Highlights
-
•
We present antibiotic-induced membrane thinning of a multi-lamellar thin film sample.
-
•
Both penicillin and sulbactam are found positioned outside the model membrane in an aqueous solution.
-
•
We demonstrate a hybrid method to study the antibiotic–membrane interaction.
Abbreviations: A. baumannii, Acinetobacter baumannii; LXD, lamellar X-ray diffraction; GUV, giant unilamellar vesicle
Keywords: Penicillin, Sulbactam, Cholesterol, GUV, LXD
Abstract
Antibiotic drug resistance is a serious issue for the treatment of bacterial infection. Understanding the resistance to antibiotics is a key issue for developing new drugs. We used penicillin and sulbactam as model antibiotics to study their interaction with model membranes. Cholesterol was used to target the membrane for comparison with the well-known insertion model. Lamellar X-ray diffraction (LXD) was used to determine membrane thickness using successive drug-to-lipid molar ratios. The aspiration method for a single giant unilamellar vesicle (GUV) was used to monitor the kinetic binding process of antibiotic–membrane interactions in an aqueous solution. Both penicillin and sulbactam are found positioned outside the model membrane, while cholesterol inserts perpendicularly into the hydrophobic region of the membrane in aqueous solution. This result provides structural insights for understanding the antibiotic–membrane interaction and the mechanism of antibiotics.
1. Introduction
Acinetobacter baumannii (A. baumannii) is an aerobic Gram-negative bacterium that is resistant to most antibiotics [1,2]. Since 1990s, it has been recognized as an important infectious bacterium within hospitals, especially in ICUs [3,4]. A survey conducted in 2007 by the Center for Disease Control in Taiwan indicated that A. baumannii ranked the first among common pathogens for nosocomial infections in ICUs. Developing a new type of antibiotic is a priority for the treatment of A. baumannii infections. There are no known antibiotics to kill A. baumannii effectively. However, it has recently been reported that a compound, sulbactam, killed A. baumannii 19606 effectively [5], but the detail mechanism of the bactericidal effect of sulbactam remains unclear. In that report the authors found that when treated with sulbactam the bacterial ATP-binding cascade (ABC) was down-regulated which inhibited the ability of bacteria to uptake nutrients and expel toxic material and subsequently caused bacterial death. We compared the interactions of penicillin, sulbactam, and cholesterol with a model membrane, giant unilamellar vesicle (GUV), by measuring the changes of membrane thickness and surface area of GUV, as well as drug-membrane kinetic binding behavior, to elucidate a possible structural insight for understanding the sulbactam-membrane interaction and the mechanism of bactericidal effect of sulbactam.
Penicillin is the most important antibacterial compound used clinically [6,7] and is historically significant because it was the first antibiotic that was efficient for treating many bacterial infections. The interaction between penicillin and its targeted penicillin binding proteins (PBPs) is the most typical example for elucidating the molecular mechanism of disrupting bacterial growth by antibiotics [8,9]. Various forms of PBP exist and bacteria usually contain many types of PBPs within them. The most common forms of PBPs are grouped into classes 1, 2, 3, and 4 [10,11]. Very early studies showed that higher concentration of antibiotics caused the death of bacteria [12] and interfered with the cell membrane synthesis [13]. Further extensive and laborious studies in 1970s and 1980s then established our present knowledge for the bacteria-killing mechanism of β-lactam antibiotics. In this mechanism, the β-lactam moiety of penicillin binds covalently with the catalytic serine residue on the active site of PBPs [8,14,15]. PBPs are constituent proteins on bacterial membranes and are involved in the synthesis of cross-linked peptidoglycan, which is the major component of bacterial cell walls. Penicillin binds with the transpeptidase C-terminal domain of PBPs and inhibits the cross-linking of peptidoglycan [16,17] and eventually leads to bacteria death. A recent crystal structure proved that a native form of PBP (PBP3) becomes covalently linked to antibiotics (carbenicillin and ceftazidime) [18]. In addition, bacteria always have higher internal osmotic pressure than the external one [19]. It has been reported that resisting the osmotic stress is crucial for cell growth and division, as well as maintaining the normal cell shapes. It was found that penicillin could be involved in the osmotic barrier and lying close to the bacteria cell surface [20,21]. Structure defects of peptidoglycan on bacterial cell wall would disrupt the integrity of cell membrane and resulted in the rupture of peptidoglycan [19]. Thus, β-lactam antibiotics such as penicillin would disrupt the osmotic pressure and expedite bacteria death. Hence, penicillin and derivatives have been widely used for treating hospital infections [22]. However, most bacteria soon developed antibiotic-resistance. Sulbactam is a β-lactam analog that is typically given in combination with another antibiotics ampicillin to form a β-lactam–β-lactamase inhibitor. Ampicillin/sulbactam is a common penicillin-derived antibiotic that is used to treat infections caused by bacteria that are resistant to β-lactam antibiotics. In recent years, researchers evaluated the clinical efficacy of ampicillin/sulbactam in the treatment of infections caused by multidrug-resistant A. baumannii [23,24]. Currently, ampicillin/sulbactam is the first-line therapy for diverse respiratory and skin infections [25].
Here, we focused on the direct interaction between penicillin, sulbactam and model membranes to clarify this mechanism of inhibition of bacterial growth [5]. Cholesterol is known to interact with membranes by inserting into their hydrophobic region to cause membrane thickening [26–28]. Therefore, cholesterol is used as a good model molecule for comparison with antibiotics.
