Abstract
Electrotaxis, directional cell movement in response to an electric potential, has been demonstrated in a wide range of cell types including lymphocytes. Exoelectrogens, microorganisms capable of generating electrical currents, have been identified in microbial fuel cells. However, no studies have investigated exoelectrogenic microbes in fresh feces or the effects of an exoelectrogenic microbiota on the host organism. Here we show that commensal gut microbial populations differ in their capacity for electrical current production by exoelectrogens and that those differences are predictive of increased lymphocyte trafficking to the gut in vivo, despite the lack of increased production of canonical lymphocyte-specific chemokines. Additionally, we demonstrate that the difference in current production between mice purchased from different commercial sources correlates reproducibly with the presence or absence of segmented filamentous bacteria, and while our data do not support a direct role for segmented filamentous bacteria in ex vivo current production, an exoelectrogenic microbiota can be transferred in vivo via mucosa-associated bacteria present in the ileum. Moreover, we detect upregulation of microbial genes associated with extracellular electron transfer in feces of mice colonized with exoelectrogenic microbiota containing segmented filamentous bacteria. While still correlative, these results suggest a novel means by which the gut microbiota modulates the recruitment of cells of the immune system to the gut.
Keywords: microbiota, segmented filamentous bacteria, exoelectrogen, electrotaxis
endogenous bioelectric fields have recognized roles in regulating cellular proliferation, migration, differentiation, regeneration, and wound healing. Electrotaxis (or galvanotaxis), cellular migration in response to electrical fields, has been demonstrated in numerous cell types including keratinocytes, fibroblasts, neural crest cells, stem cells (10), and most recently, lymphocytes (18, 19). The best-characterized scenario in which electrotaxis plays a physiological role is wound-healing. Following a penetrating injury, the constitutive transepithelial potential (TEP), maintained at all epithelial surfaces via directional ion transport (Cl− secretion/Na+ absorption) and the resistance provided by tight junctions is disrupted, resulting in a relative decrease in potential at the site of injury compared with surrounding epithelium (39). The resulting laterally oriented electric field recruits surrounding keratinocytes in a sodium channel- and cAMP-dependent manner (28, 36).
The role of electrotaxis in gut epithelial restitution or recruitment of other cell types is unclear, and its study is complicated by several factors. Homeostatic regulation of ion transport across the gastrointestinal (GI) epithelium is complex and varies along the length of the GI tract, along the crypt axis, and between species (15). Moreover, ionic balance in the gut can vary based on diet and the microbiota (4, 8), as well as other factors. Early studies of the TEP across intestinal mucosa confirmed that, similar to the TEP maintained at dermal surfaces, it is driven by ionic balance, with Na+ and Cl− being the strongest determinants (2), and mucosal injury induces a significant decrease in potential (29). There is however a gap in our understanding of the interactions between the TEP, the gut microbiota, and the recruitment of epithelial cells and other cell types that are responsive to electric fields.
Physical contact between microbes capable of penetrating the mucus barrier and pathogen recognition receptors expressed on underlying intestinal epithelial cells and dendritic cells leads to the induction of chemoattractant molecules (i.e., chemokines) and subsequent recruitment of leukocytes. To the authors' knowledge, the contribution of electric fields in this context has not been investigated. Providing evidence of a physiological role for endogenous electrical fields, lymphocytes exposed to both chemotactic and electrotactic gradients demonstrate comparable migration along either axis (18). Exoelectrogens, microorganisms capable of generating electrical currents via oxidation of organic material and electron transfer to an anode, have been identified in microbial fuel cells (MFC), often using wastewater as a source (20). Notably, the majority of exoelectrogenic microbes identified in MFCs belong to taxa commonly found in the GI tracts of mammals, including microbes in the classes Deltaproteobacteria and Gammaproteobacteria (20). However, no prior work has investigated exoelectrogenic microbes in animal fecal samples or explored the effects of an electrochemically active microbiota on its host. We sought to assess the ability of intestinal bacterial communities, herein referred to as gut microbiota (GM), to generate an electrical current and to determine if this process has a physiological role in lymphocyte trafficking in the host organism.
MATERIALS AND METHODS
Mice.
All experiments were performed in accordance with the Guide for the Care and Use of Laboratory Animals and were approved by the University of Missouri Institutional Animal Care and Use Committee. A/J, BALB/cJ, C57BL/6J, C3H/HeJ, C.129S7(B6)-Rag1tm1Mom/J, and C57BL/6-Tg(UBC-GFP)30Scha/J (Jackson Laboratory, Bar Harbor, ME); C57BL/6NHsd, A/JOlaHsd, BALB/cAnNHsd, C3H/HeNHsd, CrlHli:CD-1, and ND4 SW (Harlan Sprague-Dawley, Indianapolis, IN); Crl:CD1 (ICR), Crl:CFW (SW) (Charles River Laboratories International, Wilmington, MA); A/JCr (Frederick Cancer Research and Development Center, Frederick, MD); and C57BL/6NTac (Taconic, Hudson, NY) mice were individually housed in microisolator cages on ventilated racks (Thoren Caging Systems, Hazleton, PA) with ad libitum access to irradiated chow (LabDiet 5058; LabDiet, St. Louis, MO) and acidified, autoclaved water. Mice were humanely euthanized via inhaled CO2 at a regulated flow rate, in accordance with the AVMA Guide to Euthanasia.
Microbial electrolysis cells.
Microbial electrolysis cells (MECs) were adapted from a previously published design (3). Briefly, sterile glass serum bottles were aseptically filled with 5 ml brain heart infusion broth (Sigma Aldrich, St. Louis, MO) supplemented with 0.1% wt/vol l-cysteine (Sigma Aldrich) and 5 mg/ml yeast extract (Fisher Scientific, Hampton, NH). Oxygen was removed, and MECs were flushed with a mixture of 80% N2/20% CO2 three times. Following inoculation with fecal homogenate, MECs were incubated at 37°C, and a potential of 0.7 V was applied via a 3645 DC power supply (Circuit Specialists, Mesa, AZ). Voltages were measured across a 10Ω resistor every 10 min with an Agilent 34972A LXI Data Acquisition/Switch Unit (Agilent Technologies, Santa Clara, CA). Current was calculated according to Ohm's law (I = V/R).
MEC sample collection and preparation.
Mice were placed in an empty autoclaved cage, and two freshly evacuated fecal pellets were collected into a 2.0 ml round-bottom tube containing 800 μl sterile phosphate-buffered saline (PBS) and a 0.5 mm diameter stainless steel bead. Fecal pellets were homogenized with a TissueLyser II (Qiagen, Venlo, Limburg, Netherlands) for 3 min at 50 oscillations per second. We then used 100 μl of the fecal homogenate to inoculate MECs.
Electron microscopy.
