR4 rs434434 GG | 29 | 77.9 (44.6, 132.8) | 0.11 | 26 (16, 38) | 0.92 | 1829 (676, 3652) | 0.99 |
| FGFR4 rs434434 GA | 18 | 107.2 (63.9,182.4) | 26 (12, 59) | 1890 (1078, 2894) |
Data show median and interquartile range.
DISCUSSION
Our study has provided several novel insights about mucosal pathobiology of IBS by means of studies of RSM biopsies from 47 IBS-D patients or controls. The clinical and biochemical characteristics of the participants in this study are consistent with those described recently in a somewhat larger cohort (n = 64) that included the same 47 patients. However, the present study could only include the 47 patients who consented to undergo rectosigmoid biopsies. One difference in the present group of patients is that fasting serum FGF19 was not significantly different between IBS-D and healthy controls, in contrast to the original report by Walters et al. (62) and our prior studies (10, 64) with larger sample sizes. We perceive that the lack of significant difference in fasting serum FGF19 between IBS-D and healthy controls represents a type 2 error: among the 17 healthy controls who had undergone rectosigmoid biopsies, only 4 had fasting serum FGF19 measurement.
The following observations were made in the present study:
First, targeted mRNA analysis by quantitative RT-PCR replicates our prior results showing generally increased expression of intestinal ion transport mechanisms and generally reduced intestinal barrier and up- or downregulation of mucosal immune mechanisms. The significance of the observed associations is limited by the relatively small number of biopsies assessed (total 64); however, we present both uncorrected P values and, more importantly, we present the significance of observations corrected for the comparison of 19 selected genes. One of the well-recognized ion transport mechanisms (PDZ) showing increased mRNA expression was also associated with increased expression of PDZD3 protein on immunohistochemistry and Western blot analysis.
Second, there were no differences in mucosal expression in IBS-D associated with degree of fecal BA excretion.
Our third general finding was that genotype was not significantly associated with the level of expression of mRNA in RSM, colonic transit, or BA parameters, with the exception of the GPBAR1 gene, which is significantly associated with mucosal expression of GPBAR1 and with colonic transit.
Fourth, the level of expression of mRNA in RSM was not associated with ascending colon emptying time.
Mucosal mRNA Expression in IBS-D
Among the intestinal secretory mechanisms, increased mRNA expression of GUCA2B and P2RY4 could all support fluid and electrolyte secretion through actions on enterocytes or submucosal neurons. On the other hand, the significantly increased expression of PDZD3 is associated with either increased sodium ion and fluid absorption (through effects on NHE3) or increased chloride ion and fluid secretion through CFTR, and borderline decreased expression of VIP (P = 0.003) may conceivably be associated with reduced intestinal secretion.
The largest fold increases in mRNA expression in our IBS-D patients were observed for the purinergic receptor, P2RY4. Purinergic receptors are divided into adenosine P1 [A(1), A(2A), A(2B), A(3)], ionotropic ATP-gated P2X receptors [P2X(1–7) that form ion channels or pores], or metabotropic P2Y(1, 2, 4, 6, 11–14) receptors. Metabotropic receptors are indirectly linked with ion channels on the plasma membrane through transduction mechanisms, often G proteins. The purinergic hypothesis is based on ATP (or a related nucleotide, e.g., ADP or AMP) release at the neurotransmitter synapses or on neuromuscular transmission, and these may involve βNAD+ and ADP ribose in neurotransmission in rodents, primates, and humans (24, 29, 30). Mechanically evoked reflex electrogenic chloride secretion in rat distal colon is triggered by endogenous nucleotides acting at P2Y1, P2Y2, and P2Y4 receptors (21). P2Y2, 4, and 6 receptors regulate Cl−, Na+, and K+ secretion in the intestinal tract, and absorption mechanisms, in particular, P2Y4 receptors, are involved in chloride secretion and potassium secretion (20, 28, 38). Thus our observation of marked increase in expression of P2RY4 is consistent with the increased expression of secretory mechanisms (in addition to GUCA2B) in IBS-D patients. In addition to the novel mechanisms potentially related to ion transport in the manifestations of IBS-D, the present findings provide the basis for further hypothesis testing and, possibly, testing novel pharmacological approaches to reverse electrolyte secretion (41) in IBS-D. These findings complement the observation of increased small intestine secretion in response to BA infusion in IBS-D (42).