In this study, we used penicillin and sulbactam as model antibiotics and studied their interactions with model membranes. Cholesterol was used in the same condition for comparison. Lamellar X-ray diffraction (LXD) was used to determine membrane thickness. Thickness changes induced by antibiotics or cholesterol binding to the membrane were extracted from the X-ray data. The giant unilamellar vesicle (GUV) experiment was used to examine the area change of a single GUV in an aqueous solution during the entire binding process.
The structural changes induced by cholesterol partitioning into the membrane were observed not only by LXD but also by GUV measurements. Furthermore, sulbactam and penicillin caused structural changes that were observed by LXD but not GUV measurements. The results of this study show that cholesterol was found perpendicularly inserted to model membrane whereas penicillin and sulbactam were found positioned outside the surface of model membrane in aqueous solution.
2. Materials and methods
2.1. Materials
2.1.1. 1,2-Dierucoyl-sn-glycero-3-phosphocholine (Di22:1PC),
1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) (ammonium salt) (Rhod PE), and cholesterol were purchased from Avanti Polar Lipids (Alabaster, AL). Penicillin was purchased from YF Chemical Corp., Taiwan. Sulbactam was purchased from TTY Biopharm, Taiwan.
2.2. Lamellar X-ray diffraction (LXD)
Sample preparation was performed as described in previous reports [29–31]. In brief, antibiotics or cholesterol and lipids of a chosen molar ratio were co-dissolved in methanol and chloroform 1:1 (v/v). A suitable amount of solution was deposited onto a clean glass surface. The sample was vacuumed to remove the remaining solvent and then incubated under high humidity for hydration and rearrangement.
For lamellar X-ray diffraction measurement [30,31], the sample was held in a temperature-humidity controlled chamber (Fig. S1 Supplementary data, schematic of chamber). The machine collected at least five diffraction peaks using the θ–2θ reflectivity mode. The 12 keV X-ray light source of BL13A at the National Synchrotron Radiation Research Center (NSRRC) was used for the measurements. Each θ–2θ scan was measured from θ = 0.5° to 7.5° with a step angle of Δθ = 0.001° and a 1-s exposure time per step. Every sample was measured at near full hydration and two lower levels of humidity for the purpose of phase determination by the swelling method [32]. Each sample was measured twice to confirm that it was in equilibrium and to prove that it was not damaged by X-ray radiation. X-ray diffraction was used to construct the relative electron density profile along the direction normal to the bilayer surface to determine membrane thickness.
2.3. Giant unilamellar vesicle (GUV) aspiration method
Di22:1PC and a 1% molar ratio of the dye-labeled lipid Rhod PE were co-dissolved in chloroform. The proper amount of solution was deposited on an indium tin oxide coating glass (ITO glass) in the production chamber. The steps of the GUV experiment were described in our previous report [30]. Briefly, GUVs were produced in 100 mM sucrose solution by the electroformation method [33–35]. Then, the isotonic glucose solution was added to the production chamber (Fig. S2(A)) so that the molar ratio of glucose to sucrose outside the GUVs was 3:4. A glass micropipette of 5–10 μm in diameter was used to slightly suck one selected GUV to transfer it to the solution containing antibiotics or cholesterol in the observation chamber (Fig. S2(B)), which was separated from the production chamber. Because cholesterol is insoluble in water, we dissolved cholesterol in methanol first [36]. The protrusion of the GUV in the glass micropipette was monitored during the entire process of the binding of the antibiotics or cholesterol to the GUV [30].
3. Results and discussion
3.1. Membrane-thinning effect examined by LXD
The diffraction patterns of all samples measured at 30 °C and near full hydration are shown in Fig. 1. Penicillin/sulbactam and lipids were mixed to form solutions with P/L molar ratios of 0, 1/50, 1/15, and 1/5. Cholesterol and lipids were mixed to form solutions with P/L molar ratios of 0, 1/50, 1/25, 1/15, 1/10, and 1/5. At least six peaks were recorded for each diffraction pattern. No peak broadening was observed, indicating that fluctuations in the measurements were negligible. To determine the phases of the diffraction peaks, each sample was measured in a series of lowering hydration levels to produce patterns at different repeating distance D values. They were normalized relative to each other by the Blaurock method [32] and plotted as a function of scattering vector Q to determine the phases on the basis of the swelling method [37–39]. A representative example is shown in Fig. 2. After phase determination, the amplitudes of the diffraction patterns were used to remodel the transbilayer electron density profiles. The electron density profiles constructed from the diffraction data are shown in Fig. 3. The peak-to-peak distance (PtP) of the trans-bilayer electron density profile was defined as the membrane thickness. The previous study shows that the repeating distances of Di22:1PC at full hydration and excess water are 69.3 Å and 70.0 Å respectively [40]. And the membrane thickness of multilamellar vesicles (MLVs) at the excess water is about 44.3 Å. As a comparison, the repeating distance and membrane thickness of our lamellar sample of Di22:1PC at 98% RH are 60.7 Å and 45.0 Å. The repeating distance is 10 Å smaller than the ones at full hydration and excess water but the membrane thickness is comparable. The structure change from near full hydration (98% RH) to excess water is to increase the thickness of water layer. Fig. 4 shows the PtP versus the concentration of antibiotics and cholesterol. For penicillin-to-lipid and sulbactam-to-lipid, the phosphate peak-to-phosphate peak distance (PtP) decreased with increasing P/L molar ratios. The observed bilayer thinning in reverse proportion to P/L was consistent with the antibiotic molecules being embedded in the headgroup region of the membrane surface. The area of expansion was generated by the displacement of water molecules during the embedding of the antibiotic molecules in the headgroup region of the membrane surface. In contrast, for cholesterol-to-lipid, the phosphate peak-to-phosphate peak distance (PtP) increased with increasing P/L molar ratios, indicating that cholesterol inserted into the membrane in a manner that decreased lipid area as the number of inserted cholesterol molecules increased.