Unless otherwise stated, all reagents were purchased from Electron Microscopy Sciences and all specimen preparation was performed at the Electron Microscopy Core Facility, University of Missouri. For transmission electron microscopy (TEM) sample preparation, primary fixation of bacteria was performed in 2% paraformaldehyde, 2% glutaraldehyde, in 100 mM sodium cacodylate buffer, pH 7.35. Next, fixed bacteria were pelleted, embedded in 10 μl of Histogel agar (Thermo Scientific Richard-Allen Scientific), and rinsed with 100 mM sodium cacodylate buffer, pH 7.35, containing 10 mM β-mercaptoethanol (Sigma Aldrich) and 130 mM sucrose (further referred to as 2-ME buffer). Using a Pelco Biowave (Ted Pella, Redding, CA), secondary fixation was performed using 1% osmium tetroxide (Ted Pella) in 2-ME buffer using a Pelco Biowave, incubated at 4°C for 1 h, then rinsed with 2-ME buffer, followed by distilled water. Using the Pelco Biowave, we performed a graded dehydration series using ethanol, transitioned into acetone, and dehydrated tissues were then infiltrated with Epon/Spurr's resin and polymerized at 60°C overnight. Sections were cut to a thickness of 85 nm with an ultramicrotome (Ultracut UCT, Leica) and a 45° diamond knife (Diatome, Hatfield, PA). Images were acquired at an accelerating voltage of 80 kV on a JEOL JEM 1400 transmission electron microscope (JEOL, Peabody, MA) equipped with a Gatan UltraScan 1000 CCD camera (Gatan, Warrensdale, PA). For scanning electron microscopy (SEM) sample preparation, fixed bacterial cultures were incubated for 1 h on poly-l-lysine-coated Thermanox coverslips. Coverslip-adherent bacteria were rinsed with 100 mM sodium cacodylate buffer, pH 7.35; secondary fixation was performed with 2% osmium tetroxide using a Pelco Biowave, incubated at 4°C for 1 h, then rinsed with 100 mM sodium cacodylate, followed by water. Using the Pelco Biowave, we performed a graded dehydration series using ethanol, and the dehydrated samples were then critically point dried with an Autosamdri-815 (Tousimis, Rockville, MD). Samples were sputter-coated (Emitech K575x; Quorum Technologies, Sussex, UK), with a 10 nm layer of platinum to ensure conductivity. Samples were imaged with either a Hitachi S4700 operated at 5 kV and 10 microAmps or a Quanta 600F environmental SEM operated at 10 kV spot 3.
Immunohistochemistry and confocal microscopy.
Immunohistochemical staining of CD3, B220, CCL21, and enhanced green fluorescent protein (eGFP) was performed at IDEXX BioResearch (Columbia, MO). Briefly, tissues were fixed overnight in 4% paraformaldehyde and processed the next day for paraffin embedding. We mounted 5 μm thick paraffin-embedded sections onto ProbeOn Plus microscope slides (Fisher Scientific, Pittsburgh, PA). Prior to immunohistochemical analysis, sections were dewaxed in xylene, rehydrated through graded concentrations of EtOH, and then rinsed in distilled water. Antigen retrieval was performed by immersing sections in heated 10 mM citrate buffer (pH 6.0) for 30 min, followed by cooling to room temperature while immersed in buffer. All subsequent incubations were performed at room temperature. Slides intended for enzyme-mediated histochemistry were treated with 3% hydrogen peroxide (H2O2) and rinsed. All slides were immersed in blocking buffer of 5% bovine serum albumin for 20 min and rinsed prior to incubation for 60 min with the following primary antibodies: anti-CD3 [1:500 dilution of a rabbit polyclonal antibody (A0452), DAKO, Carpinteria, CA]; anti-B220 [1:1,000 dilution of a rat monoclonal antibody (clone Ra3-6b2, RM2600), Invitrogen, Grand Island, NY]; anti-CCL21 [1:80 dilution of a goat polyclonal antibody (AF452), R&D Systems, Minneapolis, MN] and anti-GFP [1:300 dilution of a chicken polyclonal antibody (ab13970), Abcam, Cambridge, MA]. Sections were then washed and incubated for 30 min with the appropriate biotinylated secondary antibody (for CD3, B220 and CCL21) or with a secondary antibody bound to Alexa Fluor 488 (for GFP). Bound biotinylated antibodies were incubated with a horseradish peroxidase-streptavidin complex and then with the substrate 3,3′-diaminobenzidine (DAB) solution (0.05% DAB with 0.015% H2O2 in PBS; DAKO). DAB-labeled sections were counterstained with Mayer's hematoxylin, dehydrated, and coverslipped. Sections for fluorescence were rinsed and coverslips were mounted with MOWIOL 4-88 medium (Sigma-Aldrich). Images for DAB-labeled slides were photographed on a Nikon Eclipse Ni. Confocal images were obtained on a TCS SPE system equipped with a DMI 4000B confocal microscope (Leica, Solms, Germany) at 488 nm excitation, with an emission filter of 503 to 563 nm for Alexa-fluor 488-conjugated IgY.
Enzyme-linked immunosorbent assay.
CCL19- and CCL25-specific enzyme-linked immunosorbent assay (ELISA) kits were purchased from Abcam; CCL21-specific ELISA kits were purchased from Cell Sciences (Canton, MA). Assays were performed according to manufacturer's instructions using 1:2 dilutions of culture supernatant as sample. All biological replicates were analyzed as two technical replicates using a Spectramax M3 plate reader (Molecular Devices, Sunnyvale, CA). Technical replicates were averaged and data reported are the mean of the three biological replicates.
Microbial DNA and RNA extraction and quantification.
Microbial DNA and RNA were extracted according to an adaptation of a previously published protocol (38). Briefly, samples were placed in 800 μl of lysis buffer [500 mM NaCl (Fisher Scientific), 50 mM Tris·HCl (Sigma Aldrich), 50 mM EDTA (Fisher Scientific), and 4% SDS (Fisher Scientific)], homogenized thoroughly, and incubated at 70°C for 20 min. The supernatant was collected, mixed with 200 μl of 10 mM ammonium acetate, incubated on ice for 5 min, and then centrifuged at room temperature for 10 min at 16,000 g. We then mixed 750 μl of supernatant with an equal volume of chilled isopropanol and incubated it for 30 min on ice. The contents of the tube were then centrifuged at 4°C for 15 min to pellet nucleic acid. The pellet was then rinsed twice with 70% EtOH and resuspended in 150 μl of Tris-EDTA (Sigma Aldrich). For DNA purification, 15 μl of proteinase-K and 200 μl of buffer AL (DNeasy kit, Qiagen) were added, and tubes were incubated at 70°C for 10 min. We then added 200 μl of 100% EtOH and transferred the entire contents of the tube to a Qiagen spin column before continuing with the manufacturer's instructions for DNA purification (DNeasy Kit, Qiagen). DNA was eluted in 200 μl of EB buffer (Qiagen). For RNA purification, 600 μl RLT buffer (RNeasy kit, Qiagen) supplemented with β-mercaptoethanol (Sigma Aldrich) was added to nucleic acid and mixed thoroughly. Following centrifugation for 3 min at 8,000 g, 350 μl of supernatant was mixed with one volume chilled 100% EtOH and mixed immediately via pipette. We then transferred 700 μl of sample to an RNeasy spin column and processed it according to the manufacturer's instructions. Yield of double-stranded DNA and single-stranded RNA were determined via fluorometry (Qubit 2.0; Life Technologies, Carlsbad, CA).
Segmented filamentous bacteria PCR.
For these assays, template DNA was extracted from feces with the Qiagen DNeasy kit, according to the manufacturer's protocol. Segmented filamentous bacteria (SFB)-specific PCR was performed according to a previously published protocol (31). PCR products were visualized on a Qiaxcel capillary electrophoresis system using a DNA high-resolution gel cartridge (Qiagen).