The specificity of the observed fold changes in mRNA expression is enhanced by the differences in the observations for IBS-D and the additional control group of IBS-C. The biological significance of the significant associations with IBS-D is discussed in the next section.
Increased mRNA expression of GUCA2B is associated with the endogenous GC-C ligand, uroguanylin, which is secreted by intestinal goblet cells (50). In the same way that guanylate cyclase C agonists are effective in the treatment of IBS-C, it is conceivable that antagonists at the guanylate cyclase receptors may impact the management of IBS-D in the future. Atrial and brain natriuretic peptides are agonists at the guanylate cyclase B receptor (66); moreover, homozygous knockout of the guanylate cyclase B receptor in mice results in a severe phenotype with gastric motor dysfunction and intestinal dilatation (49). These data suggest that further study of expression of guanylate cyclase receptors or GC-C ligands (guanylin and uroguanylin) in IBS may yield important insights into pathobiology or treatment of IBS.
We observed borderline increased mRNA expression of FXR in colonic mucosa in the whole IBS-D group (P = 0.0081, relative to significance value with FDR, P = 0.00263); this was also observed in both IBS-D subgroups. FXR activation prevents chemically induced intestinal inflammation, with improvement of colitis symptoms, inhibition of epithelial permeability, and reduced goblet cell loss in a mouse model of inflammatory bowel disease (27). FXR expression is decreased in colonic mucosa of patients with primary sclerosing cholangitis [PSC (54)], and PSC is associated with higher circulating levels of the conjugated primary BAs (3). These data are consistent with a potential role of colonic mucosal FXR in protecting the colonic mucosal integrity in patients with IBS-D. Increased expression of C4BPA and reduced expression of CCL20 reflect changes in immune functions that may ultimately lead to the immune activation observed in IBS-D (45).
Decreased expression of several barrier proteins, especially claudin-1 and fibronectin-1, may reflect the observed increase in intestinal mucosal permeability in IBS-D (14). The observed borderline increased expression of RBP-2 may be a compensatory change to correct the decreased expression of the other tight junction proteins.
On the other hand, there were no significant differences (see Table 3) in the mucosal expression of any of the genes of interest among IBS-D patients with high compared with normal fecal BA excretion. The significance of the increased mucosal expression in P2RY4 and VIP in IBS-C is unclear, although their increased expression may be related to muscle function (e.g., relaxation) rather than ion secretion. The increased OCLN in mucosa from IBS-C patients may reflect greater mucosal barrier functions in these patients; prior work (5) had documented the preservation of occludin and claudin expression in IBS-C in contrast to IBS-D.
There are only a few other papers in the published literature that describe alterations in mucosal mRNA expression of several mechanisms by next-generation sequencing methods in patients with IBS-D. Our previous study used next-generation RNA sequencing and RT-PCR of RSM in nine patients with IBS-D and nine healthy controls; we identified differential expression of secretory and barrier genes (11). In the present study in IBS-D patients, we confirmed the mRNA fold changes for 8 of the 10 genes evaluated in the prior study: FN1, IFIT3, PDZD3, TFF1, GUCA2B, RBP2, C4BPA, and P2RY4. The Barcelona group used combinations of microarray and PCR, focused their studies on mucosal tight junction expression and immune activation in jejunal mucosal biopsies, and identified reduced expression of ZO-1 in IBS-D at both gene and protein levels (37), as well as higher mucosal immune activity in IBS-D, with upregulation of germline transcripts and immunoglobulin genes (58). A recent combined study from Helsinki and Nottingham explored the association of host rectal mucosal expression and the fecal microbiome in different subgroups of IBS, including IBS-D and postinfectious IBS (31), rather than the differences in mucosal expression between IBS subgroups and healthy controls. However, one of the strongest associations between microbial populations and rectal mucosal mRNA expression in IBS patients pertains to NR1H4 or FXR, which was a change of borderline significance (P = 0.0081) in our present study.
Relation of mRNA expression and genotype and phenotype in IBS-D.
Among the genotypes studied, the only one that is significantly associated with colonic transit is GPBAR1 genotype. We had previously observed this relationship in studies in several hundred patients (17), but we present it now in only 47 patients with IBS-D and present this unique statistically significant finding in the context of a broad spectrum of genes associated with absorption, feedback regulation, and action of BAs, and selected immune function and serotonin transporter genes. None of the other genes of interest was associated with alteration in colonic transit. Importantly, we did not observe association with variation in ASBT, which has been reported rarely in familial or sporadic diarrhea (39, 40).