Fig. 1.
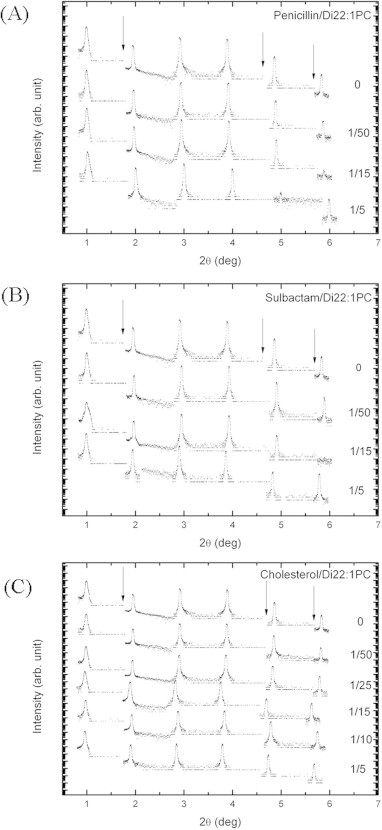
Diffraction patterns of the samples with different concentrations at 30 °C and near full hydration. (A) Penicillin was mixed with Di22:1PC bilayers at molar ratios of 0, 1/50, 1/15, and 1/5. (B) Sulbactam was mixed with Di22:1PC bilayers at molar ratios of 0, 1/50, 1/15, and 1/5. (C) Cholesterol was mixed with Di22:1PC bilayers at molar ratios of 0, 1/50, 1/25, 1/15, 1/10, and 1/5. The discontinuity in the diffraction patterns indicated by the arrow is due to the change of attenuator.
Fig. 2.
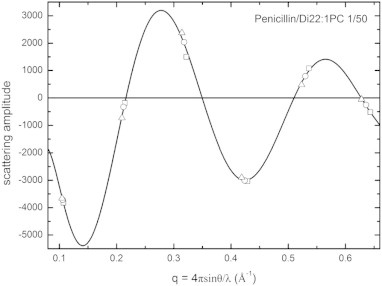
Phase diagram for the X-ray diffraction of Di22:1PC containing penicillin at a 1/50 molar ratio as an example of the swelling method. The repeating distance D was 58.6 (opened square), 59.2 (opened circle) and 60.0 (opened triangle) Å at 90.0%, 94.0% and 98.0% RH, respectively. The black curve shows Fourier-constructed from the diffraction amplitudes measured at one humidity level. All other samples had similar phase diagrams.
Fig. 3.
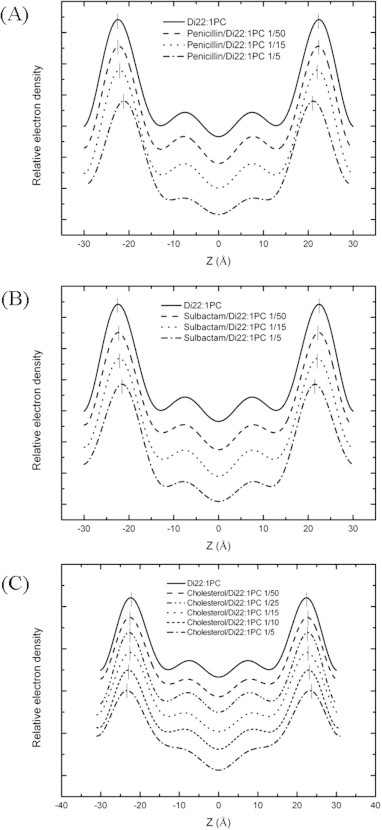
Electron density profiles of pure Di22:1PC and antibiotics or cholesterol in Di22:1PC at various molar ratios. (A) Penicillin was mixed with Di22:1PC bilayers at molar ratios of 0, 1/50, 1/15, and 1/5. (B) Sulbactam was mixed with Di22:1PC bilayers at molar ratios of 0, 1/50, 1/15, and 1/5. (C) Cholesterol was mixed with Di22:1PC bilayers at molar ratios of 0, 1/50, 1/25, 1/15, 1/10, and 1/5. The profiles were not normalized and were displaced for clarity. The short vertical bars indicate the positions of the peaks.
Fig. 4.
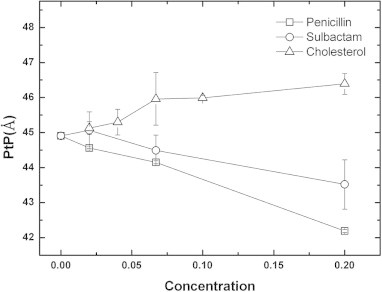
Membrane thicknesses (PtP) determined by X-ray diffraction change with penicillin, sulbactam, and cholesterol concentrations. The Di22:1PC bilayers were mixed with penicillin (opened square) and sulbactam (opened circle) at molar ratios of 0, 1/50, 1/15, and 1/5 and with cholesterol (opened triangle) at molar ratios of 0, 1/50, 1/25, 1/15, 1/10, and 1/5. The error bars are determined from the average of more than three repeated measurements.