Reverse transcription.
We reverse-transcribed 5 μg of total RNA using oligo(dT) primers according to the manufacturer's protocol (Superscript II; Invitrogen, Carlsbad, CA). cDNA was diluted with ddH2O to a final concentration of 10 ng/μl.
Real-time PCR.
Real-time PCR to measure microbial expression of MtrC, OmcA, and GyrB was performed on a Rotor-Gene Q (Qiagen) with previously published primers (6, 24).
Gastric gavage.
For gavage of fecal material, fecal samples were collected in PBS and mechanically homogenized with a TissueLyser II (Qiagen). Fecal homogenates were centrifuged at 300 g for 5 min, and the supernatant collected for gavage. Recipient mice received 0.5 ml supernatant via gastric gavage. For gavage of ileal mucosal scrapes, material obtained from mucosal scrapes was suspended in 1 ml sterile PBS, and 0.5 ml was administered via gastric gavage.
Adoptive transfer.
Donor spleens were collected aseptically, minced, and forced through a 100 μm nylon mesh filter. Erythrocytes were lysed using BD Pharm Lyse (BD Biosciences, San Jose, CA), and remaining cells were diluted to 1 × 107 cells/ml. Mice were anaesthetized via inhaled isoflurane, and ∼5 × 105 cells were administered intravenously per mouse. Mice were euthanized 48 h later, and intestinal tissue and spleens were collected for immunohistochemical staining.
Cell culture preparation.
Immediately after euthanasia, 4 cm of proximal colon adjacent to the ileocecocolic junction was excised, rinsed thoroughly with sterile PBS, minced, and placed in sterile RPMI culture media (RPMI 1640; Mediatech, Manassas, VA) supplemented with 0.6 mM HEPES (Sigma Aldrich), 10% sterile filtered fetal bovine serum (Sigma Aldrich), and a combination of penicillin, streptomycin, and amphotericin-B (EMD Millipore, Billerica, MA). Tissue was washed repeatedly in supplemented RPMI and then digested in 25 mg/ml collagenase-D (Sigma Aldrich) for 1 h at 37°C in a shaking incubator (C24KC refrigerated shaker incubator; New Brunswick Scientific, Edison, NJ). Digested tissue was then filtered through a 70 μm nylon mesh filter (Becton Dickinson, Franklin Lakes, NJ), and the flow-through collected. Cells were then washed repeatedly and filtered through progressively smaller filter pore sizes to remove clumps of epithelial cells. Cells in suspension were counted with a hemocytometer on an inverted light microscope and plated at 200,000 cells/ml as three biological replicates per animal in 96-well plates (Falcon, Becton Dickinson). Cells were incubated for 36 h at 37°C, and the supernatant was then collected for ELISA.
Metagenomic library preparation and sequencing.
Sequencing of the V4 region of the 16S rRNA gene was performed on the Illumina MiSeq platform. Bacterial 16S ribosomal DNA amplicons were constructed by amplification of the V4 hypervariable region of the 16s rRNA gene with primers flanked by Illumina standard adapter sequences. Universal primers (U515F/806R) previously developed against the V4 region were used for generating amplicons (5, 34). Oligonucleotide sequences were obtained at proBase (21). A single forward primer and reverse primers with unique 12-base indexes were used in all reactions. Extracted DNA was quantitated by Qubit fluorometer using the quant-iT HS dsDNA reagent kit (Invitrogen). PCR reactions (50 μl) contained 100 ng of genomic DNA, forward and reverse primers (0.2 μM each), dNTPs (200 μM each), and Phusion High-Fidelity DNA Polymerase (1 U). PCR amplification was performed as follows: 98°C(3:00) + [98°C(0:15) + 50°C(0:30) + 72°C(0:30)] × 25 cycles + 72°C(7:00). Amplified product (5 μl) from each reaction was combined and thoroughly mixed; pooled amplicons were purified by addition of 50 μl Axygen AxyPrep MagPCR Clean-up beads (Axygen Biosciences, Union City, CA) to an equal volume of amplicons and incubated at room temperature for 15 min. Products were washed multiple times with 80% EtOH, and the dried pellet resuspended in Qiagen EB buffer (32.5 μl), incubated at room temperature for 2 min, and then placed on a magnetic stand for 5 min. Supernatant (30 μl) was transferred to a low binding microcentrifuge tube for storage. The final amplicon pool was evaluated using the Advanced Analytical Fragment Analyzer (Advanced Analytical Technologies, Ames, IA) automated electrophoresis system, quantified with the Qubit fluorometer using the quant-iT HS dsDNA reagent kit, and diluted according to the manufacturer's protocol for sequencing on the MiSeq.
Informatics analysis.
Assembly, binning, and annotation of DNA sequences were performed at the University of Missouri Informatics Research Core Facility. Briefly, contiguous sequences of DNA were assembled using FLASH software (23), and Qiime v1.7 (14) was used to select representative operational taxonomic units (OTUs). Taxonomy was assigned to selected OTUs using BLAST (1) against the Greengenes database (7) of 16S rRNA sequences and taxonomy.
Statistical analysis.
All statistical analyses were performed with SigmaPlot 12.0. Groups were compared by repeated-measures analysis of variance (MEC data), Mann-Whitney nonparametric analysis (differences between CD3+, B220+, and eGFP+ cell numbers) due to unequal variance, Student's t-test (differences between real-time RT-PCR threshold cycles), and comparative CT analysis (30) (fold change between groups in gene expression). Significance was assigned to P ≤ 0.05.
RESULTS
Current production by GM.