We found no relation between genotype and expression for all the candidate genes of interest, and, similarly, we did not find significant associations of BA-related genotypes with serum C4, serum FGF19, and fecal BA excretion. This suggests that posttranslational modification may be a more relevant mechanism controlling these phenotypic parameters of BA control and expression than genotype.
Finally, there was no relation between mRNA expression and ascending colon emptying or transit; we focused on the latter parameter of transit, since it appears to be more closely related to the state of secretion within the colon. Thus ascending colon emptying is accelerated in carcinoid diarrhea [a classical secretory diathesis (61)], and proximal colon emptying is positively correlated with stool weight (55).
Strengths and Limitations
We have studied more than five times as many IBS-D patients in the present study compared with the prior pilot study, and we evaluated mRNA expression in RSM for 19 genes; the statistical analysis was corrected for multiple comparisons. We also replicated the numerically increased or decreased mRNA fold changes (identified as FDR uncorrected changes in the prior study) in 8 of the first 10 genes of interest. Overall, these findings support the potential role of altered mucosal functions in IBS-D.
The limitations include the following: First, we restricted our study to 19 candidates, for which there was a strong biological rationale for each biological process (e.g., ion transport, immune mechanisms, barrier proteins) in IBS. However, several different potential mechanisms were explored and, therefore, we did not bias the study, other than using the prior pilot study conducted with RNA sequencing and RT-PCR confirmation of the main findings (11) as a starting point. In designing such studies, one has to balance the need to explore as many potentially relevant mechanisms with the sample size available and the need to correct for FDR.
Second, although genetic variation in GPBAR1 was associated with colonic transit (with P < 0.05 for colonic transit at 48 h), the association of genotype with colonic transit appraised nine variants in seven genes [SLC6A4, GPBAR1, TNFSF15, KLB, FGFR4, FXR, SLC10A2 (ASBT)] and three measurements of colonic transit. Therefore, the observation should be regarded as hypothesis generating and not definitely proven in this study of 64 people. Nevertheless, the present sample of patients is much smaller than the ∼650 IBS patients and healthy controls that were used to demonstrate in prior studies the association of GPBAR1 (17, 18) and other genes [e.g., KLB, FAAH (summarized in Ref. 12)] with small bowel or colonic transit.
Third, we do not know that the parallel expression (mRNA and protein) of PDZD3 is necessarily a good “representation” for the other mRNA changes.
Conclusion
In conclusion, the present data demonstrate that mucosal ion transport mechanisms (mRNA for several factors, and PDZD3 protein) are generally upregulated and barrier genes downregulated in IBS-D compared with healthy controls, independent of genotype. There are no differences in gene expression in IBS-D with high compared with normal fecal BA excretion. These pathobiological mechanisms deserve further study to further advance the understanding of pathophysiological mechanisms in patients with IBS and diarrhea.
GRANTS
This research was funded by RO1-DK92179 grant from National Institutes of Health to M. Camilleri.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
M.C. conception and design of research; M.C. analyzed data; M.C. interpreted results of experiments; M.C. prepared figures; M.C. drafted manuscript; M.C., P.C., A.A., and I.A.B. edited and revised manuscript; M.C., P.C., A.A., and I.A.B. approved final version of manuscript; P.C., A.A., and I.A.B. performed experiments.
ACKNOWLEDGMENTS
The authors thank Cindy Stanislav for excellent secretarial assistance.