Fig. 4 shows that the phosphate-to-phosphate distance (PtP) of di22:1PC decreased from 44.9 Å to 42.1 Å with a 0.2 penicillin-to-lipid molar ratio and from 44.9 Å to 43.5 Å with a sulbactam-to-lipid molar ratio of 0.2. In both cases, the PtP distance decreased linearly as the drug concentrations increased. In contrast, when the multilamellar bilayer was mixed with a solution with a cholesterol-to-lipid molar ratio of 0.2, the PtP distance increased to 46.1 Å. This indicated that the interactions of the antibiotics and cholesterol with the multilamellar bilayer were different. Our previous reports indicated that cholesterol molecules inserted into the lipid bilayer [41,42] and increased the PtP distance [43]. Other studies by us also indicated that peptides lay horizontally on the membrane outer surface, which shortens the PtP distance [44,45]. Similarly, Fig. 4 confirms that penicillin and sulbactam both lay horizontally outside the bilayer surface. Thus, penicillin and sulbactam do not tend to enter into bacterial cell walls. A recent report showed that sulbactam killed A. baumannii by inhibiting bacterial ABC transporters [5]. Thus, penicillin and sulbactam very likely exert their bactericidal effects at the cell wall. This result is consistent with the fact that penicillin binds to the polysaccharide chain of the bacterial cell wall [16,46].
The mechanism of peptide-induced membrane thinning was investigated in a previous study [45]. To recognize the interaction between antibiotics and the membrane, we used the same approach as was used previously to quantitatively analyze our X-ray data. Due to the volume conservation of the hydrophobic region of the lipid bilayer, we were able calculate the normalized area expansion by the relation, −Δh/h = ΔA/A, where h is the thickness of the hydrophobic region and A is the membrane area. The h value was determined by subtracting twice the length of the glycerol region, i.e., ∼10 Å, from the PtP [47,48]. With the assumption that the area expansion was due to the partitioning of the antibiotics into the interface of the lipid bilayer, the following equation was used: ΔA/A ≈ (Na/Nl)Aa/Al, where (Na/Nl) is the antibiotic-to-lipid molar ratio defined as the concentration of antibiotic, Aa is the area of expansion induced by one antibiotic and Al is the cross-sectional area of the lipids. Fig. 5 shows ΔA/A vs. concentration of antibiotic, and the data presented in Fig. 5 was converted from the data presented in Fig. 4. Using Al ∼ 69.3 Å2 and the slopes of linear fitting in Fig. 5, we obtained Aa values of 26.9 ± 1.5 Å2 and 16.5 ± 2.1 Å2 for penicillin and sulbactam, respectively. The molecular sizes of penicillin and sulbactam are 6.1 Å * 4.5 Å * 11.2 Å and 7.0 Å * 4.5 Å * 8.2 Å. The larger Aa value found for penicillin was most likely due its larger molecular size. Furthermore, the antibiotics need to remove water molecules around the headgroups of lipid and then pull the headgroups away to case the area expansion. We deduced the simple relation according to the geometrical argument: Σa = Aa + Aw. Here Σa is the cross-sectional area of antibiotics, Aa is the area expansion induced by one antibiotic binding and Aw is the area occupied by the water molecules around the headgroup of lipid. At the near hydration range, the area occupied by the water molecules per lipid is about 10 Å2 [49] and there are approximately 2–4 lipids surrounding one antibiotic. Thus Aw is approximately 20–40 Å2 for one binding penicillin and sulbactam. The Σa of penicillin and sulbactam are 47–67 Å2 and 37–57 Å2 respectively. The above quantitative analysis confirms that penicillin and sulbactam both lay horizontally on the interface of bilayer, again (Fig. S3).
Fig. 5.
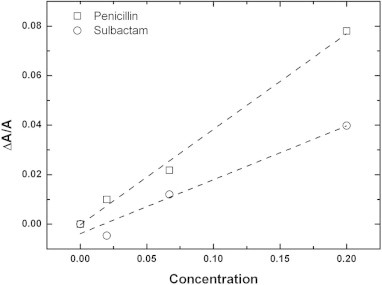
The normalized area expansion, ΔA/A, versus the concentration of penicillin (square) or sulbactam (circle). ΔA/A is equal to −Δh/h, as calculated from Fig. 1 using the relation h = PtP − 10 Å. The dashed lines indicate the linear fitting of the data.
3.2. Aspiration method for GUV
The aspiration method for GUV was developed by E. Evan and used to determine the elasticity of a membrane [50,51]. Our previous studies [30,52] applied this method to the measurement of the area expansion induced by peptide binding to a membrane. When the volume of a GUV is constant [53], the increase in the aspirated projection length LP inside the micropipette, ΔLP, can be converted to area expansion, ΔA, by the following approximate relation: ΔLP: ΔA = πDP (1 − DP/DV) ΔLP [51]. DP and DV are the inner diameter of the micropipette and the diameter of the GUV, respectively. The increase in the aspirated projection length LP inside the micropipette provides a direct measurement of the membrane area expansion induced by drug or cholesterol binding. Fig. 6 shows the time evolution fluorescence images of GUV interacting with penicillin, sulbactam, and cholesterol. We calculated the normalized area expansion, ΔA/A, from the images using the geometrical relation mentioned above. The normalized area expansions, ΔA/A, of a GUV exposed to antibiotics and cholesterol in different concentrations are shown in Fig. 7. All of the concentrations were measured 3–5 times to determine the statistical average. There is no significant change in area for a GUV exposed to penicillin or sulbactam. Combined the results of X-ray diffraction and GUV experiment, antibiotics target to membranes but don’t cause area expansion, i.e. Aa ∼ 0. The area occupied by the water molecules around the headgroup of lipid (Aw) in the excess water is much larger than in near full hydration so that Aa ∼ 0. Conversely, the percentage of expansion of the area of a GUV exposed to high-concentration cholesterol solutions is 2%. In the GUV system, there are two competitive effects on the change in area. One is the “condensing effect” on lipid cross-sectional area, which decreases the area of the membrane in the presence of low cholesterol concentrations and achieves saturation in the presence of high cholesterol concentrations [43]. The other effect is the “insertion effect” on total membrane area, which increases the membrane area as the number of inserted cholesterols increases. Consequently, the two effects cancel out in the presence of low cholesterol concentrations, and the insertion effect dominates the change in area in the presence of high cholesterol concentrations. The cholesterol induced effects on membrane area we observed in this study is agree with the previous report [54].