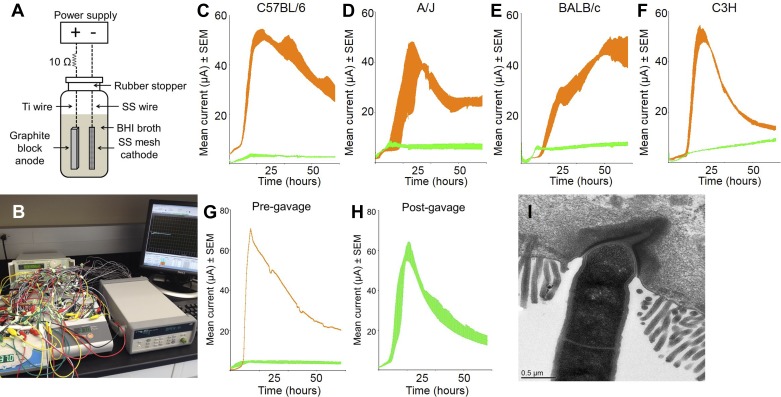
To measure the electrical current generated by GM, MECs fabricated according to the design of Call and Logan (3) were inoculated with murine GM. Briefly, a closed system containing an anode and cathode immersed in a growth medium was flushed to remove oxygen, inoculated with freshly prepared fecal homogenates, and subjected to an applied potential of 0.7 V (Fig. 1, A and B). Recognizing the diversity of the GM due to host genetics and environment, we surveyed the exoelectrogenic capacity of fecal samples from several genetic backgrounds of mice purchased from multiple vendors. MECs inoculated with murine GM produced an electrical current within 8–12 h. Unexpectedly, we detected a significant vendor-specific difference in the exoelectrogenic capacity of the GM used to inoculate MECs. Specifically, MECs inoculated with fecal homogenates collected from four different inbred strains of mice (A/J, BALB/c, C57BL/6, and C3H) purchased at Harlan Laboratories (HSD) consistently generated electrical currents peaking at ∼50 μA, while MECs inoculated with fecal homogenates from the same strains of mice purchased from The Jackson Laboratory (Jax) generated substantially less current, peaking at between 5 and 15 μA (Fig. 1, C–F). These currents correspond with current densities per area (IA; A/cm2) of 10.9 μA/cm2 and ∼2.17 μA/cm2, respectively [using anode-specific surface area of 92 cm2/cm3 based on its dimensions and normalized for 5 ml media volume (3)]. These measurements are comparable to physiologically relevant endogenous sources of electrical currents (25); epithelial wounds, for example, produce outward currents of 4–8 μA/cm2 (40). Fecal samples from two outbred stocks of mice (CD-1 and Swiss Webster) purchased from both HSD and Charles River Laboratories consistently produced electrical currents peaking between 50 and 100 μA, similar to the inbred strains from HSD (data not shown). Similarly, samples collected from A/JCr mice and C57BL/6NTac mice, obtained from the National Cancer Institute and Taconic respectively, both produced currents comparable to those from HSD (data not shown). Thus, the GM of mice from Jax differed from the GM of all mice tested from other vendors in its ability to produce an electrical current, suggesting some difference in the composition of the GM in mice from Jax. To determine whether the exoelectrogenic capacity of the GM was an intrinsic function of the GM or the host, we transferred an exoelectrogenic GM collected from an HSD mouse to Jax mice via gastric gavage. Two weeks after receiving a fecal slurry prepared from C57BL/6NHsd donor mice, fecal samples collected from C57BL/6J recipients were capable of producing an electrical current equivalent to that seen in donor mice (Fig. 1, G and H), demonstrating that the exoelectrogenic property was transferrable via the GM.
Fig. 1.
Current production by murine gut microbiota varies according to commercial source of mouse and is transferrable via feces. Schematic diagram (A) and photograph (B) of microbial electrolysis cells (MECs) used to measure current production. Ti, titanium; SS, stainless steel; BHI, brain heart infusion. Current production was measured in MECs inoculated with fecal samples collected from C57BL/6J and C57BL/6NHsd (C), A/J and A/JOlaHsd (D), BALB/cJ and BALB/cAnNHsd (E), and C3H/HeJ and C3H/HeNHsd (F) mice. Current (μA) is shown as mean ± SE from 4 identically treated MECs per group, and data from Jackson and Harlan mice are shown in green and orange, respectively. G: current production by feces from C57BL/6J recipients prior to gavage and 1 C57BL/6NHsd donor mouse. H: current production by feces from C57BL/6J recipient mice postgavage. I: transmission electron microscopic image of SFB adherent to ileal mucosa.
SFB (Candidatus Arthromitus) are commensal microbes that adhere to the ileal epithelium of mice, rats, and many other species including humans (37). SFB have gained recent attention due to their ability to induce the proliferation or recruitment of Th17 (12) and αβTCR-bearing intraepithelial lymphocytes (32) in the gut. SFB are generally considered refractory to in vitro culture, although a recent report demonstrated successful cultivation with heavily supplemented media (30a). Notably, SFB were detected via 16S rRNA-specific PCR in all mice purchased from HSD, Charles River Laboratories, Taconic, and the National Cancer Institute, but was absent in all mice purchased from Jax (data not shown). Direct visualization of SFB via TEM revealed an electron-dense region adjacent to the site of SFB attachment to the ileal mucosa (Fig. 1I).
Transfer of exoelectrogenicity.
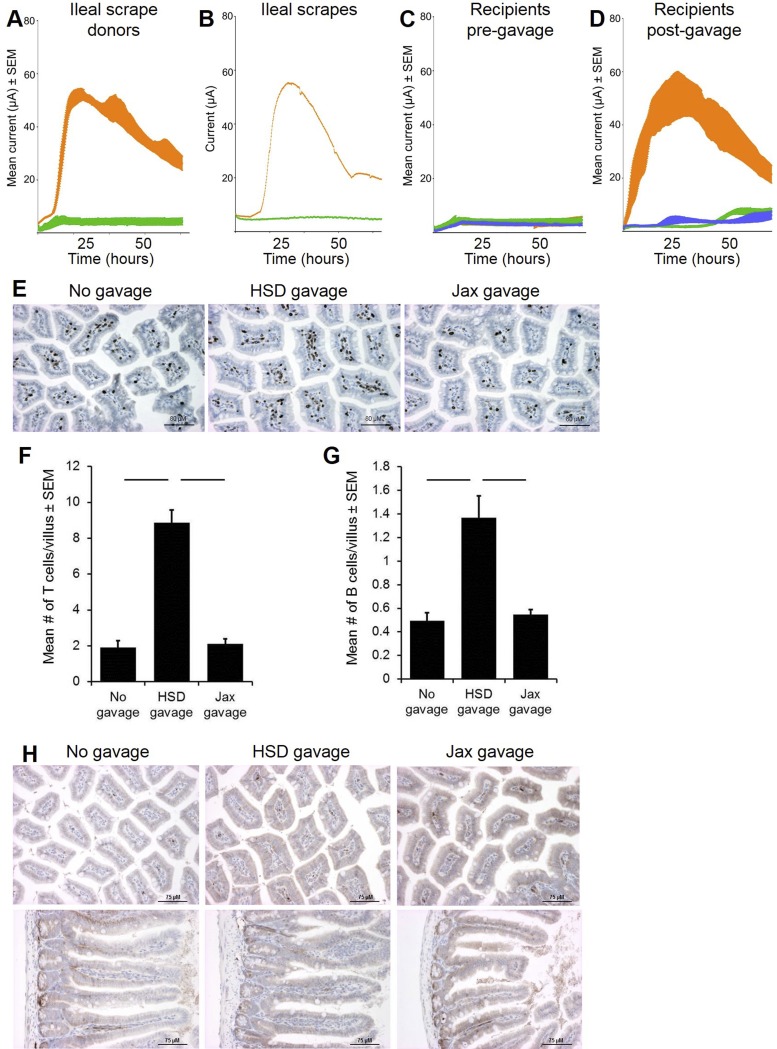
To more precisely determine the role of SFB in the differential current production of mice from different vendors, we transferred SFB to C57BL/6J recipient mice via gastric gavage of an ileal mucosal scrape collected from C57BL/6NHsd mice. Briefly, the ilea of donor mice were rinsed thoroughly with sterile PBS to remove all visible luminal contents, the mucosa was vigorously scraped with a scalpel blade and rinsed in PBS, and the rinsed material administered via gavage. SFB-specific PCR revealed colonization of SFB in all mice receiving ileal mucosal scrapes collected from HSD donors (data not shown). With this technique, the exoelectrogenic property was again transferred to Jax mice, suggesting that SFB or other bacteria adherent to the ileal mucosa were responsible for the ability of the GM to produce an electrical current (Fig. 2, A–D). To assess whether the introduction of an exoelectrogenic GM resulted in increased recruitment of lymphocytes to the gut, ileal T cells and B cells were quantified in Jax mice that had received an ileal mucosal scrape from either an HSD or Jax donor, using immunohistochemistry for CD3 and B220 respectively. Accordingly, there were significantly greater numbers of CD3+ and B220+ cells in the ileal lamina propria of mice that had received material obtained via mucosal scrape from an HSD donor 6 wk earlier (Fig. 2, E–G). Additionally, immunohistochemical staining was performed for CCL21, a canonical chemokine in the recruitment of naïve and activated lymphocytes to secondary lymphoid tissue in the gut (22). Moderate diffuse staining was seen near the basement membrane underlying the villous epithelium in all mice (Fig. 2H). No difference was detected however between mice that received no gavage or ileal mucosal scrapes collected from HSD or Jax mice, as determined by two blinded pathologists. Collectively, these data suggest that mucosa-associated microbes present in the ileum are capable of increasing lymphocyte recruitment to the gut.