REFERENCES
- 1.Aerssens J, Camilleri M, Talloen W, Thielemans L, Göhlmann HW, Van Den Wyngaert I, Thielemans T, De Hoogt R, Andrews CN, Bharucha AE, Carlson PJ, Busciglio I, Burton DD, Smyrk T, Urrutia R, Coulie B. Alterations in mucosal immunity identified in the colon of patients with irritable bowel syndrome. Clin Gastroenterol Hepatol 6: 194–205, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altman DG, Bland JM. The normal distribution. Br Med J 310: 298, 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bell LN, Wulff J, Comerford M, Vuppalanchi R, Chalasani N. Serum metabolic signatures of primary biliary cirrhosis and primary sclerosing cholangitis. Liver Int 35: 263–274, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belmonte L, Beutheu Youmba S, Bertiaux-Vandaële N, Antonietti M, Lecleire S, Zalar A, Gourcerol G, Leroi AM, Déchelotte P, Coëffier M, Ducrotté P. Role of toll like receptors in irritable bowel syndrome: differential mucosal immune activation according to the disease subtype. PLoS One 7: e42777–e42786, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertiaux-Vandaële N, Youmba SB, Belmonte L, Lecleire S, Antonietti M, Gourcerol G, Leroi AM, Déchelotte P, Ménard JF, Ducrotté P, Coëffier M. The expression and the cellular distribution of the tight junction proteins are altered in irritable bowel syndrome patients with differences according to the disease subtype. Am J Gastroenterol 106: 2165–2173, 2011. [DOI] [PubMed] [Google Scholar]
- 6.Brydon WG, Nyhlin H, Eastwood MA, Merrick MV. Serum 7 alpha-hydroxy-4-cholesten-3-one and selenohomocholyltaurine (SeHCAT) whole body retention in the assessment of bile acid induced diarrhoea. Eur J Gastroenterol Hepatol 8: 117–123, 1996. [DOI] [PubMed] [Google Scholar]
- 7.Camilleri M. Perspective: Intestinal secretory mechanisms in irritable bowel syndrome-diarrhea. Clin Gastroenterol Hepatol. 2014. July 17 pii: S1542-3565(14)01049-0. doi: 10.1016/j.cgh.2014.07.020 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Camilleri M. Physiological underpinnings of irritable bowel syndrome: neurohormonal mechanisms. J Physiol 592: 2967–2980, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Camilleri M, Andrews CN, Bharucha AE, Carlson PJ, Ferber I, Stephens D, Smyrk TC, Urrutia R, Aerssens J, Thielemans L, Göhlmann H, van den Wyngaert I, Coulie B. Alterations in expression of p11 and SERT in mucosal biopsy specimens of patients with irritable bowel syndrome. Gastroenterology 132: 17–25, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Camilleri M, Busciglio I, Acosta A, Shin A, Carlson P, Burton D, Ryks M, Rhoten D, Lamsam J, Lueke A, Donato LJ, Zinsmeister AR. Effect of increased bile acid synthesis or fecal excretion in irritable bowel syndrome-diarrhea. Am J Gastroenterol 109: 1621–1630, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Camilleri M, Carlson P, Acosta A, Busciglio I, Nair AA, Gibbons SJ, Farrugia G, Klee EW. RNA sequencing shows transcriptomic changes in rectosigmoid mucosa in patients with irritable bowel syndrome-diarrhea: a pilot case-control study. Am J Physiol Gastrointest Liver Physiol 306: G1089–G1098, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Camilleri M, Katzka DA. Irritable bowel syndrome: methods, mechanisms, and pathophysiology. Genetic epidemiology and pharmacogenetics in irritable bowel syndrome. Am J Physiol Gastrointest Liver Physiol 302: G1075–G1084, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Camilleri M, Klee EW, Shin A, Carlson P, Li Y, Grover M, Zinsmeister AR. Irritable bowel syndrome-diarrhea: characterization of genotype by exome sequencing, and phenotypes of bile acid synthesis and colonic transit. Am J Physiol Gastrointest Liver Physiol 306: G13–G26, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Camilleri M, Lasch K, Zhou W. Irritable bowel syndrome: methods, mechanisms, and pathophysiology. The confluence of increased permeability, inflammation, and pain in irritable bowel syndrome. Am J Physiol Gastrointest Liver Physiol 303: G775–G785, 2012. [DOI] [PubMed] [Google Scholar]