Fig. 6.
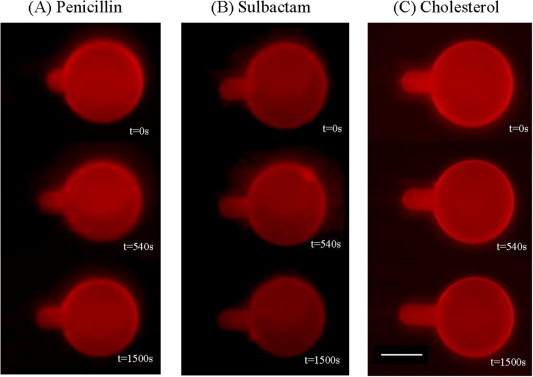
Time evolution fluorescence images of GUVs exposed to penicillin, sulbactam, and cholesterol. The typical GUV diameter (DV) is 35–50 μm. The inner diameter of the pipette (DP) is 7–10 μm. GUVs are exposed to solutions with (A) 1.72 × 10−3 M penicillin (B) 1.11 × 10−3 M sulbactam and (C) 5.56 × 10−5 M cholesterol. The scale bar in the image is 20 μm.
Fig. 7.
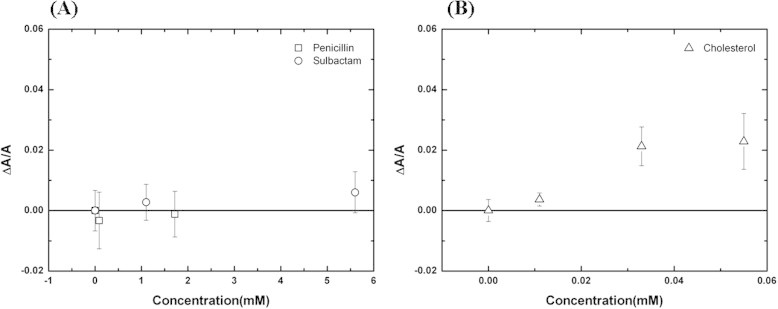
Concentration dependence of the normalized area expansion induced by (A) penicillin and sulbactam or (B) cholesterol at t = 540 s. After an exposure time of 540 s, the area expansions are saturated and no longer change in all cases. The error bars are determined from the average of more than three repeated measurements.
4. Conclusions
Our previous study showed that the protein expression of ABC transporter was decreased after treating A. baumannii with sulbactam [5]. The biological function of ABC transporter is to import nutrient into the bacteria and also to export toxic material. We compared the protein sequences of transpeptidase domain of penicillin binding proteins (PBP) 1a, 1b (class A), and 2, 3 (class B), with the ATPase and permease components (APC) of ABC transporter of A. baumannii. Table S1 shows the sequence alignment of these four transpeptidases of PBPs and APC. As can be seen, the homology within PBPs is greater, whereas with APC the homology is poor.
Further quantitative analysis is shown in Table S2. ClustalW score indicated the close genetic relationship between PBP 1a and 1b, score 36.26; PBP 2 and 3, score 28.67. On the contrary, APC shows poor genetic relationship with PBP 1a, 1b, 2, and 3, with scores ranging from 2.17 to 8.79, all less than 10. A genetic phylogram and distance between the protein sequences is also shown in Table S2. In this phylogram, the transpeptidase domain of PBP 1a and 1b, PBP 2 and 3 are each genetically related, whereas APC domain of ABC transporter is not related with PBPs.
In summary, the bactericidal effect of sulbactam against A. baumannii involves down-regulating the ABC transporter [5]. According to the protein sequence and phylographic analyses, ATPase and permease components, the active site of ABC transporter, has low relationship with the transpeptidase domain of PBP classes. Therefore, the mechanism of the bactericidal effect of sulbactam against A. baumannii is different from that of penicillin (with penicillin binding proteins). The bactericidal effect of sulbactam against A. baumannii is a new phenomenon.
On the other hand, we studied the interaction between antibiotics and membranes using not only X-ray diffraction in equilibrium but also kinetic experiments involving a single GUV in solution. The result of X-ray diffraction indicates penicillin and sulbactam target to membrane and lay horizontally on the interface of bilayer rather than insert into membrane like cholesterol. The GUV experiment suggests that neither penicillin nor sulbactam is able to disturb the structure of membrane and that cholesterol inserts perpendicularly into a membrane in an aqueous solution. The less perturbation means less deformation energy cost of antibiotic-membrane binding. Due to the hydrophobic interaction, penicillin and sulbactam prefer to bind on membrane rather dissolve in the solution. It implies that the antibiotics have more chances to “meet” the receptor protein or disturb the osmotic pressure to kill the bacteria. The hypothesis provides the new insight on the membrane mediated enhancement of bacteria inhibition.