Fig. 2.
Fecal electrical current production is transferrable via material adherent to ileal mucosa and correlates with lymphocyte recruitment. Current production was measured in MECs inoculated with fecal samples (A, n = 4) or ileal mucosal scrapes (B, n = 1) collected from C57BL/6J or C57BL/6NHsd mice. Data from Jackson (Jax) and Harlan (HSD) mice are shown in green and orange, respectively. C: current production by fecal samples collected from 12 C57BL/6J recipients prior to gavage. D: current production in MECs inoculated with feces collected from recipients 48 h postgavage with ileal scrape material collected from C57BL/6NHsd (orange) or C57BL/6J (green) mice or following no gavage (blue). Current (μA) is shown as mean ± SE. E: immunohistochemical staining of ileal tissue for CD3 was performed in recipient mice 6 wk postgavage with ileal scrape material. Mean (± SE) number of CD3+ (F) and B220+ (G) cells per villus. Bars denote P ≤ 0.05 as determined via Mann-Whitney nonparametric analysis. H: immunohistochemical staining for CCL21 in the ileum of recipient mice 6 wk postgavage.
Recruitment of exogenous lymphocytes.
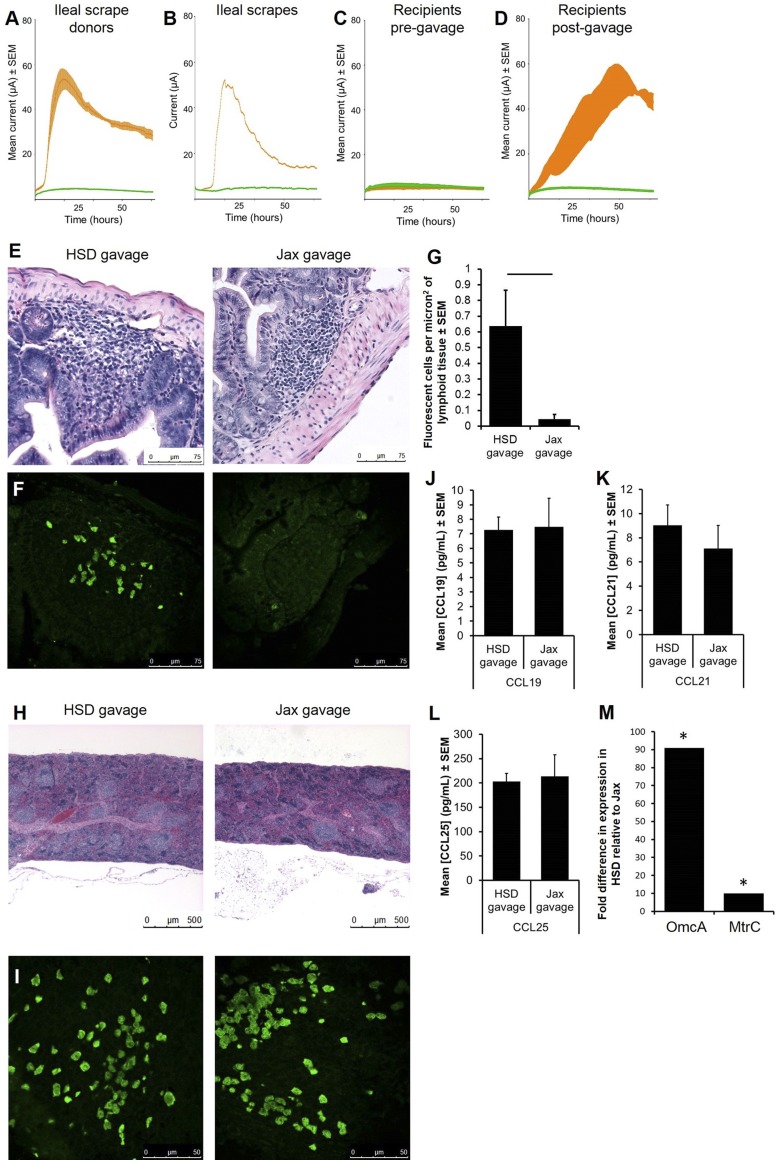
To confirm that the difference in the number of gut lymphocytes was due to differential recruitment and not proliferation of endogenous lymphocytes, SFB-negative Rag1−/− mice were gavaged with an ileal mucosal scrape from HSD or Jax mice, followed 2 wk later by adoptive transfer of eGFP+ splenocytes. As anticipated, the exoelectrogenic property was transferred to Rag1−/− mice receiving gavage material from HSD donors (Fig. 3, A–D). While Rag1−/− mice lack mature lymphocytes and normal mucosa-associated lymphoid tissue, they still possess small lymphoid organs containing immature lymphoid cells (26) (Fig. 3E). Confocal microscopy revealed rare eGFP+ lymphocytes in the immature lymphoid aggregates in the ileum of mice that had received a mucosal scrape from SFB-negative Jax mice. Conversely, eGFP+ lymphocytes were found scattered throughout the mucosal lymphoid aggregates of all mice that had received a mucosal scrape from SFB-positive HSD mice (Fig. 3, F and G). Staining of splenic tissue confirmed successful transfer of cells in both groups (Fig. 3, H and I). To evaluate the contribution of chemokine expression in the increased recruitment of transferred cells, cells were isolated from adjacent colonic tissue of Rag1−/− mice that had received an ileal mucosal scrape from HSD and Jax donors 2 days earlier, and cultured ex vivo for 36 h. Production of the three principal gut-associated chemokines in the supernatant of those cultures was determined via ELISA. No difference was detected between groups in the production of CCL19, CCL21, or CCL25 (Fig. 3, J–L), suggesting that differential production of these canonical gut-specific chemokines was not responsible for the difference seen in lymphocyte recruitment to the gut.
Fig. 3.
Gavage with exoelectrogenic material results in increased lymphocyte recruitment to the gut. Current production was measured in MECs inoculated with feces (A, n = 4) or ileal mucosal scrapes (B, n = 1) collected from C57BL/6J or C57BL/6NHsd mice. Current (μA) is shown as mean ± SE, and data from Jax and HSD mice are shown in green and orange, respectively. C: current production by Rag1−/− recipients (n = 8) prior to gavage of material collected from ileal mucosal scrape. D: current production by recipients postgavage with ileal mucosal scrape. Data from Jax and HSD mice are shown in green and orange, respectively. E: photomicrographs of hematoxylin and eosin (H&E)-stained sections of ileal lymphoid follicles in Rag1−/− recipients following adoptive transfer of eGFP+ lymphocytes. F: confocal microscopic images of the regions shown in E. G: mean number (± SE) of eGFP+ cells detected in Rag1−/− recipients. Bar denotes P ≤ 0.05 as determined via t-test. H&E-stained (H) and confocal microscopic (I) images of spleens from recipient mice in E and F. Production of CCL19 (J), CCL21 (K), and CCL25 (L) in colonocytes collected from Rag1−/− recipients 48 h postgavage and cultured ex vivo for 36 h. M: fold difference in expression of OmcA and MtrC, normalized to that of the housekeeping gene GyrB, in feces of Rag1−/− recipients as determined via comparative CT analysis.*Significant difference (P < 0.05).