- 15.Camilleri M, McKinzie S, Busciglio I, Low PA, Sweetser S, Burton D, Baxter K, Ryks M, Zinsmeister AR. Prospective study of motor, sensory, psychologic, and autonomic functions in patients with irritable bowel syndrome. Clin Gastroenterol Hepatol 6: 772–781, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Camilleri M, Nadeau A, Tremaine WJ, Lamsam J, Burton D, Odunsi S, Sweetser S, Singh R. Measurement of serum 7α-hydroxy-4-cholesten-3-one (or 7αC4), a surrogate test for bile acid malabsorption in health, ileal disease and irritable bowel syndrome using liquid chromatography-tandem mass spectrometry. Neurogastroenterol Motil 21: 734–e43, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Camilleri M, Shin A, Busciglio I, Carlson P, Acosta A, Bharucha AE, Burton D, Lamsam J, Lueke A, Donato LJ, Zinsmeister AR. Genetic variation in GPBAR1 predisposes to quantitative changes in colonic transit and bile acid excretion. Am J Physiol Gastrointest Liver Physiol 307: G508–G516, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Camilleri M, Vazquez-Roque MI, Carlson P, Burton D, Wong BS, Zinsmeister AR. Association of bile acid receptor TGR5 variation and transit in health and lower functional gastrointestinal disorders. Neurogastroenterol Motil 23: 995–999, e458, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chadwick VS, Gaginella TS, Carlson GL, Debongnie JC, Phillips SF, Hofmann AF. Effect of molecular structure on bile acid-induced alterations in absorptive function, permeability, and morphology in the perfused rabbit colon. J Lab Clin Med 94: 661–674, 1979. [PubMed] [Google Scholar]
- 20.Christofi FL. Purinergic receptors and gastrointestinal secretomotor function. Purinergic Signal 4: 213–236, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Christofi FL, Wunderlich J, Yu JG, Wang YZ, Xue J, Guzman J, Javed N, Cooke H. Mechanically evoked reflex electrogenic chloride secretion in rat distal colon is triggered by endogenous nucleotides acting at P2Y1, P2Y2, and P2Y4 receptors. J Comp Neurol 469: 16–36, 2004. [DOI] [PubMed] [Google Scholar]
- 22.Coates MD, Mahoney CR, Linden DR, Sampson JE, Chen J, Blaszyk H, Crowell MD, Sharkey KA, Gershon MD, Mawe GM, Moses PL. Molecular defects in mucosal serotonin content and decreased serotonin reuptake transporter in ulcerative colitis and irritable bowel syndrome. Gastroenterology 126: 1657–1664, 2004. [DOI] [PubMed] [Google Scholar]
- 23.Deiteren A, Camilleri M, Bharucha AE, Burton D, McKinzie S, Rao AS, Zinsmeister AR. Performance characteristics of scintigraphic colon transit measurement in health and irritable bowel syndrome and relationship to bowel functions. Neurogastroenterol Motil 22: 415–423, e95, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Durnin L, Hwang SJ, Ward SM, Sanders KM, Mutafova-Yambolieva VN. Adenosine 5′-diphosphate-ribose is a neural regulator in primate and murine large intestine along with β-NAD+. J Physiol 590: 1921–1941, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.El-Salhy M, Gundersen D, Hatlebakk JG, Gilja OH, Hausken T. Abnormal rectal endocrine cells in patients with irritable bowel syndrome. Regul Pept 188: 60–65, 2014. [DOI] [PubMed] [Google Scholar]
- 26.El-Salhy M, Wendelbo I, Gundersen D. Serotonin and serotonin transporter in the rectum of patients with irritable bowel disease. Mol Med Rep 8: 451–455, 2013. [DOI] [PubMed] [Google Scholar]
- 27.Gadaleta RM, van Erpecum KJ, Oldenburg B, Willemsen EC, Renooij W, Murzilli S, Klomp LW, Siersema PD, Schipper ME, Danese S, Penna G, Laverny G, Adorini L, Moschetta A, van Mil SW. Farnesoid X receptor activation inhibits inflammation and preserves the intestinal barrier in inflammatory bowel disease. Gut 60: 463–472, 2011. [DOI] [PubMed] [Google Scholar]