Acknowledgments
This study was supported by Taiwan Ministry of Science and Technology Contracts MOST 103-2112-M-213-003 and the National Synchrotron Radiation Research Center (NSRRC), Taiwan.
Appendix A. Supplementary data
Supplementary Fig. S1.
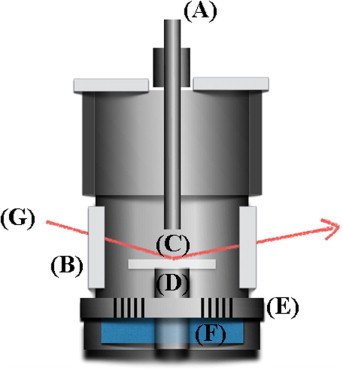
Schematic of the temperature-humidity controlled chamber. (A) Temperature-humidity sensor (B) mylar window (C) sample (D) temperature sensor (E) isolated layer (F) water cap (G) incident X-ray.
Supplementary Fig. S2.
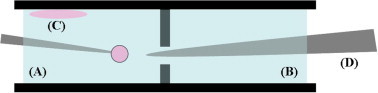
Schematic of the side view of the cell used in the GUV experiment. (A) Production chamber (B) observation chamber (C) lipid film (D) glass tip used to align the micropipette and to isolate two chambers before transferring the GUV. In brief, a 2-μl drop of a lipid solution was deposited on the upper side of the conductive ITO-coated glasses of the production chamber (A). The vesicles were produced in the production region containing a 4:3 mixture of sucrose/glucose sugar solutions. A micropipette (internal diameter 7–10 μm) was used to select a GUV (35–50 μm in diameter). Then, the micropipette with the selected GUV was aligned to the glass tip, (D) and the GUV was transferred to the solution with antibiotic or cholesterol in the observation chamber (B). The protrusion length inside the micropipette was monitored with time.
Supplementary Fig. S3.
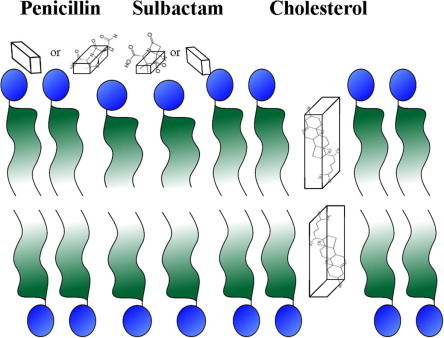
The cartoon represents the possible orientations of penicillin, sulbactam and cholesterol in membranes.
References
- 1.Perez F., Hujer A.M., Hujer K.M., Decker B.K., Rather P.N., Bonomo R.A. Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 2007;51:3471–3484. doi: 10.1128/AAC.01464-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maragakis L.L., Perl T.M. Acinetobacter baumannii: epidemiology, antimicrobial resistance, and treatment options. Clin. Infect. Dis. 2008;46:1254–1263. doi: 10.1086/529198. [DOI] [PubMed] [Google Scholar]
- 3.Lin N.T., Chiou P.Y., Chang K.C., Chen L.K., Lai M.J. Isolation and characterization of phi AB2: a novel bacteriophage of Acinetobacter baumannii. Res. Microbiol. 2010;161:308–314. doi: 10.1016/j.resmic.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 4.Peleg A.Y., Seifert H., Paterson D.L. Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 2008;21:538–582. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin C.H., Su S.C., Ho K.H., Hsu Y.W., Lee K.R. Bactericidal effect of sulbactam against Acinetobacter baumannii ATCC 19606 studied by 2D-DIGE and mass spectrometry. Int. J. Antimicrob. Agents. 2014;44:38–46. doi: 10.1016/j.ijantimicag.2014.03.004. [DOI] [PubMed] [Google Scholar]
- 6.Jager S.A., Shapovalova I.V., Jekel P.A., Alkema W.B., Svedas V.K., Janssen D.B. Saturation mutagenesis reveals the importance of residues alphaR145 and alphaF146 of penicillin acylase in the synthesis of beta-lactam antibiotics. J. Biotechnol. 2008;133:18–26. doi: 10.1016/j.jbiotec.2007.08.039. [DOI] [PubMed] [Google Scholar]
- 7.Medeiros A.A. Evolution and dissemination of beta-lactamases accelerated by generations of beta-lactam antibiotics. Clin. Infect. Dis. 1997;24(Suppl. 1):S19–S45. doi: 10.1093/clinids/24.supplement_1.s19. [DOI] [PubMed] [Google Scholar]
- 8.Waxman D.J., Strominger J.L. Penicillin-binding proteins and the mechanism of action of beta-lactam antibiotics. Annu. Rev. Biochem. 1983;52:825–869. doi: 10.1146/annurev.bi.52.070183.004141. [DOI] [PubMed] [Google Scholar]
- 9.Ghuysen J.M. Serine beta-lactamases and penicillin-binding proteins. Annu. Rev. Microbiol. 1991;45:37–67. doi: 10.1146/annurev.mi.45.100191.000345. [DOI] [PubMed] [Google Scholar]
- 10.Scheffers D.J., Pinho M.G. Bacterial cell wall synthesis: new insights from localization studies. Microbiol. Mol. Biol. Rev. MMBR. 2005;69:585–607. doi: 10.1128/MMBR.69.4.585-607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Massova I., Mobashery S. Kinship and diversification of bacterial penicillin-binding proteins and beta-lactamases. Antimicrob. Agents Chemother. 1998;42:1–17. doi: 10.1128/aac.42.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gardner A.D. Morphological effects of penicillin on bacteria. Nature. 1940;146:837–838. [Google Scholar]
- 13.Duguid J.P. The sensitivity of bacteria to the action of penicillin. Edinburgh Med. J. 1946;53:401–412. [PMC free article] [PubMed] [Google Scholar]
- 14.Waxman D.J., Strominger J.L. Sequence of active site peptides from the penicillin-sensitive d-alanine carboxypeptidase of Bacillus subtilis. Mechanism of penicillin action and sequence homology to beta-lactamases. J. Biol. Chem. 1980;255:3964–3976. [PubMed] [Google Scholar]