Upregulation of microbial electron transfer genes.
Exoelectrogenic microbes transfer electrons to other materials via soluble shuttles, conductive nanowires, or cytochromes. Absence of functional genes encoding the c-type decaheme cytochromes MtrC and OmcA results in poor electrical conductivity (11). To evaluate the role of these genes in the differential exoelectrogenic capacity of the gut microbiota, RNA was extracted from feces of Rag1−/− mice that had received an ileal mucosal scrape from HSD or Jax mice, and used as template to generate cDNA for use in real-time PCR. A standardized amount of cDNA was amplified for 50 cycles, and fold difference was calculated by the comparative CT method (30) after normalization to the housekeeping gene GyrB. Significantly greater levels of mRNA specific for MtrC and OmcA were seen in the feces of mice that received an ileal mucosal scrape from HSD donors compared with mice that received material collected from Jax donors (Fig. 3M), suggesting that the exoelectrogenic properties of the GM observed in MECs were also occurring in vivo and were not an artifact of the culture system.
Characterization of microbes associated with exoelectrogenicity.
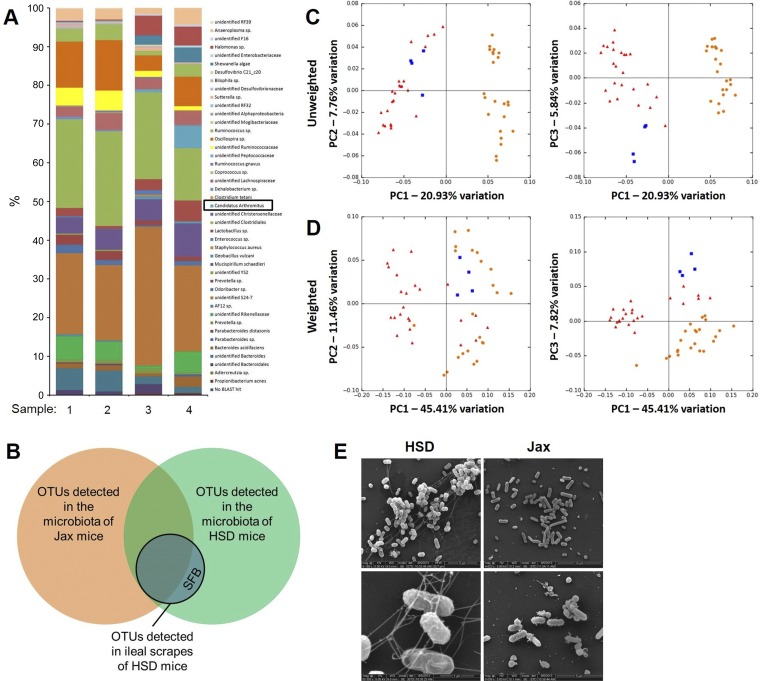
To better define which microbes were involved in the exoelectrogenic properties observed, we performed bar-coded sequencing of the V4 region of microbial 16S rRNA genes of mucosal scrapes from HSD mice capable of transferring exoelectrogenicity. Using a 97% nucleotide identity OTU, we detected 45 distinct OTUs in the mucosal scrapes, including SFB (i.e., Candidatus Arthomitus) (Fig. 4A). We also performed sequencing on the feces of C57BL/6J mice before and after they received a mucosal scrape. While there was collateral transfer of several other microbial species with SFB, we reasoned that microbes involved in production of electrical currents would consistently be present in the feces of mice from HSD and absent in the feces of mice from Jax. Sequencing of the feces from eight C57BL/6, eight A/J, and eight BALB/c mice from each vendor resulted in a total of 94 and 82 consistently detected OTUs from HSD and Jax mice, respectively, with 70 of those OTUs being shared between vendors. Compared with OTUs present in an exoelectrogenic mucosal scrape, SFB was the only OTU that had not previously been detected in the GM of Jax mice (Fig. 4B). That said, several OTUs were resolved to the taxonomic level of family (or higher), and sequences annotated to those levels might represent completely different species whose sequences vary at regions of the 16S rRNA gene other than that used in the current study. Principal component analysis based on either unweighted or weighted UniFrac distances reflected a clear separation between the GM present in mice from the two vendors, with bacteria detected in exoelectrogenic mucosal scrapes clustering with samples from HSD mice (Fig. 4, C and D). The unweighted UniFrac distance between two samples is calculated based on the agreement regarding the presence or absence of each OTU; weighted UniFrac distances also take into account how closely samples agree based on the relative abundance of shared OTUs. In either case, distances of 0 and 1 indicate identical or mutually exclusive communities, respectively. The mean unweighted UniFrac distance between all possible pairwise combinations of a mucosal scrape capable of conferring exoelectrogenicity and the GM of HSD mice was significantly lower than comparable pairings of the same mucosal scrapes and the GM of Jax mice (P < 0.001, t-test). Surprisingly, there was also a significant difference in the mean weighted UniFrac distance although, in this case, the GM of Jax mice was significantly closer to the mucosal scrapes (P < 0.001, t-test). Collectively, these data suggest that while the mucosal scrapes are in closer accord to the GM of HSD mice with regard to which taxa are actually present, they agree more closely with the GM of Jax mice with regard to the abundance of those OTUs shared between the GM of Jax mice and the mucosal scrapes.
Fig. 4.
The presence of segmented filamentous bacteria (SFB) in feces correlates with current production and the presence of microbial nanowires in MECs. A: relative abundance of operational taxonomic units (OTUs) in mucosal scrapes collected from C57BL/6NHsd mice; legend at right. B: Venn diagram depicting overlap between OTUs in feces of HSD mice (C57BL/6NHsd, A/JOlaHsd, and BALB/cAnNHsd mice, n = 8/strain); Jax mice (C57BL/6J, A/J, and BALB/cJ mice, n = 8/strain); and the ileal mucosal scrapes depicted in A. Unweighted (C) and weighted (D) principal component analysis of the fecal microbiota of C57BL/6, A/J, and BALB/c mice from Jackson (orange circles) and Harlan (red triangles) Labs (n = 8 mice/strain/vendor) and mucosal scrape material (blue squares) collected from C57BL/6NHsd mice (n = 4). E: scanning electron microscopic images of microbes present in MECs inoculated with feces from C57BL/6NHsd (HSD) or C57BL/6J (Jax) mice.