- 28.Ghanem E, Robaye B, Leal T, Leipziger J, Van Driessche W, Beauwens R, Boeynaems JM. The role of epithelial P2Y2 and P2Y4 receptors in the regulation of intestinal chloride secretion. Br J Pharmacol 146: 364–369, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hwang SJ, Blair PJ, Durnin L, Mutafova-Yambolieva V, Sanders KM, Ward SM. P2Y1 purinoreceptors are fundamental to inhibitory motor control of murine colonic excitability and transit. J Physiol 590: 1957–1972, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hwang SJ, Durnin L, Dwyer L, Rhee PL, Ward SM, Koh SD, Sanders KM, Mutafova-Yambolieva VN. β-Nicotinamide adenine dinucleotide is an enteric inhibitory neurotransmitter in human and nonhuman primate colons. Gastroenterology 140: 608–617, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jalanka-Tuovinen J, Salojärvi J, Salonen A, Immonen O, Garsed K, Kelly FM, Zaitoun A, Palva A, Spiller RC, de Vos WM. Faecal microbiota composition and host-microbe cross-talk following gastroenteritis and in postinfectious irritable bowel syndrome. Gut 63: 1737–1745, 2014. [DOI] [PubMed] [Google Scholar]
- 32.Kalia N, Hardcastle J, Keating C, Grasa L, Keating C, Pelegrin P, Bardhan KD, Grundy D. Intestinal secretory and absorptive function in Trichinella spiralis mouse model of postinfective gut dysfunction: role of bile acids. Gut 57: 41–49, 2008. [DOI] [PubMed] [Google Scholar]
- 33.Kerckhoffs AP, ter Linde JJ, Akkermans LM, Samsom M. SERT and TPH-1 mRNA expression are reduced in irritable bowel syndrome patients regardless of visceral sensitivity state in large intestine. Am J Physiol Gastrointest Liver Physiol 302: G1053–G1060, 2012. [DOI] [PubMed] [Google Scholar]
- 34.Kim HJ, Camilleri M, Carlson PJ, Cremonini F, Ferber I, Stephens D, McKinzie S, Zinsmeister AR, Urrutia R. Association of distinct α2 adrenoceptor and serotonin-transporter polymorphisms associated with constipation and somatic symptoms in functional gastrointestinal disorders. Gut 53: 829–837, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lamprecht G, Seidler U. The emerging role of PDZ adapter proteins for regulation of intestinal ion transport. Am J Physiol Gastrointest Liver Physiol 291: G766–G777, 2006. [DOI] [PubMed] [Google Scholar]
- 36.Martínez C, Lobo B, Pigrau M, Ramos L, González-Castro AM, Alonso C, Guilarte M, Guilá M, de Torres I, Azpiroz F, Santos J, Vicario M. Diarrhoea-predominant irritable bowel syndrome: an organic disorder with structural abnormalities in the jejunal epithelial barrier. Gut 62: 1160–1168, 2013. [DOI] [PubMed] [Google Scholar]
- 37.Martínez C, Vicario M, Ramos L, Lobo B, Mosquera JL, Alonso C, Sánchez A, Guilarte M, Antolín M, de Torres I, González-Castro AM, Pigrau M, Saperas E, Azpiroz F, Santos J. The jejunum of diarrhea-predominant irritable bowel syndrome shows molecular alterations in the tight junction signaling pathway that are associated with mucosal pathobiology and clinical manifestations. Am J Gastroenterol 107: 736–746, 2012. [DOI] [PubMed] [Google Scholar]
- 38.Matos JE, Robaye B, Boeynaems JM, Beauwens R, Leipziger J. K+ secretion activated by luminal P2Y2 and P2Y4 receptors in mouse colon. J Physiol 564: 269–279, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Montagnani M, Abrahamsson A, Gälman C, Eggertsen G, Marschall HU, Ravaioli E, Einarsson C, Dawson PA. Analysis of ileal sodium/bile acid cotransporter and related nuclear receptor genes in a family with multiple cases of idiopathic bile acid malabsorption. World J Gastroenterol 12: 7710–7714, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Montagnani M, Love MW, Rössel P, Dawson PA, Qvist P. Absence of dysfunctional ileal sodium-bile acid cotransporter gene mutations in patients with adult-onset idiopathic bile acid malabsorption. Scand J Gastroenterol 36: 1077–1080, 2001. [DOI] [PubMed] [Google Scholar]