- 15.Yocum R.R., Rasmussen J.R., Strominger J.L. The mechanism of action of penicillin. Penicillin acylates the active site of Bacillus stearothermophilusd-alanine carboxypeptidase. J. Biol. Chem. 1980;255:3977–3986. [PubMed] [Google Scholar]
- 16.Wise E.M., Jr., Park J.T. Penicillin: its basic site of action as an inhibitor of a peptide cross-linking reaction in cell wall mucopeptide synthesis. Proc. Natl. Acad. Sci. U.S.A. 1965;54:75–81. doi: 10.1073/pnas.54.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tipper D.J., Strominger J.L. Mechanism of action of penicillins: a proposal based on their structural similarity to acyl-d-alanyl-d-alanine. Proc. Natl. Acad. Sci. U.S.A. 1965;54:1133–1141. doi: 10.1073/pnas.54.4.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sainsbury S., Bird L., Rao V., Shepherd S.M., Stuart D.I., Hunter W.N., Owens R.J., Ren J. Crystal structures of penicillin-binding protein 3 from Pseudomonas aeruginosa: comparison of native and antibiotic-bound forms. J. Mol. Biol. 2011;405:173–184. doi: 10.1016/j.jmb.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koch A.L. How bacteria grow and divide in spite of internal hydrostatic pressure. Can. J. Microbiol. 1985;31:1071–1084. doi: 10.1139/m85-204. [DOI] [PubMed] [Google Scholar]
- 20.Cooper P.D. The site of action of penicillin. III. Effect of surface-active substances on penicillin uptake by Staphylococcus aureus. Biochim. Biophys. Acta. 1954;13:433–438. doi: 10.1016/0006-3002(54)90350-9. [DOI] [PubMed] [Google Scholar]
- 21.Cooper P.D. Site of action of radiopenicillin. Bacteriol. Rev. 1956;20:28–48. doi: 10.1128/br.20.1.28-48.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barber M. Hospital infection yesterday and today. J. Clin. Pathol. 1961;14:2–10. doi: 10.1136/jcp.14.1.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karlowsky J.A., Draghi D.C., Jones M.E., Thornsberry C., Friedland I.R., Sahm D.F. Surveillance for antimicrobial susceptibility among clinical isolates of Pseudomonas aeruginosa and Acinetobacter baumannii from hospitalized patients in the United States, 1998 to 2001. Antimicrob. Agents Chemother. 2003;47:1681–1688. doi: 10.1128/AAC.47.5.1681-1688.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levin A.S., Levy C.E., Manrique A.E., Medeiros E.A., Costa S.F. Severe nosocomial infections with imipenem-resistant Acinetobacter baumannii treated with ampicillin/sulbactam. Int. J. Antimicrob. Agents. 2003;21:58–62. doi: 10.1016/s0924-8579(02)00276-5. [DOI] [PubMed] [Google Scholar]
- 25.Lode H.M. Rational antibiotic therapy and the position of ampicillin/sulbactam. Int. J. Antimicrob. Agents. 2008;32:10–28. doi: 10.1016/j.ijantimicag.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 26.Samsonov A.V., Mihalyov I., Cohen F.S. Characterization of cholesterol-sphingomyelin domains and their dynamics in bilayer membranes. Biophys. J. 2001;81:1486–1500. doi: 10.1016/S0006-3495(01)75803-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sessa G., Freer J.H., Colacicco G., Weissmann G. Interaction of alytic polypeptide, melittin, with lipid membrane systems. J. Biol. Chem. 1969;244:3575–3582. [PubMed] [Google Scholar]
- 28.Needham D., Nunn R.S. Elastic deformation and failure of lipid bilayer membranes containing cholesterol. Biophys. J. 1990;58:997–1009. doi: 10.1016/S0006-3495(90)82444-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ludtke S., He K., Huang H. Membrane thinning caused by magainin 2. Biochemistry. 1995;34:16764–16769. doi: 10.1021/bi00051a026. [DOI] [PubMed] [Google Scholar]
- 30.Lee M.T., Hung W.C., Chen F.Y., Huang H.W. Mechanism and kinetics of pore formation in membranes by water-soluble amphipathic peptides. Proc. Natl. Acad. Sci. U.S.A. 2008;105:5087–5092. doi: 10.1073/pnas.0710625105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hung W.C., Chen F.Y., Lee C.C., Sun Y., Lee M.T., Huang H.W. Membrane-thinning effect of curcumin. Biophys. J. 2008;94:4331–4338. doi: 10.1529/biophysj.107.126888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blaurock A.E. Structure of the nerve myelin membrane: proof of the low-resolution profile. J. Mol. Biol. 1971;56:35–52. doi: 10.1016/0022-2836(71)90082-9. [DOI] [PubMed] [Google Scholar]
- 33.Okumura Y., Oana S. Effect of counter electrode in electroformation of giant vesicles. Membranes. 2011;1:345–353. doi: 10.3390/membranes1040345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhelev D.V., Needham D. Tension-stabilized pores in giant vesicles: determination of pore size and pore line tension. Biochim. Biophys. Acta. 1993;1147:89–104. doi: 10.1016/0005-2736(93)90319-u. [DOI] [PubMed] [Google Scholar]