Lastly, we performed electron microscopy of actively exoelectrogenic bacteria collected from an MEC. SEM images of the bacteria present in MECs inoculated with an ileal mucosal scrape collected from an HSD mouse revealed an extensive network of pilus-like structures resembling nanowires reported in other electrically conductive microbes (16) (Fig. 4E). These structures were not visible in MEC contents inoculated with similar material from a Jax mouse. Notably, however, no microbes morphologically similar to SFB were detected in the MECs capable of producing current. These data were interpreted to suggest that SFB serve as biomarkers or hallmarks of exoelectrogenic microbial communities, and other microbial taxa, unresolved in 16S rRNA amplicon sequencing, are likely responsible for the observed phenomenon. Alternatively, SFB may have an indirect role by conferring the exoelectrogenic property to other microbes present in the GM.
DISCUSSION
Here we show that the exoelectrogenic activity of resident gut bacteria correlates with the recruitment of lymphocytes to the gut, suggesting a role for electric fields in intestinal immune cell recruitment. The exoelectrogenic capacity of the microbiota had field magnitudes (μV/cm2) equivalent to those previously shown to induce lymphocyte electrotaxis in vitro and in vivo (19). Moreover, direct comparisons between electrotaxis and chemotaxis of activated T cells have shown equal or greater orientation toward electric fields (18). Electrotaxis in physiological fields of similar magnitude has been shown in re-epithelialization of wounds (40) and stem cell migration (17); both cell types have prominent roles in maintaining intestinal homeostasis, raising questions about what other tissues besides lymphocytes may be affected by bioelectric fields at the intestinal barrier. The presence of structures resembling conductive pili or nanowires on rod-shaped microbes present in current-producing MECs suggests that the electron transfer occurs via direct contact, as opposed to soluble shuttles, although this was not directly tested. That said, microbially derived B vitamins are enriched in the gut and are not only key factors for animal health, but also soluble shuttles in extracellular electron transfer among exoelectrogenic microbes (13, 33), suggesting that the host-microbe interface merits consideration as a unique redox boundary.
While the presence of SFB correlated perfectly with the production of electrical current in all experiments, we were repeatedly unable to detect SFB in any of the MECs inoculated via electron microscopy. While PCR with SFB-specific primers detected SFB-specific DNA in MECs inoculated with feces from HSD mice, attempts to document proliferation of SFB via real-time PCR analysis of longitudinal samples were unsuccessful, implying that the previously detected DNA did not represent viable bacteria, an expected finding considering the difficulty in ex vivo cultivation of SFB. While purely speculative, it is possible that SFB creates a favorable niche for unidentified exoelectrogens or that it triggers morphologic changes and electrogenic metabolism in other gut constituents that have, in other experimental contexts (primarily bioenergy applications), been shown to have exoelectrogenic abilities. The morphology of SFB resembles that of Desulfobulbaceae, which have been shown to couple reduction of oxygen at marine sediment surfaces to sulfide oxidation in deeper anoxic zones via long-distance electron transport (27). The unique attachment of SFB to its host raises the question of whether host oxygen could be exploited in a manner similar to Desulfobulbaceae.
Another key difference in the GM of mice from the different commercial sources used in the current study is overall microbial richness and alpha diversity. The GM of mice used in biomedical research is greatly simplified relative to that of feral mice (35), and the GM of mice purchased from the Jackson Laboratory exhibits a substantially lower microbial richness than that of HSD mice (9). Considering the limited resolution provided by 16S rRNA amplicon sequencing, there is the possibility that there are undefined taxa present within OTUs that annotated to higher taxonomic levels such as family or order involved in the differential current production. A whole genome shotgun sequencing approach would provide greater resolution of the microbial communities present in the fecal samples tested but was cost-prohibitive in the present studies. Similarly, a more comprehensive analysis of host chemokine expression in mice receiving SFB-positive and negative mucosal scrapes could be accomplished by an RNA-Seq approach, and future studies are warranted.
In conclusion, our findings introduce an additional level of complexity with regard to pH and redox status in the gut, the composition of the GM, and the overall effect on host physiology. Moreover, these data illuminate the GM as a heterogeneous electroactive microbial system that can influence host immunity and underscore the need for interdisciplinary teams in biomedical and bioengineering research. Future studies are merited to investigate the influence of these processes in the context of exposure to pathogenic bacteria and other organisms.
GRANTS
This work was partially funded by National Institutes of Health Grant #5U42OD-010918-15 to the University of Missouri Mutant Mouse Resource and Research Center (MMRRC).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: A.C.E. and C.E.H. conception and design of research; A.C.E., D.J.D., and C.E.H. performed experiments; A.C.E., C.L.F., and C.E.H. analyzed data; A.C.E. and C.E.H. interpreted results of experiments; A.C.E. and D.J.D. prepared figures; A.C.E. drafted manuscript; A.C.E. and C.E.H. edited and revised manuscript; A.C.E., D.J.D., C.L.F., and C.E.H. approved final version of manuscript.
ACKNOWLEDGMENTS
We thank Sherri Neff for assistance with adoptive transfers; Tommi A. White for technical help with scanning and transmission electron microscopy; Cindy Besch-Williford and Jill Gruenkemeyer for technical help with immunohistochemistry; Karen Clifford for assistance formatting figures; William Spollen for technical help with bioinformatics analysis of next-generation sequencing data; and Ronnie Marshall for animal care.
REFERENCES
- 1.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402, 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Archampong EQ, Edmonds CJ. Effect of luminal ions on the transepithelial electrical potential difference of human rectum. Gut 13: 559–565, 1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Call DF, Logan BE. A method for high throughput bioelectrochemical research based on small scale microbial electrolysis cells. Biosens Bioelectron 26: 4526–4531, 2011. [DOI] [PubMed] [Google Scholar]
- 4.Campbell JM, Fahey GC Jr, Wolf BW. Selected indigestible oligosaccharides affect large bowel mass, cecal and fecal short-chain fatty acids, pH and microflora in rats. J Nutr 127: 130–136, 1997. [DOI] [PubMed] [Google Scholar]
- 5.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA 108, Suppl 1: 4516–4522, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coursolle D, Baron DB, Bond DR, Gralnick JA. The Mtr respiratory pathway is essential for reducing flavins and electrodes in Shewanella oneidensis. J Bacteriol 192: 467–474, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72: 5069–5072, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duncan SH, Louis P, Thomson JM, Flint HJ. The role of pH in determining the species composition of the human colonic microbiota. Environ Microbiol 11: 2112–2122, 2009. [DOI] [PubMed] [Google Scholar]