- 41.Ochoa-Cortes F, Liñán-Rico A, Jacobson KA, Christofi FL. Potential for developing purinergic drugs for gastrointestinal diseases. Inflamm Bowel Dis 20: 1259–1287, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oddsson E, Rask-Madsen J, Krag E. A secretory epithelium of the small intestine with increased sensitivity to bile acids in irritable bowel syndrome associated with diarrhoea. Scand J Gastroenterol 13: 408–416, 1978. [PubMed] [Google Scholar]
- 43.Odunsi-Shiyanbade ST, Camilleri M, McKinzie S, Burton D, Carlson P, Busciglio IA, Lamsam J, Singh R, Zinsmeister AR. Effects of chenodeoxycholate and a bile acid sequestrant, colesevelam, on intestinal transit and bowel function. Clin Gastroenterol Hepatol 8: 159–165, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O'Hara SP, Bogert PS, Trussoni CE, Chen X, LaRusso NF. TLR4 promotes Cryptosporidium parvum clearance in a mouse model of biliary cryptosporidiosis. J Parasitol 97: 813–821, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ohman L, Simrén M. Pathogenesis of IBS: role of inflammation, immunity and neuroimmune interactions. Nat Rev Gastroenterol Hepatol 7: 163–173, 2010. [DOI] [PubMed] [Google Scholar]
- 46.Ohman L, Stridsberg M, Isaksson S, Jerlstad P, Simrén M. Altered levels of fecal chromogranins and secretogranins in IBS: relevance for pathophysiology and symptoms? Am J Gastroenterol 107: 440–447, 2012. [DOI] [PubMed] [Google Scholar]
- 47.Raimondi F, Santoro P, Barone MV, Pappacoda S, Barretta ML, Nanayakkara M, Apicella C, Capasso L, Paludetto R. Bile acids modulate tight junction structure and barrier function of Caco-2 monolayers via EGFR activation. Am J Physiol Gastrointest Liver Physiol 294: G906–G913, 2008. [DOI] [PubMed] [Google Scholar]
- 48.Shin A, Camilleri M, Vijayvargiya P, Busciglio I, Burton D, Ryks M, Rhoten D, Lueke A, Saenger A, Girtman A, Zinsmeister AR. Bowel functions, fecal unconjugated primary and secondary bile acids, and colonic transit in patients with irritable bowel syndrome. Clin Gastroenterol Hepatol 11: 1270–1275, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sogawa C, Abe A, Tsuji T, Koizumi M, Saga T, Kunieda T. Gastrointestinal tract disorder in natriuretic peptide receptor B gene mutant mice. Am J Pathol 177: 822–828, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Steinbrecher KA, Mann EA, Giannella RA, Cohen MB. Increases in guanylin and uroguanylin in a mouse model of osmotic diarrhea are guanylate cyclase C-independent. Gastroenterology 121: 1191–1202, 2001. [DOI] [PubMed] [Google Scholar]
- 51.Swan C, Duroudier NP, Campbell E, Zaitoun A, Hastings M, Dukes GE, Cox J, Kelly FM, Wilde J, Lennon MG, Neal KR, Whorwell PJ, Hall IP, Spiller RC. Identifying and testing candidate genetic polymorphisms in the irritable bowel syndrome (IBS): association with TNFSF15 and TNFα. Gut 62: 985–994, 2013. [DOI] [PubMed] [Google Scholar]
- 52.Tagliacozzi D, Mozzi AF, Casetta B, Bertucci P, Bernardini S, Di Ilio C, Urbani A, Federici G. Quantitative analysis of bile acids in human plasma by liquid chromatography-electrospray tandem mass spectrometry: a simple and rapid one-step method. Clin Chem Lab Med 41: 1633–1641, 2003. [DOI] [PubMed] [Google Scholar]
- 53.Talley NJ, Phillips SF, Wiltgen CM, Zinsmeister AR, Melton LJ 3rd. Assessment of functional gastrointestinal disease: the bowel disease questionnaire. Mayo Clin Proc 65: 1456–1479, 1990. [DOI] [PubMed] [Google Scholar]
- 54.Torres J, Bao X, Iuga AC, Chen A, Harpaz N, Ullman T, Cohen BL, Pineton de Chambrun G, Asciutti S, Odin JA, Sachar DB, Gaskins HR, Setchell K, Colombel JF, Itzkowitz SH. Farnesoid X receptor expression is decreased in colonic mucosa of patients with primary sclerosing cholangitis and colitis-associated neoplasia. Inflamm Bowel Dis 19: 275–282, 2013. [DOI] [PubMed] [Google Scholar]
- 55.Vassallo M, Camilleri M, Phillips SF, Brown ML, Chapman NJ, Thomforde GM. Transit through the proximal colon influences stool weight in the irritable bowel syndrome. Gastroenterology 102: 102–108, 1992. [DOI] [PubMed] [Google Scholar]