- 35.Angelova M.I., Soleau S., Meleard P., Faucon J.F., Bothorel P. Preparation of giant vesicles by external AC electric fields. Kinetics and applications. Prog. Colloid Polym. Sci. 1992;89:127–131. [Google Scholar]
- 36.Lllana K.B., Nkslrn G., Sara S., Raphael B. Solubilities of cholesterol, sitosterol, and cholesteryl acetate in polar organic solvents. J. Chem. Eng. Data. 1984;29:441–443. [Google Scholar]
- 37.Wilkins M.H., Blaurock A.E., Engelman D.M. Bilayer structure in membranes. Nature: New Biol. 1971;230:72–76. doi: 10.1038/newbio230072a0. [DOI] [PubMed] [Google Scholar]
- 38.Torbet J., Wilkins M.H. X-ray diffraction studies of lecithin bilayers. J. Theor. Biol. 1976;62:447–458. doi: 10.1016/0022-5193(76)90129-6. [DOI] [PubMed] [Google Scholar]
- 39.Perutz M.F. The structure of haemoglobin. III. Direct determination of the molecular transform. R. Soc. 1954;225:264–286. [Google Scholar]
- 40.Kučerka N., Tristram-Nagle S., Nagle J. Structure of fully hydrated fluid phase lipid bilayers with monounsaturated chains. J. Membr. Biol. 2006;208:193–202. doi: 10.1007/s00232-005-7006-8. [DOI] [PubMed] [Google Scholar]
- 41.Brzustowicz M.R., Cherezov V., Caffrey M., Stillwell W., Wassall S.R. Molecular organization of cholesterol in polyunsaturated membranes: microdomain formation. Biophys. J. 2002;82:285–298. doi: 10.1016/S0006-3495(02)75394-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harroun T.A., Katsaras J., Wassall S.R. Cholesterol hydroxyl group is found to reside in the center of a polyunsaturated lipid membrane. Biochemistry. 2006;45:1227–1233. doi: 10.1021/bi0520840. [DOI] [PubMed] [Google Scholar]
- 43.Hung W.C., Lee M.T., Chen F.Y., Huang H.W. The condensing effect of cholesterol in lipid bilayers. Biophys. J. 2007;92:3960–3967. doi: 10.1529/biophysj.106.099234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee M.T., Chen F.Y., Huang H.W. Energetics of pore formation induced by membrane active peptides. Biochemistry. 2004;43:3590–3599. doi: 10.1021/bi036153r. [DOI] [PubMed] [Google Scholar]
- 45.Chen F.Y., Lee M.T., Huang H.W. Evidence for membrane thinning effect as the mechanism for peptide-induced pore formation. Biophys. J. 2003;84:3751–3758. doi: 10.1016/S0006-3495(03)75103-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakae T., Nikaido H. Outer membrane as a diffusion barrier in Salmonella typhimurium. Penetration of oligo- and polysaccharides into isolated outer membrane vesicles and cells with degraded peptidoglycan layer. J. Biol. Chem. 1975;250:7359–7365. [PubMed] [Google Scholar]
- 47.Nagle J.F., Tristram-Nagle S. Structure of lipid bilayers. Biochim. Biophys. Acta. 2000;1469:159–195. doi: 10.1016/s0304-4157(00)00016-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hung W.C., Chen F.Y. The hydrophobic–hydrophilic interface of phospholipid membranes studied by lamellar X-ray diffraction. Chin. J. Phys. 2003;41:85–91. [Google Scholar]
- 49.Lee M.T., Hung W.C., Chen F.Y., Huang H.W. Many-body effect of antimicrobial peptides: on the correlation between lipid’s spontaneous curvature and pore formation. Biophys. J. 2005;89:4006–4016. doi: 10.1529/biophysj.105.068080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kwok R., Evans E. Thermoelasticity of large lecithin bilayer vesicles. Biophys. J. 1981;35:637–652. doi: 10.1016/S0006-3495(81)84817-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Olbrich K., Rawicz W., Needham D., Evans E. Water permeability and mechanical strength of polyunsaturated lipid bilayers. Biophys. J. 2000;79:321–327. doi: 10.1016/S0006-3495(00)76294-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee M.T., Sun T.L., Hung W.C., Huang H.W. Process of inducing pores in membranes by melittin. Proc. Natl. Acad. Sci. U.S.A. 2013;110:14243–14248. doi: 10.1073/pnas.1307010110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rawicz W., Olbrich K.C., McIntosh T., Needham D., Evans E. Effect of chain length and unsaturation on elasticity of lipid bilayers. Biophys. J. 2000;79:328–339. doi: 10.1016/S0006-3495(00)76295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Olsen B.N., Schlesinger P.H., Baker N.A. Perturbations of membrane structure by cholesterol and cholesterol derivatives are determined by sterol orientation. J. Am. Chem. Soc. 2009;131:4854–4865. doi: 10.1021/ja8095224. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