- 9.Ericsson AC, Davis JW, Spollen W, Bivens N, Givan S, Hagan CE, McIntosh M, Franklin CL. Effects of vendor and genetic background on the composition of the fecal microbiota of inbred mice. PLoS One 10: e0116704, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Funk RH, Monsees T, Ozkucur N. Electromagnetic effects - From cell biology to medicine. Progr Histochem Cytochem 43: 177–264, 2009. [DOI] [PubMed] [Google Scholar]
- 11.Gorby YA, Yanina S, McLean JS, Rosso KM, Moyles D, Dohnalkova A, Beveridge TJ, Chang IS, Kim BH, Kim KS, Culley DE, Reed SB, Romine MF, Saffarini DA, Hill EA, Shi L, Elias DA, Kennedy DW, Pinchuk G, Watanabe K, Ishii S, Logan B, Nealson KH, Fredrickson JK. Electrically conductive bacterial nanowires produced by Shewanella oneidensis strain MR-1 and other microorganisms. Proc Natl Acad Sci USA 103: 11358–11363, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, Tanoue T, Imaoka A, Itoh K, Takeda K, Umesaki Y, Honda K, Littman DR. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139: 485–498, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kotloski NJ, Gralnick JA. Flavin electron shuttles dominate extracellular electron transfer by Shewanella oneidensis. mBio 4: 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuczynski J, Stombaugh J, Walters WA, Gonzalez A, Caporaso JG, Knight R. Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr Protoc Microbiol 10: Unit 1E.5, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kunzelmann K, Mall M. Electrolyte transport in the mammalian colon: mechanisms and implications for disease. Physiol Rev 82: 245–289, 2002. [DOI] [PubMed] [Google Scholar]
- 16.Leung KM, Wanger G, El-Naggar MY, Gorby Y, Southam G, Lau WM, Yang J. Shewanella oneidensis MR-1 bacterial nanowires exhibit p-type, tunable electronic behavior. Nano Lett 13: 2407–2411, 2013. [DOI] [PubMed] [Google Scholar]
- 17.Levin M. Bioelectric mechanisms in regeneration: unique aspects and future perspectives. Sem Cell Dev Biol 20: 543–556, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J, Nandagopal S, Wu D, Romanuik SF, Paul K, Thomson DJ, Lin F. Activated T lymphocytes migrate toward the cathode of DC electric fields in microfluidic devices. Lab Chip 11: 1298–1304, 2011. [DOI] [PubMed] [Google Scholar]
- 19.Lin F, Baldessari F, Gyenge CC, Sato T, Chambers RD, Santiago JG, Butcher EC. Lymphocyte electrotaxis in vitro and in vivo. J Immunol 181: 2465–2471, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Logan BE. Exoelectrogenic bacteria that power microbial fuel cells. Nat Rev Microbiol 7: 375–381, 2009. [DOI] [PubMed] [Google Scholar]
- 21.Loy A, Maixner F, Wagner M, Horn M. probeBase–an online resource for rRNA-targeted oligonucleotide probes: new features 2007. Nucleic Acids Res 35: D800–D804, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luther SA, Bidgol A, Hargreaves DC, Schmidt A, Xu Y, Paniyadi J, Matloubian M, Cyster JG. Differing activities of homeostatic chemokines CCL19, CCL21, and CXCL12 in lymphocyte and dendritic cell recruitment and lymphoid neogenesis. J Immunol 169: 424–433, 2002. [DOI] [PubMed] [Google Scholar]
- 23.Magoc T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27: 2957–2963, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martens M, Dawyndt P, Coopman R, Gillis M, De Vos P, Willems A. Advantages of multilocus sequence analysis for taxonomic studies: a case study using 10 housekeeping genes in the genus Ensifer (including former Sinorhizobium). Int J Syst Evol Microbiol 58: 200–214, 2008. [DOI] [PubMed] [Google Scholar]
- 25.McCaig CD, Rajnicek AM, Song B, Zhao M. Controlling cell behavior electrically: current views and future potential. Physiol Rev 85: 943–978, 2005. [DOI] [PubMed] [Google Scholar]
- 26.Mombaerts P, Iacomini J, Johnson RS, Herrup K, Tonegawa S, Papaioannou VE. RAG-1-deficient mice have no mature B and T lymphocytes. Cell 68: 869–877, 1992. [DOI] [PubMed] [Google Scholar]
- 27.Pfeffer C, Larsen S, Song J, Dong M, Besenbacher F, Meyer RL, Kjeldsen KU, Schreiber L, Gorby YA, El-Naggar MY, Leung KM, Schramm A, Risgaard-Petersen N, Nielsen LP. Filamentous bacteria transport electrons over centimetre distances. Nature 491: 218–221, 2012. [DOI] [PubMed] [Google Scholar]
- 28.Pullar CE, Isseroff RR. Cyclic AMP mediates keratinocyte directional migration in an electric field. J Cell Sci 118: 2023–2034, 2005. [DOI] [PubMed] [Google Scholar]
- 29.Scheppach W, Dusel G, Kuhn T, Loges C, Karch H, Bartram HP, Richter F, Christl SU, Kasper H. Effect of L-glutamine and n-butyrate on the restitution of rat colonic mucosa after acid induced injury. Gut 38: 878–885, 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3: 1101–1108, 2008. [DOI] [PubMed] [Google Scholar]
- 30a.Schnupf P, Gaboriau-Routhiau V, Gros M, Friedman R, Moya-Nilges M, Nigro G, Cerf-Bensussan N, Sansonetti PJ. Growth and host interaction of mouse segmented filamentous bacteria in vitro. Nat Lett 520: 99–103, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Snel J, Heinen PP, Blok HJ, Carman RJ, Duncan AJ, Allen PC, Collins MD. Comparison of 16S rRNA sequences of segmented filamentous bacteria isolated from mice, rats, and chickens and proposal of “Candidatus Arthromitus”. Int J Syst Bacteriol 45: 780–782, 1995. [DOI] [PubMed] [Google Scholar]
- 32.Umesaki Y, Okada Y, Matsumoto S, Imaoka A, Setoyama H. Segmented filamentous bacteria are indigenous intestinal bacteria that activate intraepithelial lymphocytes and induce MHC class II molecules and fucosyl asialo GM1 glycolipids on the small intestinal epithelial cells in the ex-germ-free mouse. Microbiol Immunol 39: 555–562, 1995. [DOI] [PubMed] [Google Scholar]
- 33.von Canstein H, Ogawa J, Shimizu S, Lloyd JR. Secretion of flavins by Shewanella species and their role in extracellular electron transfer. Appl Environ Microbiol 74: 615–623, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walters WA, Caporaso JG, Lauber CL, Berg-Lyons D, Fierer N, Knight R. PrimerProspector: de novo design and taxonomic analysis of barcoded polymerase chain reaction primers. Bioinformatics 27: 1159–1161, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilson KH, Brown RS, Andersen GL, Tsang J, Sartor B. Comparison of fecal biota from specific pathogen free and feral mice. Anaerobe 12: 249–253, 2006. [DOI] [PubMed] [Google Scholar]
- 36.Yang HY, Charles RP, Hummler E, Baines DL, Isseroff RR. The epithelial sodium channel mediates the directionality of galvanotaxis in human keratinocytes. J Cell Sci 126: 1942–1951, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yin Y, Wang Y, Zhu L, Liu W, Liao N, Jiang M, Zhu B, Yu HD, Xiang C, Wang X. Comparative analysis of the distribution of segmented filamentous bacteria in humans, mice and chickens. ISME J 7: 615–621, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu Z, Morrison M. Improved extraction of PCR-quality community DNA from digesta and fecal samples. Biotechniques 36: 808–812, 2004. [DOI] [PubMed] [Google Scholar]
- 39.Zhao M. Electrical fields in wound healing-An overriding signal that directs cell migration. Sem Cell Dev Biol 20: 674–682, 2009. [DOI] [PubMed] [Google Scholar]
- 40.Zhao M, Song B, Pu J, Wada T, Reid B, Tai G, Wang F, Guo A, Walczysko P, Gu Y, Sasaki T, Suzuki A, Forrester JV, Bourne HR, Devreotes PN, McCaig CD, Penninger JM. Electrical signals control wound healing through phosphatidylinositol-3-OH kinase-gamma and PTEN. Nature 442: 457–460, 2006. [DOI] [PubMed] [Google Scholar]