- 56.Vazquez-Roque MI, Camilleri M, Smyrk T, Murray JA, O'Neill J, Carlson P, Lamsam J, Eckert D, Janzow D, Burton D, Ryks M, Rhoten D, Zinsmeister AR. Association of HLA-DQ gene with bowel transit, barrier function and inflammation in irritable bowel syndrome with diarrhea. Am J Physiol Gastrointest Liver Physiol 303: G1262–G1269, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vazquez-Roque MI, Camilleri M, Smyrk T, Murray JA, Marietta E, O'Neill J, Carlson P, Lamsam J, Janzow D, Eckert D, Burton D, Zinsmeister AR. A controlled trial of gluten-free diet in irritable bowel syndrome-diarrhea: effect on bowel frequency and intestinal functions. Gastroenterology 144: 903–911, e3, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vicario M, González-Castro AM, Martínez C, Lobo B, Pigrau M, Guilarte M, de Torres I, Mosquera JL, Fortea M, Sevillano-Aguilera C, Salvo-Romero E, Alonso C, Rodiño-Janeiro BK, Söderholm JD, Azpiroz F, Santos J. Increased humoral immunity in the jejunum of diarrhoea-predominant irritable bowel syndrome associated with clinical manifestations. Gut. 2014. September 10 pii: gutjnl-2013-306236. doi: 10.1136/gutjnl-2013-306236 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 59.Vijayvargiya P, Camilleri M, Shin A, Saenger A. Methods for diagnosis of bile acid malabsorption in clinical practice. Clin Gastroenterol Hepatol 11: 1232–1239, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Villani AC, Lemire M, Thabane M, Belisle A, Geneau G, Garg AX, Clark WF, Moayyedi P, Collins SM, Franchimont D, Marshall JK. Genetic risk factors for post-infectious irritable bowel syndrome following a waterborne outbreak of gastroenteritis. Gastroenterology 138: 1502–1513, 2010. [DOI] [PubMed] [Google Scholar]
- 61.von der Ohe M, Camilleri M, Kvols LK, Thomforde GM. Motor dysfunction of the small bowel and colon in patients with the carcinoid syndrome and diarrhea. N Engl J Med 329: 1073–1078, 1993. [DOI] [PubMed] [Google Scholar]
- 62.Walters JR, Tasleem AM, Omer OS, Brydon WG, Dew T, le Roux CW. A new mechanism for bile acid diarrhea: defective feedback inhibition of bile acid biosynthesis. Clin Gastroenterol Hepatol 7: 1189–1194, 2009. [DOI] [PubMed] [Google Scholar]
- 63.Wang YM, Chang Y, Chang YY, Cheng J, Li J, Wang T, Zhang QY, Liang DC, Sun B, Wang BM. Serotonin transporter gene promoter region polymorphisms and serotonin transporter expression in the colonic mucosa of irritable bowel syndrome patients. Neurogastroenterol Motil 24: 560–565, e254–e255, 2012. [DOI] [PubMed] [Google Scholar]
- 64.Wong BS, Camilleri M, Carlson P, McKinzie S, Busciglio I, Bondar O, Dyer RB, Lamsam J, Zinsmeister AR. Increased bile acid biosynthesis is associated with irritable bowel syndrome with diarrhea. Clin Gastroenterol Hepatol 10: 1009–1015, e3, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wouters MM, Lambrechts D, Knapp M, Cleynen I, Whorwell P, Agréus L, Dlugosz A, Schmidt PT, Halfvarson J, Simrén M, Ohlsson B, Karling P, Van Wanrooy S, Mondelaers S, Vermeire S, Lindberg G, Spiller R, Dukes G, D'Amato M, Boeckxstaens G. Genetic variants in CDC42 and NXPH1 as susceptibility factors for constipation and diarrhoea predominant irritable bowel syndrome. Gut 63: 1103–1111, 2014. [DOI] [PubMed] [Google Scholar]
- 66.Wunder F, Woermann A, Geerts A, Milde M. Pharmacological characterization of receptor guanylyl cyclase reporter cell lines. Eur J Pharmacol 698: 131–136, 2013. [DOI] [PubMed] [Google Scholar]
- 67.Zigmond AS, Snaith RP. The hospital anxiety and depression scale. Acta Psychiatr Scand 67: 361–370, 1983. [DOI] [PubMed] [Google Scholar]
- 68.Zucchelli M, Camilleri M, Nixon Andreasson A, Bresso F, Dlugosz A, Halfvarson J, Törkvist L, Schmidt PT, Karling P, Ohlsson B, Duerr RH, Simren M, Lindberg G, Agreus L, Carlson P, Zinsmeister AR, D'Amato M. Association of TNFSF15 polymorphism with irritable bowel syndrome. Gut 60: 1671–1677, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]